Submitted:
10 August 2025
Posted:
11 August 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. HPV-DNA Test and Genotyping
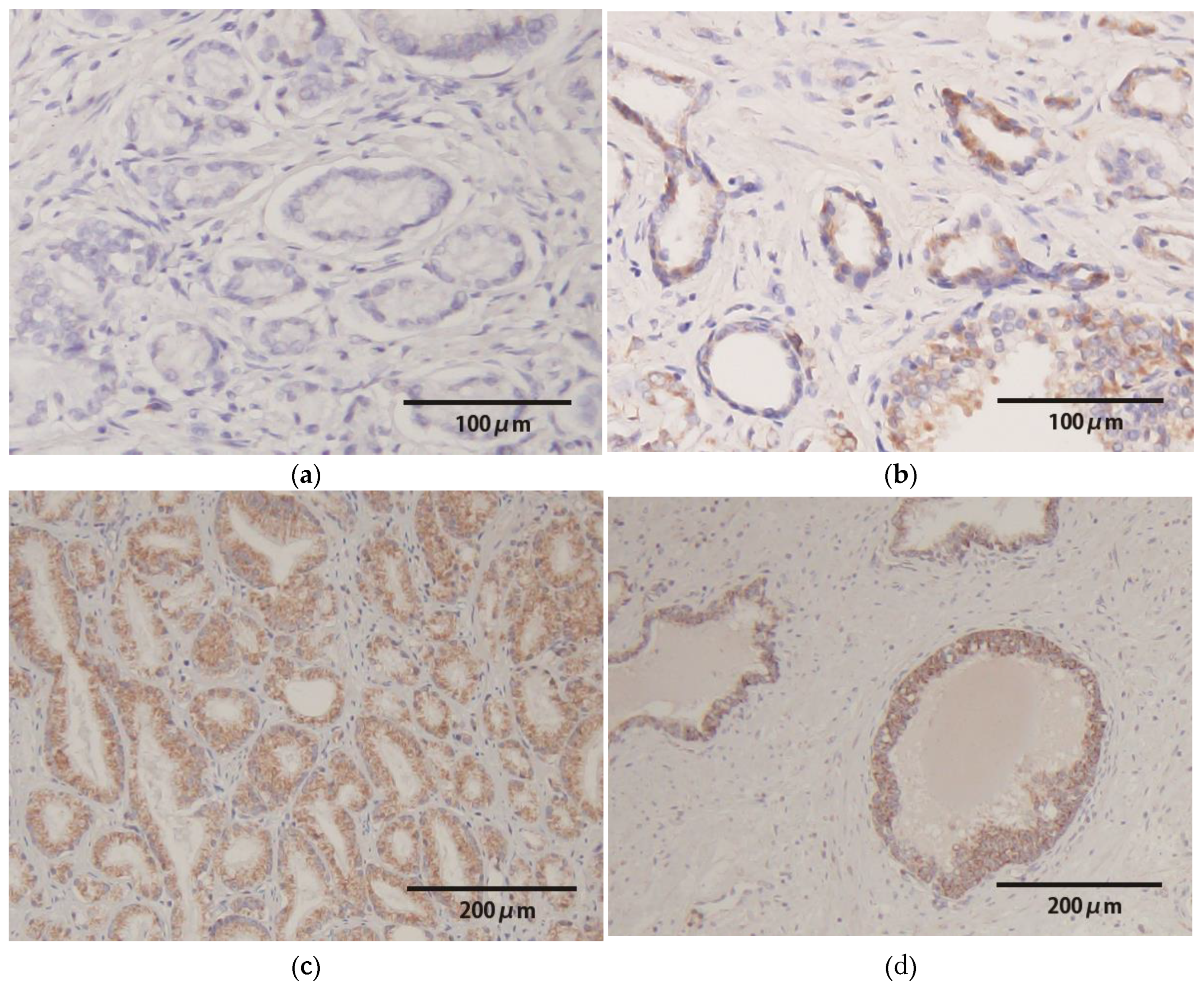
2.3. HPV-DNA In Situ Hybridization
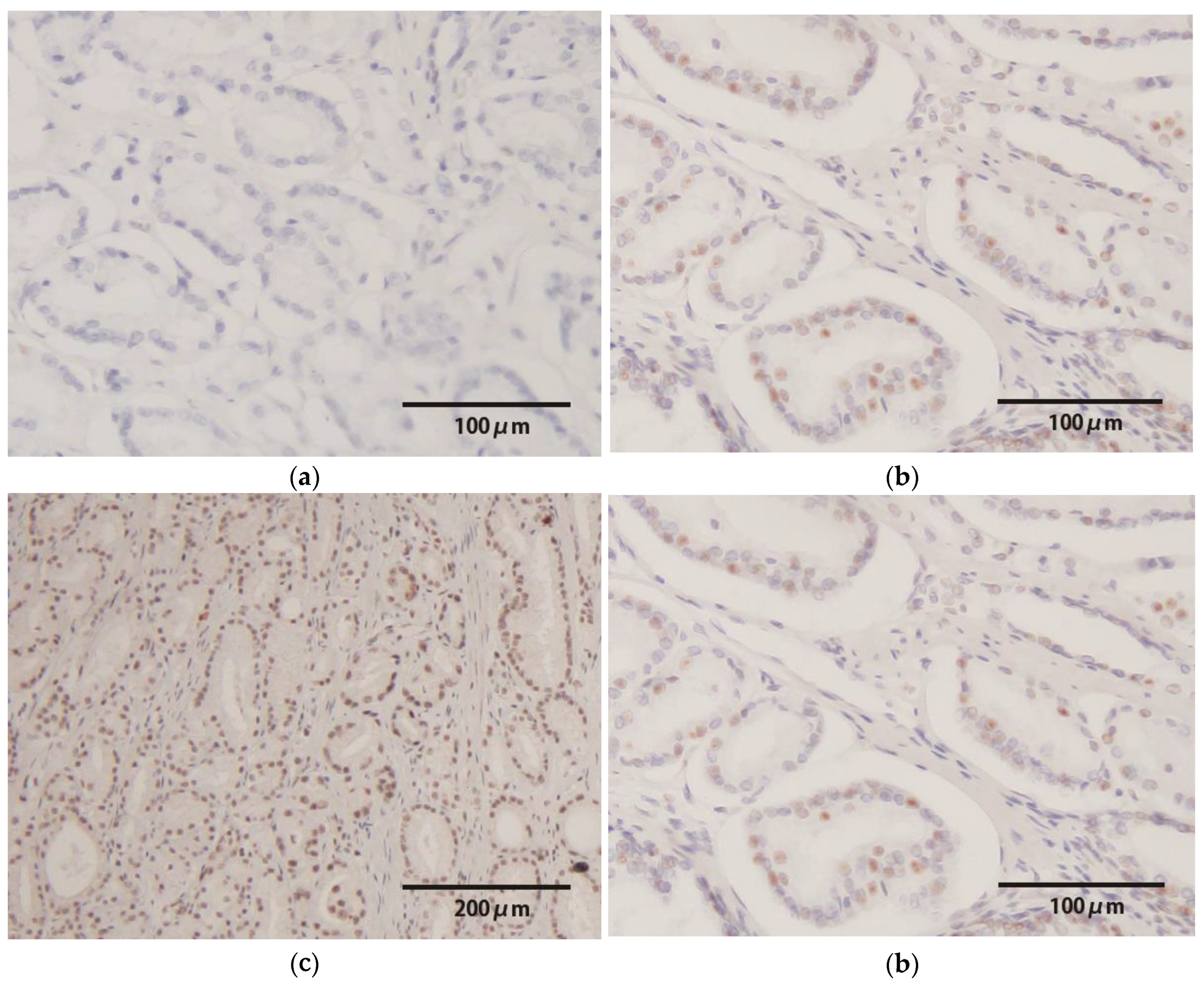
2.4. HPV-E6/E7 mRNA In Situ Hybridization
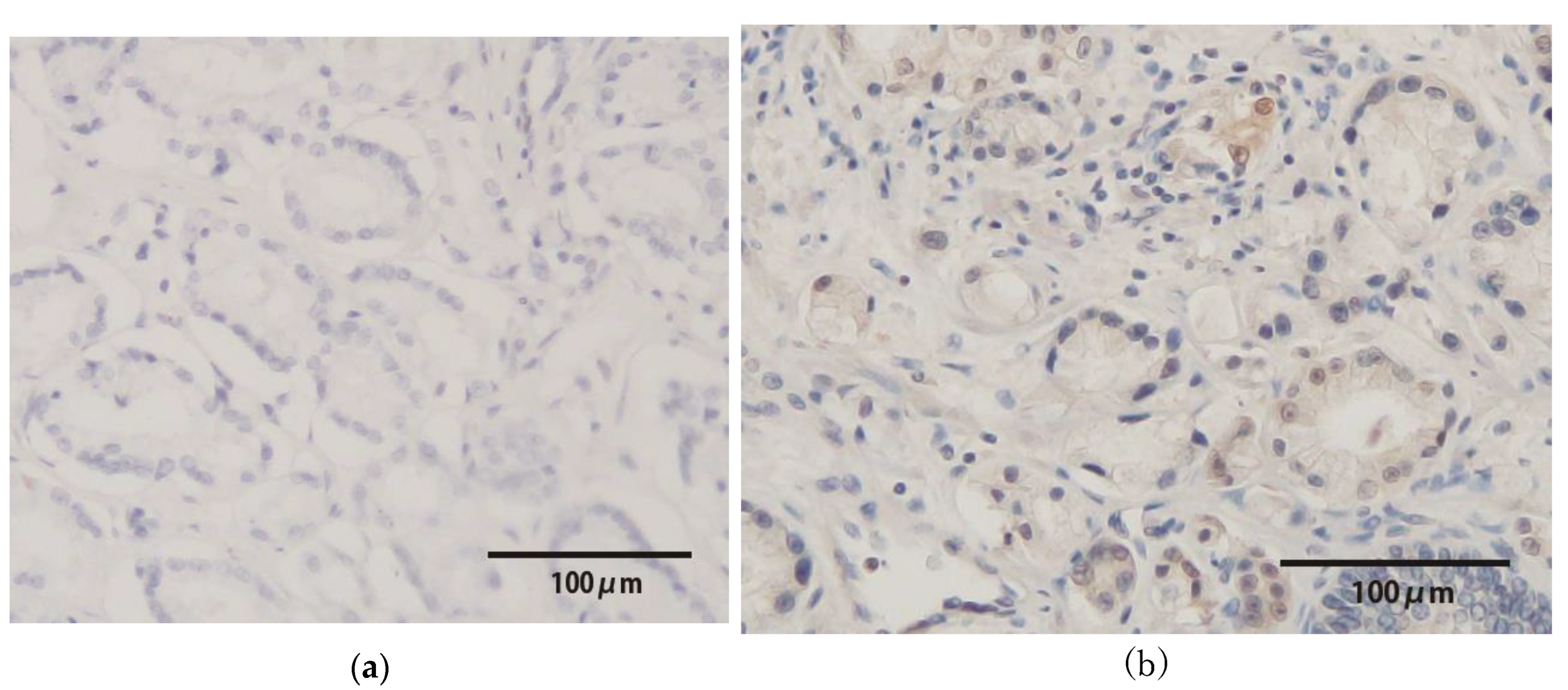
2.5. Immunohistochemistry of p16INK4a, a Surrogate Marker of the E7 Protein
2.6. Statistical Analysis
3. Results
3.1. Patient Characteristics and HPV Prevalence
3.2. HPV-DNA In Situ Hybridization
3.3. HPV-E6/E7 mRNA Expression
3.4. p16INK4a Protein Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dunne, E.F.; Nielson, C.M.; Stone, K.M.; Markowitz, L.E.; Giuliano, A.R. Prevalence of HPV infection among men: A systematic review of the literature. J Infect Dis 2006, 194, 1044–1057. [Google Scholar] [CrossRef]
- Giuliano, A.R.; Nielson, C.M.; Flores, R.; et al. The optimal anatomic sites for sampling heterosexual men for human papillomavirus (HPV) detection: The HPV detection in men study. J Infect Dis 2007, 196, 1146–1152. [Google Scholar] [CrossRef] [PubMed]
- Shigehara, K.; Sasagawa, T.; Namiki, M. Human papillomavirus infection and pathogenesis in urothelial cells: A mini-review. J Infect Chemother 2014, 20, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Laprise, C.; Trottier, H.; Monnier, P.; Coutlée, F.; Mayrand, M.H. Prevalence of human papillomaviruses in semen: A systematic review and meta-analysis. Hum Reprod 2014, 29, 640–651. [Google Scholar] [CrossRef]
- Kato, Y.; Shigehara, K.; Nakagawa, T.; et al. Human papillomavirus detected in sperm of Japanese infertile males affects reproductive parameters. Int J Infect Dis 2021, 112, 294–299. [Google Scholar] [CrossRef]
- Jenson, A.B.; Geyer, S.; Sundberg, J.P.; Ghim, S. Human papillomavirus and skin cancer. J Investig Dermatol Symp Proc 2001, 6, 203–206. [Google Scholar] [CrossRef]
- Fonsêca, T.C.; Jural, L.A.; Marañón-Vásquez, G.A.; et al. Global prevalence of human papillomavirus-related oral and oropharyngeal squamous cell carcinomas: A systematic review and meta-analysis. Clin Oral Investig 2023, 28, 62. [Google Scholar] [CrossRef]
- Sakamoto, J.; Shigehara, K.; Nakashima, K.; et al. Etiological role of human papillomavirus infection in the development of penile cancer. Int J Infect Dis 2019, 78, 148–154. [Google Scholar] [CrossRef]
- zur Hausen, H. Papillomaviruses and cancer: From basic studies to clinical application. Nat Rev Cancer 2002, 2, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Tsydenova, I.A.; Ibragimova, M.K.; Tsyganov, M.M.; Litviakov, N.V. Human papillomavirus and prostate cancer: Systematic review and meta-analysis. Sci Rep 2023, 13, 16597. [Google Scholar] [CrossRef]
- de Roda Husman, A.M.; Walboomers, J.M.; van den Brule, A.J.; et al. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J Gen Virol 1995, 76, 1057–1062. [Google Scholar] [CrossRef]
- Shigehara, K.; Sasagawa, T.; Kawaguchi, S.; et al. Prevalence of human papillomavirus infection in the urinary tract of men with urethritis. Int J Urol 2010, 17, 563–568. [Google Scholar] [CrossRef]
- Nakagawa, S. ; Yoshikawa. H.; Yasugi, T.; et al. Ubiquitous presence of E6 and E7 transcripts in human papillomavirus-positive cervical carcinomas regardless of its type. J Med Virol 2000, 62, 251–258. [Google Scholar] [CrossRef]
- Schmitt, A.; Harry, J.B.; Rapp, B.; et al. Comparison of the properties of the E6 and E7 genes of low- and high-risk cutaneous papillomaviruses reveals strongly transforming and high Rb-binding activity for the E7 protein of the low-risk human papillomavirus type 1. J Virol 1994, 68, 7051–7059. [Google Scholar] [CrossRef] [PubMed]
- Darragh, T.M.; Colgan, T.J.; Thomas Cox, J.; et al. The Lower Anogenital Squamous Terminology Standardization project for HPV-associated lesions: Background and consensus recommendations. Int J Gynecol Pathol 2013, 32, 76–115. [Google Scholar] [CrossRef] [PubMed]
- Rotola, A.; Monini, P.; Di Luca, D.; et al. Presence and physical state of HPV DNA in prostate and urinary-tract tissues. Int J Cancer 1992, 52, 359–365. [Google Scholar] [CrossRef]
- Ahmed, M.Y.; Salman, N.A.; Sandhu, S.; et al. Detection of high-risk Human Papillomavirus in prostate cancer from a UK based population. Sci Rep 2023, 13, 7633. [Google Scholar] [CrossRef]
- Noda, T.; Sasagawa, T.; Dong, Y.; et al. Detection of human papillomavirus (HPV) DNA in archival specimens of benign prostatic hyperplasia and prostatic cancer using a highly sensitive nested PCR method. Urol Res 1998, 26, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Komiya, A.; Aida, S.; et al. Detection of human papillomavirus DNA and p53 gene mutations in human prostate cancer. Prostate 1996, 28, 318–324. [Google Scholar] [CrossRef]
- Bruni, L.; Albero, G.; Rowley, J.; et al. Global and regional estimates of genital human papillomavirus prevalence among men: A systematic review and meta-analysis. Lancet Glob Health 2023, 11, e1345–e1362. [Google Scholar] [CrossRef]
- Moghoofei, M.; Keshavarz, M.; Ghorbani, S.; et al. Association between human papillomavirus infection and prostate cancer: A global systematic review and meta-analysis. Asia Pac J Clin Oncol 2019, 15, e59–e67. [Google Scholar] [CrossRef]
- Nakashima, K.; Shigehara, K.; Kitamura, T.; et al. Risk factors for human papillomavirus detection in urine samples of heterosexual men visiting urological clinics in Japan. J Infect Chemother 2018, 24, 713–717. [Google Scholar] [CrossRef]
- Singh, N.; Hussain, S.; Kakkar, N.; et al. Implication of high risk human papillomavirus (HR-HPV) infection in prostate cancer in Indian population: A pioneering case-control analysis. Sci Rep 2015, 5, 7822. [Google Scholar] [CrossRef]
- Pascale, M.; Pracella, D.; Barbazza, R.; et al. Is human papillomavirus associated with prostate cancer survival? Dis Markers 2013, 35, 607–613. [Google Scholar] [CrossRef]
- Shigehara, K.; Sasagawa, T.; Kawaguchi, S.; et al. Etiologic role of human papillomavirus infection in bladder carcinoma. Cancer 2011, 117, 2067–2076. [Google Scholar] [CrossRef] [PubMed]
- Abumsimir, B.; Mrabti, M.; Laraqui, A.; et al. Molecular characterization of human papillomavirus and mouse mammary tumor virus-like infections in prostate cancer tissue and relevance with tumor characteristics. Mol Clin Oncol. 2022, 16, 16. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.K.; Panaiyadiyan, S.; Kurra, S.; et al. Association of human papillomavirus in penile cancer: A single-center analysis. Indian J Urol 2022, 38, 210–215. [Google Scholar] [CrossRef]
- Yin, J.; Li, Y.; Song, C.; et al. Evaluation of an E6/E7 PCR-capillary electrophoresis fragment analysis in the genotyping of human papillomavirus in archival FFPE samples of oropharyngeal cancer. J Med Virol 2024, 96, e29716. [Google Scholar] [CrossRef]
- Phanuphak, N.; Teeratakulpisarn, N.; Keelawat, S.; et al. Use of human papillomavirus DNA, E6/E7 mRNA, and p16 immunocytochemistry to detect and predict anal high-grade squamous intraepithelial lesions in HIV-positive and HIV-negative men who have sex with men. PLoS ONE 2013, 8, e78291. [Google Scholar] [CrossRef] [PubMed]
- Bello, R.O.; Willis-Powell, L.; James, O.; et al. Does human papillomavirus play a causative role in prostate cancer? A systematic review using Bradford Hill's criteria. Cancers (Basel) 2023, 15, 3897. [Google Scholar] [CrossRef]
- Mehanna, H.; Taberna, M.; von Buchwald, C.; et al. Prognostic implications of p16 and HPV discordance in oropharyngeal cancer (HNCIG-EPIC-OPC): A multicentre, multinational, individual patient data analysis. Lancet Oncol 2023, 24, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Zupancic, M.; Kostopoulou, O.N.; Holzhauser, S.; et al. Human papillomavirus (HPV) load is higher in HPV-DNA/p16 positive than in HPV-DNA positive/p16 negative oropharyngeal squamous cell carcinoma but does not differ significantly between various subsites or correlate to survival. Oral Oncol 2024, 151, 106749. [Google Scholar] [CrossRef] [PubMed]
- Murphy, T.M.; Sullivan, L.; Lane, C.; et al. In silico analysis and DHPLC screening strategy identifies novel apoptotic gene targets of aberrant promoter hypermethylation in prostate cancer. Prostate 2011, 71, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Medel-Flores, O.; Valenzuela-Rodríguez, V.A.; Ocadiz-Delgado, R.; et al. Association between HPV infection and prostate cancer in a Mexican population. Genet Mol Biol 2018, 41, 781–789. [Google Scholar] [CrossRef]
| Characteristics | n = 157 | HPV(+) (n = 15) | HPV(−) (n = 142) | p |
|---|---|---|---|---|
| Age (median, range) | 68 (48–76) | 70 (48–76) | 65 (56–76) | 0.981 |
| PSA, ng/ml (median, range) | 7.05 (2.42–74.2) | 6.24 (3.00–16.54) | 7.06 (2.42–74.2) | 0.375 |
| Grade Group (n, %) | ||||
| 1 | 19 (12.1%) | 0 (0%) | 19 (13.4%) | |
| 2–3 | 118 (75.2%) | 10 (66.7%) | 108 (76.1%) | |
| 4–5 | 20 (12.7%) | 5 (33.3%) | 15 (10.6%) | 0.0214 |
| T category (n, %) | ||||
| pT0 | 2 (1.3%) | 0 | 2 (1.4%) | |
| pT2 | 136 (86.6%) | 12 (80%) | 124 (87.3%) | 0.455 |
| pT3 | 15 (9.6%) | 3 (20%) | 12 (8.5%) | |
| pT4 | 4 (2.5%) | 0 | 4 (2.8%) |
| PCR results. | Prostate Cancer Tissue | Seminal Vesicle Tissue |
| Any HPV | 15 (9.6%) | 0 (0%) |
| HR-HPV | 7 (4.5%) | 0 (0%) |
| The HPV genotype | N | |
| 31 | 2 | |
| 44 | 5 | |
| 52 | 2 | |
| 58 | 2 | |
| 66 | 1 | |
| Unknown 1 | 6 |
| No | Age | GG | T | Genotype | HPV Risk | HPV-DNA ISH | |
| Normal Lesion | Cancer Lesion | ||||||
| 1 | 48 | 3 | 2c | 31 | HR | ++ | − |
| 2 | 63 | 2 | 3a | 31 | HR | ++ | ++ |
| 3 | 74 | 5 | 3b | 44 | LR | + | ++ |
| 4 | 59 | 2 | 2c | 44 | LR | ++ | ++ |
| 5 | 59 | 5 | 3b | 52 | HR | − | + |
| 6 | 71 | 2 | 2c | UK | Unknown | + | − |
| 7 | 72 | 4 | 2a | 58 | HR | ++ | ++ |
| 8 | 76 | 2 | 2c | 66 | HR | − | ++ |
| 9 | 72 | 3 | 2b | 52/44 | HR/LR | ++ | ++ |
| 10 | 63 | 4 | 2a | 58/44 | HR/LR | + | − |
| 11 | 64 | 4 | 2c | 44 | LR | − | − |
| 12 | 70 | 3 | 2c | UK | Unknown | ++ | ++ |
| 13 | 72 | 2 | 2c | UK | Unknown | − | − |
| 14 | 72 | 2 | 2b | UK | Unknown | + | + |
| 15 | 65 | 2 | 2c | UK | Unknown | − | − |
| No | Genotype | HPV risk | E6/E7 oncogenic protein | |
|---|---|---|---|---|
| E6/E7 mRNA ISH | p16INK4a protein | |||
| 1 | 31 | HR | ++ | + |
| 2 | 31 | HR | ++ | − |
| 5 | 52 | HR | ++ | − |
| 7 | 58 | HR | − | + |
| 8 | 66 | HR | ++ | + |
| 9 | 52/44 | HR/LR | ++ | + |
| 10 | 58/44 | HR/LR | ++ | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





