Submitted:
31 July 2025
Posted:
31 July 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Key Pathophysiological Basis and Signaling Molecules of HLHS
2.1. Key Signaling Pathways for Heart Development and HLHS
2.2. Transcription Factors Underpinning Cardiac Development and HLHS
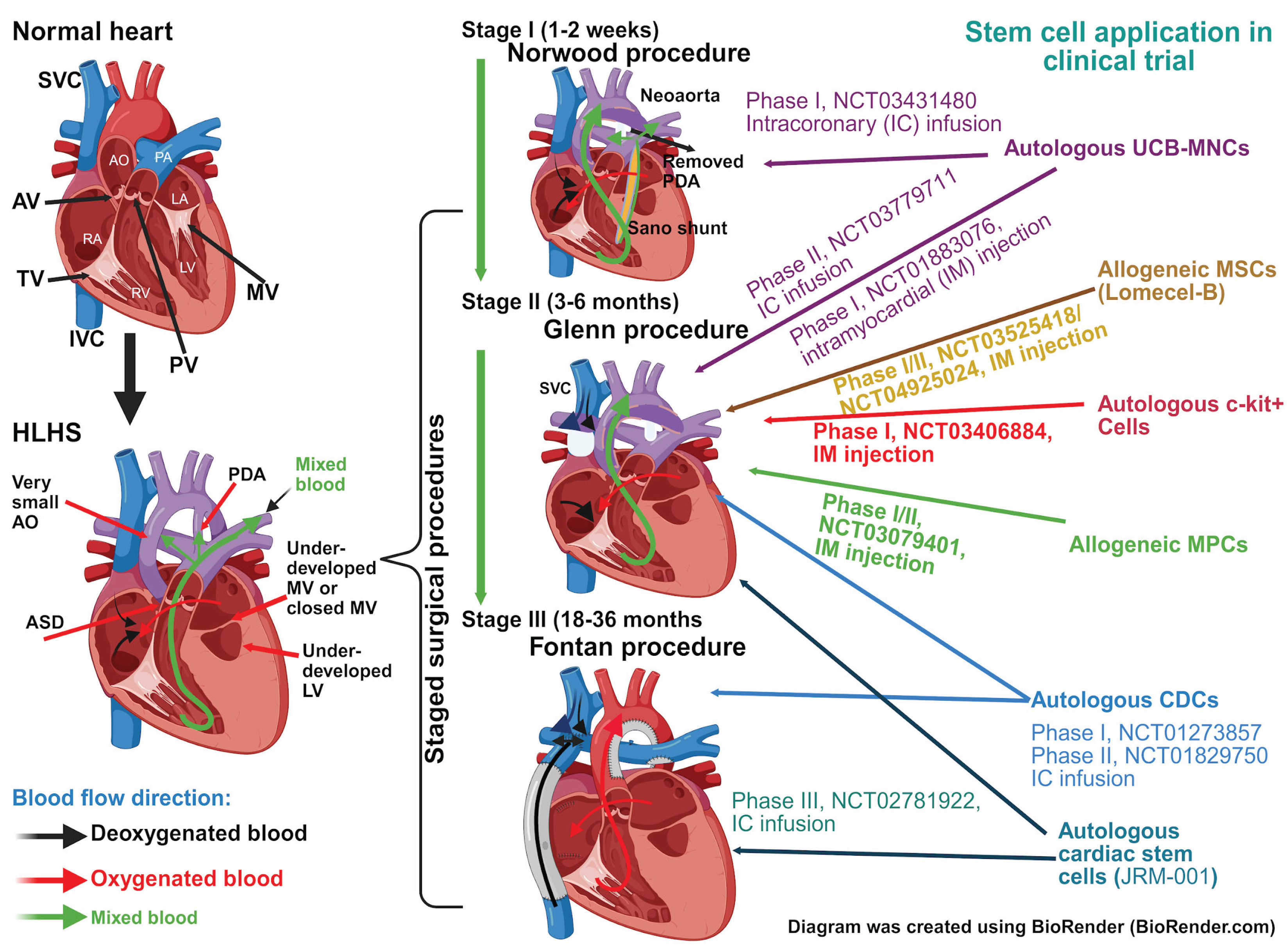
3. Application of Stem Cell in HLHS as an Adjunct Treatment
3.1. Umbilical Cord Derived Stem Cells
3.2. Bone Marrow Stem Cells
3.3. Cardiac Stem and Cardiosphere-Derived Cells
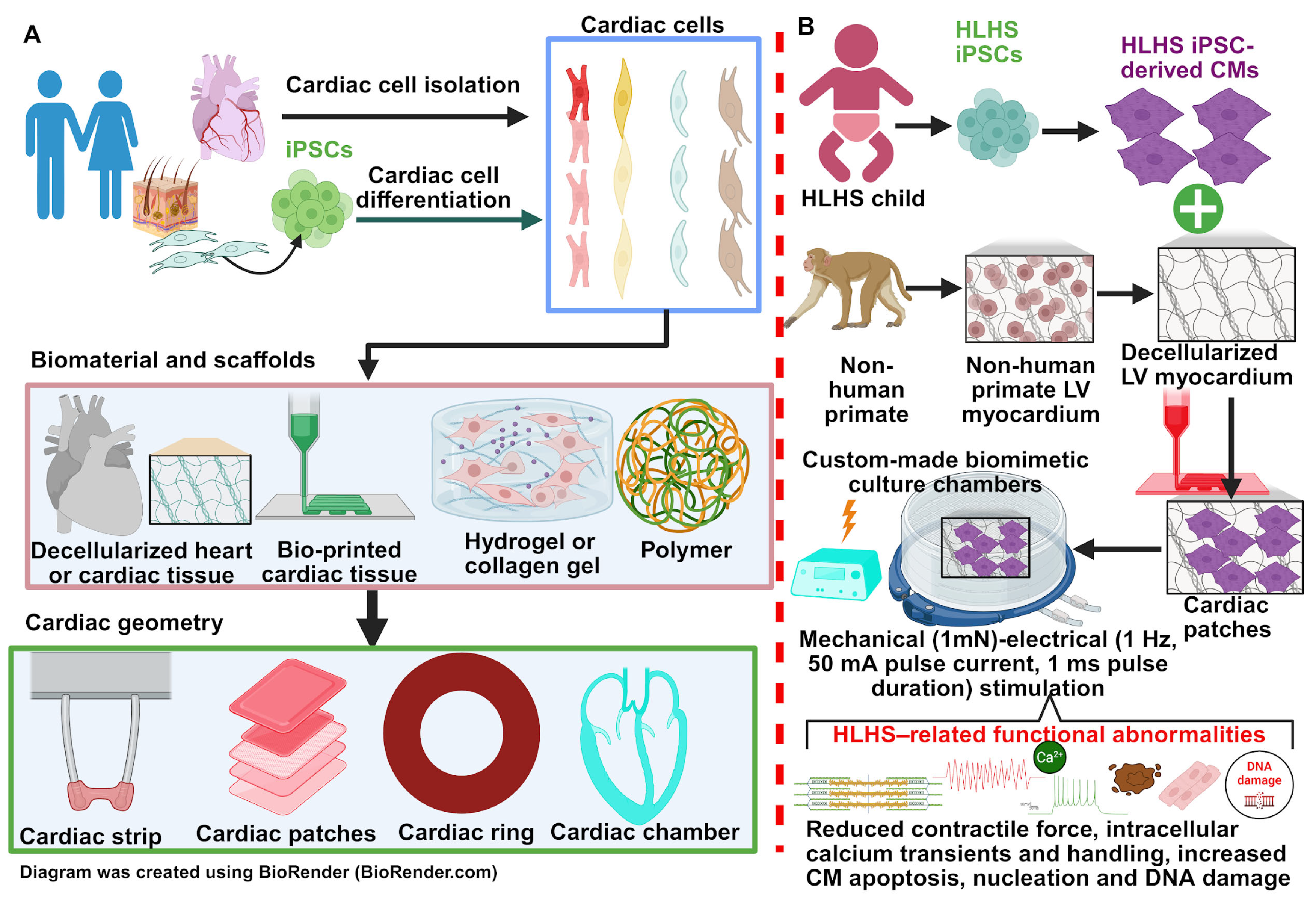
4. PSC Derived Cardiomyocytes for Studying HLHS
4.1. PSC-Derived Cardiomyocyte from HLHS Patients
4.2. Human iPSC-Derived Cardiomyocytes as Disease Modelling for HLHS
4.3. HLHS iPSC-CMs: A Platform for Drug Discovery and Evaluating Drug Toxicity
4.4. Limitations of iPSC-CMs 2D Model
5. Implications of hiPSC-Derived 3D Cardiac Patches for Treating HLHS
6. Future Directions
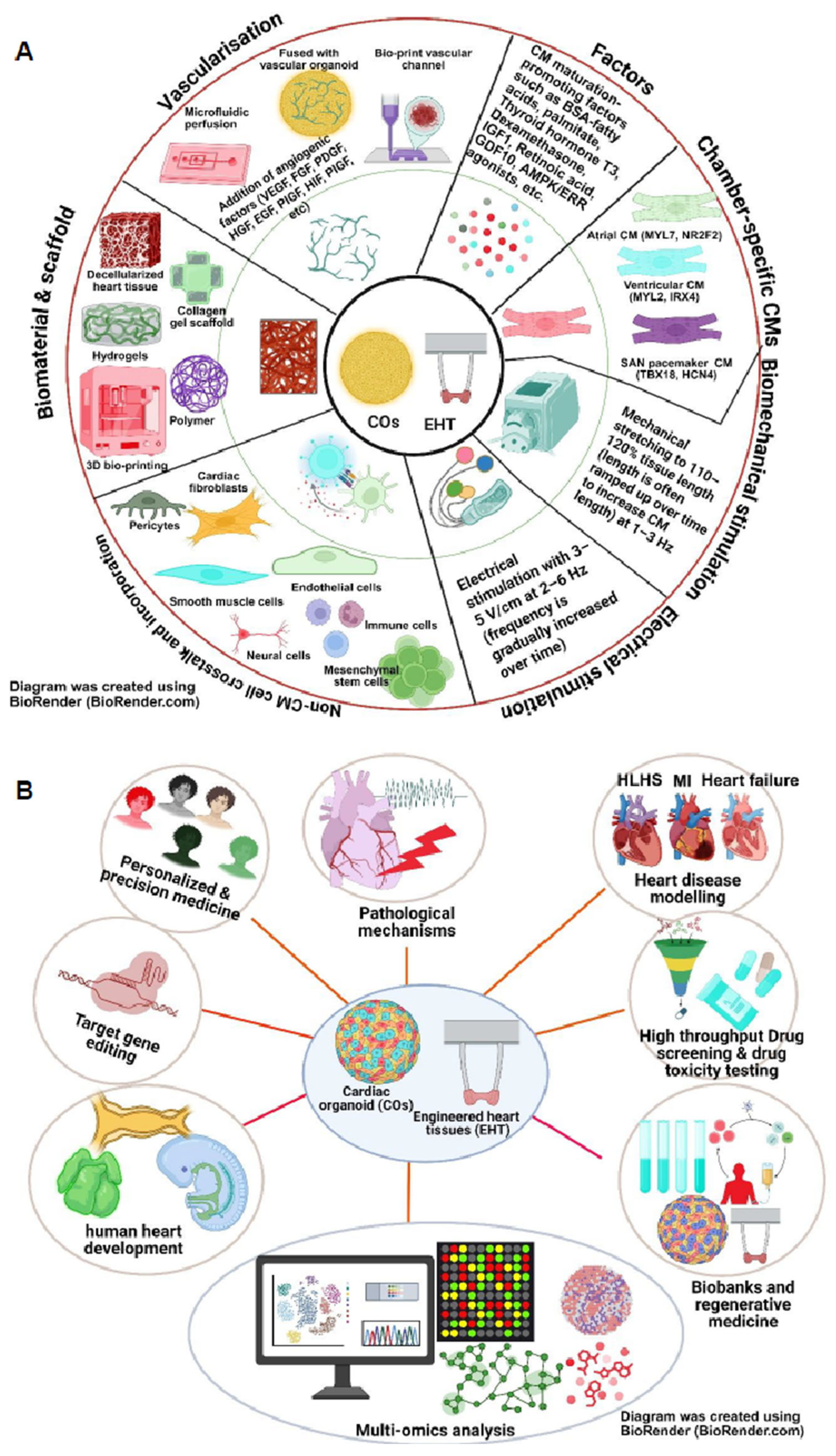
6.1. Cardiac Organoids: Better Model for HLHS?
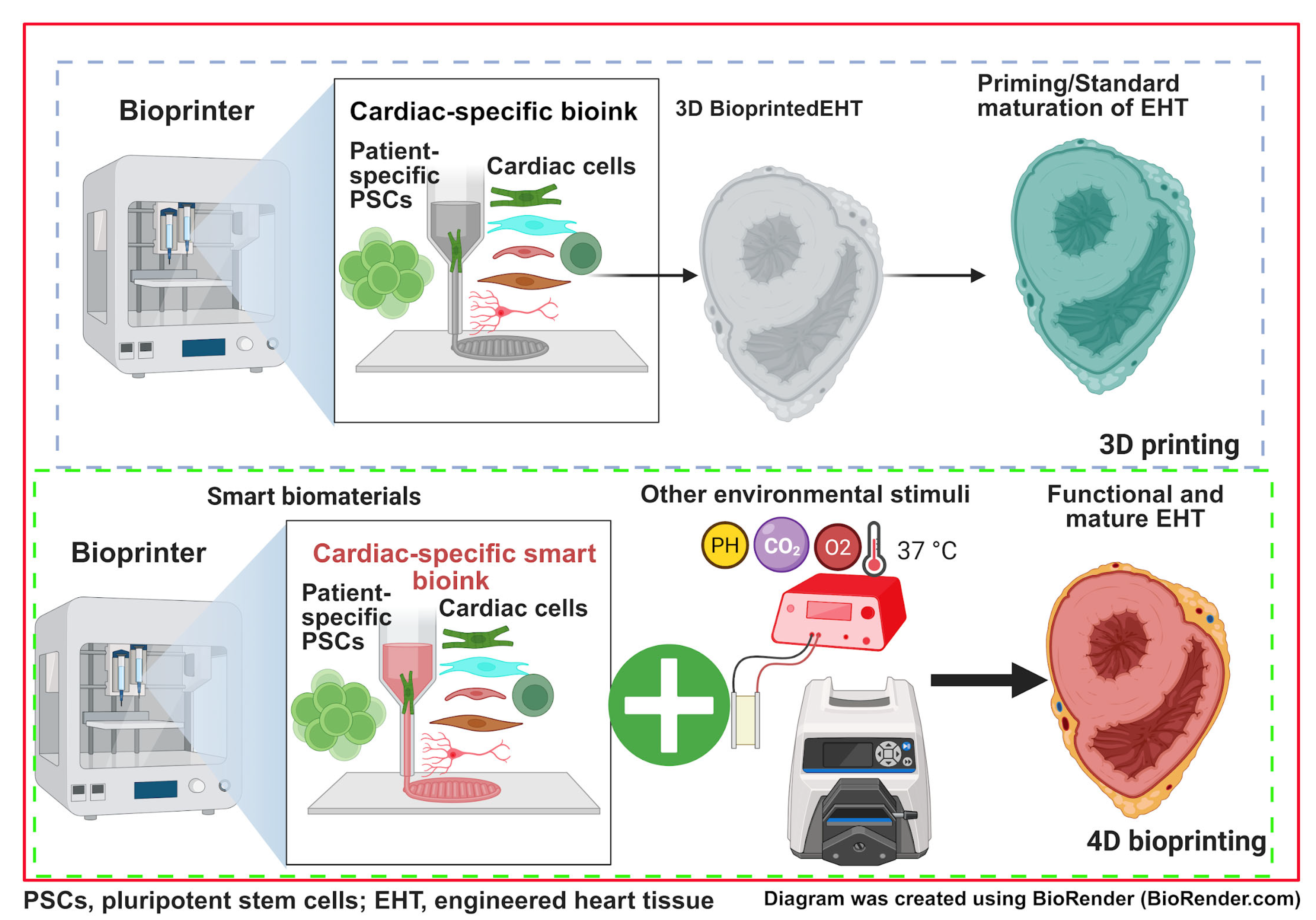
6.2. 3D/4D Bioprinting PSC-Derived Cardiac Tissues for Studying HLHS
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| HLHS | hypoplastic left heart syndrome |
| PSC | pluripotent stem cell |
| iPSC | induced pluripotent stem cell |
| CHD | congenital heart disease |
| CM | cardiomyocyte |
| RV | right ventricle |
| LV | left ventricle |
| FHF | first heart field |
| SHF | second heart field |
| EF | ejection fraction |
| BM-MSC | bone marrow-derived mesenchymal stem cell |
| BM-MNC | bone marrow-derived mononuclear cells |
| CDC | cardiosphere-derived cells |
| UCB-MSC | umbilical cord blood-derived mesenchymal stem cells |
| UCB-MNC | umbilical cord blood-derived mononuclear cells |
| NKX2 | NK homeobox 2 |
References
- Stallings, E.B.; Isenburg, J.L.; Rutkowski, R.E.; Kirby, R.S.; Nembhard, W.N.; Sandidge, T.; Villavicencio, S.; Nguyen, H.H.; McMahon, D.M.; Nestoridi, E. , et al. National population-based estimates for major birth defects, 2016-2020. Birth Defects Res 2024, 116, e2301. [Google Scholar] [CrossRef]
- Feinstein, J.A.; Benson, D.W.; Dubin, A.M.; Cohen, M.S.; Maxey, D.M.; Mahle, W.T.; Pahl, E.; Villafane, J.; Bhatt, A.B.; Peng, L.F. , et al. Hypoplastic left heart syndrome: current considerations and expectations. Journal of the American College of Cardiology 2012, 59, S1–42. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, K.; Ogawa, K.; Hishitani, T.; Kitazawa, R.; Uehara, R. Hypoplastic left heart syndrome: duration of survival without surgical intervention. Am Heart J 1999, 137, 535–542. [Google Scholar] [CrossRef]
- Alphonso, N.; Angelini, A.; Barron, D.J.; Bellsham-Revell, H.; Blom, N.A.; Brown, K.; Davis, D.; Duncan, D.; Fedrigo, M.; Galletti, L. , et al. Guidelines for the management of neonates and infants with hypoplastic left heart syndrome: The European Association for Cardio-Thoracic Surgery (EACTS) and the Association for European Paediatric and Congenital Cardiology (AEPC) Hypoplastic Left Heart Syndrome Guidelines Task Force. Eur J Cardiothorac Surg 2020, 58, 416–499. [Google Scholar] [CrossRef]
- Ohye, R.G.; Schranz, D.; D'Udekem, Y. Current Therapy for Hypoplastic Left Heart Syndrome and Related Single Ventricle Lesions. Circulation 2016, 134, 1265–1279. [Google Scholar] [CrossRef]
- Norwood, W.I.; Lang, P.; Casteneda, A.R.; Campbell, D.N. Experience with operations for hypoplastic left heart syndrome. J Thorac Cardiovasc Surg 1981, 82, 511–519. [Google Scholar] [CrossRef]
- Lee, M.; Geoffrion, T.R. Norwood Procedure. In StatPearls, Treasure Island (FL) ineligible companies. Disclosure: Tracy Geoffrion declares no relevant financial relationships with ineligible companies., 2025.
- Glenn, W.W. Circulatory bypass of the right side of the heart. IV. Shunt between superior vena cava and distal right pulmonary artery; report of clinical application. The New England journal of medicine 1958, 259, 117–120. [Google Scholar] [CrossRef]
- Fontan, F.; Baudet, E. Surgical repair of tricuspid atresia. Thorax 1971, 26, 240–248. [Google Scholar] [CrossRef]
- Steele, M.M.; Zahr, R.A.; Kirshbom, P.M.; Kopf, G.S.; Karimi, M. Quality of Life for Historic Cavopulmonary Shunt Survivors. World J Pediatr Congenit Heart Surg 2016, 7, 630–634. [Google Scholar] [CrossRef] [PubMed]
- Marshall, K.H.; D'Udekem, Y.; Sholler, G.F.; Opotowsky, A.R.; Costa, D.S.J.; Sharpe, L.; Celermajer, D.S.; Winlaw, D.S.; Newburger, J.W.; Kasparian, N.A. Health-Related Quality of Life in Children, Adolescents, and Adults With a Fontan Circulation: A Meta-Analysis. Journal of the American Heart Association 2020, 9, e014172. [Google Scholar] [CrossRef] [PubMed]
- Dib, N.; Poirier, N.; Samuel, M.; Hermann Honfo, S.; Zaidi, A.; Opotowsky, A.R.; Mongeon, F.P.; Mondesert, B.; Kay, J.; Ibrahim, R. , et al. Cardiovascular Outcomes Associated With Hypoplastic Left Heart Syndrome Versus Other Types of Single Right Ventricle: A Multicenter Study. Journal of the American Heart Association 2024, 13, e034757. [Google Scholar] [CrossRef] [PubMed]
- Yimer, M.A.; Tisma-Dupanovic, S.; Malloy-Walton, L.; Connelly, D.; Noel-Macdonnell, J.; J, O.B.; Papagiannis, J. Post-operative arrhythmias in patients with hypoplastic left heart syndrome and anatomic variants: incidence, type, and course. Cardiol Young 2021, 31, 1412–1418. [Google Scholar] [CrossRef] [PubMed]
- Poh, C.L.; d'Udekem, Y. Life After Surviving Fontan Surgery: A Meta-Analysis of the Incidence and Predictors of Late Death. Heart, lung & circulation 2018, 27, 552–559. [Google Scholar] [CrossRef]
- Chrisant, M.R.; Naftel, D.C.; Drummond-Webb, J.; Chinnock, R.; Canter, C.E.; Boucek, M.M.; Boucek, R.J.; Hallowell, S.C.; Kirklin, J.K.; Morrow, W.R. , et al. Fate of infants with hypoplastic left heart syndrome listed for cardiac transplantation: a multicenter study. The Journal of heart and lung transplantation : the official publication of the International Society for Heart Transplantation 2005, 24, 576–582. [Google Scholar] [CrossRef]
- Pittenger, M.F.; Mackay, A.M.; Beck, S.C.; Jaiswal, R.K.; Douglas, R.; Mosca, J.D.; Moorman, M.A.; Simonetti, D.W.; Craig, S.; Marshak, D.R. Multilineage potential of adult human mesenchymal stem cells. Science 1999, 284, 143–147. [Google Scholar] [CrossRef]
- Zuk, P.A.; Zhu, M.; Mizuno, H.; Huang, J.; Futrell, J.W.; Katz, A.J.; Benhaim, P.; Lorenz, H.P.; Hedrick, M.H. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng 2001, 7, 211–228. [Google Scholar] [CrossRef]
- Wang, H.S.; Hung, S.C.; Peng, S.T.; Huang, C.C.; Wei, H.M.; Guo, Y.J.; Fu, Y.S.; Lai, M.C.; Chen, C.C. Mesenchymal stem cells in the Wharton's jelly of the human umbilical cord. Stem cells 2004, 22, 1330–1337. [Google Scholar] [CrossRef]
- Bittle, G.J.; Morales, D.; Deatrick, K.B.; Parchment, N.; Saha, P.; Mishra, R.; Sharma, S.; Pietris, N.; Vasilenko, A.; Bor, C. , et al. Stem Cell Therapy for Hypoplastic Left Heart Syndrome: Mechanism, Clinical Application, and Future Directions. Circulation research 2018, 123, 288–300. [Google Scholar] [CrossRef]
- Kaushal, S.; Hare, J.M.; Hoffman, J.R.; Boyd, R.M.; Ramdas, K.N.; Pietris, N.; Kutty, S.; Tweddell, J.S.; Husain, S.A.; Menon, S.C. , et al. Intramyocardial cell-based therapy with Lomecel-B during bidirectional cavopulmonary anastomosis for hypoplastic left heart syndrome: the ELPIS phase I trial. Eur Heart J Open 2023, 3, oead002. [Google Scholar] [CrossRef]
- Lian, X.; Hsiao, C.; Wilson, G.; Zhu, K.; Hazeltine, L.B.; Azarin, S.M.; Raval, K.K.; Zhang, J.; Kamp, T.J.; Palecek, S.P. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proceedings of the National Academy of Sciences of the United States of America 2012, 109, E1848–1857. [Google Scholar] [CrossRef]
- van Mil, A.; Balk, G.M.; Neef, K.; Buikema, J.W.; Asselbergs, F.W.; Wu, S.M.; Doevendans, P.A.; Sluijter, J.P.G. Modelling inherited cardiac disease using human induced pluripotent stem cell-derived cardiomyocytes: progress, pitfalls, and potential. Cardiovascular research 2018, 114, 1828–1842. [Google Scholar] [CrossRef] [PubMed]
- Caspi, O.; Huber, I.; Gepstein, A.; Arbel, G.; Maizels, L.; Boulos, M.; Gepstein, L. Modeling of arrhythmogenic right ventricular cardiomyopathy with human induced pluripotent stem cells. Circulation. Cardiovascular genetics 2013, 6, 557–568. [Google Scholar] [CrossRef]
- Yazawa, M.; Hsueh, B.; Jia, X.; Pasca, A.M.; Bernstein, J.A.; Hallmayer, J.; Dolmetsch, R.E. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature 2011, 471, 230–234. [Google Scholar] [CrossRef]
- Lan, F.; Lee, A.S.; Liang, P.; Sanchez-Freire, V.; Nguyen, P.K.; Wang, L.; Han, L.; Yen, M.; Wang, Y.; Sun, N. , et al. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell stem cell 2013, 12, 101–113. [Google Scholar] [CrossRef]
- Wolfe, J.T.; He, W.; Kim, M.S.; Liang, H.L.; Shradhanjali, A.; Jurkiewicz, H.; Freudinger, B.P.; Greene, A.S.; LaDisa, J.F., Jr.; Tayebi, L. , et al. 3D-bioprinting of patient-derived cardiac tissue models for studying congenital heart disease. Front Cardiovasc Med 2023, 10, 1162731. [Google Scholar] [CrossRef]
- Alonzo, M.; Contreras, J.; Bering, J.; Zhao, M.T. In Vivo and In Vitro Approaches to Modeling Hypoplastic Left Heart Syndrome. Curr Cardiol Rep 2024, 26, 1221–1229. [Google Scholar] [CrossRef] [PubMed]
- Van Vliet, P.; Wu, S.M.; Zaffran, S.; Puceat, M. Early cardiac development: a view from stem cells to embryos. Cardiovascular research 2012, 96, 352–362. [Google Scholar] [CrossRef]
- Brand, T. Heart development: molecular insights into cardiac specification and early morphogenesis. Dev Biol 2003, 258, 1–19. [Google Scholar] [CrossRef]
- Kirby, M.L.; Waldo, K.L. Neural crest and cardiovascular patterning. Circulation research 1995, 77, 211–215. [Google Scholar] [CrossRef]
- Gitler, A.D.; Lu, M.M.; Jiang, Y.Q.; Epstein, J.A.; Gruber, P.J. Molecular markers of cardiac endocardial cushion development. Developmental dynamics : an official publication of the American Association of Anatomists 2003, 228, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Grossfeld, P.; Nie, S.; Lin, L.; Wang, L.; Anderson, R.H. Hypoplastic Left Heart Syndrome: A New Paradigm for an Old Disease? J Cardiovasc Dev Dis 2019, 6. [Google Scholar] [CrossRef]
- Datta, S.; Cao, W.; Skillman, M.; Wu, M. Hypoplastic Left Heart Syndrome: Signaling & Molecular Perspectives, and the Road Ahead. International journal of molecular sciences 2023, 24. [Google Scholar] [CrossRef]
- Niessen, K.; Karsan, A. Notch signaling in cardiac development. Circulation research 2008, 102, 1169–1181. [Google Scholar] [CrossRef]
- Kerstjens-Frederikse, W.S.; van de Laar, I.M.; Vos, Y.J.; Verhagen, J.M.; Berger, R.M.; Lichtenbelt, K.D.; Klein Wassink-Ruiter, J.S.; van der Zwaag, P.A.; du Marchie Sarvaas, G.J.; Bergman, K.A. , et al. Cardiovascular malformations caused by NOTCH1 mutations do not keep left: data on 428 probands with left-sided CHD and their families. Genet Med 2016, 18, 914–923. [Google Scholar] [CrossRef]
- McBride, K.L.; Riley, M.F.; Zender, G.A.; Fitzgerald-Butt, S.M.; Towbin, J.A.; Belmont, J.W.; Cole, S.E. NOTCH1 mutations in individuals with left ventricular outflow tract malformations reduce ligand-induced signaling. Human molecular genetics 2008, 17, 2886–2893. [Google Scholar] [CrossRef]
- Rahman, A.; DeYoung, T.; Cahill, L.S.; Yee, Y.; Debebe, S.K.; Botelho, O.; Seed, M.; Chaturvedi, R.R.; Sled, J.G. A mouse model of hypoplastic left heart syndrome demonstrating left heart hypoplasia and retrograde aortic arch flow. Dis Model Mech 2021, 14. [Google Scholar] [CrossRef] [PubMed]
- Durbin, M.D.; Cadar, A.G.; Williams, C.H.; Guo, Y.; Bichell, D.P.; Su, Y.R.; Hong, C.C. Hypoplastic Left Heart Syndrome Sequencing Reveals a Novel NOTCH1 Mutation in a Family with Single Ventricle Defects. Pediatric cardiology 2017, 38, 1232–1240. [Google Scholar] [CrossRef] [PubMed]
- Winnier, G.; Blessing, M.; Labosky, P.A.; Hogan, B.L. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes & development 1995, 9, 2105–2116. [Google Scholar] [CrossRef]
- Beppu, H.; Kawabata, M.; Hamamoto, T.; Chytil, A.; Minowa, O.; Noda, T.; Miyazono, K. BMP type II receptor is required for gastrulation and early development of mouse embryos. Dev Biol 2000, 221, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Jiao, K.; Kulessa, H.; Tompkins, K.; Zhou, Y.; Batts, L.; Baldwin, H.S.; Hogan, B.L. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes & development 2003, 17, 2362–2367. [Google Scholar] [CrossRef]
- Wang, J.; Greene, S.B.; Martin, J.F. BMP signaling in congenital heart disease: new developments and future directions. Birth Defects Res A Clin Mol Teratol 2011, 91, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Ricci, M.; Mohapatra, B.; Urbiztondo, A.; Birusingh, R.J.; Morgado, M.; Rodriguez, M.M.; Lincoln, J.; Vatta, M. Differential changes in TGF-beta/BMP signaling pathway in the right ventricular myocardium of newborns with hypoplastic left heart syndrome. J Card Fail 2010, 16, 628–634. [Google Scholar] [CrossRef] [PubMed]
- Stefanovic, S.; Zaffran, S. Mechanisms of retinoic acid signaling during cardiogenesis. Mechanisms of development 2017, 143, 9–19. [Google Scholar] [CrossRef]
- Vermot, J.; Niederreither, K.; Garnier, J.M.; Chambon, P.; Dolle, P. Decreased embryonic retinoic acid synthesis results in a DiGeorge syndrome phenotype in newborn mice. Proceedings of the National Academy of Sciences of the United States of America 2003, 100, 1763–1768. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Choudhary, B.; Merki, E.; Chien, K.R.; Maxson, R.E.; Sucov, H.M. Normal fate and altered function of the cardiac neural crest cell lineage in retinoic acid receptor mutant embryos. Mechanisms of development 2002, 117, 115–122. [Google Scholar] [CrossRef]
- Horitani, K.; Shiojima, I. Wnt signaling in cardiac development and heart diseases. In Vitro Cell Dev Biol Anim 2024, 60, 482–488. [Google Scholar] [CrossRef]
- Lorenzon, A.; Calore, M.; Poloni, G.; De Windt, L.J.; Braghetta, P.; Rampazzo, A. Wnt/beta-catenin pathway in arrhythmogenic cardiomyopathy. Oncotarget 2017, 8, 60640–60655. [Google Scholar] [CrossRef]
- Briggs, L.E.; Burns, T.A.; Lockhart, M.M.; Phelps, A.L.; Van den Hoff, M.J.; Wessels, A. Wnt/beta-catenin and sonic hedgehog pathways interact in the regulation of the development of the dorsal mesenchymal protrusion. Developmental dynamics : an official publication of the American Association of Anatomists 2016, 245, 103–113. [Google Scholar] [CrossRef]
- Washington Smoak, I.; Byrd, N.A.; Abu-Issa, R.; Goddeeris, M.M.; Anderson, R.; Morris, J.; Yamamura, K.; Klingensmith, J.; Meyers, E.N. Sonic hedgehog is required for cardiac outflow tract and neural crest cell development. Dev Biol 2005, 283, 357–372. [Google Scholar] [CrossRef]
- Nanni, L.; Ming, J.E.; Bocian, M.; Steinhaus, K.; Bianchi, D.W.; Die-Smulders, C.; Giannotti, A.; Imaizumi, K.; Jones, K.L.; Campo, M.D. , et al. The mutational spectrum of the sonic hedgehog gene in holoprosencephaly: SHH mutations cause a significant proportion of autosomal dominant holoprosencephaly. Human molecular genetics 1999, 8, 2479–2488. [Google Scholar] [CrossRef]
- Wang, J.; Liu, S.; Heallen, T.; Martin, J.F. The Hippo pathway in the heart: pivotal roles in development, disease, and regeneration. Nat Rev Cardiol 2018, 15, 672–684. [Google Scholar] [CrossRef]
- Zhou, Q.; Li, L.; Zhao, B.; Guan, K.L. The hippo pathway in heart development, regeneration, and diseases. Circulation research 2015, 116, 1431–1447. [Google Scholar] [CrossRef] [PubMed]
- Yagi, H.; Xu, X.; Gabriel, G.C.; Lo, C. Molecular Pathways and Animal Models of Hypoplastic Left Heart Syndrome. Advances in experimental medicine and biology 2024, 1441, 947–961. [Google Scholar] [CrossRef]
- Khosravi, F.; Ahmadvand, N.; Bellusci, S.; Sauer, H. The Multifunctional Contribution of FGF Signaling to Cardiac Development, Homeostasis, Disease and Repair. Front Cell Dev Biol 2021, 9, 672935. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chang, J.Y.; Huang, Y.; Lin, X.; Luo, Y.; Schwartz, R.J.; Martin, J.F.; Wang, F. The FGF-BMP signaling axis regulates outflow tract valve primordium formation by promoting cushion neural crest cell differentiation. Circulation research 2010, 107, 1209–1219. [Google Scholar] [CrossRef]
- Reuter, M.S.; Sokolowski, D.J.; Javier Diaz-Mejia, J.; Keunen, J.; de Vrijer, B.; Chan, C.; Wang, L.; Ryan, G.; Chiasson, D.A.; Ketela, T. , et al. Decreased left heart flow in fetal lambs causes left heart hypoplasia and pro-fibrotic tissue remodeling. Commun Biol 2023, 6, 770. [Google Scholar] [CrossRef]
- Han, Z.; Yi, P.; Li, X.; Olson, E.N. Hand, an evolutionarily conserved bHLH transcription factor required for Drosophila cardiogenesis and hematopoiesis. Development 2006, 133, 1175–1182. [Google Scholar] [CrossRef] [PubMed]
- Riley, P.; Anson-Cartwright, L.; Cross, J.C. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nature genetics 1998, 18, 271–275. [Google Scholar] [CrossRef]
- Reamon-Buettner, S.M.; Ciribilli, Y.; Inga, A.; Borlak, J. A loss-of-function mutation in the binding domain of HAND1 predicts hypoplasia of the human hearts. Human molecular genetics 2008, 17, 1397–1405. [Google Scholar] [CrossRef]
- Kobayashi, J.; Yoshida, M.; Tarui, S.; Hirata, M.; Nagai, Y.; Kasahara, S.; Naruse, K.; Ito, H.; Sano, S.; Oh, H. Directed differentiation of patient-specific induced pluripotent stem cells identifies the transcriptional repression and epigenetic modification of NKX2-5, HAND1, and NOTCH1 in hypoplastic left heart syndrome. PloS one 2014, 9, e102796. [Google Scholar] [CrossRef]
- Akazawa, H.; Komuro, I. Cardiac transcription factor Csx/Nkx2-5: Its role in cardiac development and diseases. Pharmacology & therapeutics 2005, 107, 252–268. [Google Scholar] [CrossRef]
- Elliott, D.A.; Kirk, E.P.; Yeoh, T.; Chandar, S.; McKenzie, F.; Taylor, P.; Grossfeld, P.; Fatkin, D.; Jones, O.; Hayes, P. , et al. Cardiac homeobox gene NKX2-5 mutations and congenital heart disease: associations with atrial septal defect and hypoplastic left heart syndrome. Journal of the American College of Cardiology 2003, 41, 2072–2076. [Google Scholar] [CrossRef] [PubMed]
- Doering, L.; Cornean, A.; Thumberger, T.; Benjaminsen, J.; Wittbrodt, B.; Kellner, T.; Hammouda, O.T.; Gorenflo, M.; Wittbrodt, J.; Gierten, J. CRISPR-based knockout and base editing confirm the role of MYRF in heart development and congenital heart disease. Dis Model Mech 2023, 16. [Google Scholar] [CrossRef]
- Qi, H.; Yu, L.; Zhou, X.; Wynn, J.; Zhao, H.; Guo, Y.; Zhu, N.; Kitaygorodsky, A.; Hernan, R.; Aspelund, G. , et al. De novo variants in congenital diaphragmatic hernia identify MYRF as a new syndrome and reveal genetic overlaps with other developmental disorders. PLoS genetics 2018, 14, e1007822. [Google Scholar] [CrossRef] [PubMed]
- Watt, A.J.; Battle, M.A.; Li, J.; Duncan, S.A. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proceedings of the National Academy of Sciences of the United States of America 2004, 101, 12573–12578. [Google Scholar] [CrossRef]
- Misra, C.; Sachan, N.; McNally, C.R.; Koenig, S.N.; Nichols, H.A.; Guggilam, A.; Lucchesi, P.A.; Pu, W.T.; Srivastava, D.; Garg, V. Congenital heart disease-causing Gata4 mutation displays functional deficits in vivo. PLoS genetics 2012, 8, e1002690. [Google Scholar] [CrossRef]
- Ang, Y.S.; Rivas, R.N.; Ribeiro, A.J.S.; Srivas, R.; Rivera, J.; Stone, N.R.; Pratt, K.; Mohamed, T.M.A.; Fu, J.D.; Spencer, C.I. , et al. Disease Model of GATA4 Mutation Reveals Transcription Factor Cooperativity in Human Cardiogenesis. Cell 2016, 167, 1734–1749. [Google Scholar] [CrossRef]
- Li, Y.; Du, J.; Deng, S.; Liu, B.; Jing, X.; Yan, Y.; Liu, Y.; Wang, J.; Zhou, X.; She, Q. The molecular mechanisms of cardiac development and related diseases. Signal Transduct Target Ther 2024, 9, 368. [Google Scholar] [CrossRef]
- Patel, C.; Silcock, L.; McMullan, D.; Brueton, L.; Cox, H. TBX5 intragenic duplication: a family with an atypical Holt-Oram syndrome phenotype. Eur J Hum Genet 2012, 20, 863–869. [Google Scholar] [CrossRef] [PubMed]
- Bruneau, B.G.; Nemer, G.; Schmitt, J.P.; Charron, F.; Robitaille, L.; Caron, S.; Conner, D.A.; Gessler, M.; Nemer, M.; Seidman, C.E. , et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 2001, 106, 709–721. [Google Scholar] [CrossRef]
- Yang, C.; Xu, Y.; Yu, M.; Lee, D.; Alharti, S.; Hellen, N.; Ahmad Shaik, N.; Banaganapalli, B.; Sheikh Ali Mohamoud, H.; Elango, R. , et al. Induced pluripotent stem cell modelling of HLHS underlines the contribution of dysfunctional NOTCH signalling to impaired cardiogenesis. Human molecular genetics 2017, 26, 3031–3045. [Google Scholar] [CrossRef]
- von Gise, A.; Zhou, B.; Honor, L.B.; Ma, Q.; Petryk, A.; Pu, W.T. WT1 regulates epicardial epithelial to mesenchymal transition through beta-catenin and retinoic acid signaling pathways. Dev Biol 2011, 356, 421–431. [Google Scholar] [CrossRef]
- Crucean, A.; Alqahtani, A.; Barron, D.J.; Brawn, W.J.; Richardson, R.V.; O'Sullivan, J.; Anderson, R.H.; Henderson, D.J.; Chaudhry, B. Re-evaluation of hypoplastic left heart syndrome from a developmental and morphological perspective. Orphanet J Rare Dis 2017, 12, 138. [Google Scholar] [CrossRef]
- Qiao, X.H.; Wang, F.; Zhang, X.L.; Huang, R.T.; Xue, S.; Wang, J.; Qiu, X.B.; Liu, X.Y.; Yang, Y.Q. MEF2C loss-of-function mutation contributes to congenital heart defects. Int J Med Sci 2017, 14, 1143–1153. [Google Scholar] [CrossRef] [PubMed]
- Barnes, R.M.; Harris, I.S.; Jaehnig, E.J.; Sauls, K.; Sinha, T.; Rojas, A.; Schachterle, W.; McCulley, D.J.; Norris, R.A.; Black, B.L. MEF2C regulates outflow tract alignment and transcriptional control of Tdgf1. Development 2016, 143, 774–779. [Google Scholar] [CrossRef]
- Gao, R.; Liang, X.; Cheedipudi, S.; Cordero, J.; Jiang, X.; Zhang, Q.; Caputo, L.; Gunther, S.; Kuenne, C.; Ren, Y. , et al. Pioneering function of Isl1 in the epigenetic control of cardiomyocyte cell fate. Cell research 2019, 29, 486–501. [Google Scholar] [CrossRef]
- Stevens, K.N.; Hakonarson, H.; Kim, C.E.; Doevendans, P.A.; Koeleman, B.P.; Mital, S.; Raue, J.; Glessner, J.T.; Coles, J.G.; Moreno, V. , et al. Common variation in ISL1 confers genetic susceptibility for human congenital heart disease. PloS one 2010, 5, e10855. [Google Scholar] [CrossRef]
- Tomita-Mitchell, A.; Stamm, K.D.; Mahnke, D.K.; Kim, M.S.; Hidestrand, P.M.; Liang, H.L.; Goetsch, M.A.; Hidestrand, M.; Simpson, P.; Pelech, A.N. , et al. Impact of MYH6 variants in hypoplastic left heart syndrome. Physiological genomics 2016, 48, 912–921. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Fleres, B.; Lovett, J.; Anfinson, M.; Samudrala, S.S.K.; Kelly, L.J.; Teigen, L.E.; Cavanaugh, M.; Marquez, M.; Geurts, A.M. , et al. Contractility of Induced Pluripotent Stem Cell-Cardiomyocytes With an MYH6 Head Domain Variant Associated With Hypoplastic Left Heart Syndrome. Front Cell Dev Biol 2020, 8, 440. [Google Scholar] [CrossRef]
- Christ, A.; Christa, A.; Klippert, J.; Eule, J.C.; Bachmann, S.; Wallace, V.A.; Hammes, A.; Willnow, T.E. LRP2 Acts as SHH Clearance Receptor to Protect the Retinal Margin from Mitogenic Stimuli. Developmental cell 2015, 35, 36–48. [Google Scholar] [CrossRef] [PubMed]
- Kantarci, S.; Al-Gazali, L.; Hill, R.S.; Donnai, D.; Black, G.C.; Bieth, E.; Chassaing, N.; Lacombe, D.; Devriendt, K.; Teebi, A. , et al. Mutations in LRP2, which encodes the multiligand receptor megalin, cause Donnai-Barrow and facio-oculo-acoustico-renal syndromes. Nature genetics 2007, 39, 957–959. [Google Scholar] [CrossRef]
- Theis, J.L.; Vogler, G.; Missinato, M.A.; Li, X.; Nielsen, T.; Zeng, X.I.; Martinez-Fernandez, A.; Walls, S.M.; Kervadec, A.; Kezos, J.N. , et al. Patient-specific genomics and cross-species functional analysis implicate LRP2 in hypoplastic left heart syndrome. eLife 2020, 9. [Google Scholar] [CrossRef]
- Britz-Cunningham, S.H.; Shah, M.M.; Zuppan, C.W.; Fletcher, W.H. Mutations of the Connexin43 gap-junction gene in patients with heart malformations and defects of laterality. The New England journal of medicine 1995, 332, 1323–1329. [Google Scholar] [CrossRef]
- Dasgupta, C.; Martinez, A.M.; Zuppan, C.W.; Shah, M.M.; Bailey, L.L.; Fletcher, W.H. Identification of connexin43 (alpha1) gap junction gene mutations in patients with hypoplastic left heart syndrome by denaturing gradient gel electrophoresis (DGGE). Mutation research 2001, 479, 173–186. [Google Scholar] [CrossRef]
- Asp, M.; Giacomello, S.; Larsson, L.; Wu, C.; Furth, D.; Qian, X.; Wardell, E.; Custodio, J.; Reimegard, J.; Salmen, F. , et al. A Spatiotemporal Organ-Wide Gene Expression and Cell Atlas of the Developing Human Heart. Cell 2019, 179, 1647–1660. [Google Scholar] [CrossRef]
- Gessert, S.; Kuhl, M. The multiple phases and faces of wnt signaling during cardiac differentiation and development. Circulation research 2010, 107, 186–199. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. Wnt/beta-catenin signaling in development and disease. Cell 2006, 127, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Tzahor, E. Wnt/beta-catenin signaling and cardiogenesis: timing does matter. Developmental cell 2007, 13, 10–13. [Google Scholar] [CrossRef] [PubMed]
- Ryckebusch, L.; Wang, Z.; Bertrand, N.; Lin, S.C.; Chi, X.; Schwartz, R.; Zaffran, S.; Niederreither, K. Retinoic acid deficiency alters second heart field formation. Proceedings of the National Academy of Sciences of the United States of America 2008, 105, 2913–2918. [Google Scholar] [CrossRef]
- Sirbu, I.O.; Zhao, X.; Duester, G. Retinoic acid controls heart anteroposterior patterning by down-regulating Isl1 through the Fgf8 pathway. Developmental dynamics : an official publication of the American Association of Anatomists 2008, 237, 1627–1635. [Google Scholar] [CrossRef]
- Ghyselinck, N.B.; Duester, G. Retinoic acid signaling pathways. Development 2019, 146. [Google Scholar] [CrossRef]
- Griffin, A.H.C.; Small, A.M.; Johnson, R.D.; Medina, A.M.; Kollar, K.T.; Nazir, R.A.; McGuire, A.M.; Schumacher, J.A. Retinoic acid promotes second heart field addition and regulates ventral aorta patterning in zebrafish. Dev Biol 2025, 522, 143–155. [Google Scholar] [CrossRef]
- Rones, M.S.; McLaughlin, K.A.; Raffin, M.; Mercola, M. Serrate and Notch specify cell fates in the heart field by suppressing cardiomyogenesis. Development 2000, 127, 3865–3876. [Google Scholar] [CrossRef] [PubMed]
- Grego-Bessa, J.; Luna-Zurita, L.; del Monte, G.; Bolos, V.; Melgar, P.; Arandilla, A.; Garratt, A.N.; Zang, H.; Mukouyama, Y.S.; Chen, H. , et al. Notch signaling is essential for ventricular chamber development. Developmental cell 2007, 12, 415–429. [Google Scholar] [CrossRef]
- Luxan, G.; D'Amato, G.; MacGrogan, D.; de la Pompa, J.L. Endocardial Notch Signaling in Cardiac Development and Disease. Circulation research 2016, 118, e1–e18. [Google Scholar] [CrossRef] [PubMed]
- MacGrogan, D.; Nus, M.; de la Pompa, J.L. Notch signaling in cardiac development and disease. Curr Top Dev Biol 2010, 92, 333–365. [Google Scholar] [CrossRef] [PubMed]
- McCulley, D.J.; Kang, J.O.; Martin, J.F.; Black, B.L. BMP4 is required in the anterior heart field and its derivatives for endocardial cushion remodeling, outflow tract septation, and semilunar valve development. Developmental dynamics : an official publication of the American Association of Anatomists 2008, 237, 3200–3209. [Google Scholar] [CrossRef]
- Chen, H.; Shi, S.; Acosta, L.; Li, W.; Lu, J.; Bao, S.; Chen, Z.; Yang, Z.; Schneider, M.D.; Chien, K.R. , et al. BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development 2004, 131, 2219–2231. [Google Scholar] [CrossRef]
- Liu, W.; Selever, J.; Wang, D.; Lu, M.F.; Moses, K.A.; Schwartz, R.J.; Martin, J.F. Bmp4 signaling is required for outflow-tract septation and branchial-arch artery remodeling. Proceedings of the National Academy of Sciences of the United States of America 2004, 101, 4489–4494. [Google Scholar] [CrossRef]
- Luna-Zurita, L.; Stirnimann, C.U.; Glatt, S.; Kaynak, B.L.; Thomas, S.; Baudin, F.; Samee, M.A.; He, D.; Small, E.M.; Mileikovsky, M. , et al. Complex Interdependence Regulates Heterotypic Transcription Factor Distribution and Coordinates Cardiogenesis. Cell 2016, 164, 999–1014. [Google Scholar] [CrossRef]
- Fahed, A.C.; Gelb, B.D.; Seidman, J.G.; Seidman, C.E. Genetics of congenital heart disease: the glass half empty. Circulation research 2013, 112, 707–720. [Google Scholar] [CrossRef]
- Bruneau, B.G. Transcriptional regulation of vertebrate cardiac morphogenesis. Circulation research 2002, 90, 509–519. [Google Scholar] [CrossRef]
- Bruneau, B.G. The developmental genetics of congenital heart disease. Nature 2008, 451, 943–948. [Google Scholar] [CrossRef]
- Olson, E.N. Gene regulatory networks in the evolution and development of the heart. Science 2006, 313, 1922–1927. [Google Scholar] [CrossRef]
- McFadden, D.G.; Barbosa, A.C.; Richardson, J.A.; Schneider, M.D.; Srivastava, D.; Olson, E.N. The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development 2005, 132, 189–201. [Google Scholar] [CrossRef]
- Oka, T.; Maillet, M.; Watt, A.J.; Schwartz, R.J.; Aronow, B.J.; Duncan, S.A.; Molkentin, J.D. Cardiac-specific deletion of Gata4 reveals its requirement for hypertrophy, compensation, and myocyte viability. Circulation research 2006, 98, 837–845. [Google Scholar] [CrossRef]
- Cai, C.L.; Liang, X.; Shi, Y.; Chu, P.H.; Pfaff, S.L.; Chen, J.; Evans, S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Developmental cell 2003, 5, 877–889. [Google Scholar] [CrossRef]
- Stennard, F.A.; Harvey, R.P. T-box transcription factors and their roles in regulatory hierarchies in the developing heart. Development 2005, 132, 4897–4910. [Google Scholar] [CrossRef]
- Lin, Q.; Schwarz, J.; Bucana, C.; Olson, E.N. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science 1997, 276, 1404–1407. [Google Scholar] [CrossRef]
- Tanaka, M.; Chen, Z.; Bartunkova, S.; Yamasaki, N.; Izumo, S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development 1999, 126, 1269–1280. [Google Scholar] [CrossRef]
- Durocher, D.; Charron, F.; Warren, R.; Schwartz, R.J.; Nemer, M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. The EMBO journal 1997, 16, 5687–5696. [Google Scholar] [CrossRef]
- Lyons, I.; Parsons, L.M.; Hartley, L.; Li, R.; Andrews, J.E.; Robb, L.; Harvey, R.P. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes & development 1995, 9, 1654–1666. [Google Scholar] [CrossRef]
- Targoff, K.L.; Colombo, S.; George, V.; Schell, T.; Kim, S.H.; Solnica-Krezel, L.; Yelon, D. Nkx genes are essential for maintenance of ventricular identity. Development 2013, 140, 4203–4213. [Google Scholar] [CrossRef]
- Pashmforoush, M.; Lu, J.T.; Chen, H.; Amand, T.S.; Kondo, R.; Pradervand, S.; Evans, S.M.; Clark, B.; Feramisco, J.R.; Giles, W. , et al. Nkx2-5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell 2004, 117, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.W.; Silberbach, G.M.; Kavanaugh-McHugh, A.; Cottrill, C.; Zhang, Y.; Riggs, S.; Smalls, O.; Johnson, M.C.; Watson, M.S.; Seidman, J.G. , et al. Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. The Journal of clinical investigation 1999, 104, 1567–1573. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.S.; Cserjesi, P.; Markham, B.E.; Molkentin, J.D. The transcription factors GATA4 and dHAND physically interact to synergistically activate cardiac gene expression through a p300-dependent mechanism. The Journal of biological chemistry 2002, 277, 24390–24398. [Google Scholar] [CrossRef]
- Risebro, C.A.; Smart, N.; Dupays, L.; Breckenridge, R.; Mohun, T.J.; Riley, P.R. Hand1 regulates cardiomyocyte proliferation versus differentiation in the developing heart. Development 2006, 133, 4595–4606. [Google Scholar] [CrossRef]
- Firulli, B.A.; George, R.M.; Harkin, J.; Toolan, K.P.; Gao, H.; Liu, Y.; Zhang, W.; Field, L.J.; Liu, Y.; Shou, W. , et al. HAND1 loss-of-function within the embryonic myocardium reveals survivable congenital cardiac defects and adult heart failure. Cardiovascular research 2020, 116, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Vincentz, J.W.; Toolan, K.P.; Zhang, W.; Firulli, A.B. Hand factor ablation causes defective left ventricular chamber development and compromised adult cardiac function. PLoS genetics 2017, 13, e1006922. [Google Scholar] [CrossRef]
- Firulli, B.A.; Toolan, K.P.; Harkin, J.; Millar, H.; Pineda, S.; Firulli, A.B. The HAND1 frameshift A126FS mutation does not cause hypoplastic left heart syndrome in mice. Cardiovascular research 2017, 113, 1732–1742. [Google Scholar] [CrossRef]
- Pierpont, M.E.; Basson, C.T.; Benson, D.W., Jr.; Gelb, B.D.; Giglia, T.M.; Goldmuntz, E.; McGee, G.; Sable, C.A.; Srivastava, D.; Webb, C.L. , et al. Genetic basis for congenital heart defects: current knowledge: a scientific statement from the American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation 2007, 115, 3015–3038. [Google Scholar] [CrossRef]
- Birla, A.K.; Brimmer, S.; Short, W.D.; Olutoye, O.O., 2nd; Shar, J.A.; Lalwani, S.; Sucosky, P.; Parthiban, A.; Keswani, S.G.; Caldarone, C.A. , et al. Current state of the art in hypoplastic left heart syndrome. Front Cardiovasc Med 2022, 9, 878266. [Google Scholar] [CrossRef]
- Ishigami, S.; Ohtsuki, S.; Tarui, S.; Ousaka, D.; Eitoku, T.; Kondo, M.; Okuyama, M.; Kobayashi, J.; Baba, K.; Arai, S. , et al. Intracoronary autologous cardiac progenitor cell transfer in patients with hypoplastic left heart syndrome: the TICAP prospective phase 1 controlled trial. Circulation research 2015, 116, 653–664. [Google Scholar] [CrossRef]
- Ishigami, S.; Ohtsuki, S.; Eitoku, T.; Ousaka, D.; Kondo, M.; Kurita, Y.; Hirai, K.; Fukushima, Y.; Baba, K.; Goto, T. , et al. Intracoronary Cardiac Progenitor Cells in Single Ventricle Physiology: The PERSEUS (Cardiac Progenitor Cell Infusion to Treat Univentricular Heart Disease) Randomized Phase 2 Trial. Circulation research 2017, 120, 1162–1173. [Google Scholar] [CrossRef]
- Burkhart, H.M.; Qureshi, M.Y.; Rossano, J.W.; Cantero Peral, S.; O'Leary, P.W.; Hathcock, M.; Kremers, W.; Nelson, T.J.; Wanek, H.C.C.P. Autologous stem cell therapy for hypoplastic left heart syndrome: Safety and feasibility of intraoperative intramyocardial injections. J Thorac Cardiovasc Surg 2019, 158, 1614–1623. [Google Scholar] [CrossRef] [PubMed]
- Brizard, C.P.; Elwood, N.J.; Kowalski, R.; Horton, S.B.; Jones, B.O.; Hutchinson, D.; Zannino, D.; Sheridan, B.J.; Butt, W.; Cheung, M.M.H. , et al. Safety and feasibility of adjunct autologous cord blood stem cell therapy during the Norwood heart operation. J Thorac Cardiovasc Surg 2023, 166, 1746–1755. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Navarro, C.; Jaggers, J.; Burkhart, H.M.; Carlo, W.F.; Morales, D.L.; Qureshi, M.Y.; Rossano, J.W.; Hagen, C.E.; Seisler, D.K.; Peral, S.C. , et al. Autologous umbilical cord blood mononuclear cell therapy for hypoplastic left heart syndrome: a nonrandomized control trial of the efficacy and safety of intramyocardial injections. Stem Cell Res Ther 2025, 16, 215. [Google Scholar] [CrossRef]
- Ren, H.; Sang, Y.; Zhang, F.; Liu, Z.; Qi, N.; Chen, Y. Comparative Analysis of Human Mesenchymal Stem Cells from Umbilical Cord, Dental Pulp, and Menstrual Blood as Sources for Cell Therapy. Stem Cells Int 2016, 2016, 3516574. [Google Scholar] [CrossRef]
- Baksh, D.; Yao, R.; Tuan, R.S. Comparison of proliferative and multilineage differentiation potential of human mesenchymal stem cells derived from umbilical cord and bone marrow. Stem cells 2007, 25, 1384–1392. [Google Scholar] [CrossRef]
- Davies, B.; Elwood, N.J.; Li, S.; Cullinane, F.; Edwards, G.A.; Newgreen, D.F.; Brizard, C.P. Human cord blood stem cells enhance neonatal right ventricular function in an ovine model of right ventricular training. The Annals of thoracic surgery 2010, 89, 585-593, 593 e581-584. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.A.; Rajamarthandan, S.; Francis, B.; O'Leary-Kelly, M.K.; Sinha, P. Update on stem cell technologies in congenital heart disease. J Card Surg 2020, 35, 174–179. [Google Scholar] [CrossRef]
- Le Blanc, K.; Mougiakakos, D. Multipotent mesenchymal stromal cells and the innate immune system. Nature reviews. Immunology 2012, 12, 383–396. [Google Scholar] [CrossRef]
- Lalu, M.M.; McIntyre, L.; Pugliese, C.; Fergusson, D.; Winston, B.W.; Marshall, J.C.; Granton, J.; Stewart, D.J.; Canadian Critical Care Trials, G. Safety of cell therapy with mesenchymal stromal cells (SafeCell): a systematic review and meta-analysis of clinical trials. PloS one 2012, 7, e47559. [Google Scholar] [CrossRef]
- Ascheim, D.D.; Gelijns, A.C.; Goldstein, D.; Moye, L.A.; Smedira, N.; Lee, S.; Klodell, C.T.; Szady, A.; Parides, M.K.; Jeffries, N.O. , et al. Mesenchymal precursor cells as adjunctive therapy in recipients of contemporary left ventricular assist devices. Circulation 2014, 129, 2287–2296. [Google Scholar] [CrossRef]
- Oswald, J.; Boxberger, S.; Jorgensen, B.; Feldmann, S.; Ehninger, G.; Bornhauser, M.; Werner, C. Mesenchymal stem cells can be differentiated into endothelial cells in vitro. Stem cells 2004, 22, 377–384. [Google Scholar] [CrossRef]
- Toma, C.; Pittenger, M.F.; Cahill, K.S.; Byrne, B.J.; Kessler, P.D. Human mesenchymal stem cells differentiate to a cardiomyocyte phenotype in the adult murine heart. Circulation 2002, 105, 93–98. [Google Scholar] [CrossRef]
- Maacha, S.; Sidahmed, H.; Jacob, S.; Gentilcore, G.; Calzone, R.; Grivel, J.C.; Cugno, C. Paracrine Mechanisms of Mesenchymal Stromal Cells in Angiogenesis. Stem Cells Int 2020, 2020, 4356359. [Google Scholar] [CrossRef] [PubMed]
- Teng, X.; Chen, L.; Chen, W.; Yang, J.; Yang, Z.; Shen, Z. Mesenchymal Stem Cell-Derived Exosomes Improve the Microenvironment of Infarcted Myocardium Contributing to Angiogenesis and Anti-Inflammation. Cell Physiol Biochem 2015, 37, 2415–2424. [Google Scholar] [CrossRef] [PubMed]
- Pu, X.; Ma, S.; Gao, Y.; Xu, T.; Chang, P.; Dong, L. Mesenchymal Stem Cell-Derived Exosomes: Biological Function and Their Therapeutic Potential in Radiation Damage. Cells 2020, 10. [Google Scholar] [CrossRef]
- Wehman, B.; Sharma, S.; Pietris, N.; Mishra, R.; Siddiqui, O.T.; Bigham, G.; Li, T.; Aiello, E.; Murthi, S.; Pittenger, M. , et al. Mesenchymal stem cells preserve neonatal right ventricular function in a porcine model of pressure overload. American journal of physiology. Heart and circulatory physiology 2016, 310, H1816–1826. [Google Scholar] [CrossRef] [PubMed]
- Liufu, R.; Shi, G.; He, X.; Lv, J.; Liu, W.; Zhu, F.; Wen, C.; Zhu, Z.; Chen, H. The therapeutic impact of human neonatal BMSC in a right ventricular pressure overload model in mice. Stem Cell Res Ther 2020, 11, 96. [Google Scholar] [CrossRef]
- Wittenberg, R.E.; Gauvreau, K.; Leighton, J.; Moleon-Shea, M.; Borow, K.M.; Marx, G.R.; Emani, S.M. Prospective randomized controlled trial of the safety and feasibility of a novel mesenchymal precursor cell therapy in hypoplastic left heart syndrome. JTCVS Open 2023, 16, 656–672. [Google Scholar] [CrossRef]
- Thomas, E.D.; Lochte, H.L., Jr.; Lu, W.C.; Ferrebee, J.W. Intravenous infusion of bone marrow in patients receiving radiation and chemotherapy. The New England journal of medicine 1957, 257, 491–496. [Google Scholar] [CrossRef]
- Gratwohl, A.; Hermans, J.; Goldman, J.M.; Arcese, W.; Carreras, E.; Devergie, A.; Frassoni, F.; Gahrton, G.; Kolb, H.J.; Niederwieser, D. , et al. Risk assessment for patients with chronic myeloid leukaemia before allogeneic blood or marrow transplantation. Chronic Leukemia Working Party of the European Group for Blood and Marrow Transplantation. Lancet 1998, 352, 1087–1092. [Google Scholar] [CrossRef]
- Burnett, A.K.; Goldstone, A.H.; Stevens, R.M.; Hann, I.M.; Rees, J.K.; Gray, R.G.; Wheatley, K. Randomised comparison of addition of autologous bone-marrow transplantation to intensive chemotherapy for acute myeloid leukaemia in first remission: results of MRC AML 10 trial. UK Medical Research Council Adult and Children's Leukaemia Working Parties. Lancet 1998, 351, 700–708. [Google Scholar] [CrossRef]
- Qureshi, M.Y.; Cabalka, A.K.; Khan, S.P.; Hagler, D.J.; Haile, D.T.; Cannon, B.C.; Olson, T.M.; Cantero-Peral, S.; Dietz, A.B.; Radel, D.J. , et al. Cell-Based Therapy for Myocardial Dysfunction After Fontan Operation in Hypoplastic Left Heart Syndrome. Mayo Clin Proc Innov Qual Outcomes 2017, 1, 185–191. [Google Scholar] [CrossRef] [PubMed]
- van Berlo, J.H.; Kanisicak, O.; Maillet, M.; Vagnozzi, R.J.; Karch, J.; Lin, S.C.; Middleton, R.C.; Marban, E.; Molkentin, J.D. c-kit+ cells minimally contribute cardiomyocytes to the heart. Nature 2014, 509, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Sultana, N.; Zhang, L.; Yan, J.; Chen, J.; Cai, W.; Razzaque, S.; Jeong, D.; Sheng, W.; Bu, L.; Xu, M. , et al. Resident c-kit(+) cells in the heart are not cardiac stem cells. Nature communications 2015, 6, 8701. [Google Scholar] [CrossRef]
- Messina, E.; De Angelis, L.; Frati, G.; Morrone, S.; Chimenti, S.; Fiordaliso, F.; Salio, M.; Battaglia, M.; Latronico, M.V.; Coletta, M. , et al. Isolation and expansion of adult cardiac stem cells from human and murine heart. Circulation research 2004, 95, 911–921. [Google Scholar] [CrossRef] [PubMed]
- Hasan, A.S.; Luo, L.; Yan, C.; Zhang, T.X.; Urata, Y.; Goto, S.; Mangoura, S.A.; Abdel-Raheem, M.H.; Zhang, S.; Li, T.S. Cardiosphere-Derived Cells Facilitate Heart Repair by Modulating M1/M2 Macrophage Polarization and Neutrophil Recruitment. PloS one 2016, 11, e0165255. [Google Scholar] [CrossRef]
- Carr, C.A.; Stuckey, D.J.; Tan, J.J.; Tan, S.C.; Gomes, R.S.; Camelliti, P.; Messina, E.; Giacomello, A.; Ellison, G.M.; Clarke, K. Cardiosphere-derived cells improve function in the infarcted rat heart for at least 16 weeks--an MRI study. PloS one 2011, 6, e25669. [Google Scholar] [CrossRef]
- Lee, S.T.; White, A.J.; Matsushita, S.; Malliaras, K.; Steenbergen, C.; Zhang, Y.; Li, T.S.; Terrovitis, J.; Yee, K.; Simsir, S. , et al. Intramyocardial injection of autologous cardiospheres or cardiosphere-derived cells preserves function and minimizes adverse ventricular remodeling in pigs with heart failure post-myocardial infarction. Journal of the American College of Cardiology 2011, 57, 455–465. [Google Scholar] [CrossRef]
- Gallet, R.; Dawkins, J.; Valle, J.; Simsolo, E.; de Couto, G.; Middleton, R.; Tseliou, E.; Luthringer, D.; Kreke, M.; Smith, R.R. , et al. Exosomes secreted by cardiosphere-derived cells reduce scarring, attenuate adverse remodelling, and improve function in acute and chronic porcine myocardial infarction. European heart journal 2017, 38, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Bittle, G.J.; Morales, D.; Pietris, N.; Parchment, N.; Parsell, D.; Peck, K.; Deatrick, K.B.; Rodriguez-Borlado, L.; Smith, R.R.; Marban, L. , et al. Exosomes isolated from human cardiosphere-derived cells attenuate pressure overload-induced right ventricular dysfunction. J Thorac Cardiovasc Surg 2021, 162, 975–986 e976. [Google Scholar] [CrossRef]
- Sano, T.; Ousaka, D.; Goto, T.; Ishigami, S.; Hirai, K.; Kasahara, S.; Ohtsuki, S.; Sano, S.; Oh, H. Impact of Cardiac Progenitor Cells on Heart Failure and Survival in Single Ventricle Congenital Heart Disease. Circulation research 2018, 122, 994–1005. [Google Scholar] [CrossRef]
- Tarui, S.; Ishigami, S.; Ousaka, D.; Kasahara, S.; Ohtsuki, S.; Sano, S.; Oh, H. Transcoronary infusion of cardiac progenitor cells in hypoplastic left heart syndrome: Three-year follow-up of the Transcoronary Infusion of Cardiac Progenitor Cells in Patients With Single-Ventricle Physiology (TICAP) trial. J Thorac Cardiovasc Surg 2015, 150, 1198–1207. [Google Scholar] [CrossRef]
- Hirai, K.; Sawada, R.; Hayashi, T.; Araki, T.; Nakagawa, N.; Kondo, M.; Yasuda, K.; Hirata, T.; Sato, T.; Nakatsuka, Y. , et al. Eight-Year Outcomes of Cardiosphere-Derived Cells in Single Ventricle Congenital Heart Disease. Journal of the American Heart Association 2024, 13, e038137. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Ye, L.; Yu, Y.; Zhao, Z.A.; Zhao, D.; Ni, X.; Wang, Y.; Fang, X.; Yu, M.; Wang, Y.; Tang, J.M. , et al. Patient-specific iPSC-derived cardiomyocytes reveal abnormal regulation of FGF16 in a familial atrial septal defect. Cardiovascular research 2022, 118, 859–871. [Google Scholar] [CrossRef]
- Thareja, S.K.; Anfinson, M.; Cavanaugh, M.; Kim, M.S.; Lamberton, P.; Radandt, J.; Brown, R.; Liang, H.L.; Stamm, K.; Afzal, M.Z. , et al. Altered contractility, Ca(2+) transients, and cell morphology seen in a patient-specific iPSC-CM model of Ebstein's anomaly with left ventricular noncompaction. American journal of physiology. Heart and circulatory physiology 2023, 325, H149–H162. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Buganim, Y.; Faddah, D.A.; Cheng, A.W.; Itskovich, E.; Markoulaki, S.; Ganz, K.; Klemm, S.L.; van Oudenaarden, A.; Jaenisch, R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell 2012, 150, 1209–1222. [Google Scholar] [CrossRef] [PubMed]
- Devalla, H.D.; Schwach, V.; Ford, J.W.; Milnes, J.T.; El-Haou, S.; Jackson, C.; Gkatzis, K.; Elliott, D.A.; Chuva de Sousa Lopes, S.M.; Mummery, C.L. , et al. Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO Mol Med 2015, 7, 394–410. [Google Scholar] [CrossRef]
- Protze, S.I.; Liu, J.; Nussinovitch, U.; Ohana, L.; Backx, P.H.; Gepstein, L.; Keller, G.M. Sinoatrial node cardiomyocytes derived from human pluripotent cells function as a biological pacemaker. Nature biotechnology 2017, 35, 56–68. [Google Scholar] [CrossRef]
- Burridge, P.W.; Matsa, E.; Shukla, P.; Lin, Z.C.; Churko, J.M.; Ebert, A.D.; Lan, F.; Diecke, S.; Huber, B.; Mordwinkin, N.M. , et al. Chemically defined generation of human cardiomyocytes. Nat Methods 2014, 11, 855–860. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Pabon, L.; Murry, C.E. Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circulation research 2014, 114, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Tohyama, S.; Hattori, F.; Sano, M.; Hishiki, T.; Nagahata, Y.; Matsuura, T.; Hashimoto, H.; Suzuki, T.; Yamashita, H.; Satoh, Y. , et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell stem cell 2013, 12, 127–137. [Google Scholar] [CrossRef]
- Hofbauer, P.; Jahnel, S.M.; Papai, N.; Giesshammer, M.; Deyett, A.; Schmidt, C.; Penc, M.; Tavernini, K.; Grdseloff, N.; Meledeth, C. , et al. Cardioids reveal self-organizing principles of human cardiogenesis. Cell 2021, 184, 3299–3317. [Google Scholar] [CrossRef]
- Jiang, Y.; Habibollah, S.; Tilgner, K.; Collin, J.; Barta, T.; Al-Aama, J.Y.; Tesarov, L.; Hussain, R.; Trafford, A.W.; Kirkwood, G. , et al. An induced pluripotent stem cell model of hypoplastic left heart syndrome (HLHS) reveals multiple expression and functional differences in HLHS-derived cardiac myocytes. Stem Cells Transl Med 2014, 3, 416–423. [Google Scholar] [CrossRef]
- Ye, S.; Wang, C.; Xu, Z.; Lin, H.; Wan, X.; Yu, Y.; Adhicary, S.; Zhang, J.Z.; Zhou, Y.; Liu, C. , et al. Impaired Human Cardiac Cell Development due to NOTCH1 Deficiency. Circulation research 2023, 132, 187–204. [Google Scholar] [CrossRef] [PubMed]
- Lewis J, L.T. , Schlegel B, Woods D, Rao K, Sentis A, Tan J, Jagannathan R, Javed Z, Boudreau AN, Nelson T, Moulik M, Hempel N, Chen BB, Shiva S, Rajasundaram D, Finkel T, Saraf A. Hypomorphic NOTCH1 Expression Alters Cardiomyocyte Cellular Architecture in Hypoplastic Left Heart Syndrome. bioRxiv, 2024; 10.1101/2024.09.30.615809. [Google Scholar] [CrossRef]
- Krane, M.; Dressen, M.; Santamaria, G.; My, I.; Schneider, C.M.; Dorn, T.; Laue, S.; Mastantuono, E.; Berutti, R.; Rawat, H. , et al. Sequential Defects in Cardiac Lineage Commitment and Maturation Cause Hypoplastic Left Heart Syndrome. Circulation 2021, 144, 1409–1428. [Google Scholar] [CrossRef]
- Hrstka, S.C.; Li, X.; Nelson, T.J.; Wanek Program Genetics Pipeline, G. NOTCH1-Dependent Nitric Oxide Signaling Deficiency in Hypoplastic Left Heart Syndrome Revealed Through Patient-Specific Phenotypes Detected in Bioengineered Cardiogenesis. Stem cells 2017, 35, 1106–1119. [Google Scholar] [CrossRef]
- Theis, J.L.; Hrstka, S.C.; Evans, J.M.; O'Byrne, M.M.; de Andrade, M.; O'Leary, P.W.; Nelson, T.J.; Olson, T.M. Compound heterozygous NOTCH1 mutations underlie impaired cardiogenesis in a patient with hypoplastic left heart syndrome. Hum Genet 2015, 134, 1003–1011. [Google Scholar] [CrossRef]
- Paige, S.L.; Galdos, F.X.; Lee, S.; Chin, E.T.; Ranjbarvaziri, S.; Feyen, D.A.M.; Darsha, A.K.; Xu, S.; Ryan, J.A.; Beck, A.L. , et al. Patient-Specific Induced Pluripotent Stem Cells Implicate Intrinsic Impaired Contractility in Hypoplastic Left Heart Syndrome. Circulation 2020, 142, 1605–1608. [Google Scholar] [CrossRef]
- Xu, X.; Jin, K.; Bais, A.S.; Zhu, W.; Yagi, H.; Feinstein, T.N.; Nguyen, P.K.; Criscione, J.D.; Liu, X.; Beutner, G. , et al. Uncompensated mitochondrial oxidative stress underlies heart failure in an iPSC-derived model of congenital heart disease. Cell stem cell 2022, 29, 840–855. [Google Scholar] [CrossRef] [PubMed]
- Alonzo, M.; Contreras, J.; Ye, S.; Lin, H.; Hernandez-Rosario, L.; McBride, K.L.; Texter, K.; Garg, V.; Zhao, M.T. Characterization of an iPSC line NCHi006-A from a patient with hypoplastic left heart syndrome (HLHS). Stem Cell Res 2022, 64, 102892. [Google Scholar] [CrossRef] [PubMed]
- Adhicary, S.; Ye, S.; Lin, H.; Texter, K.; Garg, V.; Zhao, M.T. Establishment of NCHi009-A, an iPSC line from a patient with hypoplastic left heart syndrome (HLHS) carrying a heterozygous NOTCH1 mutation. Stem Cell Res 2023, 66, 103013. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, C.; Ye, S.; Qin, H.; Alonzo, M.; Onorato, A.; Argall, A.; Texter, K.; Ma, Q.; Garg, V. , et al. Common and divergent cellular aetiologies underlying hypoplastic left heart syndrome and hypoplastic right heart syndrome. European heart journal 2025, 46, 1946–1949. [Google Scholar] [CrossRef]
- Kussauer, S.; David, R.; Lemcke, H. hiPSCs Derived Cardiac Cells for Drug and Toxicity Screening and Disease Modeling: What Micro- Electrode-Array Analyses Can Tell Us. Cells 2019, 8. [Google Scholar] [CrossRef]
- Sinnecker, D.; Laugwitz, K.L.; Moretti, A. Induced pluripotent stem cell-derived cardiomyocytes for drug development and toxicity testing. Pharmacology & therapeutics 2014, 143, 246–252. [Google Scholar] [CrossRef]
- Liu, S.; Fang, C.; Zhong, C.; Li, J.; Xiao, Q. Recent advances in pluripotent stem cell-derived cardiac organoids and heart-on-chip applications for studying anti-cancer drug-induced cardiotoxicity. Cell Biol Toxicol 2023, 39, 2527–2549. [Google Scholar] [CrossRef]
- Colatsky, T.; Fermini, B.; Gintant, G.; Pierson, J.B.; Sager, P.; Sekino, Y.; Strauss, D.G.; Stockbridge, N. The Comprehensive in Vitro Proarrhythmia Assay (CiPA) initiative - Update on progress. J Pharmacol Toxicol Methods 2016, 81, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Li, B.; Wang, S. Establishment and validation of a torsade de pointes prediction model based on human iPSC-derived cardiomyocytes. Exp Ther Med 2023, 25, 61. [Google Scholar] [CrossRef]
- Blinova, K.; Dang, Q.; Millard, D.; Smith, G.; Pierson, J.; Guo, L.; Brock, M.; Lu, H.R.; Kraushaar, U.; Zeng, H. , et al. International Multisite Study of Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes for Drug Proarrhythmic Potential Assessment. Cell reports 2018, 24, 3582–3592. [Google Scholar] [CrossRef] [PubMed]
- Bedut, S.; Seminatore-Nole, C.; Lamamy, V.; Caignard, S.; Boutin, J.A.; Nosjean, O.; Stephan, J.P.; Coge, F. High-throughput drug profiling with voltage- and calcium-sensitive fluorescent probes in human iPSC-derived cardiomyocytes. American journal of physiology. Heart and circulatory physiology 2016, 311, H44–53. [Google Scholar] [CrossRef]
- Huang, C.Y.; Nicholson, M.W.; Wang, J.Y.; Ting, C.Y.; Tsai, M.H.; Cheng, Y.C.; Liu, C.L.; Chan, D.Z.H.; Lee, Y.C.; Hsu, C.C. , et al. Population-based high-throughput toxicity screen of human iPSC-derived cardiomyocytes and neurons. Cell reports 2022, 39, 110643. [Google Scholar] [CrossRef]
- Goversen, B.; van der Heyden, M.A.G.; van Veen, T.A.B.; de Boer, T.P. The immature electrophysiological phenotype of iPSC-CMs still hampers in vitro drug screening: Special focus on I(K1). Pharmacology & therapeutics 2018, 183, 127–136. [Google Scholar] [CrossRef]
- Koivumaki, J.T.; Naumenko, N.; Tuomainen, T.; Takalo, J.; Oksanen, M.; Puttonen, K.A.; Lehtonen, S.; Kuusisto, J.; Laakso, M.; Koistinaho, J. , et al. Structural Immaturity of Human iPSC-Derived Cardiomyocytes: In Silico Investigation of Effects on Function and Disease Modeling. Front Physiol 2018, 9, 80. [Google Scholar] [CrossRef]
- Vuckovic, S.; Dinani, R.; Nollet, E.E.; Kuster, D.W.D.; Buikema, J.W.; Houtkooper, R.H.; Nabben, M.; van der Velden, J.; Goversen, B. Characterization of cardiac metabolism in iPSC-derived cardiomyocytes: lessons from maturation and disease modeling. Stem Cell Res Ther 2022, 13, 332. [Google Scholar] [CrossRef] [PubMed]
- Tu, C.; Chao, B.S.; Wu, J.C. Strategies for Improving the Maturity of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Circulation research 2018, 123, 512–514. [Google Scholar] [CrossRef]
- Ronaldson-Bouchard, K.; Ma, S.P.; Yeager, K.; Chen, T.; Song, L.; Sirabella, D.; Morikawa, K.; Teles, D.; Yazawa, M.; Vunjak-Novakovic, G. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 2018, 556, 239–243. [Google Scholar] [CrossRef]
- Parikh, S.S.; Blackwell, D.J.; Gomez-Hurtado, N.; Frisk, M.; Wang, L.; Kim, K.; Dahl, C.P.; Fiane, A.; Tonnessen, T.; Kryshtal, D.O. , et al. Thyroid and Glucocorticoid Hormones Promote Functional T-Tubule Development in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Circulation research 2017, 121, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Rodriguez, M.L.; Leonard, A.; Sun, L.; Fischer, K.A.; Wang, Y.; Ritterhoff, J.; Zhao, L.; Kolwicz, S.C., Jr.; Pabon, L. , et al. Fatty Acids Enhance the Maturation of Cardiomyocytes Derived from Human Pluripotent Stem Cells. Stem cell reports 2019, 13, 657–668. [Google Scholar] [CrossRef]
- Li, W.; Luo, X.; Strano, A.; Arun, S.; Gamm, O.; Poetsch, M.S.; Hasse, M.; Steiner, R.P.; Fischer, K.; Poche, J. , et al. Comprehensive promotion of iPSC-CM maturation by integrating metabolic medium with nanopatterning and electrostimulation. Nature communications 2025, 16, 2785. [Google Scholar] [CrossRef] [PubMed]
- Carey, A.S.; Liang, L.; Edwards, J.; Brandt, T.; Mei, H.; Sharp, A.J.; Hsu, D.T.; Newburger, J.W.; Ohye, R.G.; Chung, W.K. , et al. Effect of copy number variants on outcomes for infants with single ventricle heart defects. Circulation. Cardiovascular genetics 2013, 6, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Mills, J.L.; Troendle, J.; Conley, M.R.; Carter, T.; Druschel, C.M. Maternal obesity and congenital heart defects: a population-based study. Am J Clin Nutr 2010, 91, 1543–1549. [Google Scholar] [CrossRef] [PubMed]
- Nicoll, R. Environmental Contaminants and Congenital Heart Defects: A Re-Evaluation of the Evidence. Int J Environ Res Public Health 2018, 15. [Google Scholar] [CrossRef]
- Roland, T.J.; Song, K. Advances in the Generation of Constructed Cardiac Tissue Derived from Induced Pluripotent Stem Cells for Disease Modeling and Therapeutic Discovery. Cells 2024, 13. [Google Scholar] [CrossRef]
- Liu, C.; Feng, X.; Li, G.; Gokulnath, P.; Xiao, J. Generating 3D human cardiac constructs from pluripotent stem cells. EBioMedicine 2022, 76, 103813. [Google Scholar] [CrossRef]
- Min, S.; Cho, S.W. Engineered human cardiac tissues for modeling heart diseases. BMB Rep 2023, 56, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Minale, C.; Nikol, S.; Hollweg, G.; Mittermayer, C.; Messmer, B.J. Clinical experience with expanded polytetrafluoroethylene Gore-Tex surgical membrane for pericardial closure: a study of 110 cases. J Card Surg 1988, 3, 193–201. [Google Scholar] [CrossRef]
- Crawford, F.A., Jr.; Sade, R.M.; Spinale, F. Bovine pericardium for correction of congenital heart defects. The Annals of thoracic surgery 1986, 41, 602–605. [Google Scholar] [CrossRef]
- Gao, L.; Gregorich, Z.R.; Zhu, W.; Mattapally, S.; Oduk, Y.; Lou, X.; Kannappan, R.; Borovjagin, A.V.; Walcott, G.P.; Pollard, A.E. , et al. Large Cardiac Muscle Patches Engineered From Human Induced-Pluripotent Stem Cell-Derived Cardiac Cells Improve Recovery From Myocardial Infarction in Swine. Circulation 2018, 137, 1712–1730. [Google Scholar] [CrossRef] [PubMed]
- Jebran, A.F.; Seidler, T.; Tiburcy, M.; Daskalaki, M.; Kutschka, I.; Fujita, B.; Ensminger, S.; Bremmer, F.; Moussavi, A.; Yang, H. , et al. Engineered heart muscle allografts for heart repair in primates and humans. Nature 2025, 639, 503–511. [Google Scholar] [CrossRef]
- Yeung, E.; Fukunishi, T.; Bai, Y.; Bedja, D.; Pitaktong, I.; Mattson, G.; Jeyaram, A.; Lui, C.; Ong, C.S.; Inoue, T. , et al. Cardiac regeneration using human-induced pluripotent stem cell-derived biomaterial-free 3D-bioprinted cardiac patch in vivo. J Tissue Eng Regen Med 2019, 13, 2031–2039. [Google Scholar] [CrossRef] [PubMed]
- Miyagawa, S.; Kawamura, T.; Ito, E.; Takeda, M.; Iseoka, H.; Yokoyama, J.; Harada, A.; Mochizuki-Oda, N.; Imanishi-Ochi, Y.; Li, J. , et al. Pre-clinical evaluation of the efficacy and safety of human induced pluripotent stem cell-derived cardiomyocyte patch. Stem Cell Res Ther 2024, 15, 73. [Google Scholar] [CrossRef] [PubMed]
- Mahle, W.T.; Hu, C.; Trachtenberg, F.; Menteer, J.; Kindel, S.J.; Dipchand, A.I.; Richmond, M.E.; Daly, K.P.; Henderson, H.T.; Lin, K.Y. , et al. Heart failure after the Norwood procedure: An analysis of the Single Ventricle Reconstruction Trial. The Journal of heart and lung transplantation : the official publication of the International Society for Heart Transplantation 2018, 37, 879–885. [Google Scholar] [CrossRef]
- Querdel, E.; Reinsch, M.; Castro, L.; Kose, D.; Bahr, A.; Reich, S.; Geertz, B.; Ulmer, B.; Schulze, M.; Lemoine, M.D. , et al. Human Engineered Heart Tissue Patches Remuscularize the Injured Heart in a Dose-Dependent Manner. Circulation 2021, 143, 1991–2006. [Google Scholar] [CrossRef] [PubMed]
- Miyagawa, S.; Kainuma, S.; Kawamura, T.; Suzuki, K.; Ito, Y.; Iseoka, H.; Ito, E.; Takeda, M.; Sasai, M.; Mochizuki-Oda, N. , et al. Case report: Transplantation of human induced pluripotent stem cell-derived cardiomyocyte patches for ischemic cardiomyopathy. Front Cardiovasc Med 2022, 9, 950829. [Google Scholar] [CrossRef]
- Liu, C.; Niu, K.; Xiao, Q. Updated perspectives on vascular cell specification and pluripotent stem cell-derived vascular organoids for studying vasculopathies. Cardiovascular research 2022, 118, 97–114. [Google Scholar] [CrossRef]
- Cheruku, G.R.W., C. V. ; Raviendran, S.; Xiao, Q. Recent Advances and Future Perspectives in Vascular Organoids and Vessel-on-Chip Organoids 2024, 3, 203–246. [Google Scholar] [CrossRef]
- Lewis-Israeli, Y.R.; Wasserman, A.H.; Gabalski, M.A.; Volmert, B.D.; Ming, Y.; Ball, K.A.; Yang, W.; Zou, J.; Ni, G.; Pajares, N. , et al. Self-assembling human heart organoids for the modeling of cardiac development and congenital heart disease. Nature communications 2021, 12, 5142. [Google Scholar] [CrossRef]
- Drakhlis, L.; Biswanath, S.; Farr, C.M.; Lupanow, V.; Teske, J.; Ritzenhoff, K.; Franke, A.; Manstein, F.; Bolesani, E.; Kempf, H. , et al. Human heart-forming organoids recapitulate early heart and foregut development. Nature biotechnology 2021, 39, 737–746. [Google Scholar] [CrossRef]
- Yang, J.; Lei, W.; Xiao, Y.; Tan, S.; Yang, J.; Lin, Y.; Yang, Z.; Zhao, D.; Zhang, C.; Shen, Z. , et al. Generation of human vascularized and chambered cardiac organoids for cardiac disease modelling and drug evaluation. Cell Prolif 2024, 57, e13631. [Google Scholar] [CrossRef]
- Song, M.; Choi, D.B.; Im, J.S.; Song, Y.N.; Kim, J.H.; Lee, H.; An, J.; Kim, A.; Choi, H.; Kim, J.C. , et al. Modeling acute myocardial infarction and cardiac fibrosis using human induced pluripotent stem cell-derived multi-cellular heart organoids. Cell Death Dis 2024, 15, 308. [Google Scholar] [CrossRef] [PubMed]
- Richards, D.J.; Li, Y.; Kerr, C.M.; Yao, J.; Beeson, G.C.; Coyle, R.C.; Chen, X.; Jia, J.; Damon, B.; Wilson, R. , et al. Human cardiac organoids for the modelling of myocardial infarction and drug cardiotoxicity. Nat Biomed Eng 2020, 4, 446–462. [Google Scholar] [CrossRef]
- Sharma, P.; Liu Chung Ming, C.; Gentile, C. In vitro modeling of myocardial ischemia/reperfusion injury with murine or human 3D cardiac spheroids. STAR Protoc 2022, 3, 101751. [Google Scholar] [CrossRef] [PubMed]
- Pocock, M.W.; Reid, J.D.; Robinson, H.R.; Charitakis, N.; Krycer, J.R.; Foster, S.R.; Fitzsimmons, R.L.; Lor, M.; Devilee, L.A.C.; Batho, C.A.P. , et al. Maturation of human cardiac organoids enables complex disease modeling and drug discovery. Nat Cardiovasc Res 2025, 4, 821–840. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, G.; Ma, T.; Simmons, C.A.; Santerre, J.P. A critical review on advances and challenges of bioprinted cardiac patches. Acta Biomater 2024, 189, 1–24. [Google Scholar] [CrossRef]
- Lu, T.Y.; Xiang, Y.; Tang, M.; Chen, S. 3D Printing Approaches to Engineer Cardiac Tissue. Curr Cardiol Rep 2023, 25, 505–514. [Google Scholar] [CrossRef]
- Faber, L.; Yau, A.; Chen, Y. Translational biomaterials of four-dimensional bioprinting for tissue regeneration. Biofabrication 2023, 16. [Google Scholar] [CrossRef] [PubMed]
- Ashammakhi, N.; Ahadian, S.; Zengjie, F.; Suthiwanich, K.; Lorestani, F.; Orive, G.; Ostrovidov, S.; Khademhosseini, A. Advances and Future Perspectives in 4D Bioprinting. Biotechnol J 2018, 13, e1800148. [Google Scholar] [CrossRef] [PubMed]

| Signaling Pathway | Normal Function | Disease secondary to defect | Association with HLHS |
|---|---|---|---|
| NOTCH1 | Valve formation; ventricular septation; Left-right patterning; Regulation of progenitor differentiation [34]; |
Bicuspid aortic valve; Right ventricular hypoplasia; VSD [35]; |
Strong–rare genetic variants (G661S,R1279H,A683T) and de novo mutations associated with HLHS found in patients and relatives [36]; Animal models with NOTCH 1 defects display HLHS associated ventricular hypoplasia and valve abnormalities [37,38] |
| Bone Morphogenetic Protein (BMP) | Mesoderm induction and regulation [39]; FHF formation; Proliferation of CMs; Development of cardiac cushion [40]; |
Pulmonary arterial Hypertension; AV canal defects [41]; |
Moderate–BMP2/4 knockouts in mouse models lead to ventricular hypoplasia [42]; Significant interactions with other pathways including Notch, HAND, Wnt, RA and SHH; BMP/TGF-beta dysregulation in human neonatal HLHS myocardial tissue [43]; |
| Retinoic Acid (RA) | Anterior-posterior patterning; FHF and SHF development; Mesoderm formation and induction [44]; |
DiGeorge Syndrome [45]; AV cushion defects; Truncus arteriosus [46] |
Emerging–biological plausibility due to critical role in anterior-posterior axis patterning, FHF and SHF development and chamber specification [44] |
| Wnt/Beta-catenin | Mesoderm induction; CM differentiation; SHF expansion and patterning [47] |
VSD; Truncus Arteriosus; ASD; Familial Exudative Vitreoretinopathy; Arrhythmogenic Cardiomyopathy [48,49] |
Moderate–dysregulated signaling of pathways in HLHS iPSC-CMs. |
| Sonic Hedgehog Pathway (SHH) | Heart tube development; SHF proliferation [50] |
OFT defects; Holoprosencephaly [51] |
Emerging–Essential for SHF and outflow tract development which contribute to normal development of LV [50]. |
| Hippo Pathway | Organ size; Regulates myocardial thickness [52] |
Abnormal heart size [53] | Emerging–Hippo-Yap signaling associated with HLHS iPSC-CMs [54] |
| Fibroblast growth factor (FGF) family | Mesoderm induction; Outflow tracts development; SHF proliferation [55] |
OFT defects; Overriding aorta [56] |
Moderate – pathway dysregulation observed in HLHS fetal lamb model [57] |
| Transcription Factors | Normal Function | Disease secondary to defect | Association with HLHS |
| HAND1 | Ventricle development; LV specification; Ventricular trabeculation [58,59]; |
LV defects; Endocardial cushion defects [60] |
Strong–mouse models with HAND1 deletion resulting in HLHS phenotype; iPSC models demonstrate reduced expression of HAND1 and downstream targets [61]; A126fs frameshift mutation seen in patient cardiac tissue [60] |
| NKX2-5 | Differentiation of CMs and ventricle formation; Purkinje fibre network, AV node and bundle branch development [62]; |
ASD, AV block, AVSD [62] | Moderate–cohort studies show genetic variant (T178M) in subset of HLHS [63]; iPSC studies show reduced expression of NKX2.5 [61] |
| MYRF | Ventricular formation, Transcriptional regulation [64] |
Congenital Diaphragmatic Hernia with CHD and Genitourinary anomalies; Hypomyelination disorders [64] |
Moderate–Associated with syndromic presentations of HLHS; Mutated MYRF in vertebrae models demonstrate HLHS phenotype [65] |
| GATA4 | Septation of chambers; CM differentiation; Valve formation [66] |
ASD, VSD, AVSD [67,68] | Moderate–GATA4 mutation leads to thin ventricular myocardium and CM proliferation [67]; significant synergism with TBX5 required for early cardiogenesis [68] |
| TBX1 [69] | Pharyngeal Arch development; OFT formation; SHF regulation of differentiation and proliferation |
DiGeorge syndrome/velocardiofacial syndrome, Conotruncal heart defects |
Emerging–biologically plausible given role in upstream SHF signaling and regulation |
| TBX5 | Cardiac septation; Conduction system; Chamber specification; |
Holt-Oram syndrome [70,71] |
Moderate – altered function and expression seen in HLHS iPSC studies [72] |
|
WT1 |
Epicardial-to-mesenchymal transition [73] | Wilms Tumor; Denys-Drash syndrome |
Emerging – biological plausibility |
| MEF2C | Heart tube looping; Ventricle formation [74] |
VSD, OFT defects [74,75] | Emerging– knockout animal models display RV and outflow tract defects [76] |
| ISL1 | SHF formation and regulation; Coronary development; |
RV defects, OFT defects, VSD [77] | Moderate– variations can increase susceptibility to CHD including HLHS [78] |
| Structural/Regulatory Proteins | Normal Function | Disease secondary to defect | Evidence for Association with HLHS |
| MYH6 | Encodes Alpha-myosin heavy chain; predominantly atrial contractile function | ASD, Late onset Hypertrophic cardiomyopathy | Moderate– multiple rare genetic variants (R443) found in HLHS cohorts [79]; HiPSC-CM models display reduced contractility [80] |
| LRP2 | Encodes endocytic receptors responsible for developmental signalling (SHH pathway) [81] | Neural tube defects, Donnai-Barrow Syndrome [82] |
Moderate–multiple rare genetic variants found in HLHS genomic and transcriptomic studies [83] |
| GJA1 (Connexin43) | Encodes gap junctions; facilitates electrical and metabolic communications between CMs | Arrhythmogenic Cardiomyopathy, Cardiac conduction disorders, Oculodentodigital Dysplasia [84] |
Emerging – altered expression of GJA1 in HLHS heart tissue [85] |
| Study name (NCT number) | Stem cell type | Study timeline | Study stage | Enrolment: total (control/treatment) | Route and timing of administration | Key findings | Limitations | Status/reference(s) |
|---|---|---|---|---|---|---|---|---|
| Transcoronary Infusion of Cardiac Progenitor Cells in Patients With Single Ventricle Physiology (TICAP) (NCT01273857) | Autologous CDC | 2011-2013 | Phase I | 14 (7/7) | IC at stage II/III surgical palliation | ↑RV function over 18 month period; ↑HF status Safety and feasibility of CDCs; Long-standing benefits on follow-up analysis; |
Non-randomised; Open-Label; Small Sample Size; Variable timing of intervention; |
Completed; Ishigami et al [124] |
| Cardiac Progenitor Cell Infusion to Treat Univentricular Heart Disease (PERSEUS) (NCT01829750) |
Autologous CDC | 2013-2016 | Phase II | 34 (17/17) | IC at stage II/III surgical palliation | ↑RV function; ↑quality of life and somatic growth; Safety and feasibility of CDCs; Long-standing benefits on follow-up analysis |
Limited long term conclusion; Open label; Single Ventricle; Disease heterogeneity |
Completed; Ishigami et al [125] |
| Safety Study of Autologous Umbilical Cord Blood Cells for Treatment of Hypoplastic Left Heart Syndrome (NCT01883076) |
Autologous UCB-MNCs | 2013-2021 | Phase I |
Phase I: 10 (0/10) |
IM during Glenn Operation (Stage II) | Preserved RV; Safety and Feasibility; |
Single centre study; no control group; Short-term follow up; Heterogeneity of UCB-MNCs |
Completed; Burkhart et al [126]; |
| Safety of Autologous Cord Blood Cells in HLHS Patients During Norwood Heart Surgery (NCT03431480) | Autologous UCB-MNCs | 2018-2022 | Phase I | 10 (0/10) | IC during Norwood procedure | Preserved RV; Safety and Feasibility; |
Open label; no control group; Heterogeneity of UCB-MNCs |
Completed; Brizard et al [127] |
| Intramyocardial Injection of Autologous Umbilical Cord Blood Derived Mononuclear Cells During Surgical Repair of Hypoplastic Left Heart Syndrome (NCT03779711) | Autologous UCB-MNCs | 2019-2026 | Phase IIb | 95 (45/50) | IM at Stage II surgical repair | An unfavorable change in longitudinal cardiac strain and a greater incidence (20%) of at least one severe adverse event in treatment group; Failed to enhance cardiac functions |
Multicenter, open-label, nonrandomized study; Negative results. | Completed; Gallego-Navarro et al [128] |
| Cardiac Stem/Progenitor Cell Infusion in Univentricular Physiology (APOLLON) (NCT02781922) | Autologous cardiac stem cells | 2016-2023 | Phase III | 40 (NR/NR) | IC After stage II/III surgical palliation | NR | NR | NR |
| Lomecel-B Injection in Patients With Hypoplastic Left Heart Syndrome: A Phase I/II Study (ELPIS) (NCT03525418/ NCT04925024) |
Allogeneic MSCs (Lomecel-B) | 2018-2025 | Phase I Phase II |
Phase I: 10 (0/10) Phase II: 20 (10/10) |
IM during Glenn Operation | Phase I: Safety and feasibility; No alloimmune sensitisation Phase II: NR |
Phase I: Open Label; No control; Small sample size; Non-Randomised |
Phase I: Completed; Kaushal, et al [20]; Phase II: ongoing; |
| Autologous Cardiac Stem Cell Injection in Patients With Hypoplastic Left Heart Syndrome: An Open Label Pilot Study (CHILD Trial) (NCT03406884) | Autologous c-kit+ | 2019-2024 | Phase I/II | 10 (phase I); 22 (phase II) |
IM during Glenn Operation | NR | Open Label; |
Completed; Pending to report; |
| Mesoblast Stem Cell Therapy for Patients With Single Ventricle and Borderline Left Ventricle (NCT03079401) |
Allogeneic MPCs | 2017-2024 | Phase I Phase II |
19 (9/10) | IM during Glenn Operation | NR | NR | Completed; Pending to report; |
| Cell source for iPSC reprogramming | Reprogramming system | Genetic mutations | Methodology (Medium, small molecular) | Key findings | Applications | Reference |
|---|---|---|---|---|---|---|
| H7/H9 ESCs; WTC iPSCs; 176/1 iPSC 176/5 iPSC 176/8 iPSC |
WiCell Research Institute; WiCell Research Institute; IMBA Stem Cell Core Facility |
HAND1 knockout or NKX2.5 knockout PSCs | Cardiac differentiation medium (CDM) with 30 ng/ml FGF2, 5 µM LY294002, 50 ng/ml Activin A, 10 ng/ml BMP4, and CHIR99021 (, 1-4 µM, Wnt-activation) for 36-40 hours; CDM with BMP4 (10ng/ml), FGF2 (8 ng/ml), insulin (10 µg/ml), IWP2 (5µM, Wnt inhibition) and retinoic acid (0.5 µM) for 4 days; CDM with BMP4 (10ng/ml), FGF2 (8 ng/ml) and insulin (10 µg/ml) for 2 days; Then CDM with insulin (10 µg/ml) for CM maintenance |
Main purpose: to screen for factors that are sufficient to stimulate intrinsic 3D cardiac structure formation and confirm that addition of laminins 521/511 before mesoderm induction resulted in intrinsic self-assembly of cells and the striking formation of hollow, beating 3D structures expressing the CM marker TNNT2 after 7 days of differentiation. Findings also confirm stage-specific regulation of HAND1 and NKX2.5 during cardiac development. | Cardioids generation, Molecular insights into cardiac cavity morphogenesis, disease modelling such as cryoinjury (mimicking myocardial infarcti) and HLHS |
Hofbauer et al [169] |
| Cardiac progenitor cells | Retroviruses to deliver OKSM factors | NR (not reported) | Matrigel-based monolayer: RPMI/B27 medium supplemented with 100 ng/ml activin A for 24 h, followed by incubation with 10 ng/ml BMP 4 for an additional 4 days. | Lower cardiomyogenic differentiation potential; decreased NKX2.5, TBX2, NOTCH/HEY signaling, and HAND1/2; reduced H3K4 dimethylation and histone H3 acetylation but increased H3K27 trimethylation. |
Provide molecular insights into complex transcriptional and epigenetic mechanisms underlying HLHS | Kobayashi et al [61] |
| Dermal fibroblasts | Polycistronic lentiviral system for OCT4, KLF4, SOX2, and MYC (OKSM) factors | NR | EB-based protocol: 1 mM ascorbic acid (AA), 4 × 10−4 M monothioglycerol (MTG), 10 ng/ml BMP4, 12.5 ng/ml bFGF, 6 ng/ml Activin A, 150ng/ml DKK1, 5 ng/ml VEGF, and 5.4 μM SB-431542. From day 7, EBs were plated on 0.1% gelatin-coated 12 well culture plates (20 EBs per well) and cultured in in StemPro-34 SFM media supplemented with VEGF and bFGF. |
Decreased number of beating clusters; myofibrillar disorganization, persistence of a fetal gene expression pattern, and changes in commitment to ventricular versus atrial lineages; different calcium transient patterns and electrophysiological responses |
NR | Jiang et al [170] |
| Peripheral blood mononuclear cells (PBMCs) | CytoTune-iPSC 2.0 Sendai Reprogramming Kit | Notch1 knockout | Matrigel-based monolayer (over 90% confluency): RPMI/B27 medium supplemented with 6 μM CHIR99021 for 48 h, RPMI/B27 medium for 24 h, RPMI/B27 medium plus 5 μM IWR-1 for 48 h, then maintain in RPMI/B27 untill use from day 7. Cell were incubated with RPMI1640 no glucose medium plus B27 supplement for 4 days to remove non-CMs. | Disruption of NOTCH1 blocks human ventricular-like CM differentiation but promotes atrial-like CM generation, defective CM proliferation; impaired cell cycle progression and mitosis; biased differentiation toward epicardial and second heart field progenitors at the expense of first heart field progenitors | Possibly modeling HLHS, gaining new insights into the comprehension of the mechanisms underlying HLHS etiology | Ye et al [171] |
| Fibroblast (ATCC) | Episomal plasmid (ND2.0, NIH CRM control iPSC line) | Isogenic hiPSC hypomorphic NOTCH1 clones | Matrigel-based monolayer (over 90% confluency): CM differentiation media A and B | Displayed abnormalities in pathways associated with mitochondrial function, actin cytoskeleton, and cardiomyocyte development; skew differentiation away from CMs and towards fibroblasts and smooth muscle cells; impaired cardiac cytoskeletal and mitochondrial architecture; decreased CM contractility and ATP production | Possibly modeling HLHS; high-throughput drug screen to identify potential HLHS drug such as auranofin. | Lewis et al [172] |
| Neonatal fibroblasts | Polycistronic vector encoding KLF4, OCT4, SOX2, as well as vectors encoding hc-Myc and hKlf4 |
Deleterious genetic variants of Notch1-4 | Embryoid body (EB) based protocol: StemPro-34 SFM (basal media) day 0-3 (10 ng/ml BMP4, 6 ng/ml Activin A, 5 ng/ml bFGF, 10 µM Y276321); day 3-5 (150 ng/ml DKK1, 10 ng/ml VEGF, 5.4 µM SB431542, 0.25 µM Droso, 10 µM Y276321); day 5-7 (150 ng/ml DKK1, 10 ng/ml VEGF, 10 µM Y276321) From day 7, EBs were plated on Matrigel-coated 12 well culture plates (20 EBs per well) and cultured in basal media plus 10 ng/ml VEGF and 5 ng/ml bFGF. |
Reduced ability to give rise to mesodermal, cardiac progenitors and mature CMs and an enhanced ability to differentiate to smooth muscle cells; lower beating rate, disorganised sarcomeres and sarcoplasmic reticulum; blunted response to isoprenaline | Possibly modeling HLHS; provide novel signalling and genetic insights into in HLHS pathogenesis | Yang et al [72] |
| Skin fibroblasts or PBMCs | CytoTune-iPSTM-iPS 2.0 Sendai Reprogramming Kit | heterozygous de-novo mutations in multiple genes including MYRF, BAI2, FGFR1, AIM1L, SYBU, MACF1, etc | Matrigel-based monolayer (over 90% confluency): Day 1: CDM3 (RPMI1640 supplemented with 500 μM/mL Oryza sativa-derived recombinant human albumin and 213 μg/mL L-ascorbic acid 2-phosphate) supplemented with 4-6 μM CHIR99021; Day 2-3: CDM3 plus 2 μM Wnt-C59; Fron day 4: CDM3 |
Unique aberrations in autophagy terms were present when directing HLHS iPSCs toward early cardiac progenitors (CPs), whereas apoptosis-associated pathways appeared solely affected in later CPs (day 6) and cardiomyocytes (day 8); dysregulated lineage-specific CM differentiation; disrupted both early CM subtype lineage specification and CM differentiation and maturation in HLHS |
Possibly modeling HLHS; provide novel signalling and genetic insights into in HLHS pathogenesis; use for 3D cardiax patches generation | Krane et al [173] |
| Dermal fibroblasts | Lentiviral transduction of OKSM factors | heterozygous NOTCH1(P1256L/P1964L) | Monolayer (over 90% confluency): Day 1: RPMI/B27 plus 40-100 ng/ml Activin A; Day 2-5: RPMI/B27 plus 5-20 ng/ml bFGF and BMP4; Fron day 5: RPMI/B27 |
Deficiency in Notch signaling pathway and a diminished capacity to generate CMs; impaired NO signaling | HLHS modeling; identification of small therapeutic molecules to compensate dysregulated NO signaling | Hrstka et al [174] Theis et al [175] |
| Dermal fibroblasts | Sendai reprogramming kits | MYH6-R443P variant | Geltrex-based monlayer: Day 0: mTeSR1 medium plus 5 μM ROCK inhibitor; Day 1-2: insulin-free RPMI/B27 plus 10 μM CHIR99021 and 10 ng/ml Activin-A; Day 3-6: insulin-free RPMI/B27 with 5 μM IWP; From day 7: RPMI/B27 with insulin |
MYH6-R443P variant CMs express beta myosin heavy chain expression (MYH7), with impaired contractility, relaxation, and CM differentiation, as well as sarcomere disorganization | HLHS modeling; study how genetic variants contribute to HLHS. | Kim et al [80]; Tomita-Mitchell et al [79] |
| NR | NR | NR | Small-molecule modulation of the canonical Wnt/β-catenin signaling pathway | Impaired contractility; upregulation in sarcomere and cytoskeletal genes and downregulation in genes involved in mitochondrial function and metabolism; reduced mitochondrial content, mitochondrial respiration and oxidative metabolism | HLHS modeling and drug screening | Paige et al [176] |
| Fibroblasts or lymphoblastoid cells | Episomal plasmids encoding OSKM | Pathogenic variants associated with mitochondrial metabolism | Matrigel-based monolayer (over 90% confluency): Day 1-2: basal CDM3 media (RPMI 1640, BSA, B27, 213 μg/ml Ascorbic acid) and 6 μM CHIR99021; Day 3-14: basal CDM3 media and 10 μM XAV939; Fro day 15: basal CDM3 media minus Ascorbic acid |
Impaired cardiomyocyte differentiation and contractile dysfunction; cell-cycle disturbance with metaphase arrest; incresed CM apoptosis; myofibrillar disarray; mitochondrial dysfunction and perturbation of mitochondrial dynamics; defects in YAP-regulated antioxidant response | Modelling of HLHS and early heart failure; drug screening; identification of potential therapeutics such as sildenafil and TUDCA | Xu et al [177] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





