1. Introduction
Mutations in the epidermal growth factor receptor (
EGFR) gene are the most frequent driver mutations in non-small cell lung cancer (NSCLC), especially in patients with East-Asian ethnicity and no history of smoking [
1]. Numerous subtypes of
EGFR mutations have been reported to date [
2], and these mutations are usually classified as common mutations (L858R point mutation or exon 19 in-frame deletions) and uncommon mutations (all other mutations). Uncommon mutations are usually detected in about 10% of patients with
EGFR mutations, irrespective of disease stage [
3]. The common versus uncommon classification is useful when considering treatment strategies in advanced-stage settings, because uncommon mutations are usually less sensitive to some of the currently available EGFR tyrosine kinase inhibitors (TKIs) [
4]. This observation has been validated in structure-based analysis; many of the uncommon
EGFR mutations are classified into the P-loop alphaC-helix compressing subtype that is usually insensitive to first- (1G) and third-generation (3G) TKIs, while some (such as L861Q/R) are classified as classical-like
EGFR mutations [
5].
In the Lux-Lung clinical trials [
6], afatinib monotherapy demonstrated a progression-free survival (PFS) of 10.7 months (95% confidence interval 5.6–14.7) in patients with NSCLC harboring uncommon
EGFR mutations (excluding exon 20 insertion- and T790M-positive groups) in the front-line setting. This finding has been validated in the phase III ACHILLES study, which reported superior PFS with afatinib (10.6 months) over platinum plus pemetrexed (5.7 months) in NSCLC patients with uncommon
EGFR mutations [
7]. Osimertinib has also shown some activity against NSCLC in these patients, with phase II studies reporting a PFS of 9.4 (3.7–15.2) months in Japanese patients [
8] and 8.2 months (5.9–10.5) in Korean patients [
9]. As a result, afatinib or osimertinib are often used in daily clinical practice to treat advanced NSCLC with uncommon
EGFR mutations. However, the PFS reported for these agents is shorter than that reported for patients harboring common
EGFR mutations; for example, a PFS of 18.9 months has been reported in patients with common mutations receiving osimertinib [
10]. Although the recent CHRYSALIS-2 study (cohort C) demonstrated promising efficacy for amivantamab plus lazertinib in patients with NSCLC harboring uncommon
EGFR mutations [
11] research into single-agent TKI regimens is still warranted because of the high toxicity associated with the amivantamab plus lazertinib combination.
Several novel 3G-TKIs are currently under clinical development, some of which may have activity against uncommon EGFR mutations. In addition, some of these new drugs may overcome the acquired resistance that can occur during treatment with currently available TKIs, including afatinib and osimertinib. Here, we used Ba/F3 cell models of NSCLC driven by uncommon EGFR mutations to evaluate the efficacy of novel 3G-TKIs in the first-line setting and the second line after afatinib or osimertinib treatment failure.
2. Materials and Methods
2.1. Data Collection from the cBioPortal Database
Data on
EGFR mutation subtypes and their frequencies in NSCLC were extracted from the cBioPortal database (
https://www.cbioportal.org) as of April 2024. We counted the total number of each mutation in exons 18–21 of
EGFR, excluding L858R point mutation, exon 19 in-frame deletions, and exon 20 in-frame insertions.
2.2. Cell Lines and Reagents
The Ba/F3 cell line was provided by Riken Bio Resource Center (Tsukuba, Japan). Ba/F3 cell lines driven by uncommon
EGFR mutations, E709K, G719A, exon 18 deletion (delE709_T710insD), and S768I, as well as wild-type human
EGFR were established in our previous studies [
12,
13]. In this study, Ba/F3 cells driven by
EGFR L861Q mutation were established as reported previously [
13,
14]. The cells were cultured in RPMI 1640 medium (Wako, Osaka, Japan) supplemented with 10% fetal bovine serum (Sigma-Aldrich, St. Louis, MO) and maintained at 37 °C in a humidified incubator with 5% CO2. Human recombinant EGF was purchased from Thermo Fisher Scientific (Waltham, MA). First-generation (1G) EGFR-TKIs (gefitinib and erlotinib), second-generation (2G) EGFR-TKI (afatinib), and 3G EGFR-TKIs (osimertinib, furmonertinib, lazertinib, almonertinib, rezivertinib, and befotertinib) were purchased from Selleck Chemicals (Houston, TX).
2.3. Establishment of Ba/F3 Cells with EGFR L861Q Mutation
Ba/F3 cells driven by the EGFR L861Q mutation were established in this study. Briefly, the pBABE retroviral vector with a full-length cDNA fragment of human EGFR with the L861Q point mutation was purchased from Addgene (Cambridge, MA). The pBABE construct was co-transfected into gpIRES-293 cells with the pVSV-G vector (Clontech, Mountain View, CA) using FuGENE6 transfection reagent (Promega, Madison, Wisconsin). Viral envelopes were generated to produce viral particles. After 48 hours of transfection, the culture medium was collected and centrifuged at 1500 × g for 45 minutes at 4°C to concentrate the virus particles. Viral pellets were resuspended in DMEM (Sigma-Aldrich) and stored at −80°C.
2.4. Growth Inhibition Assay
Ba/F3 cells were seeded at a density of 2500 cells/well in 96-well plates. After 24-h incubation, cells were exposed to each TKI at the concentrations determined based on the ranges of clinically achievable drug concentration. After 72 h, 10 µL of the tetrazolium salt WST-8 from a Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan) was added to each well, and the plates were incubated for an additional 1.5–3 h. The absorbance was read at 450 nm using a multiplate reader (Tecan, Mannedorf, Switzerland), and the growth inhibitory effect was calculated by comparing with DMSO-treated control cells. After calculating the half-maximal inhibitory concentration (IC50) values, the sensitivity index (SI), defined as the IC50 value divided by the trough concentration of each drug at the recommended dose (IC50/Ctrough × 100), was calculated. We also evaluated the selectivity index, which was defined as the SI divided by the SI of Ba/F3 cells with wild-type EGFR.
2.5. Establishment of Resistant Clones to Afatinib and Osimertinib
The N-ethyl-N-nitrosourea (ENU, Sigma-Aldrich, St. Louis, MO) mutagenesis technique was used to accelerate the emergence of afatinib- and osimertinib-resistant cells, as previously described [
15,
16,
17]. Ba/F3 parental cells were initially exposed to 100 mg/mL ENU for 24 h. After washing twice with RPMI 1640 medium, cells were cultured for 24 h and then plated in 96-well plates (10,000 cells/well) with 10 nM afatinib or 100 nM osimertinib. These drug concentrations were selected to be between the IC
50 values of uncommon mutations and that of wild-type
EGFR. We cultured the cells for 14–28 days, with a change of medium every 3 to 5 days. After establishing resistant cells, DNA was extracted using a DNeasy Blood & Tissue Kit (250) (QIAGEN, Venlo, the Netherlands), and secondary
EGFR mutations were detected by direct sequencing as previously described [
18]. We examined all wells with regrowth or randomly selected 12 wells if there were 13 or more wells with confluent cells. If no cells grew in the plate, the drug concentration was reduced by orders of magnitude (5 nM and 2.5 nM for afatinib, and 50 nM and 25 nM for osimertinib), and the same experiments were repeated.
3. Results
3.1. Frequency of Uncommon EGFR Mutations in cBioPortal Database
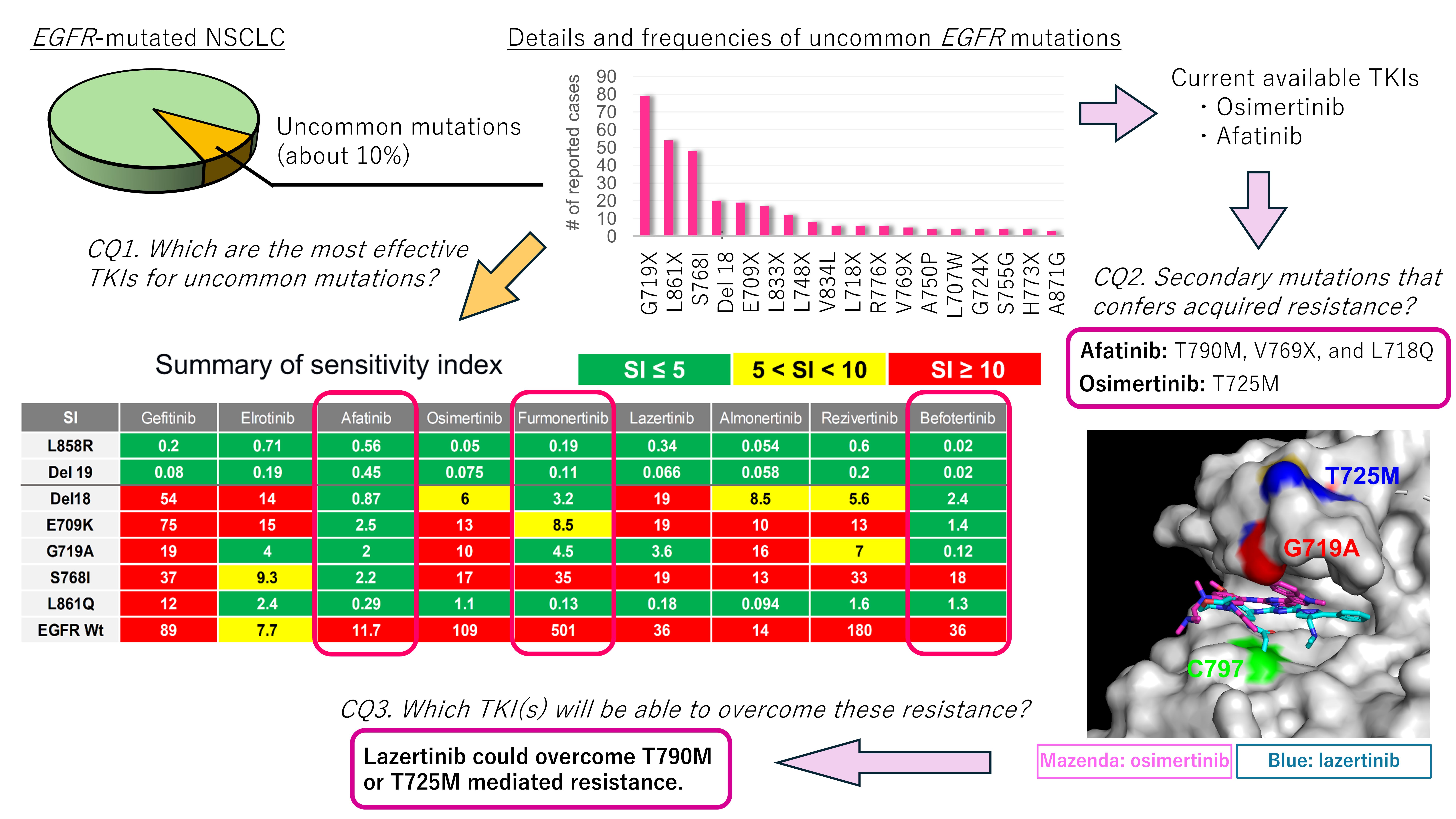
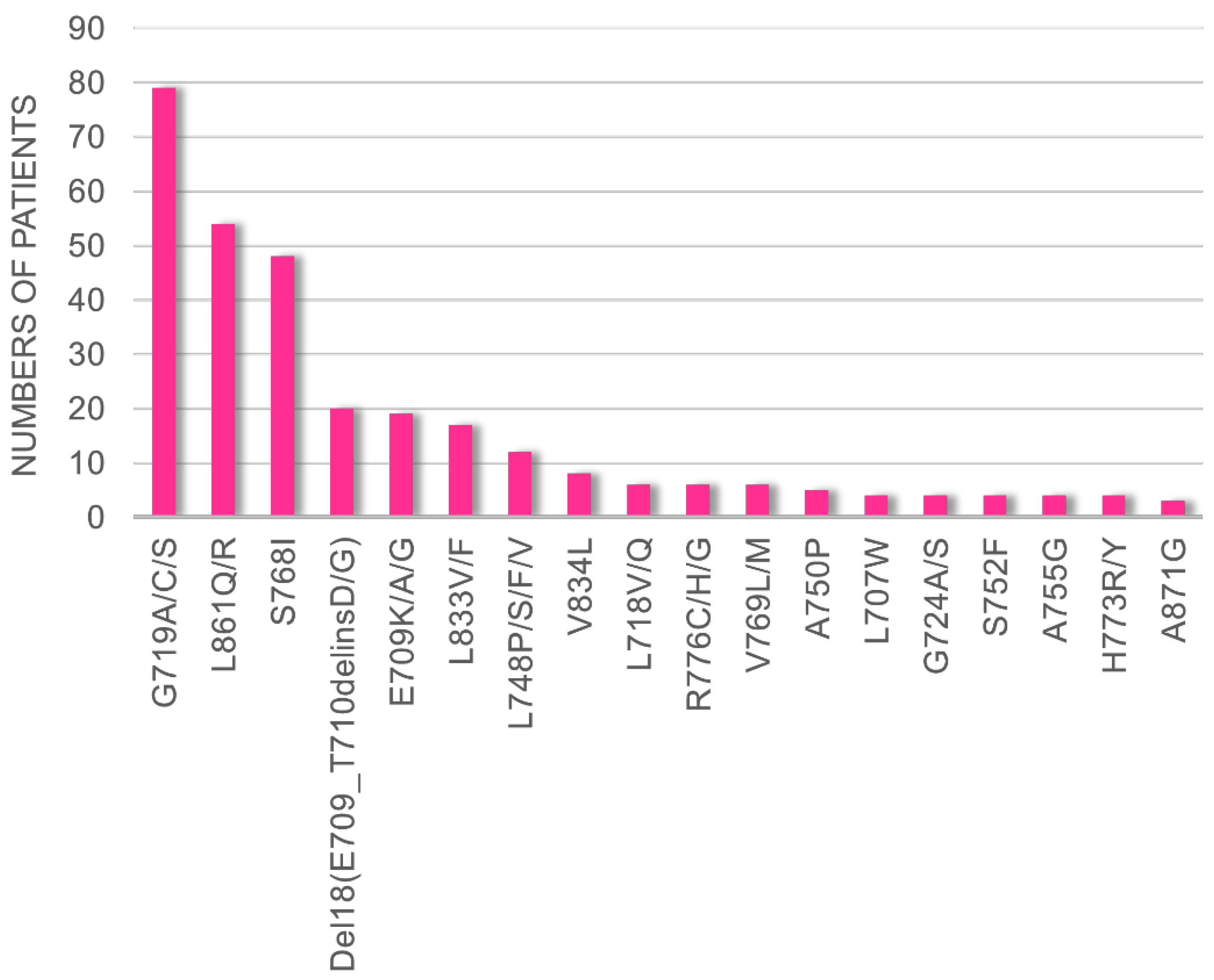
By evaluating data obtained from cBioPortal, we found that G719X, L861X, S768I, Del18 (delE709_T710insD), and E709X were the five most frequent uncommon
EGFR mutations in patients with NSCLC (
Figure 1). Therefore, we decided to evaluate the efficacies of the novel EGFR-TKIs using Ba/F3 cells harboring these five mutations.
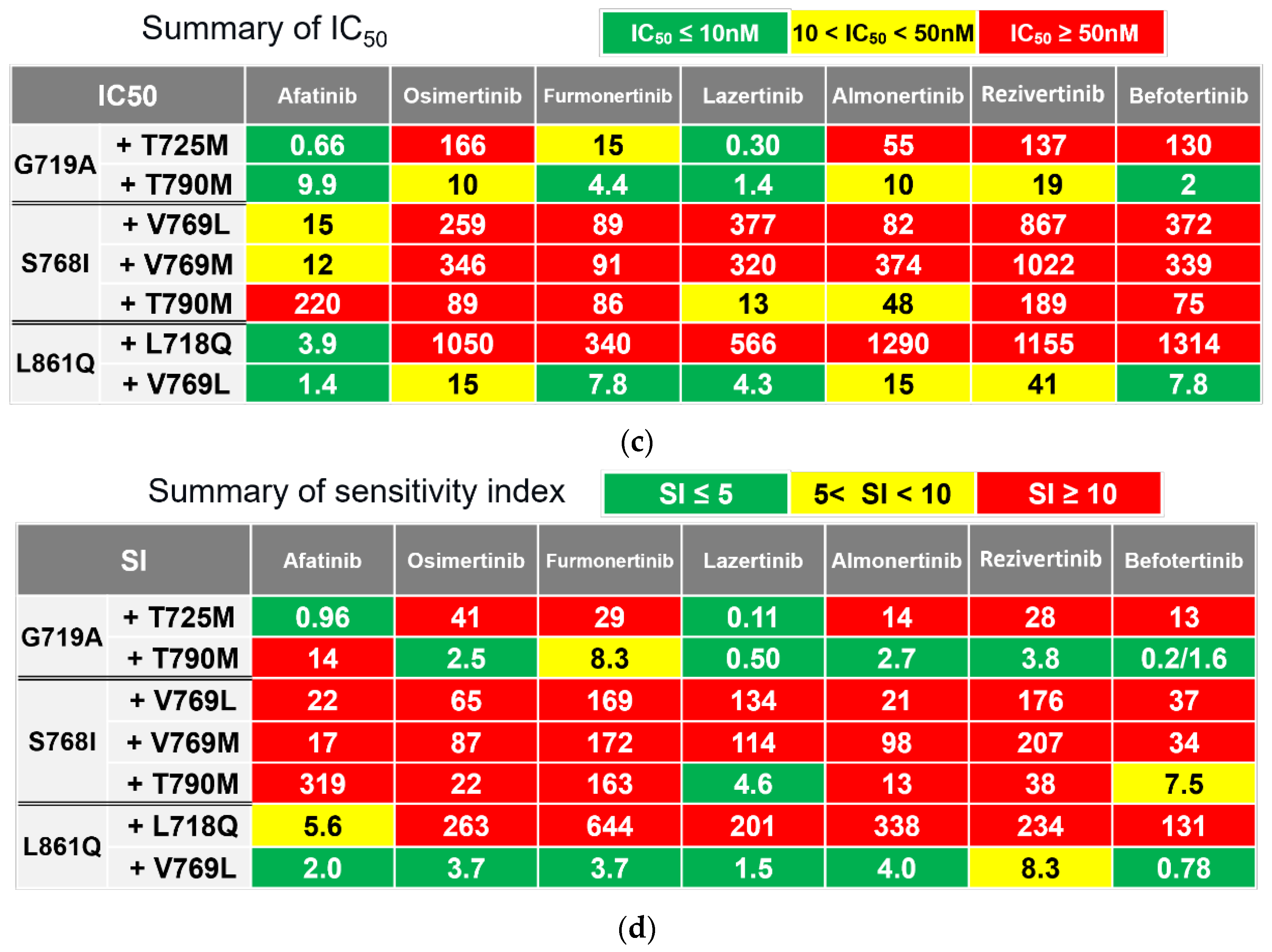
3.2. Efficacy of Novel TKIs Against Uncommon EGFR Mutations
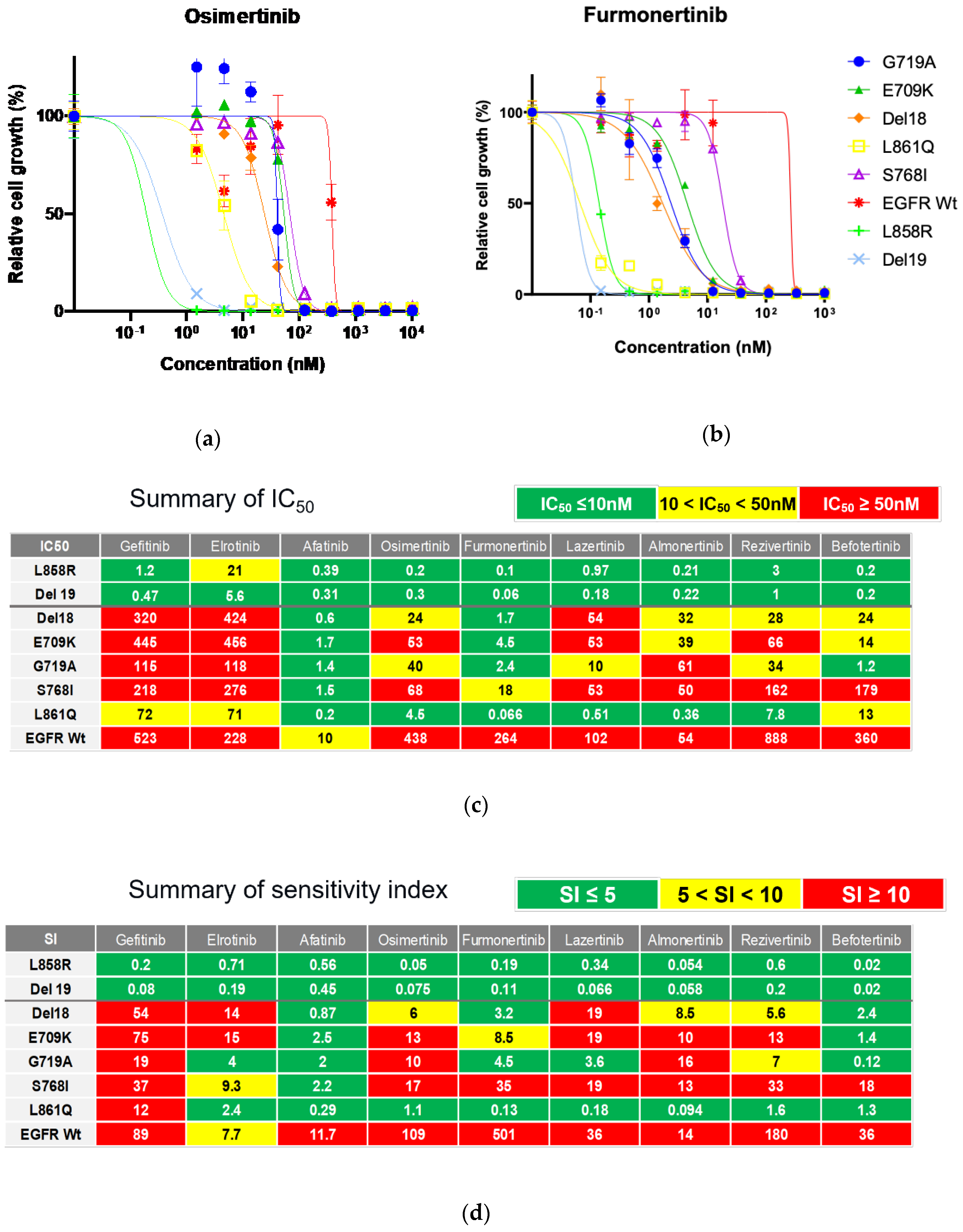
We evaluated the inhibitory effects of 1G-TKIs (gefitinib and erlotinib), 2G-TKIs (afatinib), and 3G-TKIs (osimertinib, almonertinib, lazertinib, furmonertinib, rezivertinib, and befotertinib) against Ba/F3 cells with uncommon
EGFR mutations to identify TKIs with activity against these mutations. In addition, the efficacies of these drugs were evaluated in Ba/F3 cells harboring wild-type
EGFR supplemented with 20 ng/ml of human recombinant EGF to assess the side effects of the drugs on non-cancerous cells. Growth inhibitory curves are shown in
Figure 2A and 2B, and in
Supplementary Figure S1. IC
50 values and SIs, which indicate efficacy of drug adjusted with clinically achievable drug concentration, are summarized in
Figures 2C and 2D, respectively.
As shown in Figures 2C and 2D, afatinib showed the greatest inhibitory effect against all tested Ba/F3 cell models with uncommon EGFR mutations. Erlotinib was active against L861Q and G719A mutations, and osimertinib was active against the L861Q mutation only when we defined being active by an SI of < 5. However, we observed that the ratios of SIs between wild-type and uncommon EGFR mutations, hereafter defined as the selectivity index, were largest in osimertinib (≥ 6.5 times), followed by afatinib (≥ 4.8 times), gefitinib (≥ 1.2 times), and erlotinib (≥ 0.50 times), suggesting that osimertinib may also work clinically, while preserving the phosphorylation of wild-type EGFR in noncancerous cells. Among novel 3G-TKIs, we found that all were active against Ba/F3 cells with the L861Q mutation, and all had low efficacy (SIs over 10) against Ba/F3 cells with the S768I mutation. Comparing between the 3G-TKIs, befotertinib was active against all uncommon EGFR mutations other than S768I, whereas furmonertinib had the highest selectivity index (14.4 times or higher) versus osimertinib and afatinib.
These results suggest that befotertinib or furmonertinib (especially for higher dosing) are potentially useful 3G-TKIs against NSCLC in patients harboring uncommon
EGFR mutations. Afatinib and osimertinib are also reasonable treatment options. The potential utility of other drugs (gefitinib, erlotinib, lazertinib, almonertinib, and rezivertinib) could be considered for each mutation variant in accordance with the results summarized in
Figure 2D.
3.3. Secondary Resistance Mutations to Osimertinib and Strategies to Overcome Resistance
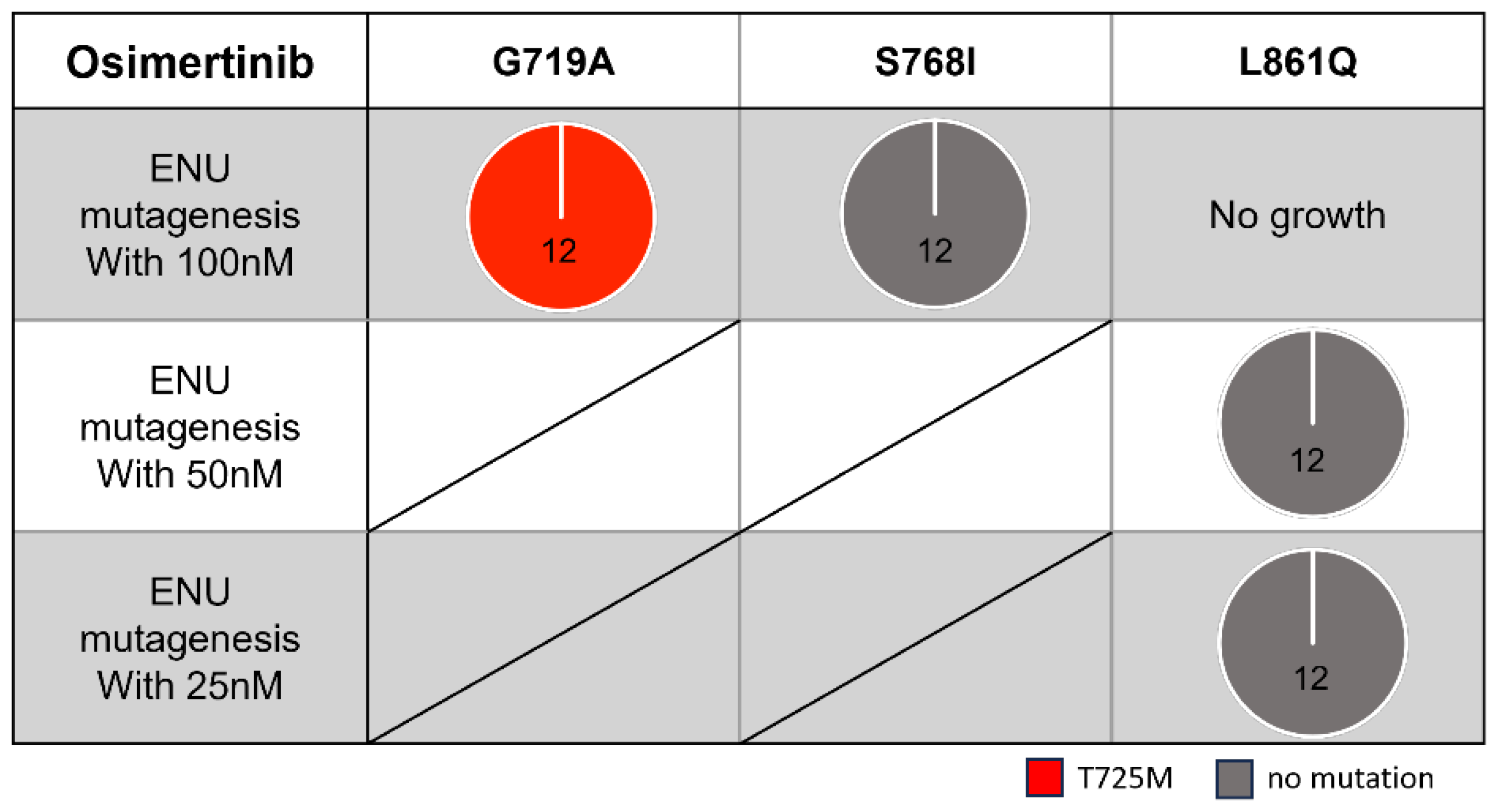
To explore useful TKIs in the second-line setting, we examined potential secondary mutations that may confer acquired resistance to simertinib using Ba/F3 cells with the three most frequent uncommon mutations (G719A, S768I, and L861Q). After exposing the Ba/F3 cells to ENU, we treated them with simertinib (starting at 100 nM) for a few weeks.
We identified the T725M secondary
EGFR mutation (
Figure 3 and
Figure 4A) in Ba/F3 cells with the G719A mutation in all viable wells tested. In contrast, we detected no secondary mutations in the kinase domain of the
EGFR gene in Ba/F3 cells with S768I or L861Q mutations after treatment with simertinib (
Figure 3).
In the growth inhibitory analysis (
Figure 4B), Ba/F3 cells with G719A/T725M mutations showed an IC
50 value of 165.6 nM for simertinib that was 4.1-fold higher than parental Ba/F3 cells with only the G719A mutation. We observed that Ba/F3 cells with G719A/T725M were insensitive to all novel 3G-TKIs except for simertini. In addition, we found that afatinib was active against Ba/F3 cells with G719A/T725M mutations (
Figure 4C and D).
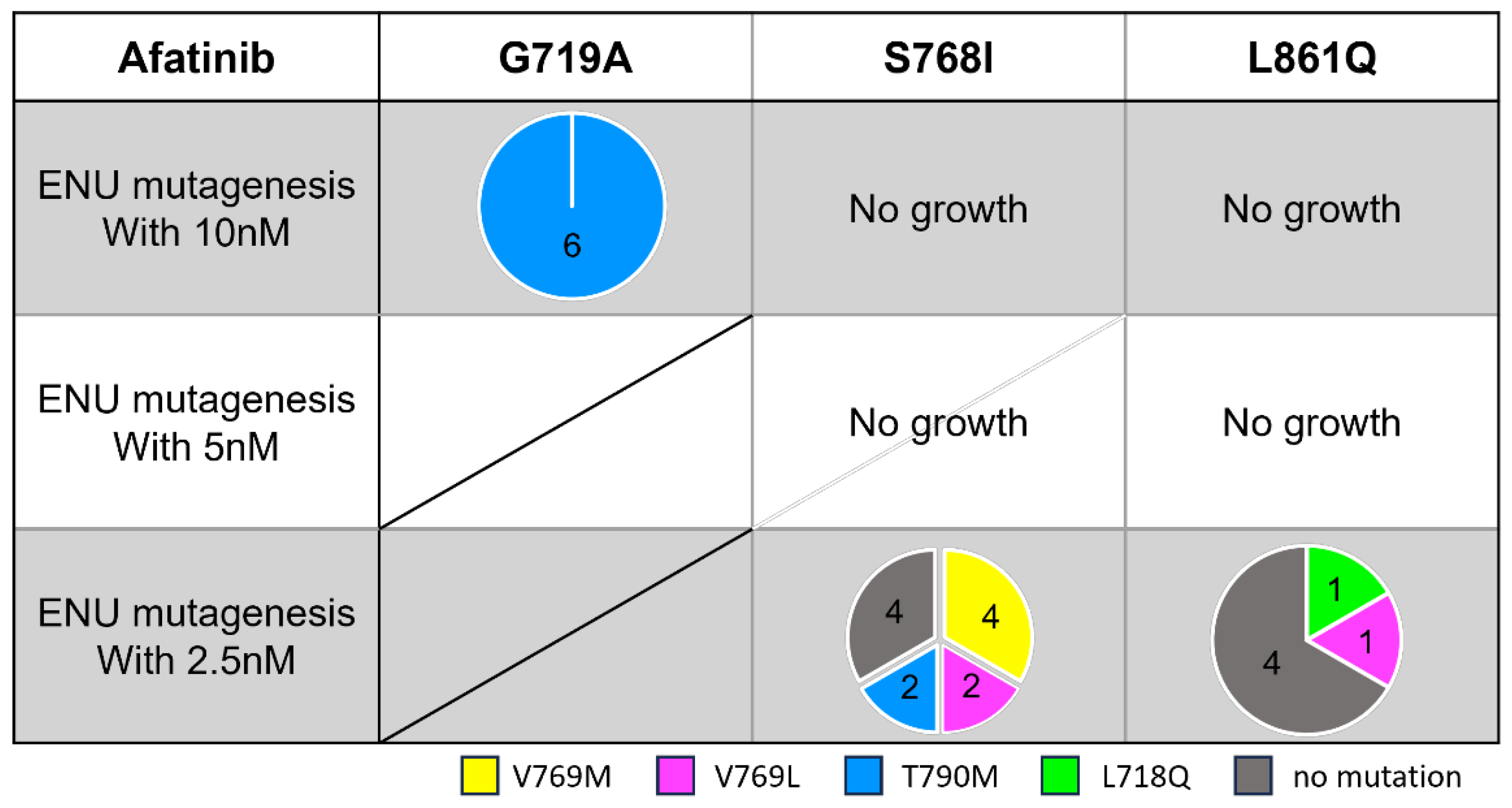
3.4. Secondary Resistance Mutations to Afatinib and Strategies to Overcome Resistance
To explore useful TKIs in the second-line setting after afatinib treatment, we examined potential secondary mutations that may confer acquired resistance to afatinib. After ENU exposure, we treated these Ba/F3 cells with afatinib (starting at 10 nM) for a few weeks.
In contrast to osimertinib-resistant cells, we detected several different secondary mutations depending on the activating
EGFR mutation subtype (
Figure 5). In the G719A model, secondary T790M mutation emerged in all six established wells. Ba/F3 cells with G719A/T790M had a 7.1-fold higher IC
50 value compared with G719A parental cells (
Figure 4C). T790M secondary mutation also emerged in cells with S768I mutation at the lowest drug concentration (2.5 nM), and Ba/F3 cells with S768I/T790M had a 146-fold higher IC
50 value compared with the parental cells. Furthermore, several different substitutions involving V769 were found in Ba/F3 cells with S768I or L861Q mutations.
To explore EGFR-TKIs that may overcome these secondary mutations induced by afatinib, we also evaluated the efficacy of osimertinib and other novel 3G-TKIs. We observed that most of the 3G-TKIs tested were active against Ba/F3 cells with G719A/T790M and S861Q/V769L; however, only lazertinib was effective against Ba/F3 cells with S768I/T790M mutations (
Figure 4D). In contrast, Ba/F3 cells with S768I/V769L or S768I/V769M mutations were refractory to all EGFR-TKIs tested.
4. Discussion
Various studies have reported that afatinib and osimertinib are effective against NSCLCs harboring uncommon
EGFR mutations; therefore, these drugs are usually the treatment of choice in clinical practice [
6,
8,
9,
19,
20,
21]. However, treatment outcomes with these TKIs are not satisfactory compared with osimertinib treatment of NSCLC in patients with common
EGFR mutations. Therefore, as a first step in this study, we screened novel 3G-TKIs to identify the most effective agent using in vitro models with uncommon
EGFR mutations. We found that afatinib was active (SI < 5) against all uncommon
EGFR mutations, but that osimertinib was less active against many of the uncommon mutations tested. This result is in agreement with a recent pooled analysis comparing the efficacy of afatinib versus osimertinib using propensity score-matching in patients with NSCLC harboring uncommon
EGFR mutations [
22]. However, in our analysis of novel 3G-TKIs, we observed that befotertinib and furmonertinib could be promising TKIs for patients with many of the uncommon
EGFR mutations, such as L861Q, G719A, or E709K, when considering IC
50 values, clinically achievable drug concentrations, and the inhibitory effect of wild-type
EGFR (selectivity index as defined in the Results section). Based on these results, we suggest that NSCLC harboring uncommon
EGFR mutations should not be treated as a single disease; rather, we should determine an appropriate TKI (and appropriate drug concentrations as reported recently [
23]) for each mutation subtype.
As the next step, we explored TKIs that can overcome acquired resistance to front-line osimertinib or afatinib. To evaluate potential secondary mutations associated with acquired resistance to osimertinib or afatinib, we used ENU mutagenesis to establish cells with acquired resistance. We detected T725M as a secondary mutation in Ba/F3 cells with G719A that acquired resistance to osimertinib. Although a machine-learning approach in a previous study has suggested the transforming ability of the
EGFR T725M mutation [
24], this mutation is very rare in clinical practice. By exploring the COSMIC database, we observed that five cases of
EGFR T725M mutation have been reported (
Supplementary Table S1). None had a concurrent G719X mutation, although some patients had concurrent L858R mutations. In addition, through a literature search, we found two studies that reported detecting T725M mutations in tumors after osimertinib treatment [
25,
26]. Therefore, it is possible that the T725M secondary mutation emerged after osimertinib exposure in our in vitro model. Our results also suggest that afatinib or lazertinib could overcome the T725M secondary mutation in
EGFR G719A-positive patients.
Furthermore, we observed that T790M or V769M/L secondary mutations emerged after afatinib exposure in Ba/F3 cell models with G719A, S768I, or L861Q mutations. We had expected the emergence of the T790M secondary mutation because it has been reported as an acquired resistance mechanism to afatinib in NSCLC among patients with common EGFR mutations. Furthermore, it is reasonable that the T790M secondary mutation could be overcome by 3G-TKIs, which were designed to overcome the T790M mutation. Our results showed that lazertinib was the most effective 3G-TKI in this setting.
Among secondary resistant mutations to afatinib, we also observed V769M and V769L mutations in models with S768I and L861Q
EGFR-activating mutations. Previous studies have reported that the
EGFR V769M mutation is a frequent
EGFR germ-line mutation [
27,
28]; however, it has also been reported that V769M/L mutations have emerged as somatic mutations, usually together with another
EGFR-activating mutation, as summarized in
Supplementary Table S2 [
29,
30]. Interestingly, V769L often co-exists with the
EGFR S768I mutation, although the molecular mechanisms responsible for this co-existence are unclear. Two of the affected patients had efficacy data for TKI treatment; neither showed a response to erlotinib or afatinib (
Supplementary Table S2). In addition to COSMIC data, additional case studies have reported TKI efficacy against NSCLCs with S768I plus V769L compound mutation; only one patient responded to full-dose afatinib [
31], whereas the other two patients showed inherent resistance to gefitinib [
32] or lower-dose afatinib [
33]. Currently, few case studies have reported the emergence of V769X as a secondary mutation [
34]. Therefore, future studies are needed to evaluate the frequency of this mutation after acquiring afatinib resistance in patients with NSCLC harboring uncommon
EGFR mutations. It should be noted that one patient with the G719A plus V769M compound mutation responded to upfront osimertinib [
35].
5. Conclusions
Our in vitro study demonstrated that afatinib showed universal activity against various uncommon EGFR mutations, while 3G-TKIs, especially furmonertinib and befotertinib, also showed high efficacy against all these mutations, except S768I. Therefore, we suggest that NSCLC with uncommon EGFR mutations should not be treated as a single disease but should be treated based on mutation subtype. Furthermore, we detected several on-target resistance mutations of EGFR, such as T725M, T790M, and V769M/L, after exposure to osimertinib or afatinib, and our results suggest that lazertinib may overcome some of these secondary mutations.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Figure S1: Growth inhibitory curves of EGFR-TKIs tested in this study; Table S1: T725M mutation reported in lung cancers in COSMIC database (accessed at January 31, 2025); Table S2: V769L/M mutations reported in lung cancers in COSMIC database (accessed at January 31, 2025).
Author Contributions
Conceptualization, K.S. and A.H.; methodology, H.O., K.S., T.F., Y.K., and T.M; validation, K.S., K.O., M.I., J.S., and Y.T; formal analysis, H.O. and K.S.; investigation, H.O., T.F., K.O., S.F., S.O., and M.I.; resources, K.S., T.F., Y.K., K.S., K.N., and T.M; writing—original draft preparation, H.O. and K.S.; writing—review and editing, all authors; visualization, H.O, K.S., and T.F.; supervision, K.S., K.N., T.M., and Y.S.; project administration, K.S., T.M., and Y.T.; funding acquisition, K.S., A.H., and J.S. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by grants-in-aid for scientific research from the Japan Society for the Promotion of Science (grants 22K07291 to Dr Suda, 23K15569 to Dr Hamada, and 22K08986 to Dr Soh).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The findings of this study are available from the corresponding author (K.S.) upon reasonable request.
Acknowledgments
The authors thank Mr. Yoshihiro Mine (Center for Instrumental Analyses, Central Research Facilities, Kindai University Faculty of Medicine) for his technical assistance. We thank Alison McTavish, MSc, from Edanz (
https://jp.edanz.com/ac) for editing a draft of this manuscript.
Conflicts of Interest
Dr. Suda has received research funding from AstraZeneca K.K. and Guardant Health Inc.; has received honoraria from Chugai Pharmaceutical Co., Ltd., Daiichi-Sankyo, Amgen, Boehringer Ingelheim, and AstraZeneca K.K. outside of the submitted work. Dr. Hamada has received research funding from AstraZeneca and Chugai Pharmaceutical Co., Ltd.; has received honoraria from AstraZeneca K.K., Chugai Pharmaceutical Co., Ltd., Ono Pharmaceuticals, Bristol-Myers Squibb, ETHICON; has undertaken an advisory board for AstraZeneca and Chugai Pharmaceutical Co., Ltd. Dr. Fujino has received research fundings from Pfizer Inc., Nuvalent Inc., Takeda Science Foundation, Eli Lilly Japan K.K., Bridge Biotherapeutics Inc., and Kinnate Biopharma Inc. outside of the submitted work; and a patent for KU220115PCT pending. Dr. Kobayashi has received honoraria from AstraZeneca K.K., Amgen, Chugai Pharmaceutical Co., Ltd., Ono Pharmaceuticals, and MSD K.K. Dr. Sakai has received honoraria from Takeda Pharmaceutical Co., Ltd., Nippon Kayaku Co.,Ltd., Qiagen, Inc., and Life Technologies Japan Ltd. outside of the submitted work. Dr. Soh has received research funding from Chugai Pharmaceutical Co., Ltd.; has received honoraria from Chugai Pharmaceutical Co., Ltd., ETHICON, Intuitive Surgical, Inc., CSL Behring K.K., Olympus Corporation, and Covidien Japan, Inc. outside of the submitted work. Dr. Nishio has received research funding from West Japan Oncology Group, Nichirei Biosciences Inc., Eli Lilly Japan K.K., Hitachi, Ltd., Nippon Boehringer Ingelheim Co., Ltd., Sysmex Corporation, Otsuka Pharmaceutical Co., Ltd., Thoracic Oncology Research Group, Okayama University, Japan Breast Cancer Research Group; has received honoraria from Chugai Pharmaceutical Co Ltd., MSD K.K., Guardant Health Inc., Daiichi-Sankyo, SymBio Pharmaceuticals Limited., Ono Pharmaceutical Co Ltd., Janssen Pharmaceutical K.K., Novartis Pharma K.K., Eli Lilly Japan K.K., Otsuka Pharmaceutical Co., Ltd., Invitae Japan, AstraZeneca K.K., Nichirei Biosciences Inc. and Maruho Co., Ltd. outside of the submitted work. Dr. Mitsudomi has received honoraria from AstraZeneca K.K., Chugai Pharmaceutical Co, Janssen Pharmaceutical K.K.; and a patent for AstraZeneca K.K. pending. Dr. Tsutani has undertaken an advisory role for AstraZeneca K.K., Bristol-Myers Squibb, Chugai Pharmaceutical Co, Ltd., MSD K.K., and Ono Pharmaceutical Co, Ltd.; received honoraria from AstraZeneca K.K., Bristol-Myers Squibb, Carenet, Chugai Pharmaceutical Co, Ltd., Covidien Japan, CSL Behring K.K., Eli Lilly, Daiichi Sankyo, HOKUTO, Japan Blood Products Organization, Johnson & Johnson, MSD K.K., MiRTeL, Nihon Medi-Physics, Nikkei BP, Novartis, Ono Pharmaceutical Co, Ltd., Olympus Corporation, Smart Hospital, Takeda Pharmaceutical Co., Ltd., and Taiho Pharmaceutical Co, Ltd.; and has received research funding from Abbott, Nippon Boehringer Ingelheim Co.,Ltd., Chugai Pharmaceutical Co, Ltd., Daiichi Sankyo, Japan Blood Products Organization, Kyowa Kirin, Medtronic, and Otsuka Pharmaceutical Co., Ltd., Taiho Pharmaceutical Co, Ltd. outside of the submitted work. The remaining authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| EGFR |
Epidermal growth factor receptor |
| ENU |
N-ethyl-N-nitrosourea |
| IC50
|
Half maximal inhibitory concentration |
| NSCLC |
Non-small cell lung cancer |
| PFS |
Progression-free survival |
| SI |
Sensitivity index |
| TKI |
Tyrosine kinase inhibitor |
| 1G |
First-generation |
| 2G |
Second-generation |
| 3G |
Third-generation |
References
- Hirsch, F.R.; Suda, K.; Wiens, J.; Bunn, P.A., Jr. New and emerging targeted treatments in advanced non-small-cell lung cancer. Lancet 2016, 388, 1012–1024. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Mitsudomi, T. Not all epidermal growth factor receptor mutations in lung cancer are created equal: Perspectives for individualized treatment strategy. Cancer Sci. 2016, 107, 1179–1186. [Google Scholar] [CrossRef]
- Suda, K.; Mitsudomi, T.; Shintani, Y.; Okami, J.; Ito, H.; Ohtsuka, T.; Toyooka, S.; Mori, T.; Watanabe, S.-I.; Asamura, H.; et al. Clinical Impacts of EGFR Mutation Status: Analysis of 5780 Surgically Resected Lung Cancer Cases. Ann. Thorac. Surg. 2021, 111, 269–276. [Google Scholar] [CrossRef]
- Gomez-Randulfe, I.; Scanlon, L.A.; Carter, M.; Moliner, L.; Cil, E.; Califano, R.; Summers, Y.; Blackhall, F.; Lindsay, C.R.; Lewis, J.; et al. First-line osimertinib compared to earlier generation TKIs in advanced EGFR-mutant NSCLC: A real-world survival analysis. Lung Cancer 2025, 200, 108084. [Google Scholar] [CrossRef] [PubMed]
- Robichaux, J.P.; Le, X.; Vijayan, R.S.K.; Hicks, J.K.; Heeke, S.; Elamin, Y.Y.; Lin, H.Y.; Udagawa, H.; Skoulidis, F.; Tran, H.; et al. Structure-based classification predicts drug response in EGFR-mutant NSCLC. Nature 2021, 597, 732–737. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.C.-H.; Sequist, L.V.; Geater, S.L.; Tsai, C.-M.; Mok, T.S.K.; Schuler, M.; Yamamoto, N.; Yu, C.-J.; I Ou, S.-H.; Zhou, C.; et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: a combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol. 2015, 16, 830–838. [Google Scholar] [CrossRef]
- S. Miura HT, T. S. Miura HT, T. Misumi. LBA66 Afatinib versus chemotherapy for treatment-naïve non-small cell lung cancer with a sensitizing uncommon epidermal growth factor receptor mutation: A phase III study (ACHILLES/TORG1834). ANNALS OF ONCOLOGY. 2023;34:S1310-S1.
- Okuma Y, Kubota K, Shimokawa M, Hashimoto K, Kawashima Y, Sakamoto T, Wakui H, Murakami S, Okishio K, Hayashihara K, et al. First-Line Osimertinib for Previously Untreated Patients With NSCLC and Uncommon EGFR Mutations: The UNICORN Phase 2 Nonrandomized Clinical Trial. JAMA Oncol. 2024;10(1):43-51.
- Cho JH, Lim SH, An HJ, Kim KH, Park KU, Kang EJ, Choi YH, Ahn MS, Lee MH, Sun JM, et al. Osimertinib for Patients With Non-Small-Cell Lung Cancer Harboring Uncommon EGFR Mutations: A Multicenter, Open-Label, Phase II Trial (KCSG-LU15-09). J Clin Oncol. 2020;38(5):488-95.
- Soria, J.-C.; Ohe, Y.; Vansteenkiste, J.; Reungwetwattana, T.; Chewaskulyong, B.; Lee, K.H.; Dechaphunkul, A.; Imamura, F.; Nogami, N.; Kurata, T.; et al. Osimertinib in Untreated EGFR-Mutated Advanced Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Catherine, A. Shu KG, Byoung Chul Cho, Frank Griesinger, James Chih-Hsin Yang, Enriqueta Felip, John Xie, Jun Chen, Janine Mahoney, Meena Thayu, Roland Elmar Knoblauch, Leonardo Trani, and Joshua Bauml. CHRYSALIS-2: A phase 1/1b study of lazertinib as monotherapy and in combination with amivantamab in patients with EGFR-mutant NSCLC. Journal of Clinical Oncology. 2021; 39.
- Banno, E.; Togashi, Y.; Nakamura, Y.; Chiba, M.; Kobayashi, Y.; Hayashi, H.; Terashima, M.; de Velasco, M.A.; Sakai, K.; Fujita, Y.; et al. Sensitivities to various epidermal growth factor receptor-tyrosine kinase inhibitors of uncommon epidermal growth factor receptor mutations L861Q and S768I: What is the optimal epidermal growth factor receptor-tyrosine kinase inhibitor? Cancer Sci. 2016, 107, 1134–1140. [Google Scholar] [CrossRef]
- Kobayashi Y, Togashi Y, Yatabe Y, Mizuuchi H, Jangchul P, Kondo C, Shimoji M, Sato K, Suda K, Tomizawa K, et al. EGFR Exon 18 Mutations in Lung Cancer: Molecular Predictors of Augmented Sensitivity to Afatinib or Neratinib as Compared with First- or Third-Generation TKIs. Clin Cancer Res. 2015;21(23):5305-13.
- Nishino, M.; Suda, K.; Kobayashi, Y.; Ohara, S.; Fujino, T.; Koga, T.; Chiba, M.; Shimoji, M.; Tomizawa, K.; Takemoto, T.; et al. Effects of secondary EGFR mutations on resistance against upfront osimertinib in cells with EGFR-activating mutations in vitro. Lung Cancer 2018, 126, 149–155. [Google Scholar] [CrossRef]
- Kobayashi Y, Azuma K, Nagai H, Kim YH, Togashi Y, Sesumi Y, Chiba M, Shimoji M, Sato K, Tomizawa K, et al. Characterization of EGFR T790M, L792F, and C797S Mutations as Mechanisms of Acquired Resistance to Afatinib in Lung Cancer. Mol Cancer Ther. 2017;16(2):357-64.
- Fujino, T.; Suda, K.; Koga, T.; Hamada, A.; Ohara, S.; Chiba, M.; Shimoji, M.; Takemoto, T.; Soh, J.; Mitsudomi, T. Foretinib can overcome common on-target resistance mutations after capmatinib/tepotinib treatment in NSCLCs with MET exon 14 skipping mutation. J. Hematol. Oncol. 2022, 15, 1–14. [Google Scholar] [CrossRef]
- Koga, T.; Suda, K.; Fujino, T.; Ohara, S.; Hamada, A.; Nishino, M.; Chiba, M.; Shimoji, M.; Takemoto, T.; Arita, T.; et al. KRAS Secondary Mutations That Confer Acquired Resistance to KRAS G12C Inhibitors, Sotorasib and Adagrasib, and Overcoming Strategies: Insights From In Vitro Experiments. J. Thorac. Oncol. 2021, 16, 1321–1332. [Google Scholar] [CrossRef]
- Suda K, Murakami I, Katayama T, Tomizawa K, Osada H, Sekido Y, Maehara Y, Yatabe Y, Mitsudomi T. Reciprocal and complementary role of MET amplification and EGFR T790M mutation in acquired resistance to kinase inhibitors in lung cancer. Clin Cancer Res. 2010;16(22):5489-98.
- Chang, L.-C.; Lim, C.-K.; Chen, K.-Y.; Shih, J.-Y.; Yu, C.-J. Non-small cell lung cancer harbouring non-resistant uncommon EGFR mutations: Mutation patterns, effectiveness of epidermal growth factor receptor-tyrosine kinase inhibitors and prognostic factors. Eur. J. Cancer 2019, 119, 77–86. [Google Scholar] [CrossRef]
- Tanaka, I.; Morise, M.; Kodama, Y.; Matsui, A.; Ozawa, N.; Ozone, S.; Goto, D.; Miyazawa, A.; Hase, T.; Hashimoto, N.; et al. Potential for afatinib as an optimal treatment for advanced non-small cell lung carcinoma in patients with uncommon EGFR mutations. Lung Cancer 2018, 127, 169–171. [Google Scholar] [CrossRef]
- Yang, J.C.-H.; Schuler, M.; Popat, S.; Miura, S.; Heeke, S.; Park, K.; Märten, A.; Kim, E.S. Afatinib for the Treatment of NSCLC Harboring Uncommon EGFR Mutations: A Database of 693 Cases. J. Thorac. Oncol. 2020, 15, 803–815. [Google Scholar] [CrossRef]
- Wang, C.; Zhao, K.; Hu, S.; Dong, W.; Gong, Y.; Xie, C. Clinical Outcomes of Afatinib Versus Osimertinib in Patients With Non-Small Cell Lung Cancer With Uncommon EGFR Mutations: A Pooled Analysis. Oncol. 2023, 28, e397–e405. [Google Scholar] [CrossRef]
- Planchard D, Le X, Yu Y, Zhao Y. FURTHER (FURMO-002): A Global, Randomized Study of Firmonertinib at Two Dose Levels in TKI-Naïve, Advanced NSCLC with EGFR PACC Mutations. 2024 World Conference on Lung Cancer. 2024:S.
- U, M.; Talevich, E.; Katiyar, S.; Rasheed, K.; Kannan, N.; Radivojac, P. Prediction and Prioritization of Rare Oncogenic Mutations in the Cancer Kinome Using Novel Features and Multiple Classifiers. PLOS Comput. Biol. 2014, 10, e1003545. [Google Scholar] [CrossRef] [PubMed]
- Lin Lin QL, Ran Cao, Qiuxiang Ou, Yutong Ma, Hua Bao, Xue Wu, Yang Shao, Zhaoxia Wang. Acquired rare recurrent EGFR mutations as mechanisms of resistance to Osimertinib in lung cancer and in silico structural modelling. American Journal of Cancer Research. 2020;2020 Nov 1.
- Janning, M.; Süptitz, J.; Albers-Leischner, C.; Delpy, P.; Tufman, A.; Velthaus-Rusik, J.-L.; Reck, M.; Jung, A.; Kauffmann-Guerrero, D.; Bonzheim, I.; et al. Treatment outcome of atypical EGFR mutations in the German National Network Genomic Medicine Lung Cancer (nNGM). Ann. Oncol. 2022, 33, 602–615. [Google Scholar] [CrossRef] [PubMed]
- Hellmann, M.D.; Hayashi, T.; Reva, B.; Yu, H.A.; Riely, G.J.; Adusumilli, P.S.; Travis, W.D.; Wilkins, O.; Bramletta, N.; Chandramohan, R.; et al. Identification and Functional Characterization of EGFR V769M, a Novel Germline Variant Associated With Multiple Lung Adenocarcinomas. JCO Precis. Oncol. 2017, 1, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Peng, M.; Chen, Q.; Yuan, M.; Chen, R.; Deng, H.; Deng, J.; Liu, O.; Weng, Y.; Chen, M.; et al. Identification of the Unique Clinical and Genetic Features of Chinese Lung Cancer Patients With EGFR Germline Mutations in a Large-Scale Retrospective Study. Front. Oncol. 2021, 11. [Google Scholar] [CrossRef]
- Huang, S.-F.; Liu, H.-P.; Li, L.-H.; Ku, Y.-C.; Fu, Y.-N.; Tsai, H.-Y.; Chen, Y.-T.; Lin, Y.-F.; Chang, W.-C.; Kuo, H.-P.; et al. High Frequency of Epidermal Growth Factor Receptor Mutations with Complex Patterns in Non–Small Cell Lung Cancers Related to Gefitinib Responsiveness in Taiwan. Clin. Cancer Res. 2004, 10, 8195–8203. [Google Scholar] [CrossRef]
- Wu, J.-Y.; Yu, C.-J.; Chang, Y.-C.; Yang, C.-H.; Shih, J.-Y.; Yang, P.-C. Effectiveness of Tyrosine Kinase Inhibitors on “Uncommon” Epidermal Growth Factor Receptor Mutations of Unknown Clinical Significance in Non–Small Cell Lung Cancer. Clin. Cancer Res. 2011, 17, 3812–3821. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Shao, Y.W.; Xia, Y. Responsiveness to Full-Dose Afatinib in a Patient With Lung Adenocarcinoma Harboring EGFR S768I and V769L Mutations. J. Thorac. Oncol. 2019, 14, e25–e27. [Google Scholar] [CrossRef] [PubMed]
- Asahina, H.; Yamazaki, K.; Kinoshita, I.; Yokouchi, H.; Dosaka-Akita, H.; Nishimura, M. Non-responsiveness to gefitinib in a patient with lung adenocarcinoma having rare EGFR mutations S768I and V769L. Lung Cancer 2006, 54, 419–422. [Google Scholar] [CrossRef]
- Niogret, J.; Coudert, B.; Boidot, R. Primary Resistance to Afatinib in a Patient with Lung Adenocarcinoma Harboring Uncommon EGFR Mutations: S768I and V769L. J. Thorac. Oncol. 2018, 13, e113–E113. [Google Scholar] [CrossRef]
- Deng, Q.; Xie, B.; Wu, L.; Ji, X.; Li, C.; Feng, L.; Fang, Q.; Bao, Y.; Li, J.; Jin, S.; et al. Competitive evolution of NSCLC tumor clones and the drug resistance mechanism of first-generation EGFR-TKIs in Chinese NSCLC patients. Heliyon 2018, 4, e01031. [Google Scholar] [CrossRef] [PubMed]
- Simionato, F.; Calvetti, L.; Cosci, M.; Scarparo, S.; Aprile, G. Case Report: A Metabolic Complete Response to Upfront Osimertinib in a Smoker Non-Small Cell Lung Cancer Patient Harbouring EGFR G719A/V769M Complex Mutation. OncoTargets Ther. 2020, 13, 12027–12031. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).