1. Introduction
Arbas cashmere, a key raw material for down products, is renowned for its warmth, lightness, and bright luster [
1]. The Arbas cashmere goat, a unique breed from Inner Mongolia, has long been globally recognized for its advantages in terms of cashmere yield, fineness, and length [
2]. The goat’s hair follicles, an appendage of the skin, play a crucial role in determining cashmere quality. Cashmere goat hair follicles are categorized into primary (hair-producing) and secondary follicles (cashmere-producing) [
3]. Research on secondary follicles is essential for improving the quality of cashmere. The development of secondary follicles comprises a typical periodic cycle with seasonal variations: anagen (April to November), catagen (December to January), and telogen (February to March) [
4,
5]. Anagen phase directly influences cashmere yield. Previous studies have shown that secondary hair follicle stem cells (SHFSCs), located in the bulge region adjacent to the arrector pili muscle at the base of the follicles, are critical for follicle growth and development. SHFSCs maintain their stemness and regulate follicle growth cycles through interactions with the surrounding microenvironment, including the dermal papilla (DP) cells, the extracellular matrix, and various signaling molecules [
6,
7,
8].
At cellular level, SHFSCs play a key role in follicular development. During the anagen phase, DP cells secrete key signals, including Wnt ligands. Activation of Wnt signaling pathway stimulates SHFSC activity, promotes proliferation, and sustains stemness. For example, WNT10b induces anagen [
9,
10], driving SHFSCs proliferation, differentiation, and initiating follicle cycle progression [
11]. During catagen phase, bone morphogenetic protein (BMP) signaling inhibits SHFSCs activation, regulating the cells into a quiescent state to prevent premature differentiation and maintain the stem cell pool, with BMP4 and BMP6 playing important roles [
10,
12,
13]. The JAK/STAT pathway modulates stem cell behavior under specific conditions, while the PI3K/AKT pathway promotes SHFSCs growth and differentiation and inhibits apoptosis, balancing quiescence and activation [
14,
15,
16]. TGF-β signaling participates in regulating inflammation and cellular responses during follicle regeneration [
17,
18,
19]. Additionally, the transcription factor RUNX1 enhances SHFSC sensitivity to activation signals, promoting the transition from telogen to anagen, while the vascular system interacts with hair stem cells to maintain follicle cycle homeostasis [
20,
21,
22]. Despite systematic studies on the regulatory mechanisms underlying SHFSC behavior, the complex follicle regulatory network requires further exploration to develop effective modulation strategies.
Quercetin, a secondary metabolite abundant in onions, grapes, and tea, exhibits antioxidant, anti-inflammatory, and anticancer properties [
23]. It enhances antioxidant capacities by scavenging free radicals and increasing the activities of enzymes such as superoxide dismutase (SOD) and glutathione peroxidase (GPx), while inhibiting the NF-κB pro-inflammatory pathway [
24]. Recent studies have indicated that quercetin can enhance chemotherapy efficacy by regulating cellular signaling pathways [
25,
26,
27], showing promise for cancer treatment. However, its application is limited due to its poor bioavailability [
28]. Notably, in hair regulation, quercetin influences follicle development through multiple pathways. It enhances DP cell viability by increasing NAD(P)H production and mitochondrial membrane potential, thereby optimizing cellular energy metabolism [
29]. Quercetin also prolongs anagen phase by upregulating the anti-apoptotic protein Bcl-2 and the proliferation gene
KI67 to regulate follicle cycle [
30]. In addition, it promotes the transcription and synthesis of growth factors such as bFGF, KGF, and VEGF, which are critical for maintaining follicle activity and periodic turnover. Mechanistically, quercetin activates the MAPK/CREB signaling pathway, inducing the phosphorylation of ERK, AKT, and CREB to regulate follicle-associated biological processes [
31]. Recent clinical studies have shown that quercetin stimulates the proliferation of keratinocytes in quiescent follicles and enhances dermal vascularization via HIF-1α activation, providing nutrients to follicles [
32].
Although the effects of quercetin on follicle development have been studied, its role in regulating follicle development through SHFSCs in cashmere goats, particularly in relation to improving cashmere yield, remains poorly understood. This study investigated the effects of quercetin in cashmere goat SHFSCs, filling research gaps and aiming to enhance cashmere production.
2. Materials and Methods
2.1. Sample Collection and Cell Culture
Three healthy adult cashmere goats were selected from the Yiwei White Goat Farm in Ordos, Inner Mongolia, China. All animals were raised under standard conditions. Skin samples (1 cm²) were collected from the back of the goats when the hair follicle cycle entered the anagen phase in September. The collected skin samples were immediately frozen in liquid nitrogen for RNA and protein extraction, RNA sequencing, and subsequent analyses.
The culture system for secondary hair follicle stem cells (SHFSCs) of Arbas cashmere goats consisted of DMEM/F-12 medium (Biological Industries, Kibbutz Beit Haemek, Israel) supplemented with 4% fetal bovine serum, 14 ng/mL epidermal growth factor (PeproTech, Rocky Hill, NJ, USA), 0.4 ng/mL hydrocortisone (Monmouth Junction, NJ, USA), and 0.5 pg/mL ITS-X (Gibco BRL, Grand Island, NY, USA). Cultures were maintained at 37 °C with 5% CO₂. The medium was changed every other day. Subsequent experiments were performed when the cells reached 70-80% confluence.
2.2. RNA Extraction and qRT-PCR Detection
Total RNA was isolated from skin samples utilizing RNAiso reagent (Takara Bio Inc., Shiga, Japan) as per the manufacturer’s protocol. The purity and concentration of RNA were determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). cDNA was synthesized using the PrimeScript FAST RT Reagent Kit with gDNA Eraser (Takara Bio Inc., Shiga, Japan). qRT-PCR was conducted using TB Green® Premix Ex Taq™ II (Takara Bio Inc., Shiga, Japan) on a CFX96 real-time PCR system (Bio-Rad Laboratories, Hercules, CA, USA). GAPDH was used as the internal reference, and the relative expression level was calculated by the 2⁻ΔΔCt method. The primer sequences are listed in Supplementary S1.docx.
2.3. Protein Extraction and Western Blot Analysis
Total protein was extracted from cell samples using a mammalian protein extraction kit (CWBIO, Beijing, China). The protein concentration was determined using a bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific, Waltham, MA, USA). Equal amounts of protein (20 μg) were loaded into each well, separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and transferred to polyvinylidene difluoride membranes. The membranes were blocked with 5% skimmed milk for 1 hour at 37 °C and then incubated with primary antibodies overnight at 4 °C. The next day, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 hour, and visualization was performed using a Tanon 5200 imaging system (Tanon, Shanghai, China). ImageJ software was used to quantify the band intensity for statistical comparison between groups. Detailed information of the antibodies used is shown in Supplementary S1.docx.
2.4. Flow Cytometry Analysis of the Cell Cycle and Cell apoptosis
Cell cycle distribution was analyzed using a cell cycle detection kit. Secondary hair follicle stem cells were cultured in 6-well plates for 48 hours. Subsequently, the cell culture medium was collected into a centrifuge tube. Adherent cells were digested with trypsin for 5 minutes, and the collected medium was added. The cells were centrifuged for 5 minutes at 1500 rcf/min and the supernatant was aspirated. Afterward, the cells were resuspended in 1 mL pre-cooled PBS and centrifuged again for 5 minutes at 1500 rcf/min. The cells were then incubated with 1 mL pre-cooled 70% ethanol for 12 hours at 4 °C, resuspended in 500 μL of propidium iodide (PI) staining buffer (Beyotime, Shanghai, China), and incubated for 30 minutes at 37 °C in the dark. The cell suspension was analyzed using a flow cytometer (FACSAria SORP, BD BioSciences, NJ, USA). Each group consisted of three independent replicates.
Cell apoptosis rate was evaluated by Annexin V-FITC/PI staining. After 48 hours of cell treatment, cells in different treatment groups were washed at least three times with 1 mL of 1× PBS (pH 7.4) and digested with trypsin. They were then collected in 15 mL centrifuge tubes, washed again with 1 mL of 1× PBS, and resuspended in 1× binding buffer (Solarbio, Beijing, China). The cells were incubated with 5 μL of Annexin V-FITC and 10 μL of PI (Solarbio, Beijing, China) for 10 minutes at 37 °C in the dark. They were then analyzed by flow cytometry (FACSAria SORP, BD BioSciences, NJ, USA).
2.5. Cell Counting Kit-8 (CCK-8) assay
Cells were seeded in 96-well plates with 100 μL of medium per well. Each treatment group had 9 independent replicates. After incubation for 48 hours at 37 °C with 5% CO₂, 10 μL of CCK-8 reagent (Biosharp, Jiangsu, China) was added to each well and incubation was continued for 4 hours. Absorbance was measured at 450 nm using a microplate reader (BioTek, Winooski, VT, USA).
2.6. EdU Detection
Detection of 5-ethynyl-2'-deoxyuridine (EdU) was performed according to the instructions of EdU Cell Proliferation Kit (RiboBio, Guangzhou, China). Before immunostaining, secondary hair follicle stem cells were first incubated with serum-free medium containing 10 μM EdU reagent for 24 hours. Each group consisted of three independent replicates. Fluorescence images were captured using a Leica fluorescence microscope (Leica, Wetzlar, Germany). ImageJ software was used to analyze the cell positivity rate to evaluate cell proliferation ability.
2.7. Transcriptome Sequencing and Bioinformatics Analysis
Total RNA was extracted using TRIzol reagent, following the product’s instructions. RNA purity and quantity were measured using a NanoDrop 2000 spectrophotometer (Thermo Scientific, USA). RNA integrity was evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The transcriptome library was constructed using the VAHTS Universal V6 RNA-seq Library Prep Kit, according to the manufacturer’s instructions. Transcriptome sequencing and analysis were performed by Shanghai OE Biotech Co., Ltd. (Shanghai, China).
The library was sequenced on an Novaseq 6000 sequencing platform (Illumina, Inc., San Diego, CA, USA), generating 150 bp paired-end reads. The fastp software was used to process the raw reads in fastq format. Clean reads for subsequent data analysis were obtained after removing low-quality reads. HISAT2 [
2] software was used to align the reads with the reference genome and to quantify gene expression. PCA analysis and plotting were performed on genes (counts) to evaluate the biological replicates of samples. DESeq2 software was used for differential expression gene analysis, where genes with q value < 0.05 or log|fold change| > 2 were defined as differentially expressed genes (DEGs). R (4.5.0) was used for analysis of DEGs, whereas ggplot2 tool was used for visualization. Enrichment analysis was performed using the tools on the GO and KEGG websites.
2.8. Statistical Analysis
All data were expressed as mean ± standard deviation. T-test was used for comparison between two groups. A p value < 0.05 was considered statistically significant.
3. Results
3.1. In Vitro Culture and Identification of Arbas Cashmere Goat SHFSCs
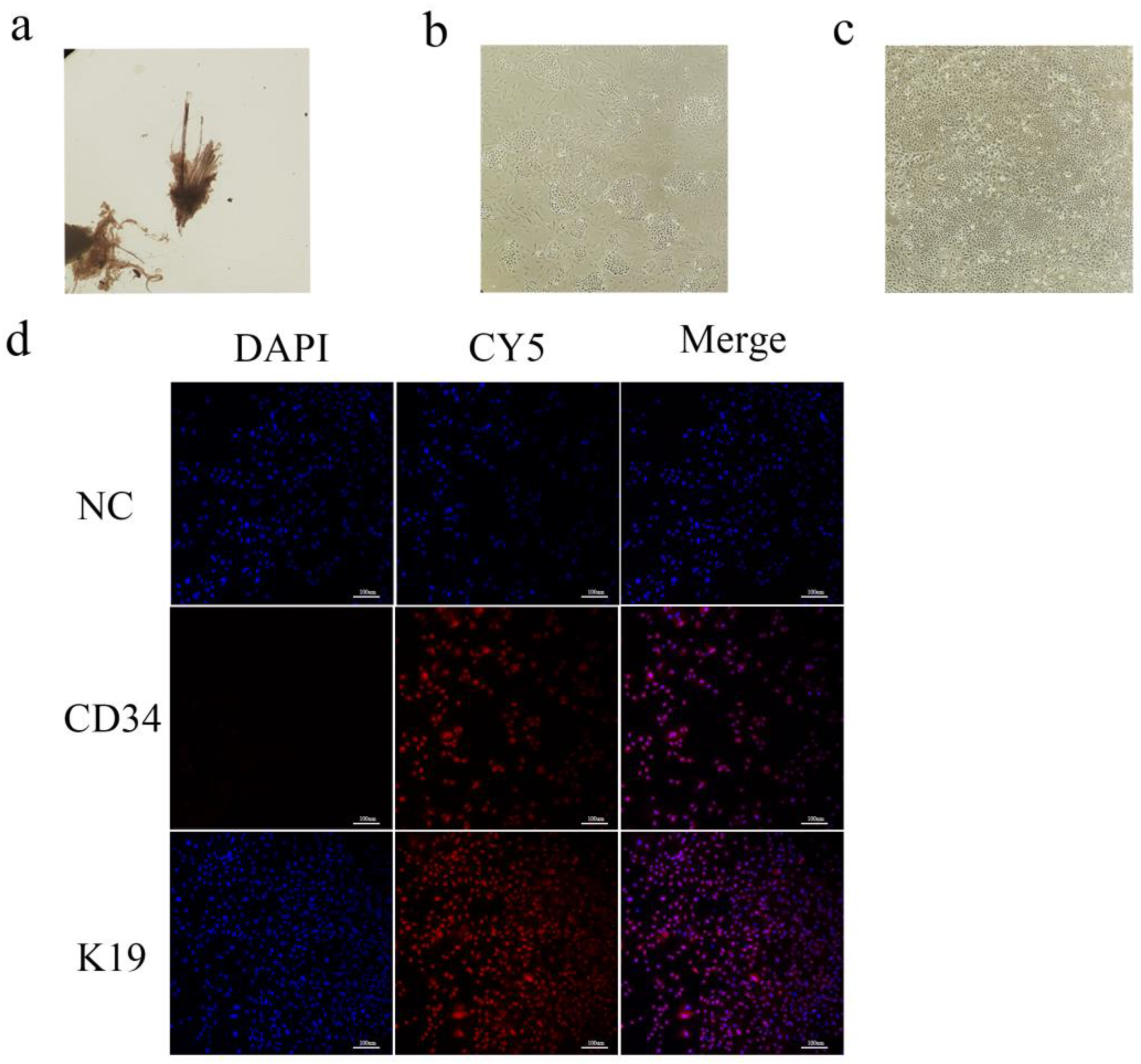
Dorsal skin samples were collected from Arbas cashmere goats during the anagen phase. Secondary follicles were isolated under a microscope (
Figure 1a) and adherent cells were obtained after primary culture (
Figure 1b). Primary and secondary follicle cells were digested with type IV collagen [
33] to obtain purified SHFSCs (
Figure 1c). To confirm their identity, immunofluorescence staining for the SHFSC surface markers CD34 and K19 was performed, with positive signals shown in red (
Figure 1d). The purified SHFSCs exhibited a cobblestone-like morphology, small size, rapid division, and high viability.
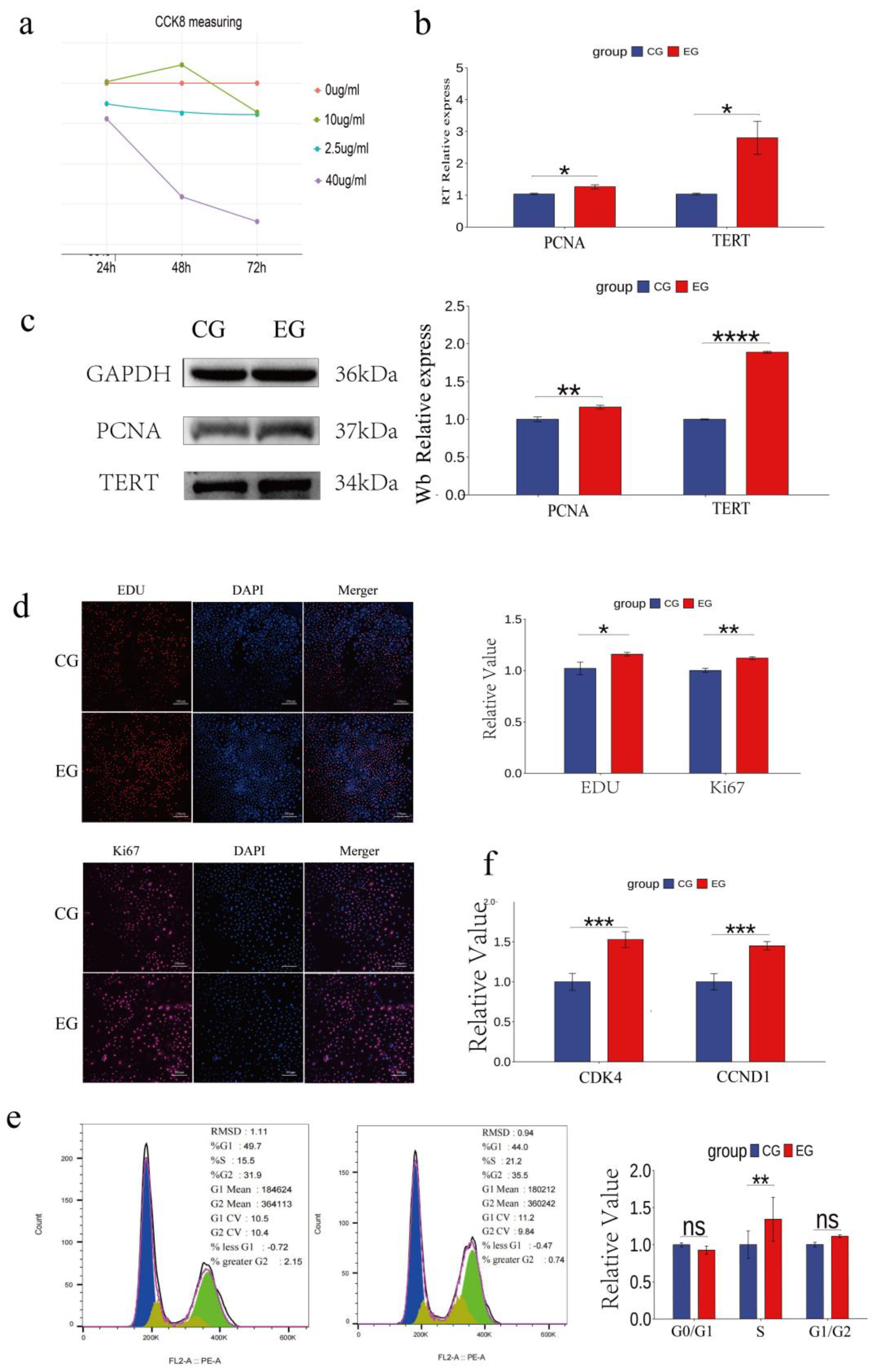
3.2. Quercetin Promotes the Proliferation of Arbas Cashmere Goat SHFSCs
Quercetin treatment at certain durations and concentrations of promoted follicle stem cell proliferation. The optimal concentration and treatment time were determined by testing different quercetin concentrations (0, 2.5, 10, and 40 μg/mL) and durations (24 h, 48 h, and 72 h). Maximum cell viability was observed after treatment with 10 μg/mL quercetin for 48 h (
Figure 2a).
Subsequent experiments used 10 μg/mL quercetin for 48 h (experimental group: EG; control group: CG). RT-qPCR analyses showed significantly increased mRNA expression levels of the proliferation markers, proliferating cell nuclear antigen (PCNA) and telomerase reverse transcriptase (TERT), in the EG compared to that in the CG (
Figure 2b). This observation was consistent with the elevated protein levels of PCNA and TERT (
Figure 2c). EdU assays revealed significantly more EdU-positive cells in the EG than in CG and confocal microscopy showed the upregulated expression of the proliferation marker KI67, further confirming enhanced proliferation (
Figure 2d). Cell cycle analyses indicated a significant increase in the number of S-phase cells in the EG (from 15.5% to 21.2%;
Figure 2e). RT-qPCR analyses also showed the upregulated expression of the cell cycle-related genes
CDK4 and
CCND1 in the EG (
Figure 2f). Collectively, these results demonstrate that quercetin promotes the proliferation of goat skin follicle stem cells.
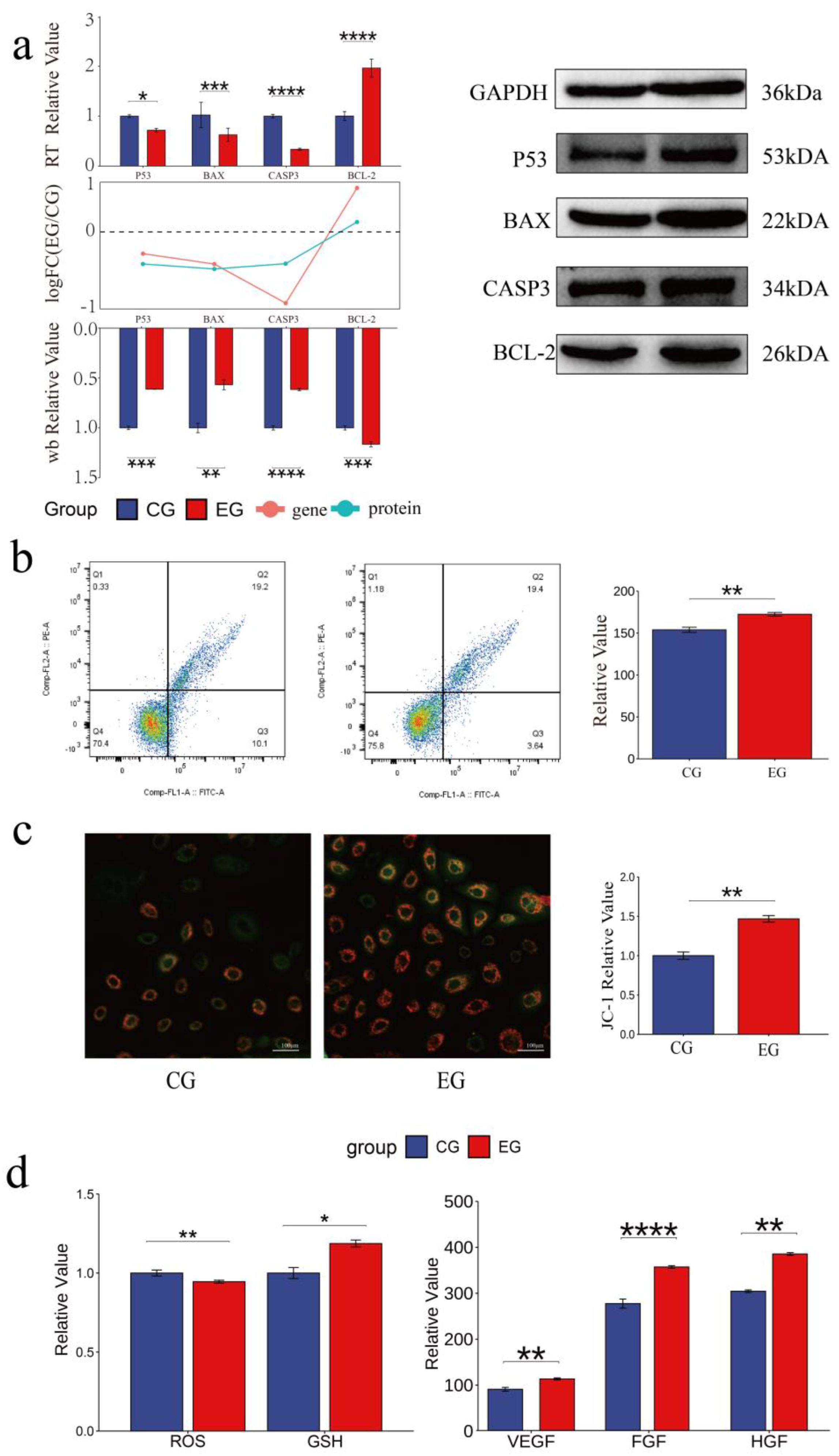
3.3. Quercetin Inhibits the Apoptosis and Modulates the Physiological Status of Arbas Cashmere Goat SHFSCs
The role of quercetin in the apoptosis of goat secondary skin follicle stem cells was further investigated. The mRNA levels of the pro-apoptotic genes
BAX,
TP53, and
CASP3 were downregulated in quercetin-treated cells, whereas the expression of the anti-apoptotic gene
BCL-
2 was upregulated. Consistently, the protein levels of BAX, p53, and
CASP3 were downregulated and that of BCL-2 was upregulated in the EG. The RNA and protein expression trends are shown as bar and line graphs in
Figure 3a. Annexin V-FITC/PI staining further showed that quercetin inhibited apoptosis and significantly reduced the proportion of late apoptotic cells (
Figure 3b), confirming its anti-apoptotic effects.
The effects of quercetin on physiological status of SHFSCs were also assessed. JC-1 staining showed increased mitochondrial membrane potential in the EG compared to that in the CG (
Figure 3c), indicating improved energy metabolism. Additionally, quercetin reduced reactive oxygen species (ROS) levels, increased the glutathione (GSH) content, and promoted the secretion of the growth factors VEGF, FGF, and HGF. These results demonstrate that quercetin enhances antioxidant capacity, sustains energy metabolism, and promotes growth factor secretion, thereby supporting SHFSC proliferation.
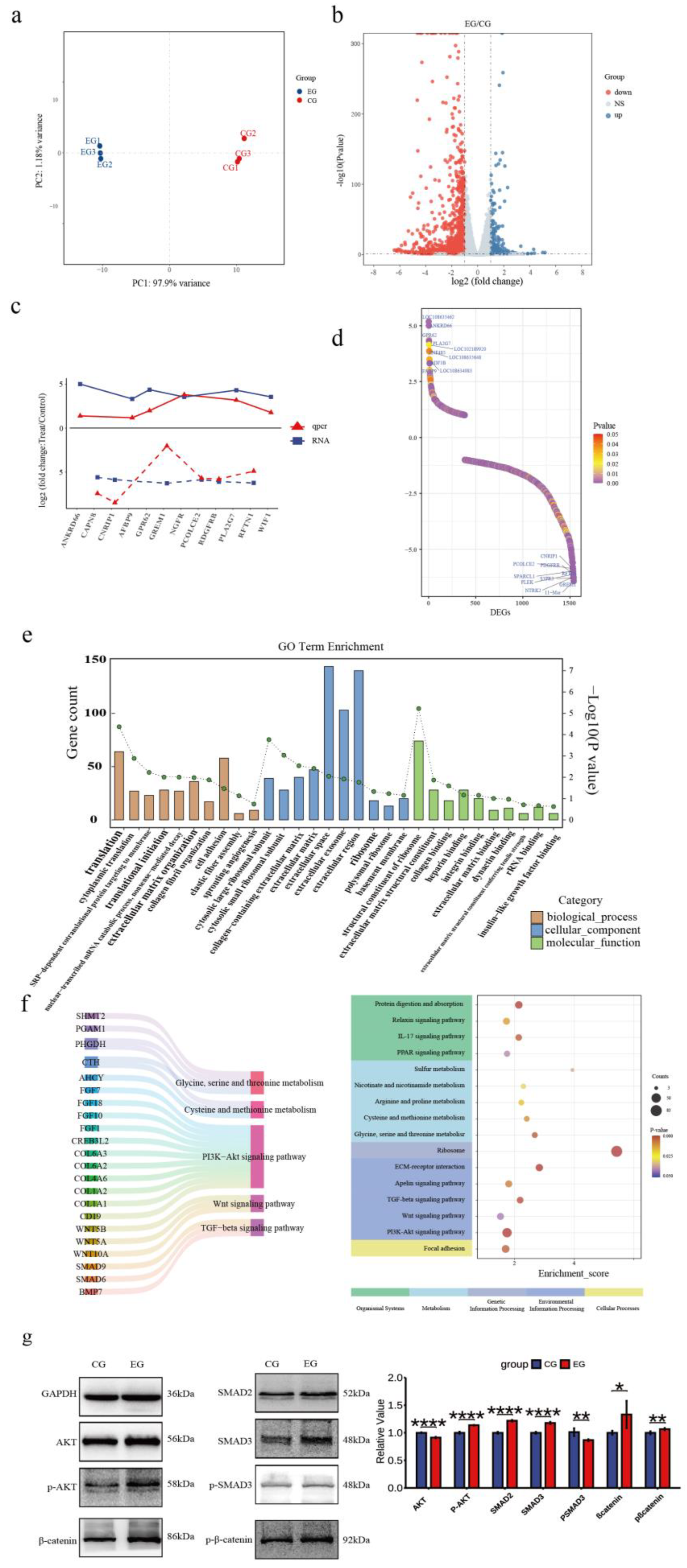
3.4. Quercetin Alters the Transcriptome of Arbas Cashmere Goat SHFSCs
To investigate quercetin’s effects on the SHFSC transcriptome, high-throughput RNA sequencing was performed on SHFSCs from EG and CG. After quality control and sequence alignment, 16,751 expressed genes were identified. A principal component analysis (PCA) distinguished gene expression profiles between the EG and CG, showing high intragroup reproducibility and confirming experimental consistency and reliability (
Figure 4a).
The DEGs were filtered using the following criteria: |logFC|≥1 and
P<0.05, identifying 1,540 DEGs in the EG vs. CG, including 1,157 upregulated and 383 downregulated genes (
Figure 4b). To validate the accuracy of the RNA-seq, six upregulated and six downregulated genes were randomly selected for qPCR verification. The logFC trends between the RNA-seq and qPCR results were consistent and visualized in a trend line comparison graph (
Figure 4c). The top 10 upregulated and downregulated DEGs were identified as potential key genes, with logFC values shown in a sorted graph (
Figure 4d). Notably,
ANKRD66 and
PLA2G7 were significantly upregulated. ANKRD66 is associated with enhanced cell proliferation in tumor cells [
34], whereas PLA2G7 catalyzes phospholipid hydrolysis to produce arachidonic acid, participating in inflammatory signaling and lipid metabolism [
35,
36]. In contrast,
GREM1 is found to be downregulated. GREM1 is a BMP antagonist that inhibits BMP signaling, which typically suppresses proliferation and promotes differentiation [
37]. Its reduced expression may enhance BMP signaling and inhibit proliferation, however, this effect may have been offset by the pro-proliferative effects of quercetin.
A GO enrichment analysis explored the biological functions associated with the DEGs, including biological processes (BPs), cellular components (CCs), and molecular functions (MFs;
Figure 4e). In BPs, “cytoplasmic translation” and “translational initiation” were enriched, indicating that quercetin promotes protein synthesis to support proliferation. In MFs, “insulin-like growth factor binding” was enriched. Insulin-like growth factors, such as IGF-1 and IGF-2, regulate proliferation via the PI3K–Akt and RAS–MAPK pathways, consistent with the observed pro-proliferative effects.
A KEGG enrichment analysis (
Figure 4f) showed that the DEGs were enriched in various pathways including Cellular Processes, Environmental Information Processing, Metabolism, Genetic Information Processing, and Organismal Systems. Key pathways included PI3K–Akt, Wnt, and TGF-β signaling, which are part of Environmental Information Processing. Additionally, under Metabolism, pathways related to cysteine/methionine metabolism and glycine/serine/threonine metabolism were enriched. These pathways are critical for follicle development and SHFSC proliferation. Key regulatory genes identified include the COL6A family and CD19 in the PI3K–Akt signaling pathway [
38,
39]; Wnt family genes in Wnt signaling; and SMAD family genes and BMP7 in TGF-β signaling. Additionally, CTH and AHCY reduced ROS levels and enhanced the antioxidant capacity through the cysteine/methionine metabolism pathway [
40,
41]. Western blot analyses of the key pathway proteins (
Figure 4g) confirmed differential expression levels consistent with the sequencing data, supporting quercetin’s role in activating the PI3K–Akt, Wnt, and TGF-β signaling pathways.
In summary, the transcriptome analysis revealed that quercetin promotes SHFSC proliferation and metabolism via multiple pathways, including PI3K–Akt, Wnt, TGF-β, and PSAT1-related pathways, clarifying its molecular mechanisms.
4. Discussion
This study found that quercetin promotes SHFSC proliferation in a concentration- and time-dependent manner, with optimal effects at treatment with 10 μg/mL quercetin for 48 h. This is consistent with previous findings in human DP cells
Error! Reference source not found.. Enhanced proliferation was revealed by an increased number of S-phase cells and upregulated PCNA and TERT expressions, suggesting that quercetin accelerates DNA replication and maintains telomere stability. Notably, upregulated Cyclin D1 and CDK4 expression—key regulators of the G1/S transition—may relieve the Rb-mediated inhibition of E2F transcription factors, activating DNA replication-related genes [
42,
43].
Transcriptome analysis and western blot showed that quercetin activates the PI3K–Akt and Wnt/β-catenin pathways. PI3K–Akt activation may inhibit β-catenin degradation via GSK3β phosphorylation, promoting nuclear translocation and the transcription of proliferation genes [
44]. This is supported by observed increase in β-catenin and phosphorylation levels. Wnt10b, a key anagen inducer, may synergize with PI3K–Akt to drive the SHFSC transition from quiescence to proliferation [
45].
Quercetin downregulated the expression of pro-apoptotic genes (
BAX,
TP53, and
CASP3) and upregulated that of
BCL-2, consistent with its anti-apoptotic effects in tumor cells [
46,
47]. Mechanistically, JC-1 staining revealed increased mitochondrial membrane potential, reduced cytochrome c release, and caspase cascade activation. As a natural antioxidant, quercetin reduces ROS levels and increases GSH levels, mitigating oxidative damage to DNA and mitochondria [
48]. An enhanced antioxidant capacity may also promote the secretion of VEGF and FGF, which further stimulates SHFSC proliferation via paracrine effects.
Transcriptome analyses identified 1,540 DEGs, with
ANKRD66 and
PLA2G7 being significantly upregulated. ANKRD66, an ankyrin repeat family member, may regulate cell proliferation via cytoskeletal dynamics, consistent with its proliferative role in cancer [
49]. PLA2G7 generates arachidonic acid, a precursor of proinflammatory and proliferative prostaglandins, aligned with an improved inflammatory microenvironment [
23]. Downregulated GREM1 expression may enhance BMP signaling (typically pro-differentiation/anti-proliferation); however, this effect may be offset by quercetin-activated pathways [
12].
KEGG enrichment analyses highlighted the PI3K–Akt, Wnt, and TGF-β pathways. The reduced phosphorylation of SMAD2/3 in TGF-β signaling may weaken the transcriptional inhibition of Cyclin D1, thereby accelerating the cell cycle [
50,
51,
52]. The activation of cysteine/methionine metabolism may provide methyl donors to support DNA/histone methylation, whereas CTH and AHCY reduce ROS, enhance the antioxidant capacity, and regulate proliferation-related genes [
53,
54]
Despite its promising results, this study had various limitations, including its reliance on in vitro experiments. This entails the need for in vivo validation in cashmere goats. Additionally, the complex SHFSC microenvironment (DP cell interactions) has not been fully recapitulated in vitro, warranting the use of 3D culture or organoid models. Furthermore, future studies should explore the association between quercetin and epigenetic modifications, such as DNA methylation and histone acetylation.
5. Conclusions
Our results demonstrate that quercetin plays a critical role in regulating follicle stem cell proliferation and apoptosis, specifically promoting proliferation and inhibiting apoptosis. It exerts these effects via multiple signaling pathways, including PI3K-Akt, Wnt, and TGF-β-related pathways, enhancing SHFSCs proliferation and metabolism. These findings provide valuable insights into developing a regulatory strategy for improving cashmere production from goats.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Supplementary S1.docx, Supplementary S2.docx
Author Contributions
Wei Lian and Guoqing Jiang: Conceptualization, Investigation, Methodology, Writing – original draft, Writing – review & editing. Xueyong Wu and Yadong Gao: Conceptualization, Project administration, Supervision, Writing – review & editing. Kun Cui and Ziyang Xu : Investigation, Methodology, Resources, Validation, Writing – review & editing. Xiao Zhang and Rui Ding : Data curation, Formal analysis, Software, Visualization, Writing – review & editing. Mingli Peng and Jiawei Wnag: Investigation, Validation, Writing – review & editing. Lei Zhu: Formal analysis, Methodology, Software, Writing – review & editing. Wei Lian: Data curation, Investigation, Writing – review & editing. Wei Lian: Validation, Visualization, Writing – review & editing. Wei Lian: Data curation, Resources, Writing – review & editing. Dongjun Liu and Fei Hao: Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
This work is financially supported by National Natural Science Foundation of China Project "Study on the Regulatory Mechanism of the edar Gene in the Growth and Development of Secondary Hair Follicles in Cashmere Goats" (Project No.: 32360811), National Natural Science Foun-dation of China Project "Study on the Mechanism of Hair Follicle Growth and Development andCashmere Trait Formation in Inner Mongolia Cashmere Goats" (Project No.: U23A20226), State KeyLaboratory of Reproductive Regulation & Breeding of Grassland Livestock (grant number.2025KYPT0081), Inner Mongolia Autonomous Region Science and Technology Plan Project "Studyon the lVechanism of Hair Follicle Growth and Development and Cashmere Trait Formation in Inner Mongolia Cashmere Goats" (Project No.: 2023KYPT0014).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.
Acknowledgments
We would like to express our gratitude to the staff of the Large Instrument Platform at Inner Mongolia University, particularly Ms. Menglan Cui , Ms. Yafei Huang and Xuan Wang, for their assistance with experimental instruments. We also thank the State Key Laboratory for providing access to the necessary experimental facilities. Additionally, we are grateful to the Yiwei White Cashmere Goat Farm in Ordos City, Inner Mongolia, China, and its staff for their valuable support in sample collection.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
| BMP |
Bone morphogenetic protein |
| BP |
Biological process |
| CC |
Cellular component |
| CCK-8 |
Cell Counting Kit-8 |
| CG |
Control group |
| DEG |
Differentially expressed gene |
| DP |
Dermal papilla |
| EdU |
5-ethynyl-2′-deoxyuridine |
| EG |
Experimental group |
| GPx |
Glutathione peroxidase |
| GO |
Gene Ontology |
| GSH |
Glutathione |
| HRP |
Horseradish peroxidase |
| KEGG |
Kyoto Encyclopedia of Genes and Genomes |
| MF |
Molecular function |
| PCA |
Principal component analysis |
| PCNA |
Proliferating cell nuclear antigen |
| PBS |
Phosphate-buffered saline |
| ROS |
Reactive oxygen species |
| SHFSC |
Secondary hair follicle stem cell |
| SOD |
Superoxide dismutase |
| TERT |
Telomerase reverse transcriptase |
References
- Dai, B., F. Hao, T. Xu, B. Zhu, L. Q. Ren, X. Y. Han, and D. J. Liu. "Thymosin Beta4 Identified by Transcriptomic Analysis from Hf Anagen to Telogen Promotes Proliferation of Shf-Dpcs in Albas Cashmere Goat." Int J Mol Sci 21, no. 7 (2020). [CrossRef]
- Gong, G., Y. Fan, W. Li, X. Yan, X. Yan, L. Zhang, N. Wang, O. Chen, Y. Zhang, R. Wang, Z. Liu, W. Jiang, J. Li, Z. Wang, Q. Lv, and R. Su. "Identification of the Key Genes Associated with Different Hair Types in the Inner Mongolia Cashmere Goat." Animals (Basel) 12, no. 11 (2022). [CrossRef]
- Nocelli, C., K. Cappelli, S. Capomaccio, L. Pascucci, F. Mercati, I. Pazzaglia, S. Mecocci, M. Antonini, and C. Renieri. "Shedding Light on Cashmere Goat Hair Follicle Biology: From Morphology Analyses to Transcriptomic Landascape." BMC Genomics 21, no. 1 (2020): 458. [CrossRef]
- Song, S., M. Yang, Y. Li, M. Rouzi, Q. Zhao, Y. Pu, X. He, J. M. Mwacharo, N. Yang, Y. Ma, and L. Jiang. "Genome-Wide Discovery of Lincrnas with Spatiotemporal Expression Patterns in the Skin of Goat during the Cashmere Growth Cycle." BMC Genomics 19, no. 1 (2018): 495. [CrossRef]
- Diao, X., L. Yao, X. Wang, S. Li, J. Qin, L. Yang, L. He, and W. Zhang. "Hair Follicle Development and Cashmere Traits in Albas Goat Kids." Animals (Basel) 13, no. 4 (2023). [CrossRef]
- Shirokova, V., L. C. Biggs, M. Jussila, T. Ohyama, A. K. Groves, and M. L. Mikkola. "Foxi3 Deficiency Compromises Hair Follicle Stem Cell Specification and Activation." Stem Cells 34, no. 7 (2016): 1896-908. [CrossRef]
- Kim, H., Y. Jang, E. H. Kim, H. Jang, H. Cho, G. Han, H. K. Song, S. H. Kim, and Y. Yang. "Potential of Colostrum-Derived Exosomes for Promoting Hair Regeneration through the Transition from Telogen to Anagen Phase." Front Cell Dev Biol 10 (2022): 815205. [CrossRef]
- Feng, C., Y. Gao, B. Dorshorst, C. Song, X. Gu, Q. Li, J. Li, T. Liu, C. J. Rubin, Y. Zhao, Y. Wang, J. Fei, H. Li, K. Chen, H. Qu, D. Shu, C. Ashwell, Y. Da, L. Andersson, X. Hu, and N. Li. "A Cis-Regulatory Mutation of Pdss2 Causes Silky-Feather in Chickens." PLoS Genet 10, no. 8 (2014): e1004576. [CrossRef]
- Sunkara, R. R., D. Mehta, R. M. Sarate, and S. K. Waghmare. "Bmp-Akt-Gsk3beta Signaling Restores Hair Follicle Stem Cells Decrease Associated with Loss of Sfrp1." Stem Cells 40, no. 9 (2022): 802-17. [CrossRef]
- Zhou, Q., Y. Song, Q. Zheng, R. Han, and H. Cheng. "Expression Profile Analysis of Dermal Papilla Cells Mrna in Response to Wnt10b Treatment." Exp Ther Med 19, no. 2 (2020): 1017-23. [CrossRef]
- Zhou, L. L., K. Yang, M. G. Xu, T. Andl, S. E. Millar, S. Boyce, and Y. H. Zhang. "Activating Β-Catenin Signaling in Cd133-Positive Dermal Papilla Cells Increases Hair Inductivity." Febs Journal 283, no. 15 (2016): 2823-35. [CrossRef]
- Wu, P., Y. Zhang, Y. Xing, W. Xu, H. Guo, F. Deng, X. Ma, and Y. Li. "The Balance of Bmp6 and Wnt10b Regulates the Telogen-Anagen Transition of Hair Follicles." Cell Commun Signal 17, no. 1 (2019): 16. [CrossRef]
- Amberg, N., M. Holcmann, G. Stulnig, and M. Sibilia. "Effects of Imiquimod on Hair Follicle Stem Cells and Hair Cycle Progression." J Invest Dermatol 136, no. 11 (2016): 2140-49. [CrossRef]
- Deng, Z., X. Lei, X. Zhang, H. Zhang, S. Liu, Q. Chen, H. Hu, X. Wang, L. Ning, Y. Cao, T. Zhao, J. Zhou, T. Chen, and E. Duan. "Mtor Signaling Promotes Stem Cell Activation Via Counterbalancing Bmp-Mediated Suppression during Hair Regeneration." J Mol Cell Biol 7, no. 1 (2015): 62-72. [CrossRef]
- Wu, Z., Y. Zhu, H. Liu, G. Liu, and F. Li. "Wnt10b Promotes Hair Follicles Growth and Dermal Papilla Cells Proliferation Via Wnt/Beta-Catenin Signaling Pathway in Rex Rabbits." Biosci Rep 40, no. 2 (2020). [CrossRef]
- Zhang, Y., Y. Xing, H. Guo, X. Ma, and Y. Li. "Immunohistochemical Study of Hair Follicle Stem Cells in Regenerated Hair Follicles Induced by Wnt10b." Int J Med Sci 13, no. 10 (2016): 765-71. [CrossRef]
- Xing, Y., X. Ma, H. Guo, F. Deng, J. Yang, and Y. Li. "Wnt5a Suppresses Beta-Catenin Signaling during Hair Follicle Regeneration." Int J Med Sci 13, no. 8 (2016): 603-10. [CrossRef]
- Lichtenberger, B. M., M. Mastrogiannaki, and F. M. Watt. "Epidermal Beta-Catenin Activation Remodels the Dermis Via Paracrine Signalling to Distinct Fibroblast Lineages." Nat Commun 7 (2016): 10537. [CrossRef]
- Hamblin, M. R. "Photobiomodulation for the Management of Alopecia: Mechanisms of Action, Patient Selection and Perspectives." Clin Cosmet Investig Dermatol 12 (2019): 669-78. [CrossRef]
- Li, K. N., P. Jain, C. H. He, F. C. Eun, S. Kang, and T. Tumbar. "Skin Vasculature and Hair Follicle Cross-Talking Associated with Stem Cell Activation and Tissue Homeostasis." Elife 8 (2019). [CrossRef]
- Martino, P., R. Sunkara, N. Heitman, M. Rangl, A. Brown, N. Saxena, L. Grisanti, D. Kohan, M. Yanagisawa, and M. Rendl. "Progenitor-Derived Endothelin Controls Dermal Sheath Contraction for Hair Follicle Regression." Nat Cell Biol 25, no. 2 (2023): 222-34. [CrossRef]
- Choi, S., B. Zhang, S. Ma, M. Gonzalez-Celeiro, D. Stein, X. Jin, S. T. Kim, Y. L. Kang, A. Besnard, A. Rezza, L. Grisanti, J. D. Buenrostro, M. Rendl, M. Nahrendorf, A. Sahay, and Y. C. Hsu. "Corticosterone Inhibits Gas6 to Govern Hair Follicle Stem-Cell Quiescence." Nature 592, no. 7854 (2021): 428-32. [CrossRef]
- Nguyen, T. L. A., and D. Bhattacharya. "Antimicrobial Activity of Quercetin: An Approach to Its Mechanistic Principle." Molecules 27, no. 8 (2022). [CrossRef]
- Kiran, F., H. K. Demirhan, O. Haliscelik, and D. Zatari. "Metabolic Profiles of Weissella Spp. Postbiotics with Anti-Microbial and Anti-Oxidant Effects." Journal of Infection in Developing Countries 17, no. 4 (2023): 507-+. [CrossRef]
- Chen, K. T. J., M. Anantha, A. W. Y. Leung, J. A. Kulkarni, G. G. C. Militao, M. Wehbe, B. Sutherland, P. R. Cullis, and M. B. Bally. "Characterization of a Liposomal Copper(Ii)-Quercetin Formulation Suitable for Parenteral Use." Drug Delivery and Translational Research 10, no. 1 (2020): 202-15. [CrossRef]
- Buchweitz, M., P. A. Kroon, G. T. Rich, and P. J. Wilde. "Quercetin Solubilisation in Bile Salts: A Comparison with Sodium Dodecyl Sulphate." Food Chemistry 211 (2016): 356-64. [CrossRef]
- Rani, N., L. P. Velan, S. Vijaykumar, and A. Arunachalam. "An Insight into the Potentially Old-Wonder Molecule-Quercetin: The Perspectives in Foresee." Chin J Integr Med (2015): 1-16. [CrossRef]
- Zhang, W., J. Sun, P. Zhang, R. Yue, Y. Zhang, F. Niu, H. Zhu, C. Ma, and S. Deng. "Design, Synthesis and Antitumor Activity of Quercetin Derivatives Containing a Quinoline Moiety." Molecules 29, no. 1 (2024). [CrossRef]
- Kim, J., S. R. Kim, Y. H. Choi, J. Y. Shin, C. D. Kim, N. G. Kang, B. C. Park, and S. Lee. "Quercitrin Stimulates Hair Growth with Enhanced Expression of Growth Factors Via Activation of Mapk/Creb Signaling Pathway." Molecules 25, no. 17 (2020). [CrossRef]
- Giuliani, C. "The Flavonoid Quercetin Induces Ap-1 Activation in Frtl-5 Thyroid Cells." Antioxidants (Basel) 8, no. 5 (2019). [CrossRef]
- Zhao, Q., Y. Zheng, D. Zhao, L. Zhao, L. Geng, S. Ma, Y. Cai, C. Liu, Y. Yan, J. C. I. Belmonte, S. Wang, W. Zhang, G. H. Liu, and J. Qu. "Single-Cell Profiling Reveals a Potent Role of Quercetin in Promoting Hair Regeneration." Protein Cell 14, no. 6 (2023): 398-415. [CrossRef]
- Bejaoui, M., M. O. Villareal, and H. Isoda. "3,4,5-Tri-O-Caffeoylquinic Acid Promoted Hair Pigmentation through Beta-Catenin and Its Target Genes." Front Cell Dev Biol 8 (2020): 175. [CrossRef]
- He, N., Z. Dong, L. Tao, S. Zhao, S. Bou, and D. Liu. "Isolation and Characterization of Hair Follicle Stem Cells from Arbas Cashmere Goat." Cytotechnology 68, no. 6 (2016): 2579-88. [CrossRef]
- Wang, T., Q. Zhen, T. Wu, L. Jin, S. Yao, Y. Feng, J. Chen, C. Chen, and Z. Huang. "Gamma-Aminobutyric Acid Type a Receptor Subunit Pi Is a Potential Chemoresistance Regulator in Colorectal Cancer." Mol Biol Rep 50, no. 4 (2023): 3167-77. [CrossRef]
- Zheng, W., Q. Lin, M. A. Issah, Z. Liao, and J. Shen. "Identification of Pla2g7 as a Novel Biomarker of Diffuse Large B Cell Lymphoma." BMC Cancer 21, no. 1 (2021): 927. [CrossRef]
- Vainio, P., L. Lehtinen, T. Mirtti, M. Hilvo, T. Seppanen-Laakso, J. Virtanen, A. Sankila, S. Nordling, J. Lundin, A. Rannikko, M. Oresic, O. Kallioniemi, and K. Iljin. "Phospholipase Pla2g7, Associated with Aggressive Prostate Cancer, Promotes Prostate Cancer Cell Migration and Invasion and Is Inhibited by Statins." Oncotarget 2, no. 12 (2011): 1176-90. [CrossRef]
- Gao, Y., S. De, and D. P. Brazil. "The Role of Gremlin1, a Bone Morphogenetic Protein Antagonist, in Cancer Stem Cell Regulation." Cells 14, no. 8 (2025). [CrossRef]
- Lettmann, S., W. Bloch, T. Maass, A. Niehoff, J. N. Schulz, B. Eckes, S. A. Eming, P. Bonaldo, M. Paulsson, and R. Wagener. "Col6a1 Null Mice as a Model to Study Skin Phenotypes in Patients with Collagen Vi Related Myopathies: Expression of Classical and Novel Collagen Vi Variants during Wound Healing." PLoS One 9, no. 8 (2014): e105686. [CrossRef]
- Ishikawa, H., N. Tsuyama, M. S. Mahmoud, R. Fujii, S. Abroun, S. Liu, F. J. Li, and M. M. Kawano. "Cd19 Expression and Growth Inhibition of Tumours in Human Multiple Myeloma." Leuk Lymphoma 43, no. 3 (2002): 613-6. [CrossRef]
- Peleli, M., I. Antoniadou, D. M. Rodrigues-Junior, O. Savvoulidou, L. Caja, A. Katsouda, D. F. J. Ketelhuth, J. Stubbe, K. Madsen, A. Moustakas, and A. Papapetropoulos. "Cystathionine Gamma-Lyase (Cth) Inhibition Attenuates Glioblastoma Formation." Redox Biol 64 (2023): 102773. [CrossRef]
- Rowland, E. C., M. D'Antuono, A. Jermakowicz, and N. G. Ayad. "Mat2a and Ahcy Inhibition Disrupts Antioxidant Metabolism and Reduces Glioblastoma Cell Survival." bioRxiv (2024).
- Hanahan, D. "Hallmarks of Cancer: New Dimensions." Cancer Discovery 12, no. 1 (2022): 31-46. [CrossRef]
- Lamb, R., S. Lehn, L. Rogerson, R. B. Clarke, and G. Landberg. "Cell Cycle Regulators Cyclin D1 and Cdk4/6 Have Estrogen Receptor-Dependent Divergent Functions in Breast Cancer Migration and Stem Cell-Like Activity." Cell Cycle 12, no. 15 (2013): 2384-94. [CrossRef]
- Fruman, D. A., H. Chiu, B. D. Hopkins, S. Bagrodia, L. C. Cantley, and R. T. Abraham. "The Pi3k Pathway in Human Disease." Cell 170, no. 4 (2017): 605-35. [CrossRef]
- Clevers, H., and R. Nusse. "Wnt/Beta-Catenin Signaling and Disease." Cell 149, no. 6 (2012): 1192-205. [CrossRef]
- Wang, Y. Y., C. Y. Chang, S. Y. Lin, J. D. Wang, C. C. Wu, W. Y. Chen, Y. H. Kuan, S. L. Liao, W. Y. Wang, and C. J. Chen. "Quercetin Protects against Cerebral Ischemia/Reperfusion and Oxygen Glucose Deprivation/Reoxygenation Neurotoxicity." J Nutr Biochem 83 (2020): 108436. [CrossRef]
- Wang, Y., Z. Z. Zhang, Y. Wu, J. J. Ke, X. H. He, and Y. L. Wang. "Quercetin Postconditioning Attenuates Myocardial Ischemia/Reperfusion Injury in Rats through the Pi3k/Akt Pathway." Braz J Med Biol Res 46, no. 10 (2013): 861-7. [CrossRef]
- Weng, C. J., M. J. Chen, C. T. Yeh, and G. C. Yen. "Hepatoprotection of Quercetin against Oxidative Stress by Induction of Metallothionein Expression through Activating Mapk and Pi3k Pathways and Enhancing Nrf2 DNA-Binding Activity." N Biotechnol 28, no. 6 (2011): 767-77. [CrossRef]
- Zhao, H., Y. Wang, Y. He, P. Zhang, C. Zeng, T. Du, Q. Shen, and S. Zhao. "Ankrd29, as a New Prognostic and Immunological Biomarker of Non-Small Cell Lung Cancer, Inhibits Cell Growth and Migration by Regulating Mapk Signaling Pathway." Biol Direct 18, no. 1 (2023): 28. [CrossRef]
- Barrientos, S., O. Stojadinovic, M. S. Golinko, H. Brem, and M. Tomic-Canic. "Growth Factors and Cytokines in Wound Healing." Wound Repair and Regeneration 16, no. 5 (2008): 585-601. [CrossRef]
- Inoue-Choi, M., N. D. Freedman, A. Etemadi, M. Hashemian, P. Brennan, G. Roshandel, H. Poustchi, P. Boffetta, F. Kamangar, T. Amiriani, A. Norouzi, S. Dawsey, R. Malekzadeh, and C. C. Abnet. "One-Carbon Metabolism Biomarkers and Upper Gastrointestinal Cancer in the Golestan Cohort Study." International Journal of Cancer 155, no. 11 (2024): 1944-57. [CrossRef]
- Ducker, G. S., and J. D. Rabinowitz. "One-Carbon Metabolism in Health and Disease." Cell Metabolism 25, no. 1 (2017): 27-42. [CrossRef]
- Liao, L. S., Z. J. Xiao, J. L. Wang, T. J. Liu, F. D. Huang, Y. P. Zhong, X. Zhang, K. H. Chen, R. L. Du, and M. Y. Dong. "A Four Amino Acid Metabolism-Associated Genes (Amgs) Signature for Predicting Overall Survival Outcomes and Immunotherapeutic Efficacy in Hepatocellular Carcinoma." Biochemical Genetics 62, no. 3 (2024): 1577-602. [CrossRef]
- Rahimi, R. A., and E. B. Leof. "Tgf-Β Signaling:: A Tale of Two Responses." Journal of Cellular Biochemistry 102, no. 3 (2007): 593-608. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).