1. Introduction
Pluripotent stem cells (PSCs), derived from the epiblast of mammalian blastocysts, have the dual capacity for unlimited self-renewal and differentiation into derivatives of all three germ layers [
1]. In rodents, PSCs can exist in two distinct states: the naïve state, represented by embryonic stem cells (ESCs) derived from the preimplantation epiblast, and the primed state, represented by epiblast stem cells (EpiSCs) derived from post-implantation epiblast tissue [
2]. These states are also recognized in human and non-human primates (NHPs), including Rhesus macaques (
Macaca mulatta), where PSCs can be stabilized in either a naïve and primed configuration depending on culture conditions and signaling inputs [
3]. Naïve and primed PSCs differ markedly in their transcriptional, epigenetic, and metabolic profiles, which in turn have a profound impact on their functional properties [
4]. Notably, only naïve PSCs in the naïve state exhibit the capacity to integrate into host preimplantation embryos and contribute to development, as demonstrated in rodents [
2], rabbits [
5], and cynomolgus monkeys [
6]. These functional differences are underpinned by distinct regulatory networks and chromatin landscapes: naïve PSCs display global DNA hypomethylation, two active X chromosomes in female cells, and reduced levels of H3K27me3 at developmental gene loci, whereas primed PSCs show higher DNA methylation, one inactive X chromosome (in females), and accumulation of repressive histone marks [
4]. Identifying the pluripotency state of a given PSC line traditionally requires multimodal analyses, including bulk RNA sequencing [
7], immunostaining [
8] for state-specific markers (e.g., KLF4, TFCP2L1 for naïve; OTX2, ZIC2 for primed), reporter-based functional assays, and epigenomic profiling. These approaches, while informative, are often labor-intensive, and not readily applicable for routine quality control. In this context, Fourier-transform infrared (FTIR) spectroscopy offers an attractive, non-invasive alternative for cell characterization [
9].
FTIR microspectroscopy analyzes vibrational energy absorption in molecular bonds, providing a composite biochemical fingerprint of cells. The technique enables rapid and label-free quantification of major cellular components—including proteins, lipids, carbohydrates, and nucleic acids—at single-cell resolution. Importantly, changes in cell state are accompanied by alterations in molecular composition and structure, which can be detected in specific IR absorption bands. For example, shifts in the amide I and II regions (1700–1500 cm⁻¹) reflect differences in protein secondary structure and abundance; CH₂ and CH₃ stretching modes (3000–2800 cm⁻¹) are indicative of membrane lipid composition and fluidity; and bands in the 1200–800 cm⁻¹ region reflect changes in nucleic acid conformation and RNA/DNA content [9-12]. Previous bio- spectroscopic studies have successfully applied FTIR to distinguish undifferentiated from differentiated cells, to assess stem cell lineage commitment, and to monitor reprogramming efficiency [
9]. However, no study has explored its application for distinguishing between naïve and primed states of pluripotency, particularly in non-human primates. Establishing such a methodology could provide a powerful tool for monitoring cell identity and quality in basic and translational stem cell research. A technique known as focal plane array (FPA) infrared imaging with multichannel detectors has recently become available for analyzing cells and tissues with allowing to measure all data points from each detector element simultaneously.
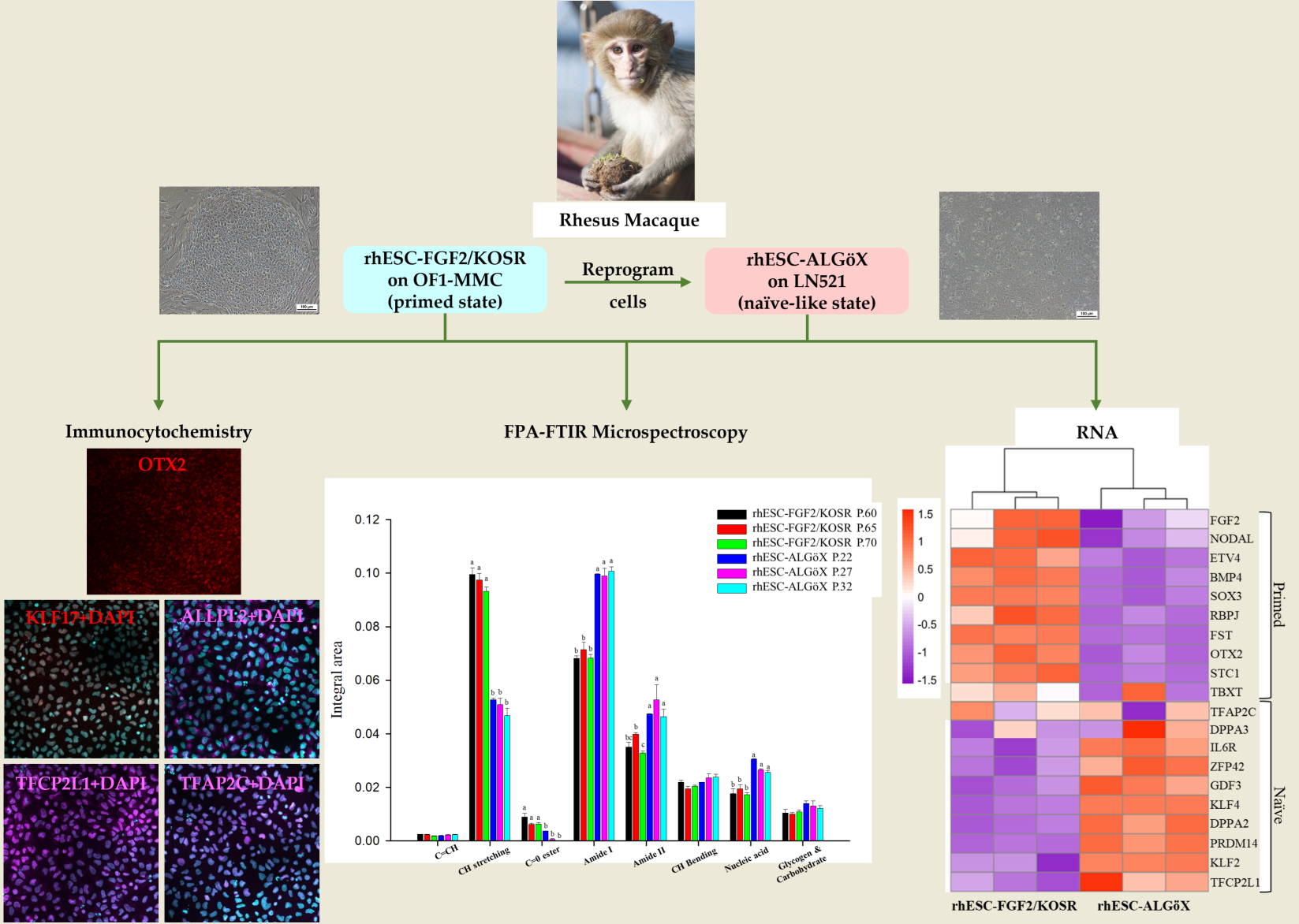
In this study, we applied FPA-FTIR microspectroscopy to rhesus macaque embryonic stem cells (rhESCs) cultured under two distinct conditions: the well-established FGF2/KOSR medium, which supports the primed state, and a novel medium we developed, ALGöX, designed to support a naïve-like pluripotent state. ALGöX consists of fibroblast-conditioned N2B27 medium supplemented with Leukemia Inhibitory Factor (LIF), Activin A, PKC inhibitor Gö6983, and tankyrase inhibitor XAV939. This combination aims to inhibit MEK/ERK and WNT signaling while maintaining key self-renewal cues, based on pathways shown to stabilize naïve pluripotency in rodent and human systems. We show that rhESCs cultured in ALGöX exhibit distinct spectral features compared to those in FGF2/KOSR.
2. Results
2.1. Characterization of rhESC-FGF2/KOSR and rhESC-ALGöX cells by Immunocytochemistry and RNA sequencing.
rhESCs were originally derived in FGF2/KOSR [
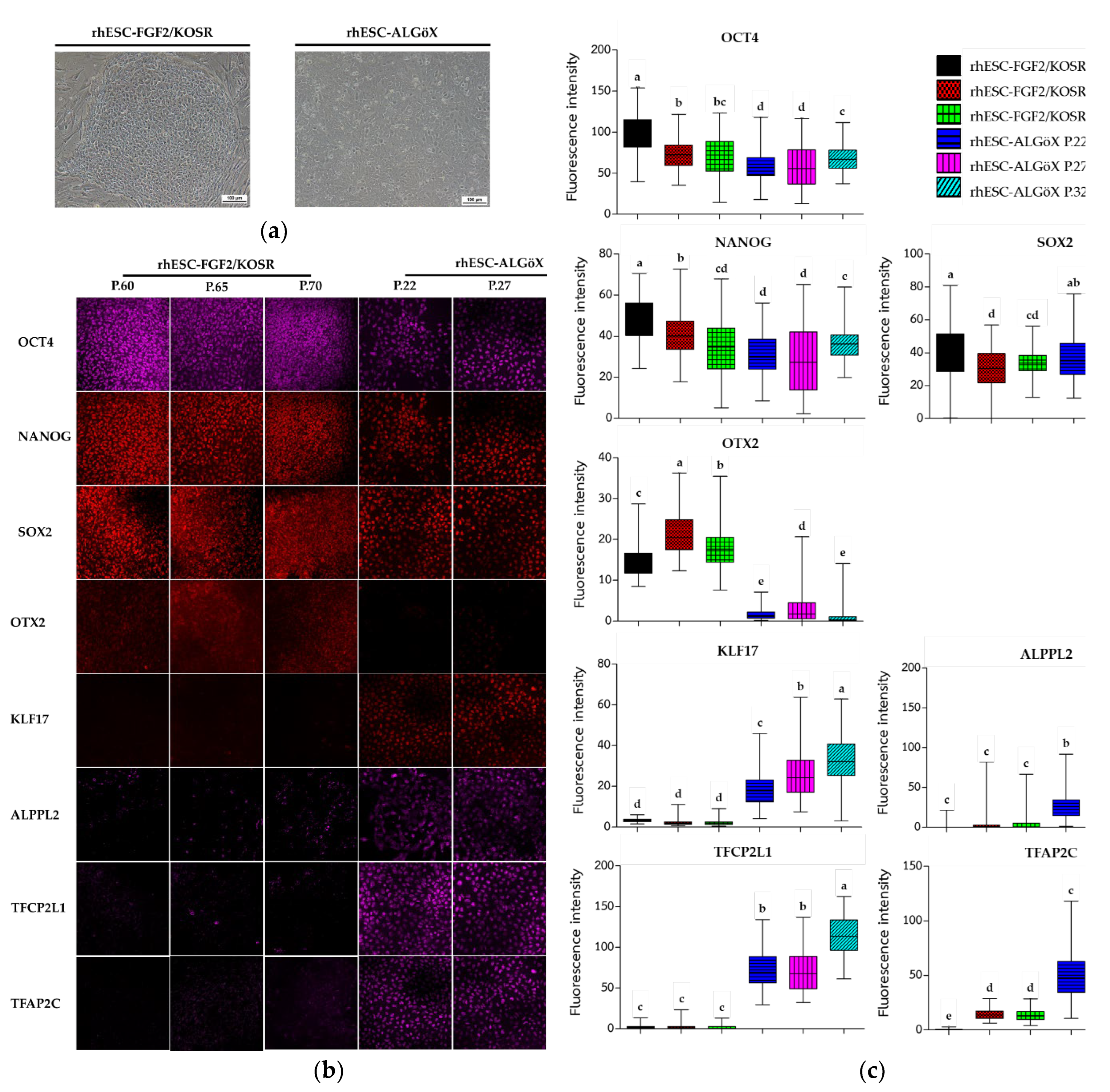
13]. Morphological changes were observed when rhESCs-FGF2/KOSR were transferred into ALGöX medium. rhESCs formed more compact colonies (
Figure 1a).
Immunolabelling confirmed that both rhESC-FGF2/KOSR and rhESCs-ALGöX retained expression of core pluripotency markers OCT4, NANOG, and SOX2 (
Figure 1b), indicating stable pluripotent identity across conditions. In rhESC-FGF2/KOSR, the intensity of these markers peaked at passage 60 and declined progressively at later passages (65 and 70). In contrast, rhESC-ALGöX maintained consistent expression levels across passages (
Figure 1c).
Notably, the primed-state marker OTX2 was detected exclusively in rhESC-FGF2/KOSR, while rhESC-ALGöX cells exhibited robust expression of KLF17, ALPPL2, TFCP2L1, and TFAP2C, consistent with a naïve pluripotency signature (
Figure 1b). Quantitative fluorescence analysis revealed statistically significant differences in marker expression over passages, with the highest levels of OTX2 in primed cells at passage 65 and peak expression of naïve markers in rhESC-ALGöX cells at passage 32 (
Figure 1c). These results indicate that rhESCs cultured in ALGöX medium maintain a naïve-like molecular profile and support stable naïve pluripotency over multiple passages.
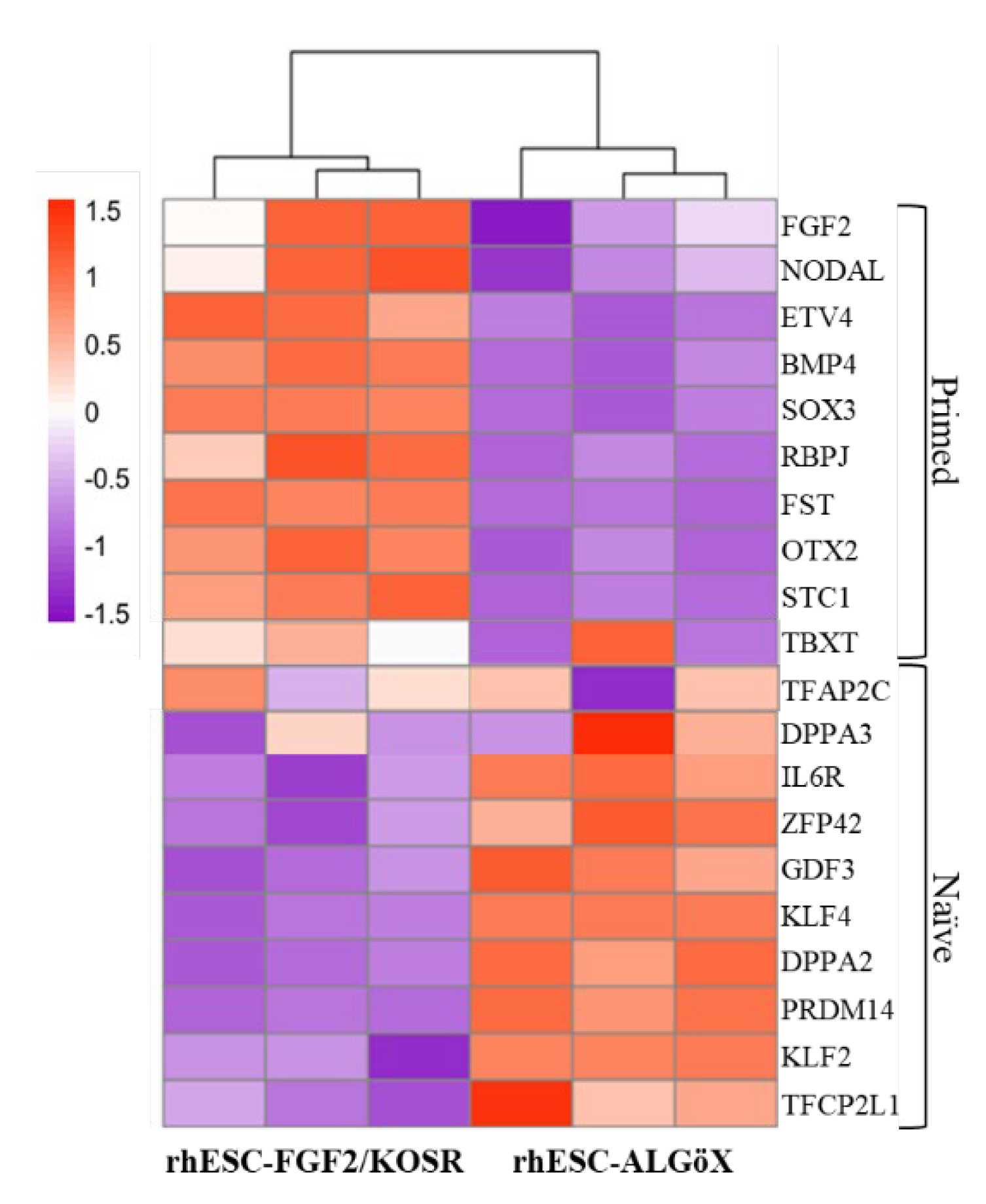
RNA-seq confirmed the transcriptional divergence between rhESC-FGF2/KOSR and rhESCs-ALGöX. Hierarchical clustering of Z-score-normalized expression profiles segregated all samples into two distinct clusters corresponding to their respective culture conditions (
Figure 2). rhESCs-FGF2/KOSR showed elevated expression of primed-state genes including NODAL, OTX2, ETV4, BMP4, FST, and SOX3, whereas rhESC-ALGöX exhibited high expression of naïve-associated genes including KLF2, DPPA2, DPPA3, ZFP42, PRDM14, and TFCP2L1. These transcriptomic data validate the immunolabelling data and strongly suggest that ALGöX conditions reprogram primed rhESCs to a molecularly distinct, naïve-like pluripotent state.
2.2. Characterization of rhESC-FGF2/KOSR and rhESC-ALGöX cells FTIR Microspectroscopy distinguishes Primed and Naïve-like rhESCs.
2.2.1. Biochemical Differences Between Cell States
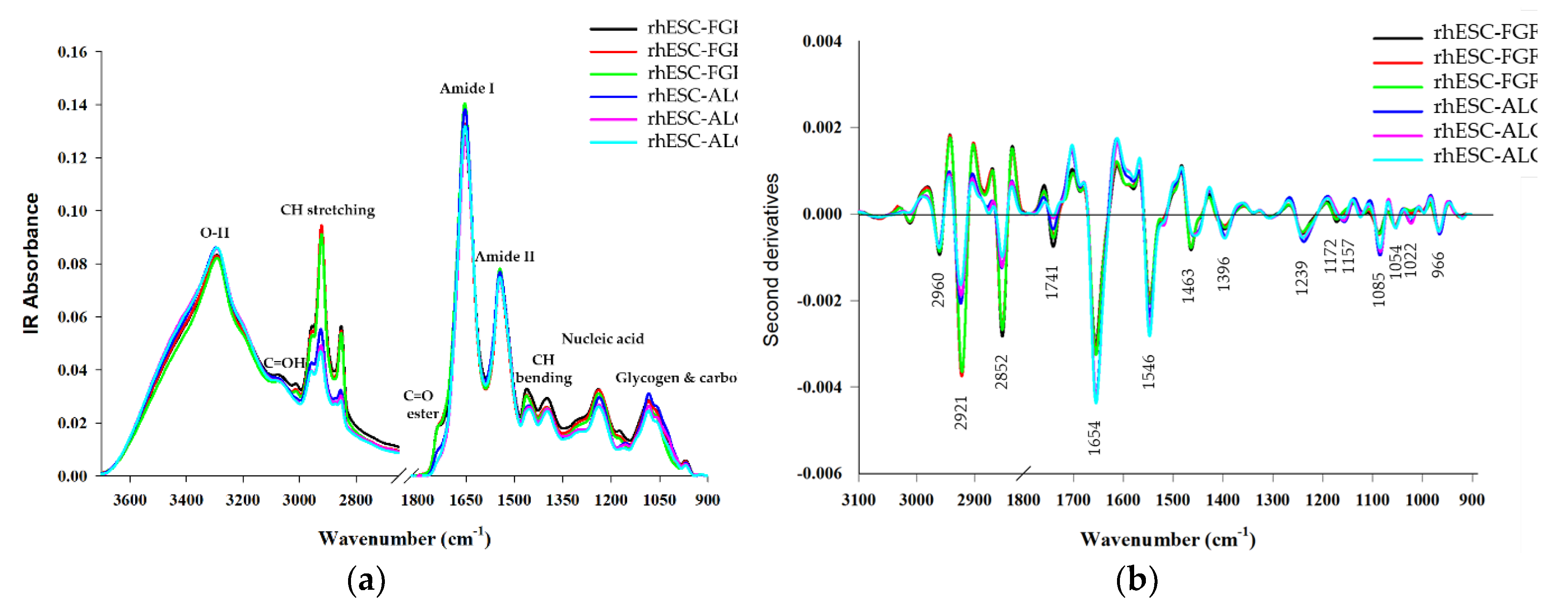
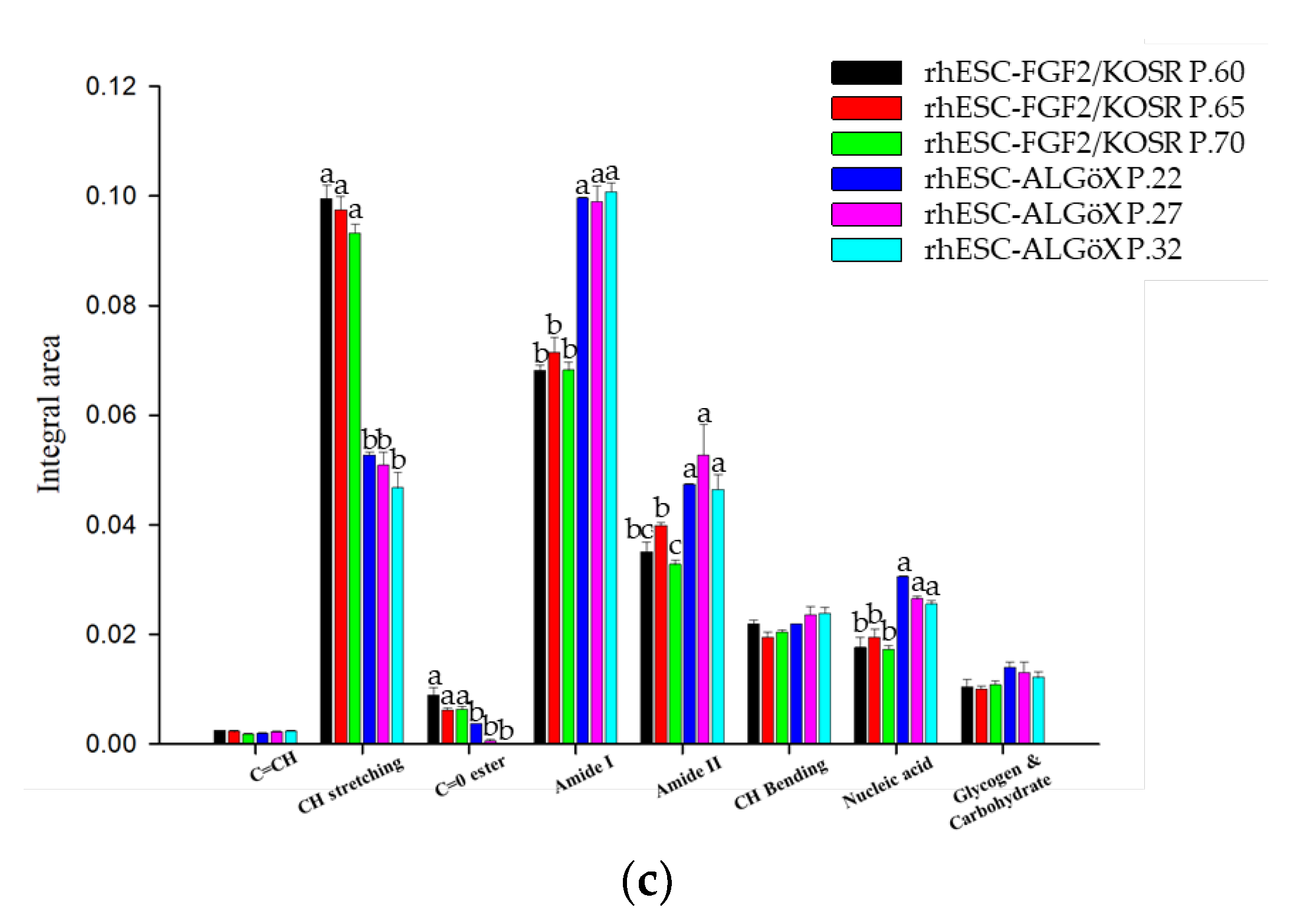
We used a FPA-based FTIR imaging system to analyze rhESC-FGF2/KOSR and rhESC-ALGöX cells at the high pixel resolution (9.6 × 9.6 μm). Three biological replicates per condition were analyzed: passages 60, 65, and 70 for primed rhESC-FGF2/KOSR, and passages 22, 27, and 32 for naïve-like rhESC-ALGöX. The amide I (1700–1600 cm⁻¹) and amide II (1600–1500 cm⁻¹) bands provide insight into protein secondary structures (
Figure 3a). The amide I band, arising primarily from C=O stretching, resolves into features attributed to α-helices (~1654 cm⁻¹), β-sheets (~1635 cm⁻¹), and β-turns (~1685 cm⁻¹) [
9]. The amide II band originates from N–H bending and C–N stretching and typically peaks around 1546 cm⁻¹. To resolve overlapping peaks, we applied second derivative spectroscopy, which enhanced the identification of individual vibrational bands. The second derivative spectra of primed and naïve-like cells are shown in
Figure 3b.
Naïve-like rhESCs-ALGöX displayed significantly higher integrated absorbance in the amide I, amide II, and nucleic acid regions compared to primed rhESC-FGF2/KOSR cells (p < 0.05, ANOVA;
Figure 3c). Notably, naïve-like cells exhibited prominant bands at 1654 and 1546 cm⁻¹, corresponding to Amide I and II, and strong nucleic acid peaks at 1239 and 1085 cm⁻¹. In contrast, primed cells showed stronger lipid-associated absorbance, particularly in the CH
₂/CH
₃ stretching region (2921–2852 cm⁻¹) and the ester carbonyl stretch at 1741 cm⁻¹ (
Figure 3b). These spectral differences suggest higher protein synthesis and nucleic acid content in naïve-like cells, while primed cells are enriched in membrane lipid signatures [14, 18-20]. No statistically significant spectral variation was observed between passages within either state (
Figure 3c).
2.2.2. Principal Component Analysis of rhESCs based on FTIR spectra.
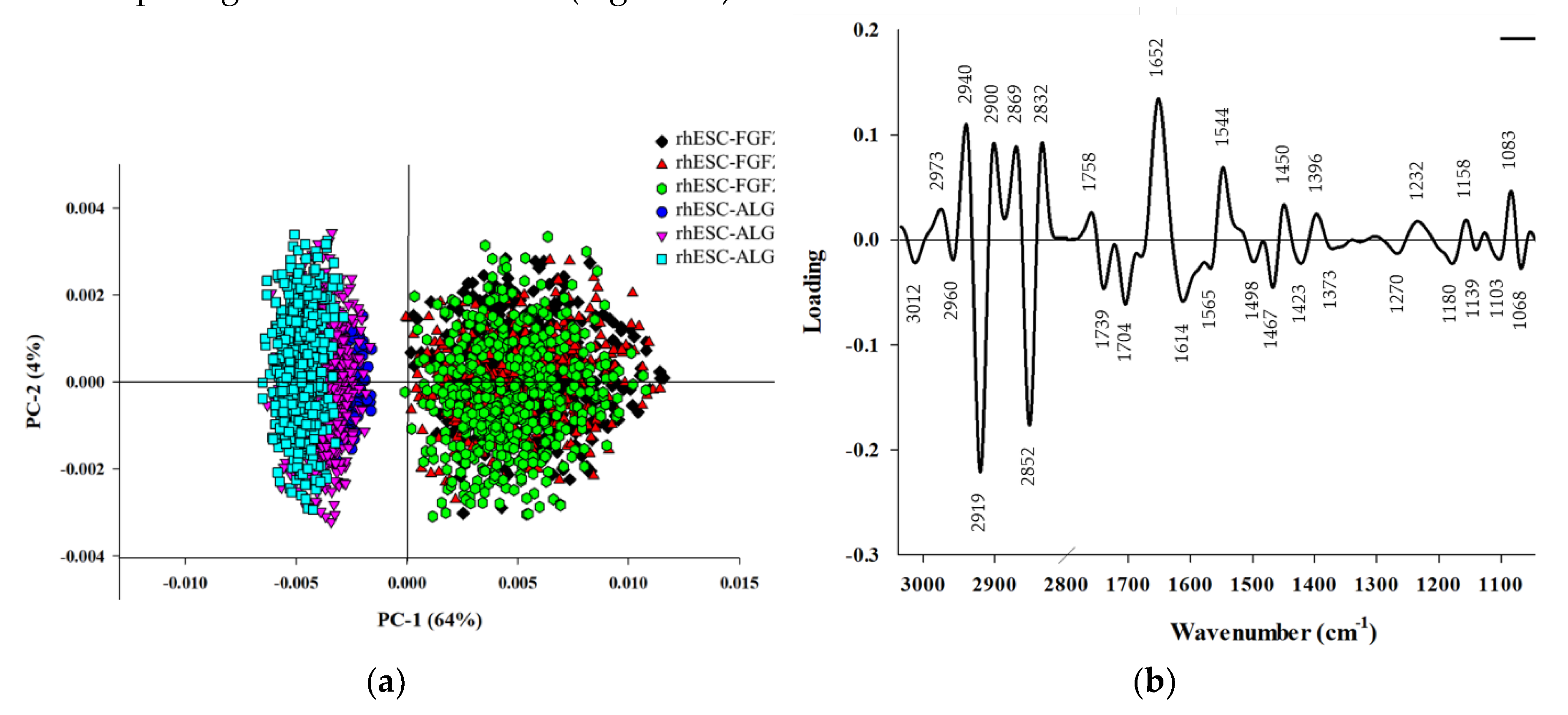
PCA was performed on the second derivative spectra from rhESC-FGF2/KOSR and rhESC-ALGöX cells in order to visualize of clustering of similar spectra within datasets in scatter plots; and identification of variables (spectral bands representing various molecular groups within the samples) in loading plots. Unsupervised PCA of the full spectral dataset revealed clear segregation between cell states. PC-1 (64% variance) and PC-2 (4% variance) of the total variance effectively separated naïve-like rhESC-ALGöX and primed rhESC-FGF2/KOSR cells (
Figure 4a). Loadings on PC-1 indicated strong contributions from lipid-associated bands (3000-2800 cm⁻¹), Lipid ester carbonyl (1750-1700 cm
-1), protein Amide I and Amide II-related bands (1700-1500 cm
-1), Phosphodiester bond from nucleic acid (1240 and 1080 cm
-1) (
Figure 4b). Primed rhESC-FGF2/KOSR cells showed positive PC-1 scores associated with negative loading from CH-stretching (2852 and 2919 cm
-1) and ester carbonyls (1739 and 1704 cm
-1), while negative score plot from naïve-like rhESC-ALGöX cells were associated with positive loading from strong amide I absorption (~1652 cm
-1), amide II absorption (~1544 cm
-1) and phosphodiester bond from nucleic acid at 1232 and 1083 cm
-1 (
Table 1).
2.2.3. Partial Least Squares-Discriminant Analysis (PLS-DA) of rhESCs based on FTIR spectra
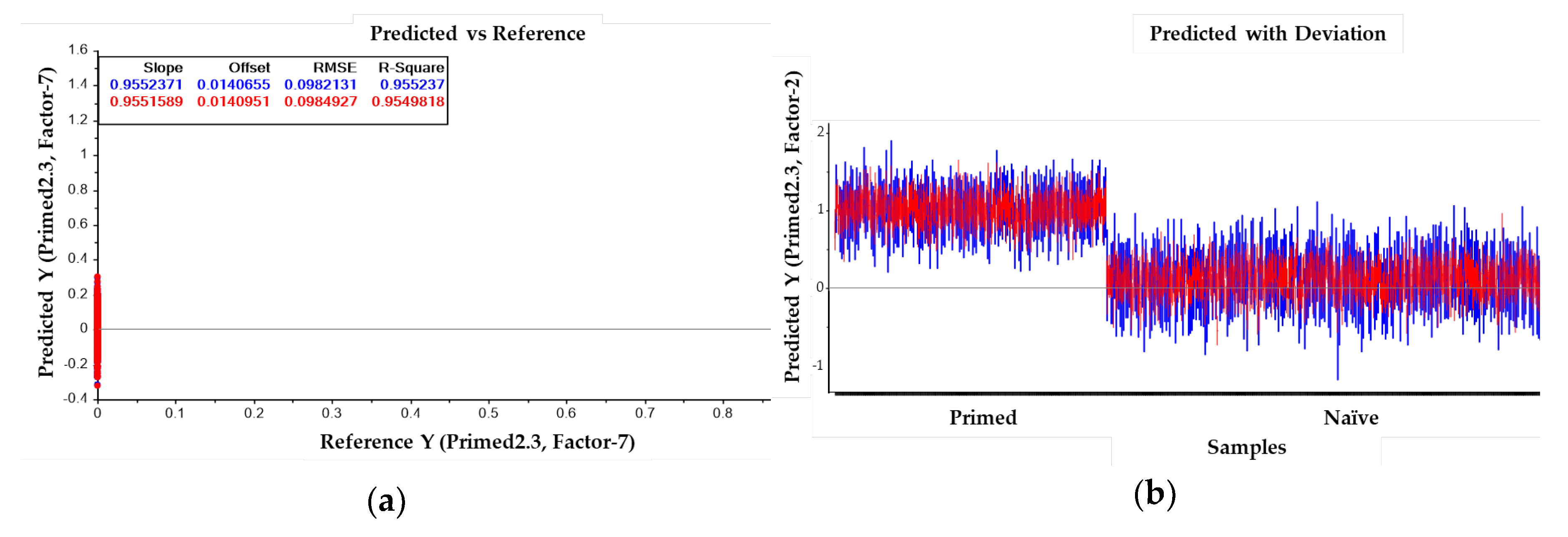
Primed or naïve-like spectra from each group were randomly separated into calibration and validation sets, comprising approximately two-thirds and one-third of spectra, respectively. A total of 1581 spectra from primed cells and 3317 spectra from naïve-like cells were used for the analysis. The calibration data matrix employed for PLS-DA consisted of the spectral dataset (multivariate X) and two Y variables with integer values of 0 or 1 coding for the each of the two modelled spectral classes. Classification of the dataset was then carried out by predicting a Y value for each spectrum in the independent validation using PLS models that had been generated from the calibration sets. Correct classification of each class was arbitrarily assigned to samples with predicted Y > 0.5 for respective spectra. The resulting model demonstrated strong predictive performance, with a correlation coefficient of
R = 0.95 (
Figure 5a, 5b). Classification accuracy reached 100% specificity and 100% sensitivity in identifying primed and naïve-like samples, respectively.
3. Discussion
Our study demonstrates that FTIR microspectroscopy is a robust, label-free, and non-invasive approach for distinguishing primed and naïve-like states in rhesus macaque ESCs. Focal plane array (FPA) detectors use multiple elements that allows for the simultaneous measurement of all data points from each detector element in the spectral interval recorded, with each detector pixel recording independently. Each detector pixel functions as an aperture and records the entire spectrum. Using FPA detector, we identified reproducible spectral signatures that correspond to characteristic differences in protein, lipid, and nucleic acid composition between the two pluripotent states [11, 17].
Naïve-like rhESCs cultured in ALGöX medium exhibited stronger absorbance in the amide I (∼1654 cm
-1) and amide II (∼1546 cm
-1) regions. Prominent peaks at 1240 and 1080 cm
-1, attributed to phosphodiester groups in nucleic acids, suggest increased transcriptional activity and molecular complexity. These biochemical features align with the molecular hallmarks of naïve pluripotency, including greater developmental plasticity and retention of an earlier embryonic identity [
18]. In contrast, primed rhESCs maintained in FGF2/KOSR medium displayed stronger lipid-associated absorbance, including CH₂/CH₃ stretching vibrations (2921-2852 cm
-1) and ester carbonyl peaks (1741 cm
-1). Similar lipid enrichment has been reported in primed human ESCs analyzed by FPA-FTIR microspectroscopy, where high CH₂/CH₃ intensity was linked to membrane remodeling and metabolic adaptations [
14]. Metabolically, naïve PSCs rely predominantly on oxidative phosphorylation, whereas primed PSCs shift toward glycolysis and increased lipid utilization, which may underlie the lipid signatures observed here [19, 20].
Multivariate analyses reinforced these findings: PCA revealed clear separation of primed and naïve-like spectra, while PLS-DA achieved 100% specificity and 100% sensitivity in classifying cell states. These FTIR-based distinctions were fully consistent with immunocytochemistry—showing mutually exclusive expression of OTX2 in primed cells and KLF17, TFCP2L1, ALPPL2, and TFAP2C in naïve-like cells—and with transcriptomic data that confirmed state-specific gene expression patterns [5, 18, 21].
This work represents, to our knowledge, the first FTIR-based biochemical profiling of primed and naïve pluripotent states in a non-human primate model. It establishes FTIR microspectroscopy as a new approach for pluripotent stem cell phenotyping at the single-cell level [11, 22]. A key question is whether this approach can reliably distinguish the naïve pluripotent state induced under diverse culture conditions and in multiple species. The discovery of a conserved and distinctive molecular signature would greatly enhance our ability to identify and validate the naïve state across experimental systems. With developing and validating PLS-DA models based on much higher sample numbers this technique might be further tested and ultimately applied as a practical tool for optimization for identifying stem cells. In our opinion, this study represents the first steps toward achieving this aim. We employed the QUASAR program [
23] for cell classification. The program was selected for its ability to analyze a large number of spectra—specifically, more than 3,000 spectra simultaneously—a significant advantage over the Unscrambler X program, which has limitations in this regard. The results demonstrated that the QUASAR program achieved 99% specificity and 99% sensitivity (Supplementary,
Figure S1), matching the performance of Unscrambler X. Furthermore, the QUASAR Program offers platform flexibility and high processing speed. For future work, we plan to compile a comprehensive database of spectra from various types of stem cells and integrate machine learning for cell classification. This approach is intended to substantially reduce the high costs, complex procedures, and extensive time currently associated with traditional analysis methods.
4. Materials and Methods
4.1. Preparation of feeder cells
Mouse embryonic fibroblasts (MEFs) were isolated from 13.5-day-old OF1 mouse embryos (Charles River) following the protocol previously described [
24]. MEFs were cultured in MEF medium consisting of Dulbecco's modified eagle medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco), 1% Non-essential amino acids (NEAA, Gibco), 1% penicillin-streptomycin-glutamine (PSG, Gibco). Cells were incubated at 37°C in a humidified atmosphere of 5% CO
2 in air. Cells were passaged with trypsin-EDTA (Gibco) and frozen at passage 2 (P2) in medium containing 10% dimethyl sulfoxide (DMSO, Sigma). Frozen-thawed MEFs were cultured to 90% confluence and mitotically inactivated by incubation in fresh MEF medium supplemented with 5 µg/ml mitomycin C (Sigma-Aldrich, M4287) at 37°C for 3 h. Cells were then washed five times with Ca
2+/Mg
2+ free PBS (Gibco), trypsinized, incubated for 5 min at 37°C, centrifuged at 400 x g for 5 min, and resuspended in MEF medium. Mitomycin-treated MEFs (2.5 x 10
5 cells) were plated in 35 mm culture dishes before seeding rhESCs.
4.2. Preparation of conditioned medium (CM)
MEFs (4 x 106) were plated in 100 mm dishes (SPL Life Sciences, Republic of South Korea) coated with 0.1% gelatin. One day after plating, MEF medium was replaced with 25 ml N2B27 basal medium containing 48.7% DMEM/F12 (Gibco), 48.7% Neurobasal medium (Gibco), 1% B27 supplement (Gibco), 0.5% N2 supplement (N2, homemade), 0.02% β-mercaptoethanol (Gibco), 1% PSG, and 20 ng/ml FGF-basic human (bFGF, Sigma). After 24 h, the N2B27 conditioned medium (N2B27-CM) was collected and replaced with an equal volume of fresh complete N2B27 medium. Conditioned medium was collected daily for three consecutive days, pooled, filtered, and stored at -20°C until use.
4.3. Culture and expansion of rhESCs-FGF2/KOSR
rhESCs-FGF2/KOSR cells were cultured on feeders in 35 mm dishes with 2 ml ESC medium containing 80% KO-DMEM (Gibco), 20% knockout serum replacement (KOSR, Gibco), 1% NEAA, 1% GlutaMAX (Gibco), 0.1mM β-mercaptoethanol (Sigma), and 5 µg/ml bFGF (Sigma). Frozen-thawed rhESCs were cultured in medium supplemented with 10 µM ROCK inhibitor (Y-27632) (TOCRIS Bioscience, UK) for the first 24 h. Cultures were maintained at 37°C, 5% CO2 and 5% O2, with daily medium changes. Cells were passaged every 3-4 days by mechanical dissociation.
4.4. Culture and expansion of rhESCs-ALGöX
rhESCs-ALGöX cells were cultured in 35 mm dishes pre-coated with 5 µg/ml laminin (LN521, STEMCELL Technologies, Sweden) for 1 h. Cells were maintained in 2 ml ALGöX medium [
5] consisting of N2B27-CM supplemented with 10 ng/ml Activin A (Peprotech, USA), 1,000 U/ml LIF (homemade), 1.25 μM Gö6983 (TOCRIS Bioscience, UK), and 2.5 μM XAV939. Cultures were maintained at 37°C, 5% CO
2 and 5% O
2, with daily medium changes until 80% confluence. Cells were passaged every 3-4 days using 1X TrypLE (Gibco) for single-cell dissociation, followed by addition of 10 µM Y-27632 for the first 24 h to promote cell survival.
4.5. Immunocytochemistry and Imaging
rhESCs-FGF2/KOSR cells and rhESCs-ALGöX cells were fixed with 4% paraformaldehyde (PFA, Sigma) in PBS for 20 min at room temperature, washed twice with PBS (5 min each), and permeabilized with PBS containing 0.5% Triton X-100. Non-specific binding sites were blocked in PBS with 10% donkey serum for 1 h at room temperature. Cells were incubated overnight at 4°C with primary antibodies (Supplementary,
Table S1). After three washes, cells were incubated with secondary antibodies (Supplementary,
Table S2) for 1 h at room temperature. Nuclei were stained with DAPI (0.5 µg/ml), and cells were mounted with M1289 mounting medium (Sigma). Confocal imaging was performed using a DM 6000 CS SP5 microscope (Leica) with a 45×/1.25 oil immersion objective (PL APO HCX, Leica) [
18].
4.6. RNA sequencing
Total RNA was extracted from 4-5 x 10
6 cells using the RNeasy mini-kit (Qiagen). Libraries were prepared from 200 ng of RNA using the NextFlex Rapid Directional mRNA-Seq kit (Bio-Scientific), and sequenced on a NextSeq500 platform (Illumina) as single-end 75 bp reads. Demultiplexing was performed with bcl2fastq (Illumina), and adapters were trimmed using Cutadapt. Sequencing depth was ~30 million reads per sample. Reads were aligned to the reference genome using HISAT2, and gene counts were generated with HTSeq. RNA-seq data from this study are available under GEO accession number
GSE146178 [20].
4.7. FPA-FTIR microspectroscopy
Cell pellets were washed three times with 0.9% NaCl, resuspended in 50 µl saline, and 5 µl aliquots were deposited onto IR-transparent 2-mm-thick barium fluoride (BaF
2) windows. Samples were air-dried and stored in a desiccator until analysis. Spectral data was acquired from an FPA-FTIR microscope (Hyperion 3000) with a FPA detector, connected to Tensor 27 FTIR spectrometer (Bruker Optics, Ettlingen, Germany) at the Synchrotron Light Research Institute (Public Organization), Nakhon Ratchasima, Thailand. The acquisition parameters were a 36X objective in transmission mode, 64 scans and all data was measured with 8 × 8 binning at a spectral resolution of 6 cm
−1 [
25]. The full area of 64 × 64 pixels equal 4,096 pixels. The field of view area using a 36X objective lens was 70.4 × 70.4 μm
2. The biochemical composition distribution was performed by OPUS 7.5 software (Bruker Optics, Ettlingen, Germany).
4.8. Multivariate data analysis of FTIR spectra
Spectral quality control was performed by visual inspection; spectra with weak absorbance (amide I band maximum absorbance <0.2), acquired from regions of the sample where there were no cells, or spectra with very high absorbance (amide I band maximum absorbance >0.8), acquired from regions where cells may have been clumped or overlaid) was reject from the analysis. Spectra from sample groups based on differentiation of cell states and passage cells were analyze using PCA. Preprocessing of the data was conduct by first performing a second derivative by the Savitzky - Golay method (13 smoothing points) and then normalized using EMSC using the spectral regions from 3000 to 2800 cm-1 and from 1800 to 800 cm-1 using The Unscrambler X 10.3 software (CAMO, Oslo, Norway). Score plots was use to visualize any clustering of the data, and the loading plot was used to determine which spectral regions contributed most to the variance in the data set, accounting for any clustering of spectra seen in scores plots. Integrate peak areas, was analyze by OPUS 7.5 software. Spectra were analyze using PLS-DA by The Unscrambler X 10.3 software. PLS-DA employed PCA models derived from calibration sets and was used to test the ability to discriminate different cell states using the independent validation set spectra. Calibration and validation of spectral data was employ using data sets that was randomly select comprising two-thirds and one-third of the spectra, respectively. The dataset can be utilized to calculate the percentages of specificity and sensitivity. FTIR microspectroscopy combined with multivariate data analysis, in particular PCA, was apply to explain biochemical changes occurring during cellular differentiation.
4.9. Statistical analysis
All statistical analyses were performed using GraphPad Prism version 5 (GraphPad Software, San Diego, CA, USA). Data are presented as mean ± standard deviation (SD). Differences among groups were analyzed using one-way analysis of variance (ANOVA), followed by Tukey-Kramer’s Honest Significant Difference (HSD) post hoc test for pairwise comparisons. A p-value of less than 0.05 was considered statistically significant. Different letters (a, b, c, d, e) above the bars indicate a statistically significant difference (p<0.05) between the groups. Identical letters indicate no significant difference. Graphs were generated using Sigma Plot version 15 (Grafiti LLC, Palo Alto, California, USA).
5. Conclusions
This work represents the first application of FTIR microspectroscopy to characterize and distinguish primed from naïve-like rhESCs. The FTIR-based findings were validated against two established methods—immunocytochemistry and RNA sequencing—which confirmed that naïve-like cells possess higher protein content, while primed cells are enriched in lipids. The three analytical approaches were complementary: immunocytochemistry localized proteins, RNA sequencing provided gene-level resolution, and FTIR offered a rapid, cost-effective overview of cellular biochemical composition.
Supplementary Materials
The following supporting information can be downloaded at:
https://www.mdpi.com/article/doi/s1, Figure S1: QUASAR: Machine Learning for cell classification.; Table S1: Primary antibodies used for Immunocytochemistry; Table S2: Secondary antibodies used for Immunocytochemistry.
Author Contributions
Conceptualization, J.S., I.A., K.T. and R.P.; methodology, J.S., A.A. and C.R.; software, J.S.; validation, I.A., K.T. and R.P.; formal analysis, J.S.; investigation, I.A., K.T. and R.P.; resources, R.P. and P.S; data curation, J.S.; writing original draft preparation, J.S., I.A. and K.T.; writing review and editing, J.S., W.S., P.A., I.A., P.S., K.T. and R.P.; visualization, J.S; supervision, I.A., P.S., K.T. and R.P.; project administration, J.S.; funding acquisition, R.P. and P.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by: National Science, Research and Innovation Fund (NSRF) through the Program Management Unit for Human Resources & Institutional Development, Research and Innovation (PMU-B), Government of Thailand, Grant Number: B16F640104; Suranaree University of Technology (SUT); Franco-Thai Mobility Programme (PHC SIAM) Year 2023-2024 to Rangsun Parnpai and Irene Aksoy; and Fondation pour la Recherche Medicale (EQU202303016295) to Pierre Savatier. Worawalan Samruan was supported by SUT full-time postdoctoral researchers (Full-time 66/26/2568); Preeyanan Anwised was supported by SUT full- time postdoctoral re-searchers (Full-time 66/17/2568); Jittanun Srisutush received One Research One Graduate (OROG) fellowship of SUT (53/2565).
Institutional Review Board Statement
The animal study protocol was approved by Ethics Committee of Institutional Animal Care and Use Committee, Suranaree University of Technology (protocol code IACUC-68-12).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
This research was supported by Suranaree University of Technology (SUT). We would like to thank Dr. Supatcharee Cael from Research Department Spectroscopy and Imaging, Leibniz Institute of Photonic Technology (Leibniz IPHT), Germany and Dr. Kanokwan Kamkajon from Synchrotron Light Research Institute (Public Organization), Nakhon Ratchasima, Thailand, for they valuable support in instructions for FTIR analysis. Special thanks also go to Miss. Chunmanus Uthaisar from Food and Agriculture Research Section (FRS), Synchrotron Research and Applications Division (SRD), Synchrotron Light Research Institute (Public Organization), Nakhon Ratchasima, Thailand, for her valuable support in set up FTIR microspectroscopy machine for analysis. During the preparation of this manuscript, the authors used ChatGPT, GPT-4o for English editing. The authors have reviewed and edited the output and take full responsibility for the content of this publication.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| PSCs |
Pluripotent Stem Cells |
| ESCs |
Embryonic Stem Cells |
| EpiSCs |
Epiblast Stem Cells |
| NHPs |
Non-Human Primates |
| FTIR |
Fourier-Transform Infrared |
| rhESCs |
Rhesus Macaque Embryonic Stem Cells |
| RNA-seq |
RNA sequencing |
| FPA |
Focal Plane Array |
| PCA |
Principal Component Analysis |
| EMSC |
Extended Multiplicative Signal Correction |
| PLS-DA |
Partial Least Squares-Discriminant Analysis |
| MEFs |
Mouse embryonic fibroblasts |
| CM |
Conditioned Medium |
| P |
Passage |
| BaF2
|
Barium Fluoride |
| C=O stretching |
Carbonyl Stretching |
| C-N stretching |
Carbon-Nitrogen Stretching |
| α-helices |
Alpha Helix |
| β-sheets |
Beta-Pleated Sheet |
| CH stretching |
Carbon-Hydrogen Stretching |
| CM-1
|
Reciprocal centimeter |
| SVM |
Support Vector Machine |
References
- Evans, M. J.; Kaufman, M. H. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981, 292, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Nichols, J.; Smith, A. Naïve and primed pluripotent states. Cell stem cell 2009, 4, 487–492. [Google Scholar] [CrossRef]
- Anwised, P.; Moorawong, R.; Samruan, W.; Somredngan, S.; Srisutush, J.; Laowtammathron, C.; Aksoy, I.; Parnpai, R.; Savatier, P. An expedition in the jungle of pluripotent stem cells of non-human primates. Stem Cell Reports 2023, 18, 2016–2037. [Google Scholar] [CrossRef] [PubMed]
- Weinberger, L.; Ayyash, M.; Novershtern, N.; Hanna, J. H. Dynamic stem cell states: naïve to primed pluripotency in rodents and humans. Nature reviews Molecular cell biology 2016, 17, 155–169. [Google Scholar] [CrossRef]
- Pham, H. T.; Perold, F.; Pijoff, Y.; Doerflinger, N.; Rival-Gervier, S.; Givelet, M.; Moulin, A.; Ressaire, M.; Da Silva Fernandes, E.; Bidault, V.; Jouneau, L.; Duranthon, V.; Wianny, F.; Pain, B.; Plotton, I.; Joly, T.; Afanassieff, M.; Savatier, P.; Beaujean, N. Efficient generation of germline chimeras in a non-rodent species using rabbit induced pluripotent stem cells. Nature Communications 2025, 16, 5165. [Google Scholar] [CrossRef]
- Li, J.; Zhu, Q.; Cao, J.; Liu, Y.; Lu, Y.; Sun, Y.; Li, Q.; Huang, Y.; Shang, S.; Bian, X.; Li, C.; Zhang, L.; Wang, Y.; Nie, Y.; Fu, J.; Li, W.; Mazid, M. A.; Jiang, Y.; Jia, W.; Wang, X.; Sun, Y.; Esteban, M. A.; Sun, Q.; Zhou, F.; Liu, Z. Cynomolgus monkey embryo model captures gastrulation and early pregnancy. Cell Stem Cell 2023, 30, 362–377. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: a revolutionary tool for transcriptomics. Nature Reviews Genetics 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Cregger, M.; Berger, A. J.; Rimm, D.L. Immunohistochemistry and Quantitative Analysis of Protein Expression. Arch Pathol Lab 2006, 130, 1026–1030. [Google Scholar] [CrossRef]
- Cao, J.; Ng, E. S.; McNaughton, D.; Stanley, E. G.; Elefanty, A. G.; Tobin, M. J.; Heraud, P. ; The characterisation of pluripotent and multipotent stem cells using Fourier transform infrared microspectroscopy. Int J Mol Sci 2013, 14, 17453–76. [Google Scholar] [CrossRef]
- Ami, D.; Neri, T.; Natalello, A.; Mereghetti, P.; Doglia, S. M.; Zanoni, M.; Zuccotti, M.; Garagna, S.; Redi, C. A. Embryonic stem cell differentiation studied by FT-IR spectroscopy. Biochimica et biophysica acta (BBA)-molecular cell research 2008, 1783, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Bassan, P.; Byrne, H. J.; Bonnier, F.; Lee, J.; Dumas, P.; Gardner, P. Resonant Mie scattering in infrared spectroscopy of biological materials–understanding the ‘dispersion artefact’. Analyst 2009, 134, 1586–1593. [Google Scholar] [CrossRef] [PubMed]
- Thumanu, K.; Tanthanuch, W.; Ye, D.; Sangmalee, A.; Lorthongpanich, C.; Parnpai, R.; Lorthongpanich, C.; Heraud, P. Spectroscopic signature of mouse embryonic stem cell–derived hepatocytes using synchrotron Fourier transform infrared microspectroscopy. Journal of Biomedical Optics 2011, 16, 057005–057005. [Google Scholar] [CrossRef] [PubMed]
- Wianny, F.; Bernat, A.; Huissoud, C.; Marcy, G.; Markossian, S.; Cortay, V.; Giroud, P.; Leviel, V.; Kennedy, H.; Savatier, P.; Dehay, C. Derivation and cloning of a novel rhesus embryonic stem cell line stably expressing tau-green fluorescent protein. Stem Cells 2008, 26, 1444–1453. [Google Scholar] [CrossRef]
- Dunkhunthod, B.; Thumanu, K.; Eumkeb, G. Application of FTIR microspectroscopy for monitoring and discrimination of the anti-adipogenesis activity of baicalein in 3T3-L1 adipocytes. Vibrational Spectroscopy 2017, 89, 92–101. [Google Scholar] [CrossRef]
- Cao, J.; Ng, E. S.; McNaughton, D.; Stanley, E. G.; Elefanty, A. G.; Tobin, M. J.; Heraud, P. Fourier transform infrared microspectroscopy reveals unique phenotypes for human embryonic and induced pluripotent stem cell lines and their progeny. Journal of Biophotonics 2014, 7, 767–781. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Rehman, S.; ur Rehman, D. I. Fourier transform infrared (FTIR) spectroscopy of biological tissues. Applied Spectroscopy Reviews 2008, 43, 134–179. [Google Scholar] [CrossRef]
- Whelan, D. R.; Bambery, K. R.; Heraud, P.; Tobin, M. J.; Diem, M.; McNaughton, D.; Wood, B. R. Monitoring the reversible B to A-like transition of DNA in eukaryotic cells using Fourier transform infrared spectroscopy. Nucleic acids research 2011, 39, 5439–5448. [Google Scholar] [CrossRef]
- Aksoy, I.; Rognard, C.; Moulin, A.; Marcy, G.; Masfaraud, E.; Wianny, F.; Cortay, V.; Ménard, A. B.; Doerflinger, N.; Dirheimer, M.; Mayère, C.; Bourillot, P. Y.; Lynch, C.; Raineteau, O.; Joly, T.; Dehay, C.; Serrano, M.; Afanassieff, M.; Savatier, P. Apoptosis, G1 phase stall, and premature differentiation account for low chimeric competence of human and rhesus monkey naïve pluripotent stem cells. Stem cell reports 2021, 16, 56–74. [Google Scholar] [CrossRef]
- Cornacchia, D.; Zhang, C.; Zimmer, B.; Chung, S.Y.; Fan, Y.; Soliman, M.A.; Tchieu, J.; Chambers, S.M.; Shah, H.; Paull, D.; Konrad, C.; Vincendeau, M.; Noggle, S.A.; Manfredi, G.; Finley, L.W.S.; Cross, J.R.; Betel, D.; Studer, L. Lipid Deprivation Induces a Stable, Naïve-to-Primed Intermediate State of Pluripotency in Human PSCs. Cell stem cell 2019, 25, 120–136. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, M.; Marashi, S.A.; Sharifi, Z.A.; Taleahmad, S. The metabolic network model of primed/naïve human embryonic stem cells underlines the importance of oxidation-reduction potential and tryptophan metabolism in primed pluripotency. Cell Biosci 2019, 9, 71. [Google Scholar] [CrossRef]
- Amzal, A.; Pijoff, Y.; Rognard, C.; Marcy, G.; Doerflinger, N.; Bréhier, C.; Anwised, P.; Srisutush, J.; Chimngam, M.; Chatdarong, K.; Cazotte, E.; Moyret, C.; Kuorome, M.; Cortay, V.; Dehay, C.; Afanassieff, M.; Joly, T.; Rougeulle, C.; Gillet, G.; Wolf, E.; Parnpai, R.; Savatier, P.; Aksoy, I. MEK and AKT signaling Determine the Potential of Pluripotent stem Cells to Colonize Heterologous Preimplantation Embryos. Nature 2025. In press. [Google Scholar]
- Heraud, P.; Ng, E. S.; Caine, S.; Yu, Q. C.; Hirst, C.; Mayberry, R.; Bruce, A.; Wood, B. R.; McNaughton, D.; Stanley, E. G.; Elefanty, A. G. Fourier transform infrared microspectroscopy identifies early lineage commitment in differentiating human embryonic stem cells. Stem Cell Research 2010, 4, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Toplak, M.; Read, S. T.; Sandt, C.; Borondics, F. Quasar: easy machine learning for biospectroscopy. Cells 2021, 10, 2300. [Google Scholar] [CrossRef] [PubMed]
- Afanassieff, M.; Tapponnier, Y.; Savatier, P. Generation of Induced Pluripotent Stem Cells in Rabbits. Induced Pluripotent Stem (iPS) Cells. Methods and Protocols, 2nd ed.; Turksen, K., Nagy, A., Eds.; Humana Press: New York, USA, 2014; Volume 1357, pp. 149–172. [Google Scholar] [CrossRef]
- Heraud, P.; Caine, S.; Sanson, G.; Gleadow, R.; Wood, B. R.; McNaughton, D. Focal plane array infrared imaging: a new way to analyse leaf tissue. New Phytologist 2007, 173, 216–225. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).