Submitted:
01 August 2025
Posted:
04 August 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
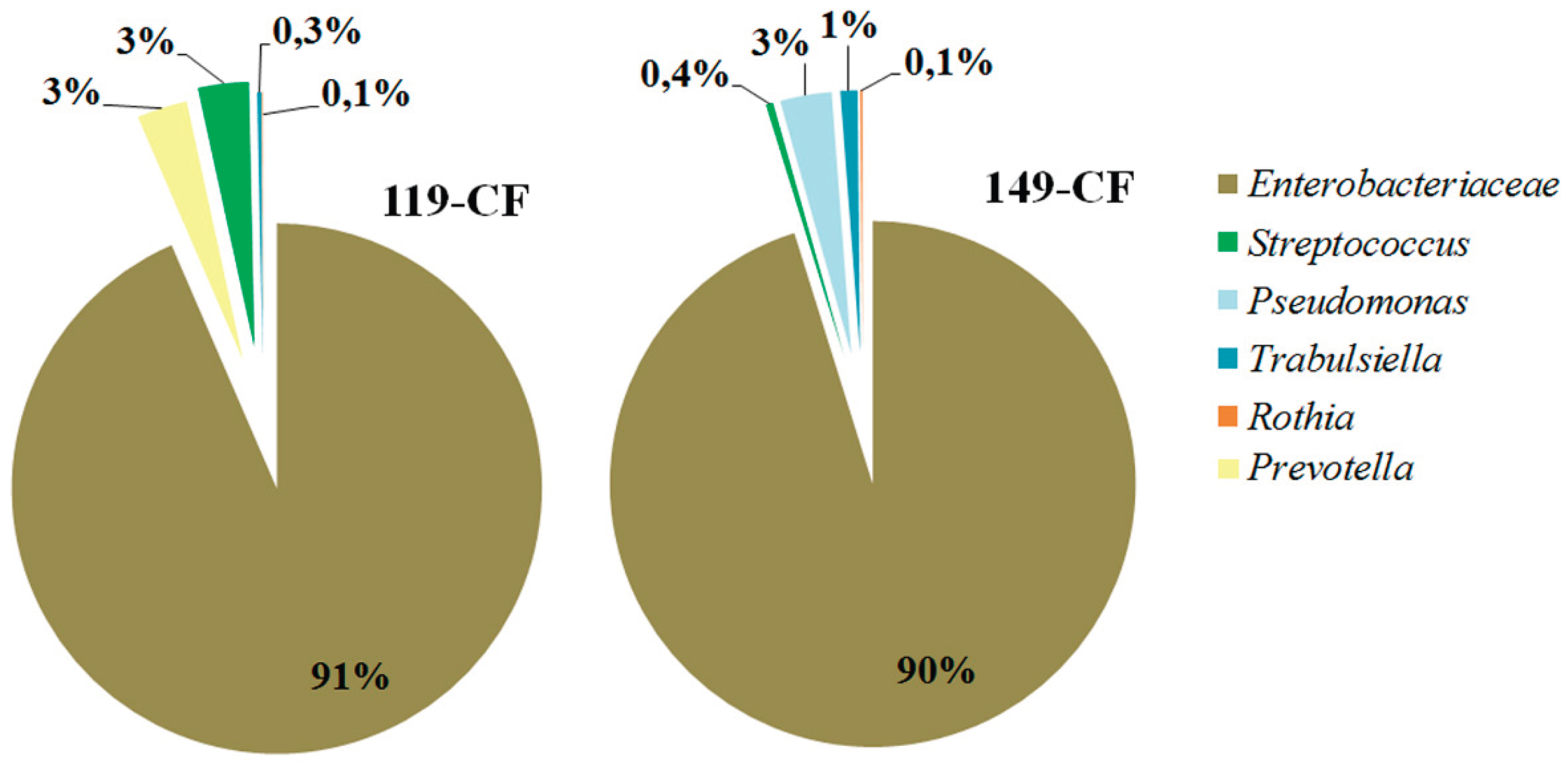
2.1. Microbiome of the Sputum Samples
2.2. Features of the Genomes of E. coli Isolates
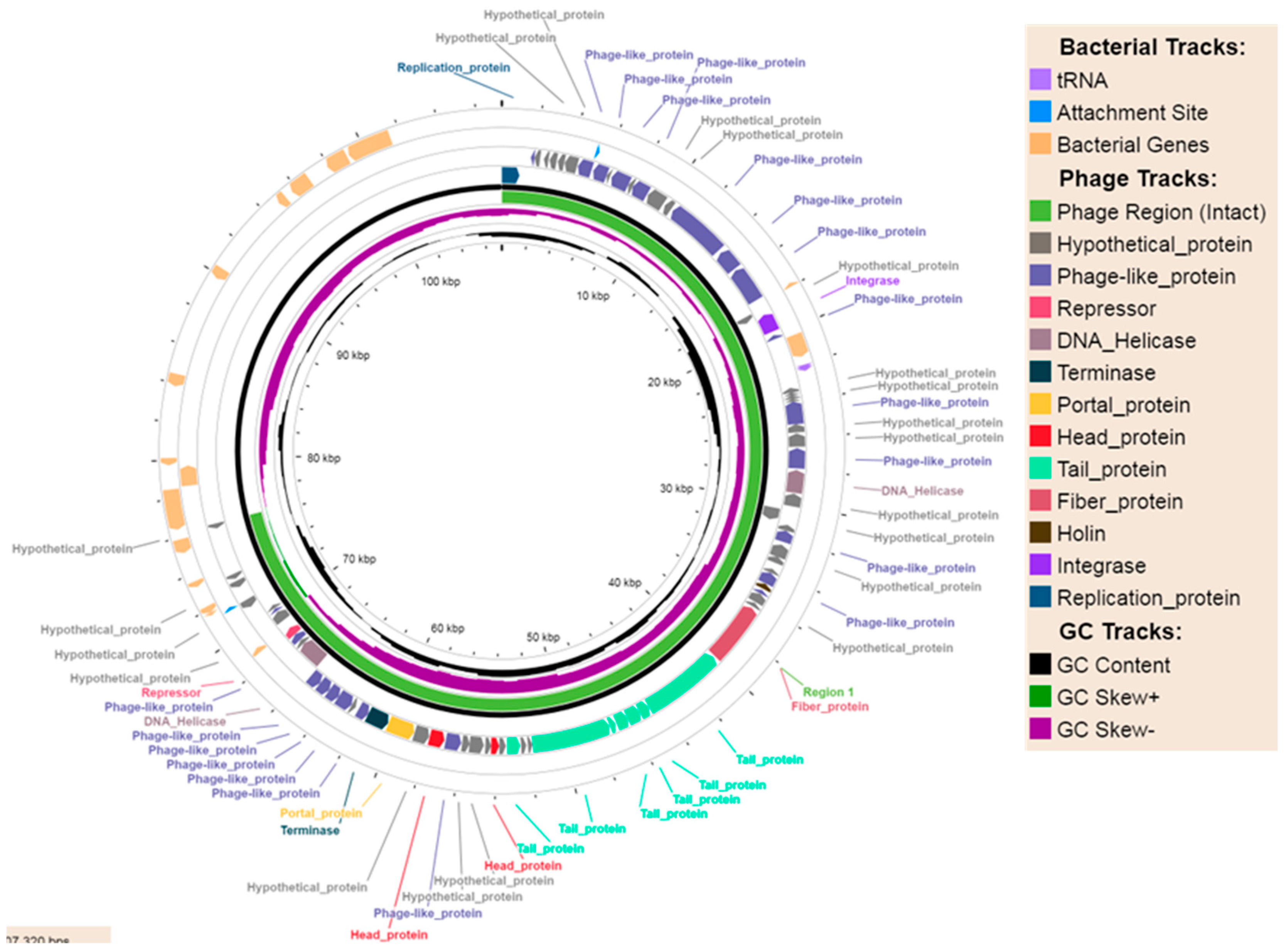
2.2.1. Phage-Plasmids
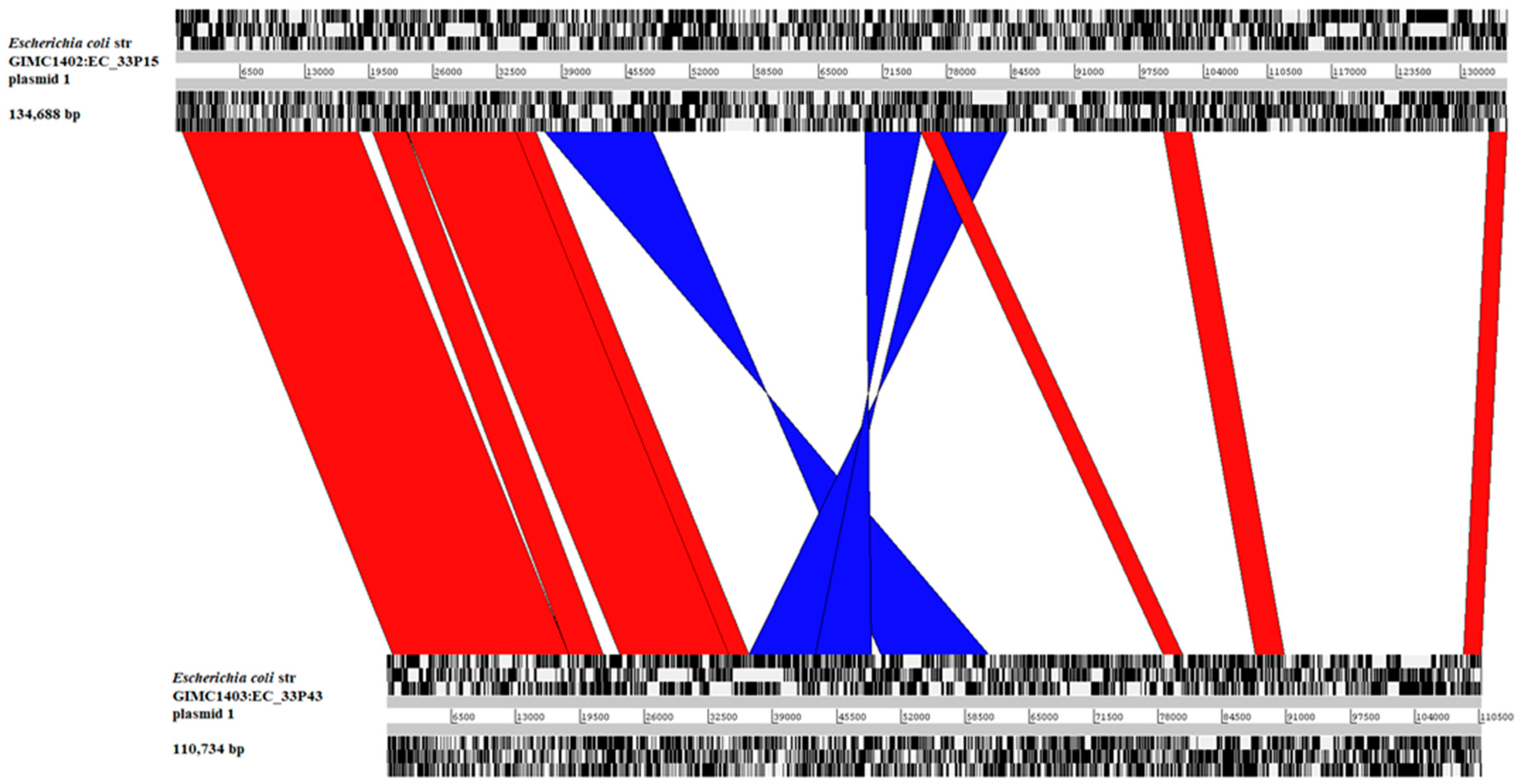
2.2.2. The Large Plasmids pEC_33P15-1 and pEC_33P43-1
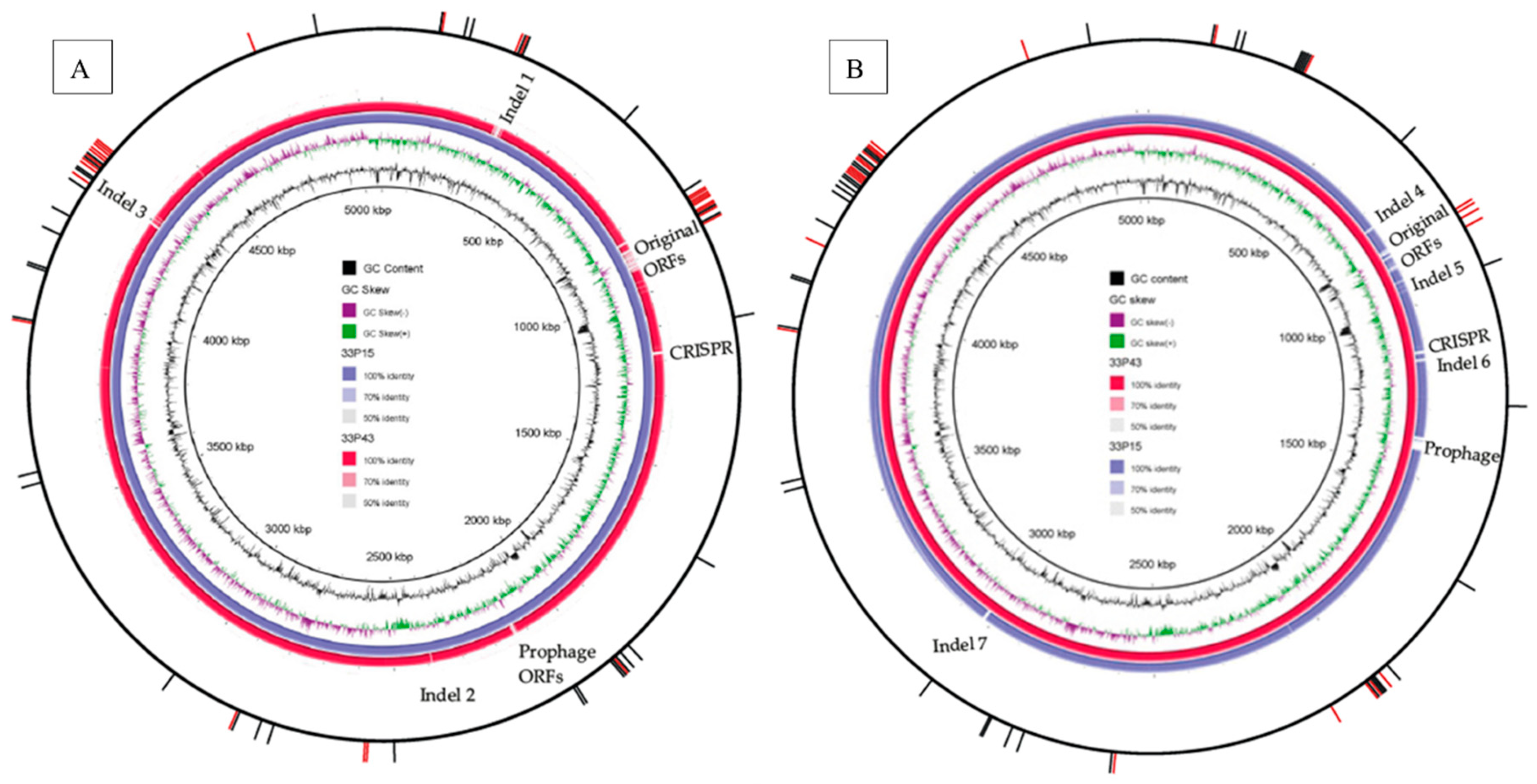
2.2.3. Comparative Analysis of Chromosomes
2.2.4. The Place of CF Isolates in the Population Structure of E. coli ST648
2.2.5. Adaptation to the Long-Term Chronic Infection
2.3. In Vitro Experiments
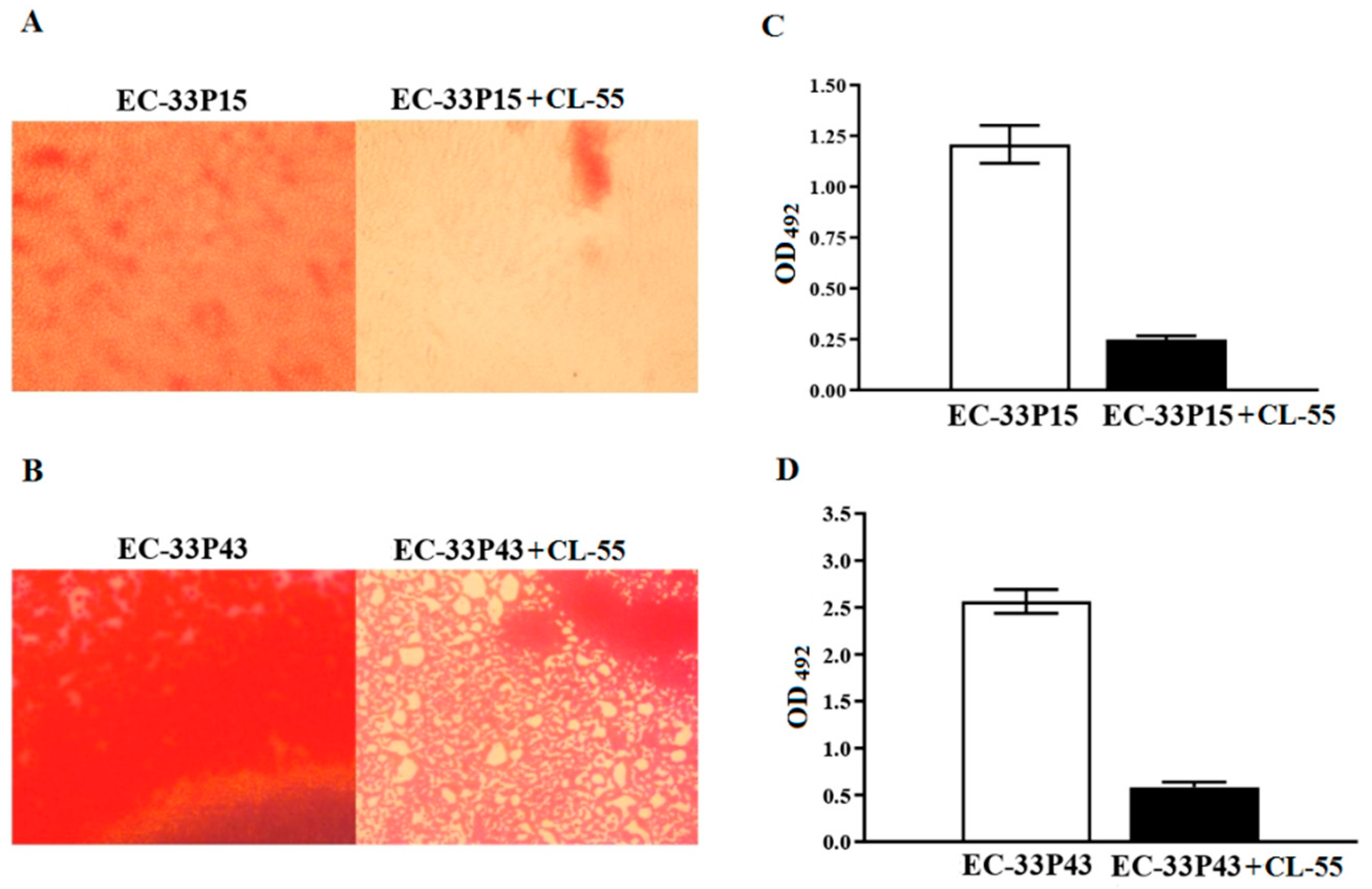
2.3.1. Biofilm Formation
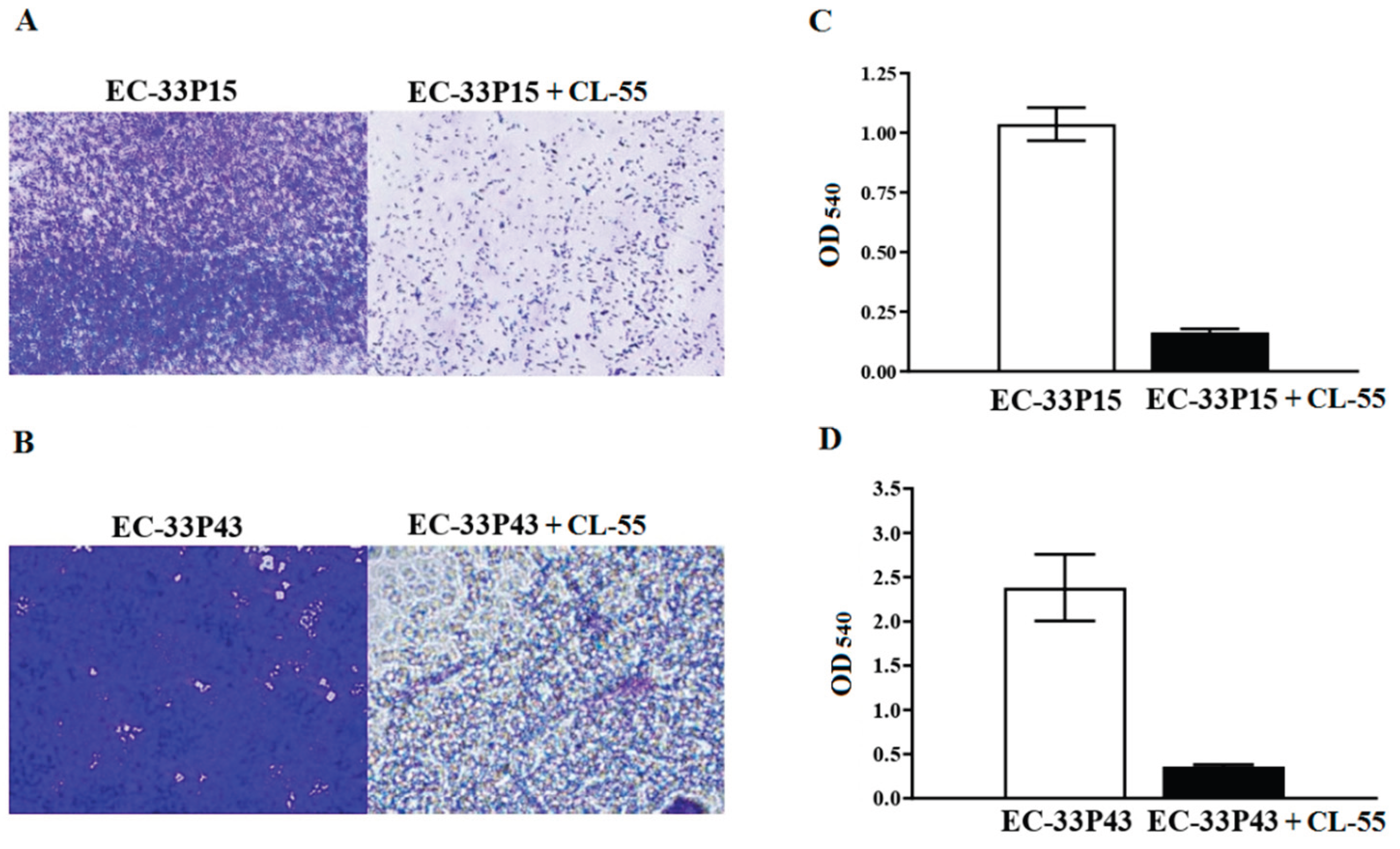
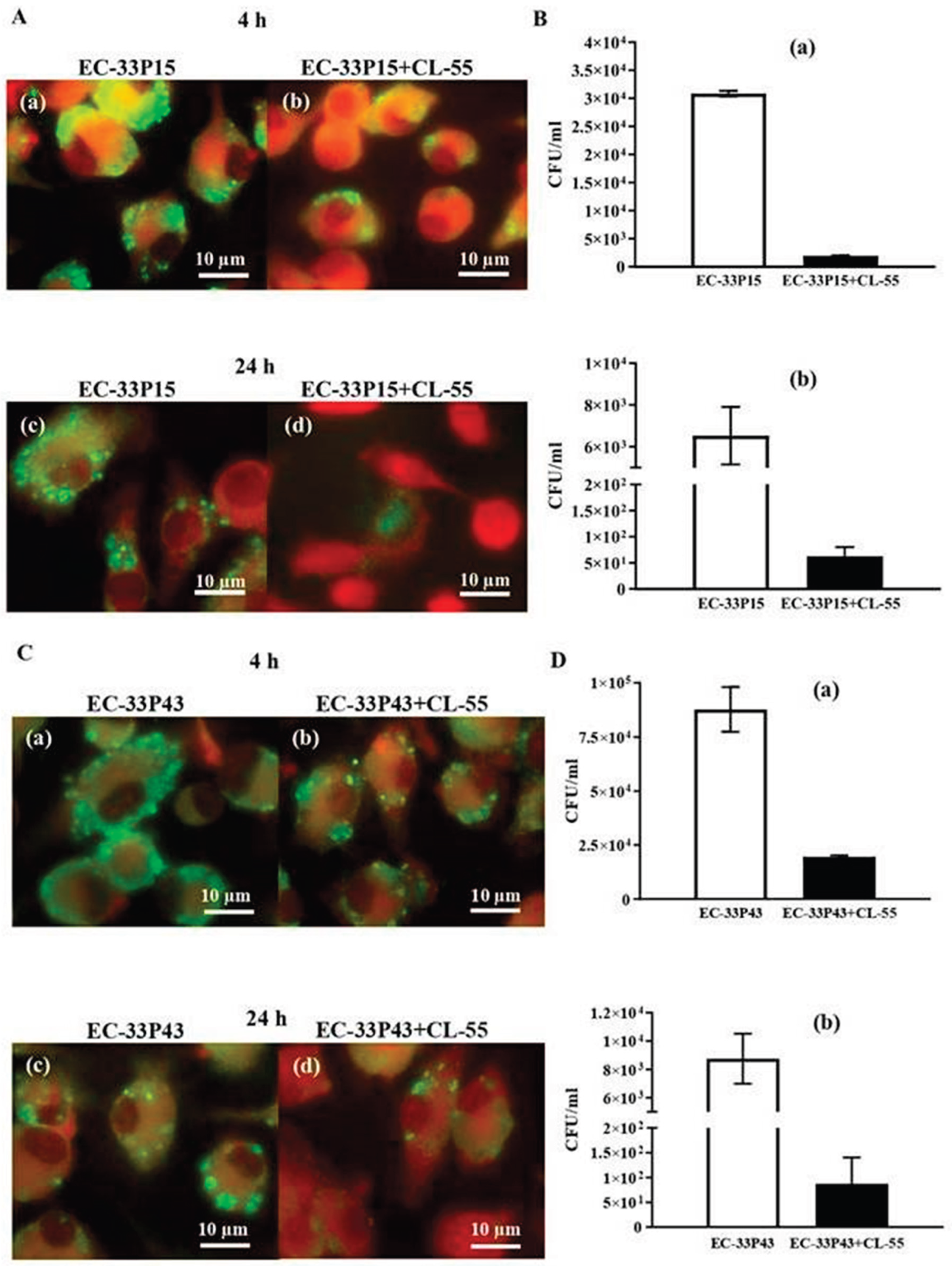
2.3.2. Macrophage Internalization and Survival
2.3.3. CL-55 Does not Inhibit Bacterial Viability
2.3.4. Effect of the Active Pharmaceutical Ingredient CL-55 on Biofilm Formation and Intracellular Survival of CF Isolates
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Methods
- Bacteria isolation and cultivation
- Isolate identification
- Microbiome analysis
- Whole genome sequencing.
- Genome analysis
- Phylogenetic analysis.
- Evaluation of the antibacterial effect of CL-55
- Biofilm assay
- Macrophage cell culture and growth conditions
- Bacterial intracellular survival in RAW264.7 macrophages.
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ExPEC | Extraintestinal pathogenic E. coli |
| CAP | Community-acquired pneumonia |
| NP | Nosocomial pneumonia |
| VAP | Ventilator-associated pneumonia |
| CF | Cystic Fibrosis |
| CPS | Capsular polysaccharide |
| FT | Fluorthiazinone |
| CL-55 | The active pharmaceutical ingredient of Fluorthiazinone |
| T3SS | Type 3 secretion system |
| T6SS | Type 6 secretion system |
| P-P | Phage-plasmid |
| ORF | Open reading frame |
| TRAP | Tripartite ATP-independent periplasmic transporter |
| ECF | energy-coupling factor |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| ESBL | Extended-spectrum beta-lactamase |
| CA | Colanic acid |
| ETT2 | E. coli type 3 secretion system 2 |
References
- Marrie, T.J.; Fine, M.J.; Obrosky, D.S.; Coley, C.; Singer, D.E.; Kapoor, W.N. Community-acquired pneumonia due to Escherichia coli. Clin. Microbiol. Infect. 1998, 4, 717–723. [Google Scholar] [CrossRef]
- La Combe, B.; Clermont, O.; Messika, J.; Eveillard, M.; Kouatchet, A.; Lasocki, S.; Corvec, S.; Lakhal, K.; Billard-Pomares, T.; Fernandes, R.; et al. Pneumonia-SpecificEscherichia coliwith Distinct Phylogenetic and Virulence Profiles, France, 2012–2014. Emerg. Infect. Dis. 2019, 25, 710–718. [Google Scholar] [CrossRef] [PubMed]
- John, T.M.; Deshpande, A.; Brizendine, K.; Yu, P.-C.; Rothberg, M.B. Epidemiology and Outcomes of Community-Acquired Escherichia coli Pneumonia. Open Forum Infect. Dis. 2021, 9, ofab597. [Google Scholar] [CrossRef] [PubMed]
- Messika, J.; Magdoud, F.; Clermont, O.; Margetis, D.; Gaudry, S.; Roux, D.; Branger, C.; Dreyfuss, D.; Denamur, E.; Ricard, J.-D. Pathophysiology of Escherichia coli ventilator-associated pneumonia: implication of highly virulent extraintestinal pathogenic strains. Intensiv. Care Med. 2012, 38, 2007–2016. [Google Scholar] [CrossRef]
- Koulenti, D.; Tsigou, E.; Rello, J. Nosocomial pneumonia in 27 ICUs in Europe: perspectives from the EU-VAP/CAP study. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 36, 1999–2006. [Google Scholar] [CrossRef]
- Stoesser, N.; Sheppard, A.E.; Pankhurst, L.; De Maio, N.; Moore, C.E.; Sebra, R.; Turner, P.; Anson, L.W.; Kasarskis, A.; Batty, E.M.; et al. Evolutionary History of the Global Emergence of the Escherichia coli Epidemic Clone ST131. mBio 2017, 7, e02162–15. [Google Scholar] [CrossRef]
- Schaufler, K.; Semmler, T.; Wieler, L.H.; Trott, D.J.; Pitout, J.; Peirano, G.; Bonnedahl, J.; Dolejska, M.; Literak, I.; Fuchs, S.; et al. Genomic and Functional Analysis of Emerging Virulent and Multidrug-Resistant Escherichia coli Lineage Sequence Type 648. Antimicrob. Agents Chemother. 2019, 63, e00243–19. [Google Scholar] [CrossRef]
- Naghavi, M.; Vollset, S.E.; Ikuta, K.S.; Swetschinski, L.R.; Gray, A.P.; E Wool, E.; Aguilar, G.R.; Mestrovic, T.; Smith, G.; Han, C.; et al. Global burden of bacterial antimicrobial resistance 1990–2021: a systematic analysis with forecasts to 2050. Lancet 2024, 404, 1199–1226. [Google Scholar] [CrossRef]
- Zemanick, E.T.; Hoffman, L.R. Cystic Fibrosis: Microbiology and Host Response. Pediatr. Clin. North Am. 2016, 63, 617–636. [Google Scholar] [CrossRef]
- Barillova, P.; Tchesnokova, V.; Dübbers, A.; Küster, P.; Peters, G.; Dobrindt, U.; Sokurenko, E.V.; Kahl, B.C. Prevalence and persistence of Escherichia coli in the airways of cystic fibrosis patients—An unrecognized CF pathogen? Int. J. Med Microbiol. 2014, 304, 415–421. [Google Scholar] [CrossRef]
- Edwards, B.D.; Somayaji, R.; Greysson-Wong, J.; Izydorczyk, C.; Waddell, B.; Storey, D.G.; Rabin, H.R.; Surette, M.G.; Parkins, M.D. Clinical Outcomes Associated With Escherichia coli Infections in Adults With Cystic Fibrosis: A Cohort Study. Open Forum Infect. Dis. 2019, 7, ofz476. [Google Scholar] [CrossRef]
- Izydorczyk, C.; Waddell, B.; Edwards, B.D.; Greysson-Wong, J.; Surette, M.G.; Somayaji, R.; Rabin, H.R.; Conly, J.M.; Church, D.L.; Parkins, M.D. Epidemiology of E. coli in Cystic Fibrosis Airways Demonstrates the Capacity for Persistent Infection but Not Patient-Patient Transmission. Front. Microbiol. 2020, 11, 475. [Google Scholar] [CrossRef]
- Registry of patients with cystic fibrosis in the Russian Federation. 2023. Editors Amelina E.L., Kashirskaya N.Yu., Kondratyeva E.I., et al.; Publishing House "MEDPRACTIKA-M", Moscow, Russia, 2025.
- Whitfield, C. Biosynthesis and Assembly of Capsular Polysaccharides in Escherichia coli. Annu. Rev. Biochem. 2006, 75, 39–68. [Google Scholar] [CrossRef]
- Xue, P.; Corbett, D.; Goldrick, M.; Naylor, C.; Roberts, I.S. Regulation of Expression of the Region 3 Promoter of the Escherichia coli K5 Capsule Gene Cluster Involves H-NS, SlyA, and a Large 5′ Untranslated Region. J. Bacteriol. 2009, 191, 1838–1846. [Google Scholar] [CrossRef] [PubMed]
- Harris, S.; Piotrowska, M.J.; Goldstone, R.J.; Qi, R.; Foster, G.; Dobrindt, U.; Madec, J.-Y.; Valat, C.; Rao, F.V.; Smith, D.G.E. Variant O89 O-Antigen of E. coli Is Associated With Group 1 Capsule Loci and Multidrug Resistance. Front. Microbiol. 2018, 9, 2026. [Google Scholar] [CrossRef]
- Cheng, H.Y.; Chen, Y.S.; Wu, C.Y.; Chang, H.Y.; Lai, Y.C.; Peng, H.L. RmpA Regulation of Capsular Polysaccharide Biosynthesis in Klebsiella pneumoniae CG43. J. Bacteriol. 2010, 192, 3144–3158. [Google Scholar] [CrossRef] [PubMed]
- Neumann, B.; Lippmann, N.; Wendt, S.; Karlas, T.; Lübbert, C.; Werner, G.; Pfeifer, Y.; Schuster, C.F. Recurrent bacteremia with a hypermucoviscous Escherichia coli isolated from a patient with perihilar cholangiocarcinoma: insights from a comprehensive genome-based analysis. Ann. Clin. Microbiol. Antimicrob. 2022, 21, 28. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-T.; Jeng, Y.-Y.; Chen, T.-L.; Fung, C.-P. Bacteremic community-acquired pneumonia due to Klebsiella pneumoniae: Clinical and microbiological characteristics in Taiwan, 2001-2008. BMC Infect. Dis. 2010, 10, 307–307. [Google Scholar] [CrossRef]
- Han, B.; Li, M.; Xu, Y.; Islam, D.; Khang, J.; Del Sorbo, L.; Lee, W.; Szaszi, K.; Zhong, N.; Slutsky, A.S.; et al. Tsr Chemoreceptor Interacts With IL-8 Provoking E. coli Transmigration Across Human Lung Epithelial Cells. Sci. Rep. 2016, 6, 31087. [Google Scholar] [CrossRef]
- Grubwieser, P.; Hoffmann, A.; Hilbe, R.; Seifert, M.; Sonnweber, T.; Böck, N.; Theurl, I.; Weiss, G.; Nairz, M. Airway Epithelial Cells Differentially Adapt Their Iron Metabolism to Infection With Klebsiella pneumoniae and Escherichia coli In Vitro. Front. Cell. Infect. Microbiol. 2022, 12, 875543. [Google Scholar] [CrossRef]
- Zhuge, X.; Sun, Y.; Jiang, M.; Wang, J.; Tang, F.; Xue, F.; Ren, J.; Zhu, W.; Dai, J. Acetate metabolic requirement of avian pathogenic Escherichia coli promotes its intracellular proliferation within macrophage. Veter- Res. 2019, 50, 31. [Google Scholar] [CrossRef]
- Zhuge, X.; Sun, Y.; Xue, F.; Tang, F.; Ren, J.; Li, D.; Wang, J.; Jiang, M.; Dai, J. A Novel PhoP/PhoQ Regulation Pathway Modulates the Survival of Extraintestinal Pathogenic Escherichia coli in Macrophages. Front. Immunol. 2018, 9, 788. [Google Scholar] [CrossRef]
- Nazareth, H.; Genagon, S.A.; Russo, T.A. Extraintestinal PathogenicEscherichia coliSurvives within Neutrophils. Infect. Immun. 2007, 75, 2776–2785. [Google Scholar] [CrossRef] [PubMed]
- Zigangirova, N.A.; Lubenec, N.L.; Beloborodov, V.B.; Sheremet, A.B.; Nelyubina, S.A.; Bondareva, N.E.; Zakharov, K.A.; Luyksaar, S.I.; Zolotov, S.A.; Levchenko, E.U.; et al. A New “Non-Traditional” Antibacterial Drug Fluorothiazinone—Clinical Research in Patients with Complicated Urinary Tract Infections. Antibiotics 2024, 13, 476. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, E.; Sousa, J.A.M.d.; Touchon, M.; Rocha, E.P.C. Bacteria have numerous distinctive groups of phage–plasmids with conserved phage and variable plasmid gene repertoires. Nucleic Acids Res. 2021, 49, 2655–2673. [Google Scholar] [CrossRef] [PubMed]
- Boyd, J.M.; Sondelski, J.L.; Downs, D.M. Bacterial ApbC Protein Has Two Biochemical Activities That Are Required for in Vivo Function. J. Biol. Chem. 2009, 284, 110–118. [Google Scholar] [CrossRef]
- Jurėnas, D.; Fraikin, N.; Goormaghtigh, F.; Van Melderen, L. Biology and evolution of bacterial toxin–antitoxin systems. Nat. Rev. Microbiol. 2022, 20, 335–350. [Google Scholar] [CrossRef]
- Akhtar, A.A.; Turner, D.P. The role of bacterial ATP-binding cassette (ABC) transporters in pathogenesis and virulence: Therapeutic and vaccine potential. Microb. Pathog. 2022, 171, 105734. [Google Scholar] [CrossRef]
- Sabri, M.; Léveillé, S.; Dozois, C.M. A SitABCD homologue from an avian pathogenic Escherichia coli strain mediates transport of iron and manganese and resistance to hydrogen peroxide. Microbiology 2006, 152, 745–758. [Google Scholar] [CrossRef]
- Hrovat, K.; Zupančič, J.Č.; Seme, K.; Avguštin, J.A. QAC Resistance Genes in ESBL-Producing E. coli Isolated from Patients with Lower Respiratory Tract Infections in the Central Slovenia Region—A 21-Year Survey. Trop Med Infect Dis. 2023, 8, 273. [Google Scholar] [CrossRef]
- Virolle, C.; Goldlust, K.; Djermoun, S.; Bigot, S.; Lesterlin, C. Plasmid Transfer by Conjugation in Gram-Negative Bacteria: From the Cellular to the Community Level. Genes 2020, 11, 1239. [Google Scholar] [CrossRef] [PubMed]
- Ghigo, J.-M. Natural conjugative plasmids induce bacterial biofilm development. Nature 2001, 412, 442–445. [Google Scholar] [CrossRef] [PubMed]
- Barrios, A.F.G.; Zuo, R.; Ren, D.; Wood, T.K. Hha, YbaJ, and OmpA regulate Escherichia coli K12 biofilm formation and conjugation plasmids abolish motility. Biotechnol. Bioeng. 2006, 93, 188–200. [Google Scholar] [CrossRef]
- Okshevsky, M.; Meyer, R.L. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit. Rev. Microbiol. 2015, 41, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Jurėnas, D.; Fraikin, N.; Goormaghtigh, F.; Van Melderen, L. Biology and evolution of bacterial toxin–antitoxin systems. Nat. Rev. Microbiol. 2022, 20, 335–350. [Google Scholar] [CrossRef]
- Boos, W. Binding protein-dependent ABC transport system for glycerol 3-phosphate of Escherichia coli. Methods Enzymol. 1998, 292, 40–51. [Google Scholar] [CrossRef]
- Karimova, G.; Davi, M.; Ladant, D. The β-Lactam Resistance Protein Blr, a Small Membrane Polypeptide, Is a Component of the Escherichia coli Cell Division Machinery. J. Bacteriol. 2012, 194, 5576–5588. [Google Scholar] [CrossRef]
- Wong, R.S.Y.; McMurry, L.M.; Levy, S.B. ‘Intergenic’blr gene in Escherichia coli encodes a 41-residue membrane protein affecting intrinsic susceptibility to certain inhibitors of peptidoglycan synthesis. Mol. Microbiol. 2000, 37, 364–370. [Google Scholar] [CrossRef]
- Davies, J.S.; Currie, M.J.; North, R.A.; Scalise, M.; Wright, J.D.; Copping, J.M.; Remus, D.M.; Gulati, A.; Morado, D.R.; Jamieson, S.A.; et al. Structure and mechanism of a tripartite ATP-independent periplasmic TRAP transporter. Nat. Commun. 2023, 14, 1120. [Google Scholar] [CrossRef]
- Domka, J.; Lee, J.; Wood, T.K. YliH (BssR) and YceP (BssS) Regulate Escherichia coli K-12 Biofilm Formation by Influencing Cell Signaling. Appl. Environ. Microbiol. 2006, 72, 2449–59. [Google Scholar] [CrossRef]
- Guiral, E.; Mendez-Arancibia, E.; Soto, S.M.; Salvador, P.; Fàbrega, A.; Gascón, J.; Vila, J. CTX-M-15–producing EnteroaggregativeEscherichia colias Cause of Travelers’ Diarrhea. Emerg. Infect. Dis. 2011, 17, 1950–1953. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Lee, S.M.; Mao, Y. Protective effect of exopolysaccharide colanic acid of Escherichia coli O157:H7 to osmotic and oxidative stress. Int. J. Food Microbiol. 2004, 93, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Makino, K.; Ohnishi, M.; Kurokawa, K.; Ishii, K.; Yokoyama, K.; Han, C.G.; Ohtsubo, E.; Nakayama, K.; Murata, T.; et al. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001, 8, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Yang, C.-H.; Kharadi, R.R.; Yuan, X.; Sundin, G.W.; Triplett, L.R.; Wang, J.; Zeng, Q.; Wang, N. Cell-length heterogeneity: a population-level solution to growth/virulence trade-offs in the plant pathogen Dickeya dadantii. PLOS Pathog. 2019, 15, e1007703. [Google Scholar] [CrossRef]
- Arredondo-Alonso, S.; Blundell-Hunter, G.; Fu, Z.; Gladstone, R.A.; Fillol-Salom, A.; Loraine, J.; Cloutman-Green, E.; Johnsen, P.J.; Samuelsen, Ø.; Pöntinen, A.K.; et al. Evolutionary and functional history of the Escherichia coli K1 capsule. Nat. Commun. 2023, 14, 3294. [Google Scholar] [CrossRef]
- Rodriguez, M.-L.; Jann, B.; Jann, K. Structure and serological characteristics of the capsular K4 antigen of Escherichia coli O5:K4:H4, a fructose-containing polysaccharide with a chondroitin backbone. Eur. J. Biochem. 1988, 177, 117–124. [Google Scholar] [CrossRef]
- Sidjabat, H.E.; Paterson, D.L.; Adams-Haduch, J.M.; Ewan, L.; Pasculle, A.W.; Muto, C.A.; Tian, G.-B.; Doi, Y. Molecular Epidemiology of CTX-M-Producing Escherichia coli Isolates at a Tertiary Medical Center in Western Pennsylvania. Antimicrob. Agents Chemother. 2009, 53, 4733–4739. [Google Scholar] [CrossRef]
- Mshana, S.E.; Imirzalioglu, C.; Hain, T.; Domann, E.; Lyamuya, E.F.; Chakraborty, T. Multiple ST clonal complexes, with a predominance of ST131, of Escherichia coli harbouring blaCTX-M-15 in a tertiary hospital in Tanzania. Clin. Microbiol. Infect. 2011, 17, 1279–1282. [Google Scholar] [CrossRef]
- Zong, Z.; Yu, R. Escherichia coli carrying the bla CTX-M-15 gene of ST648. J. Med Microbiol. 2010, 59, 1536–1537. [Google Scholar] [CrossRef]
- Sherchan, J.B.; Hayakawa, K.; Miyoshi-Akiyama, T.; Ohmagari, N.; Kirikae, T.; Nagamatsu, M.; Tojo, M.; Ohara, H.; Tandukar, S. Clinical Epidemiology and Molecular Analysis of Extended-Spectrum-β-Lactamase-Producing Escherichia coli in Nepal: Characteristics of Sequence Types 131 and 648. Antimicrob. Agents Chemother. 2015, 59, 3424–3432. [Google Scholar] [CrossRef]
- Ewers, C.; Bethe, A.; Semmler, T.; Guenther, S.; Wieler, L. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin. Microbiol. Infect. 2012, 18, 646–655. [Google Scholar] [CrossRef]
- Piekar, M.; Álvarez, V.E.; Knecht, C.; Leguina, C.; Allende, N.G.; Páez, L.C.; Gambino, A.S.; Machuca, A.G.; Campos, J.; Fox, B.; et al. Genomic data reveals the emergence of the co-occurrence of blaKPC-2 and blaCTX-M-15 in an Escherichia coli ST648 strain isolated from rectal swab within the framework of hospital surveillance. J. Glob. Antimicrob. Resist. 2023, 32, 108–112. [Google Scholar] [CrossRef]
- He, X.; Xu, L.; Dai, H.; Ge, M.; Zhu, J.; Fu, H.; Zhu, S.; Shao, J. Genomic Characteristics of a Multidrug-Resistant ST648 Escherichia coli Isolate Co-Carrying blaKPC-2 and blaCTX-M-15 Genes Recovered from a Respiratory Infection in China. Infect. Drug Resist. 2023, 16, 3535–3540. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.-S.; Feng, Y.; Lv, X.-Y.; Duan, J.-H.; Chen, J.; Fang, L.-X.; Xia, J.; Liao, X.-P.; Sun, J.; Liu, Y.-H. Emergence of NDM-5- and MCR-1-Producing Escherichia coli Clones ST648 and ST156 from a Single Muscovy Duck (Cairina moschata). Antimicrob. Agents Chemother. 2016, 60, 6899–6902. [Google Scholar] [CrossRef] [PubMed]
- Palmeira, J.D.; Ferreira, H.; Madec, J.-Y.; Haenni, M. Pandemic Escherichia coli ST648 isolate harbouring fosA3 and blaCTX-M-8 on an IncI1/ST113 plasmid: A new successful combination for the spread of fosfomycin resistance? J. Glob. Antimicrob. Resist. 2018, 15, 254–255. [Google Scholar] [CrossRef] [PubMed]
- Ewbank, A.C.; Fuentes-Castillo, D.; Sacristán, C.; Esposito, F.; Fuga, B.; Cardoso, B.; Godoy, S.N.; Zamana, R.R.; Gattamorta, M.A.; Catão-Dias, J.L.; et al. World Health Organization critical priority Escherichia coli clone ST648 in magnificent frigatebird (Fregata magnificens) of an uninhabited insular environment. Front. Microbiol. 2022, 13, 940600. [Google Scholar] [CrossRef]
- Pea, F. Intracellular Pharmacokinetics of Antibacterials and Their Clinical Implications. Clin. Pharmacokinet. 2018, 57, 177–189. [Google Scholar] [CrossRef]
- Pea, F.; Viale, P.; Furlanut, M. Antimicrobial therapy in critically ill patients: a review of pathophysiological conditions responsible for altered disposition and pharmacokinetic variability. Clin. Pharmacokinet. 2005, 44, 1009–1034. [Google Scholar] [CrossRef]
- Zigangirova, N.A.; Zayakin, E.S.; Kapotina, L.N.; Kost, E.A.; Didenko, L.V.; Davydova, D.Y.; Rumyanceva, J.P.; Gintsburg, A.L. Development of Chlamydial Type III Secretion System Inhibitors for Suppression of Acute and Chronic Forms of Chlamydial Infection. Acta Naturae 2012, 4, 87–97. [Google Scholar] [CrossRef]
- Savitskii, M.V.; Moskaleva, N.E.; Brito, A.; Markin, P.A.; Zigangirova, N.A.; Soloveva, A.V.; Sheremet, A.B.; Bondareva, N.E.; Lubenec, N.L.; Tagliaro, F.; et al. Pharmacokinetics, tissue distribution, bioavailability and excretion of the anti-virulence drug Fluorothiazinon in rats and rabbits. J. Antibiot. 2024, 77, 382–388. [Google Scholar] [CrossRef]
- Zigangirova, N.A.; Nesterenko, L.N.; Sheremet, A.B.; Soloveva, A.V.; Luyksaar, S.I.; Zayakin, E.S.; Balunets, D.V.; Gintsburg, A.L. Fluorothiazinon, a small-molecular inhibitor of T3SS, suppresses salmonella oral infection in mice. J. Antibiot. 2021, 74, 244–254. [Google Scholar] [CrossRef]
- Garcia-Medina, R.; Dunne, W.M.; Singh, P.K.; Brody, S.L. Pseudomonas aeruginosa Acquires Biofilm-Like Properties within Airway Epithelial Cells. Infect. Immun. 2005, 73, 8298–8305. [Google Scholar] [CrossRef]
- Høiby, N. A short history of microbial biofilms and biofilm infections. APMIS 2017, 125, 272–275. [Google Scholar] [CrossRef]
- Høiby, N.; Axelsen, N.H. Identification and quantitation of precipitins against Pseudomonas aeruginosa in patients with cystic fibrosis by means of crossed immunoelectrophoresis with intermediate gel. Acta Pathol. Microbiol. Scand. Sect. B Microbiol. Immunol. 1973, 81B, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Burmølle, M.; Thomsen, T.R.; Fazli, M.; Dige, I.; Christensen, L.; Homøe, P.; Tvede, M.; Nyvad, B.; Tolker-Nielsen, T.; Givskov, M.; et al. Biofilms in chronic infections – a matter of opportunity – monospecies biofilms in multispecies infections. FEMS Immunol. Med Microbiol. 2010, 59, 324–336. [Google Scholar] [CrossRef] [PubMed]
- Staudinger, B.J.; Muller, J.F.; Halldórsson, S.; Boles, B.; Angermeyer, A.; Nguyen, D.; Rosen, H.; Baldursson, Ó.; Gottfredsson, M.; Guðmundsson, G.H.; et al. Conditions Associated with the Cystic Fibrosis Defect Promote Chronic Pseudomonas aeruginosa Infection. Am. J. Respir. Crit. Care Med. 2014, 189, 812–824. [Google Scholar] [CrossRef] [PubMed]
- Voronina, O.L.; Kunda, M.S.; Ryzhova, N.N.; Aksenova, E.I.; Semenov, A.N.; Lasareva, A.V.; Amelina, E.L.; Chuchalin, A.G.; Lunin, V.G.; Gintsburg, A.L. The Variability of the Order Burkholderiales Representatives in the Healthcare Units. BioMed Res. Int. 2015, 2015, 680210. [Google Scholar] [CrossRef]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.J.; Ochman, H.; et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef]
- Voronina, O.L.; Kunda, M.S.; Ryzhova, N.N.; Aksenova, E.I.; Sharapova, N.E.; Semenov, A.N.; Amelina, E.L.; Chuchalin, A.G.; Gintsburg, A.L. On Burkholderiales order microorganisms and cystic fibrosis in Russia. BMC Genom. 2018, 19, 74. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; O’nEill, K.R.; Haft, D.H.; DiCuccio, M.; Chetvernin, V.; Badretdin, A.; Coulouris, G.; Chitsaz, F.; Derbyshire, M.K.; Durkin, A.S.; et al. RefSeq: expanding the Prokaryotic Genome Annotation Pipeline reach with protein family model curation. Nucleic Acids Res. 2021, 49, D1020–D1028. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; A Wlodarski, M.; Edalatmand, A.; Petkau, A.; A Syed, S.; Tsang, K.K.; et al. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef]
- Olson, R.D.; Assaf, R.; Brettin, T.; Conrad, N.; Cucinell, C.; Davis, J.J.; Dempsey, D.M.; Dickerman, A.; Dietrich, E.M.; Kenyon, R.W.; et al. Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): a resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023, 51, D678–D689. [Google Scholar] [CrossRef]
- Naas, T.; Oueslati, S.; Bonnin, R.A.; Dabos, M.L.; Zavala, A.; Dortet, L.; Retailleau, P.; Iorga, B.I. Beta-lactamase database (BLDB)–structure and function. J. Enzym. Inhib. Med. Chem. 2017, 32, 917–919. [Google Scholar] [CrossRef]
- Ross, K.; Varani, A.M.; Snesrud, E.; Huang, H.; Alvarenga, D.O.; Zhang, J.; Wu, C.; McGann, P.; Chandler, M.; Gottesman, S. TnCentral: a Prokaryotic Transposable Element Database and Web Portal for Transposon Analysis. mBio 2021, 12, e0206021. [Google Scholar] [CrossRef]
- Johansson, M.H.K.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garcìa-Fernandez, A.; Larsen, M.; Lund, O.; Voldby Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids. Antimicrob using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Wishart, D.S.; Han, S.; Saha, S.; Oler, E.; Peters, H.; Grant, J.R.; Stothard, P.; Gautam, V. PHASTEST: faster than PHASTER, better than PHAST. Nucleic Acids Res. 2023, 51, W443–W450. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef] [PubMed]
- Alikhan, N.-F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.J.; Rutherford, K.M.; Berriman, M.; Rajandream, M.-A.; Barrell, B.G.; Parkhill, J. ACT: the Artemis comparison tool. Bioinformatics 2005, 21, 3422–3423. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylogroups. Environ Microbiol Rep. 2013, 5, 58–65, doi: 10.1111/1758-2229.12019. Tamura K, Stecher G, Kumar S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol. 2021, 38, 3022-3027. doi: 10.1093/molbev/msab120. [Google Scholar]
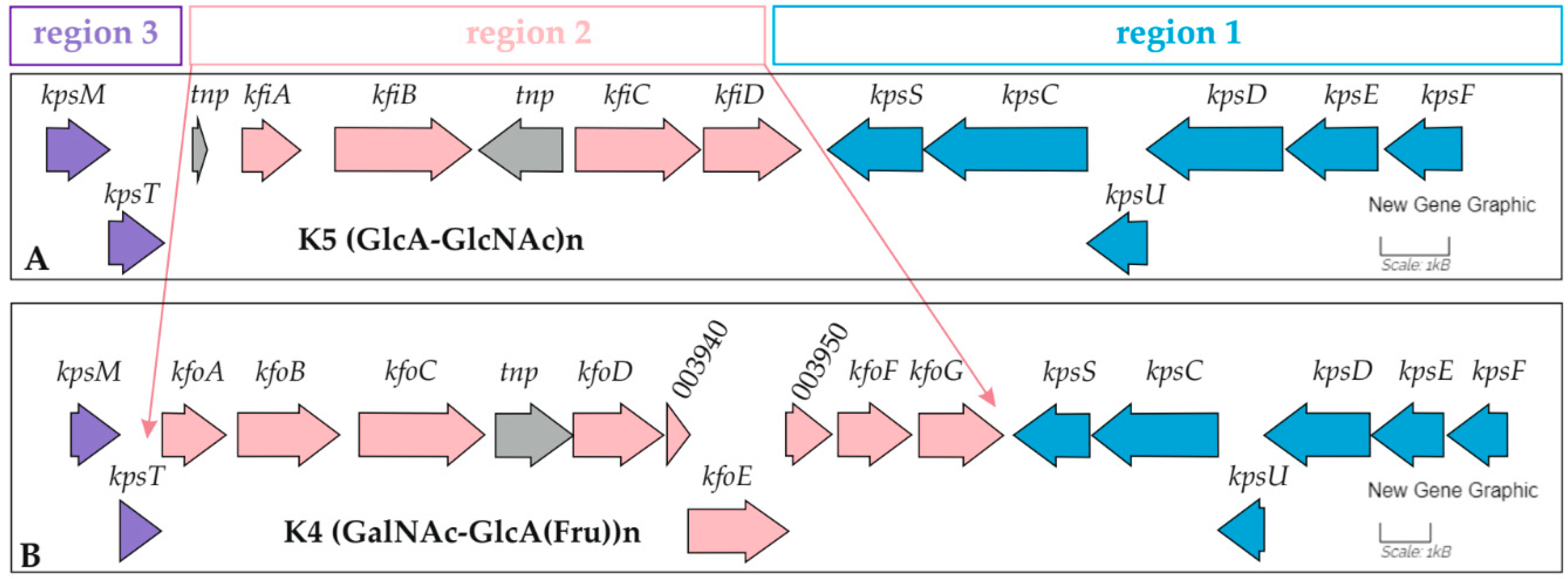

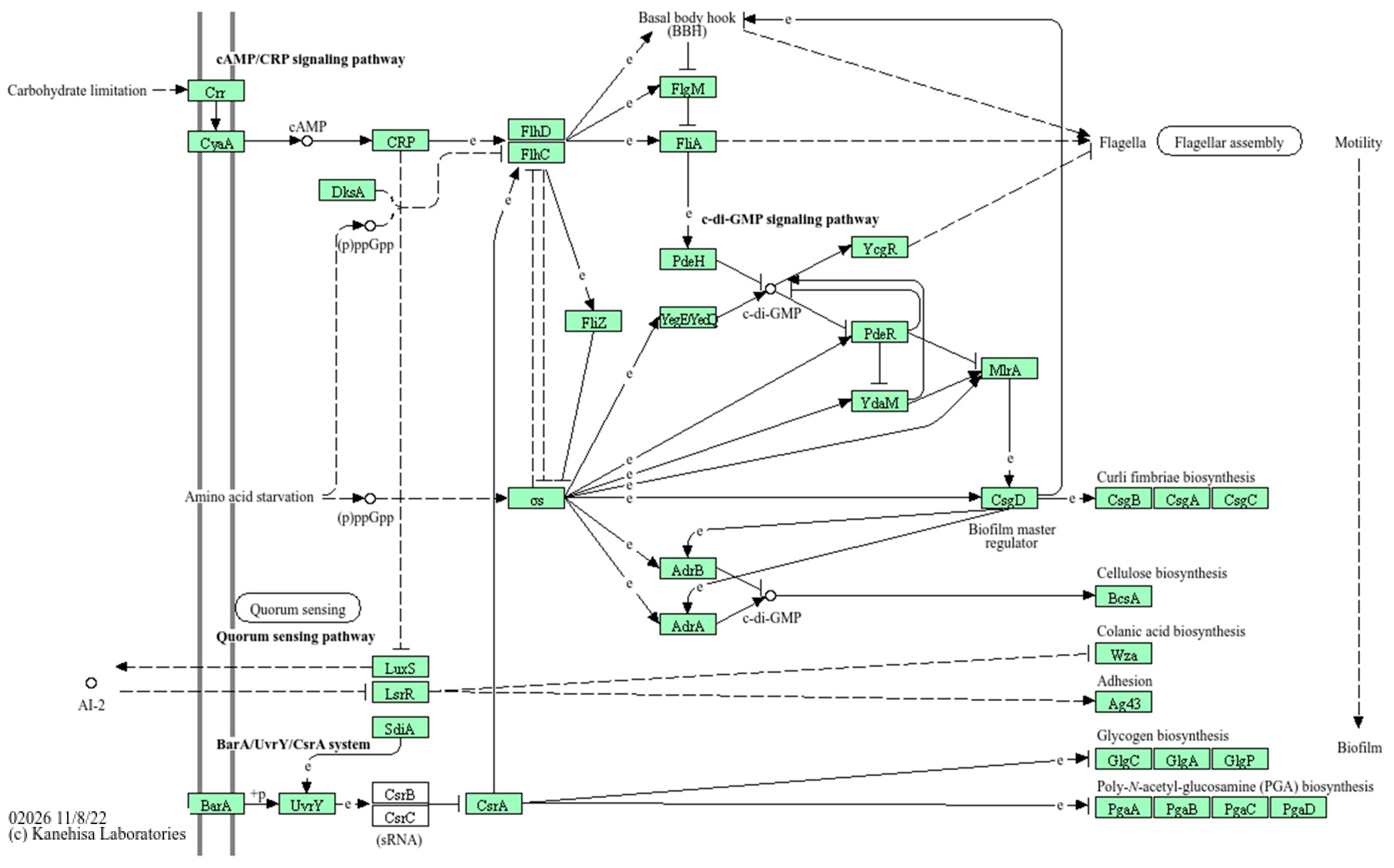
| Features | GIMC1402:EC_33P15 | GIMC1403:EC_33P43 |
| Chromosome, bp. | 5115804 | 5032495 |
| Genes (total) | 5292 | 5154 |
| CDSs (total) | 5171 | 5032 |
| Genes (RNA) | 121 | 122 |
| tRNAs | 94 | 95 |
| Pseudo Genes (total) | 206 | 202 |
| CRISPR Arrays | 2 | 2 |
| Plasmid | pEC_33P15-1 (134688 bp) | pEC_33P43-1 (110734 bp) |
| pEC_33P15-2 (4237 bp) | pEC_33P43-2 (7176 bp) | |
| pEC_33P15-3 (4072 bp) | pEC_33P43-3 (2091 bp) | |
| pEC_33P43-4 (1459 bp) | ||
| Phage-Plasmid | p-pEC_33P15 (108306 bp) | p-pEC_33P43 (107320 bp) |
| Сategory | Function | GIMC1402:EC_33P15, pl1 | GIMC1403:EC_33P43, pl1 | |
| Product (genome position) | Product (genome position) | |||
| Resistance | Tetracycline resistance | tetracycline efflux MFS transporter Tet(B) (50012..51217) | – | |
| transcriptional repressor TetR(B) (51299..51922) | – | |||
| Chloramphenicol resistance | CatA1, chloramphenicol O-acetyltransferase (54209..54868) | – | ||
| Macrolide resistance | Mph(A), family macrolide 2'-phosphotransferase (59909..60814) | – | ||
| Mrx(A), macrolide resistance MFS transporter (60811..62049) | – | |||
| Sulfonamide resistance | Sul1 sulfonamide-resistant dihydropteroate synthase (66125..66964) | – | ||
| Aminoglycoside resistance | AadA5, ANT(3'')-Ia family aminoglycoside nucleotidyltransferase (67511..68299) | – | ||
| Trimethoprim resistance | Dft17, trimethoprim-resistant dihydrofolate reductase (68430..68903) | – | ||
| Quaternary ammonium compound resistance | QacE, QAC efflux SMR transporter (66958..67305) | – | ||
| Virulence factors | Protection against the macroorganism's compliment system; participation in the biofilm formation | the F-type transfer system (24233 – 34796; 79117 – 84085) | the F-type transfer system (22132 - 41597) | |
| Colonization and survival under conditions of Fe2+, Pb2+, Zn2+ and Mn2+ deficiency | iucABCD, iutA, aerobactin (110608..119908) | – | ||
| Fe2+ ABC-transporter (38945..43430) | Fe2+ ABC-transporter (59262..54777) | |||
| Fe2+/Pb2+ ILT-transporter (43534..45988) | Fe2+/Pb2+ ILT-transporter (54673..52237) | |||
| SitABCD ABC-transporter (105156..108605) | – | |||
| – | TonB-dependent transport system (68082..71288; 81969..83939) | |||
| – | YncE protein (71357..72532) | |||
| Toxin-antitoxin systems (TA) | Selective advantage of a clone in a bacterial population, formation of a persistent cell population | Type I* | Mok/Hok TA (20002..20218) | Mok/Hok TA (18537..18753) |
| Hok/Gef TA (77769..77903) | Hok/Gef TA (42811..42945) | |||
| Type II** | – | Phd_YefM/ Fic_DOC TA (98859..99460) | ||
| VapB/VapC TA (123886..124529; 128749..129392) | – | |||
| CcdA/CcdB TA (133356..133881) | CcdA/CcdB TA (109402..109927) | |||
| PemL/PemK TA (71821..72412) | PemL/PemK TA (46343..46934) | |||
| GIMC1402:EC_33P15 | GIMC1403:EC_33P43 | |||
|---|---|---|---|---|
| Region | Gene | Product | Gene | Product |
| 1 | 351708..350878 | TEM-1 | ||
| 351852..352556 | IS6-Tnp | 331206..331280 | IS6-Tnp-pseudo | |
| 352700..353254 | AAC(6')-Ib-cr | 331424..331978 | AAC(6')-Ib-cr | |
| 353385..354215 | OXA-1 | 332109..332939 | OXA-1 | |
| 354353..354793 | CatB3-pseudo | 333077..333517 | CatB3-pseudo | |
| complement(354847..355551) | IS6-Tnp | complement(333571..334275) | IS6-Tnp | |
| 355658..356518 | AAC(3)-IIa | |||
| 356531..357073 | tmrB | |||
| 357165..358213 | IS3-Tnp | |||
| complement(358267..358971) | IS6-Tnp | |||
| 2 | complement(359039..361267) | Tn3-Tnp-pseudo | complement(334343..336571) | Tn3-Tnp-pseudo |
| complement(361672..362547) | CTX-M-15 | complement(336976..337851) | CTX-M-15 | |
| complement(362803..364065) | IS1380-Tnp | complement(338107..339369) | IS1380-Tnp | |
| Position | CRISPR Length | Consensus_Repeat | Repeat ID (CRISPRdb) | Spacers Nb | Evidence Level |
|---|---|---|---|---|---|
| GIMC1402:EC_33P15 | |||||
| 1154150…1155279 | 1129 | GTGTTCCCCGCGCCAGCGGGGATAAACCG | R6121 | 18 | 4 |
| 1180826…1181648 | 822 | GAGTTCCCCGCGCCAGCGGGGATAAACCG | R3946 | 13 | 4 |
| GIMC1403:EC_33P43 | |||||
| 1115575…1116213 | 638 | GAGTTCCCCGCGCCAGCGGGGATAAACCG | R3946 | 10 | 4 |
| 1143351…1143925 | 574 | GTGTTCCCCGCGCCAGCGGGGATAAA | Unknown | 9 | 4 |
| Strain, Accession number | GIMC1402:EC_33P15, CP181181.1 | GIMC1403:EC_33P43, CP181392.1 | NA023, JSXK000000000.1 | 32 - 2823 ED, DABAXP000000000.1 | VB 962116, DABAMI000000000.1 | F_30_1_R8, PIIR000000000.1 |
| clade | 1 | 2 | 3 | 4 | ||
| GIMC1402:EC_33P15 | - | 99,61 | 99,49 | 99,54 | 98,92 | 99,08 |
| GIMC1403:EC_33P43 | 95,65 | - | 99,39 | 99,36 | 98,84 | 98,98 |
| NA023, clade 1 | 92,45 | 91,54 | - | 99,51 | 98,98 | 98,98 |
| 32 - 2823 ED, clade 2 | 89,32 | 89,96 | 90,07 | - | 99,33 | 99,43 |
| VB 962116, clade 3 | 88,07 | 87,82 | 98,98 | 90,01 | - | 99,72 |
| F_30_1_R8, clade 4 | 90,63 | 89,98 | 89,95 | 89,76 | 90,87 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





