Submitted:
31 July 2025
Posted:
31 July 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Background
2.1. Adhesion Molecules in CNS
2.1.1. Classic Cadherins
2.1.2. Proto-Cadherins
2.1.3. Nectins
2.1.4. Nectin-Like Molecules (Necls)
2.1.5. NCAM (Neural Cell Adhesion Molecule)
2.1.6. Integrins
2.1.7. NgCAM (Neuron-Glia Cell Adhesion Molecule)
2.1.8. Contactins
2.1.9. TAG-1 (Transient Axonal Glycoprotein-1)
2.1.10. SYG-1 and SYG-2
2.1.11. Sidekicks
2.1.12. Neuroligins and Neurexins
2.2. The Na⁺,K⁺-ATPase in Neuron-Astrocyte Interactions
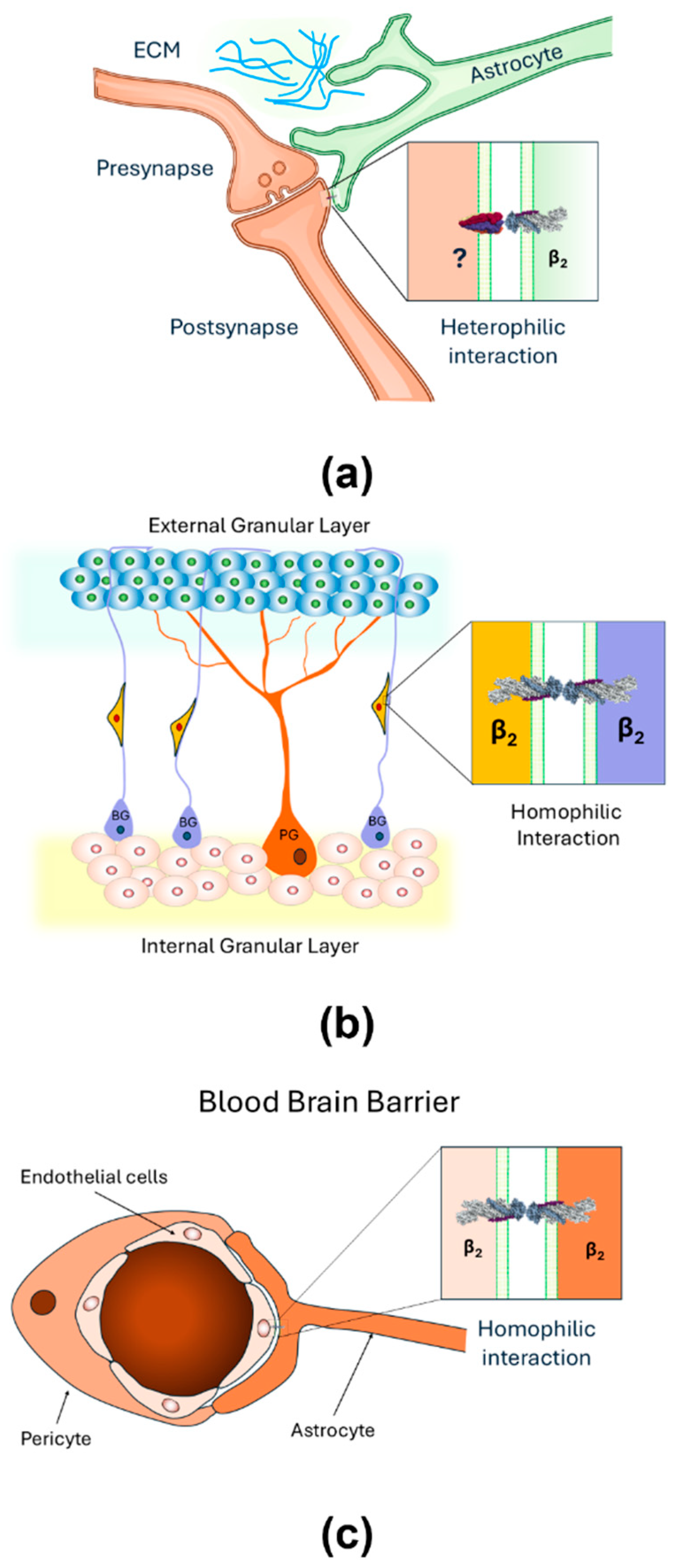
2.3. AMOG as a Heterophilic Adhesion Molecule
3. Current Understanding and Knowledge Gaps
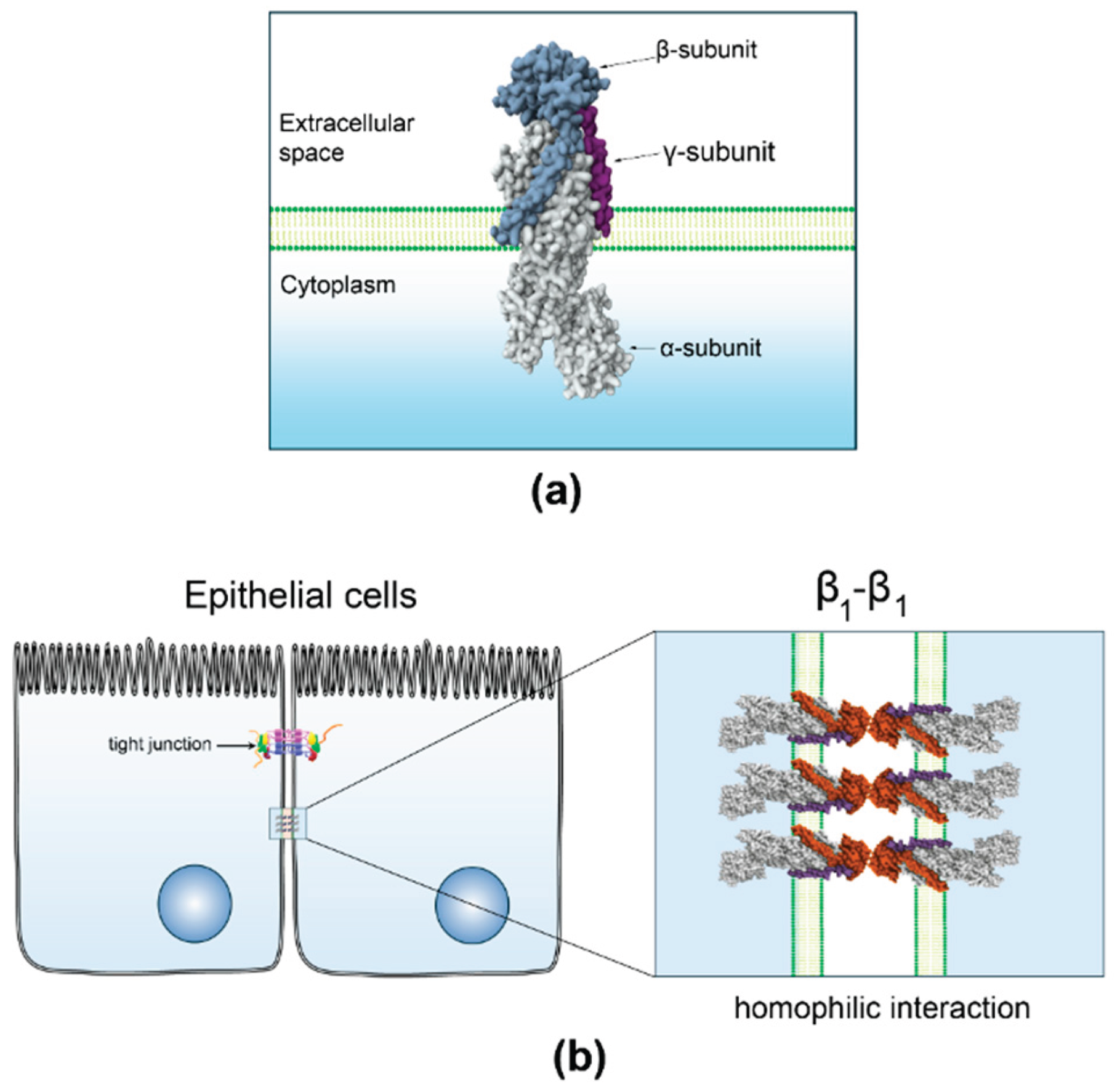
3.1. β1-Subunit as a Homophilic Adhesion Molecule in Epithelia
3.2. Gaps and Unresolved Questions
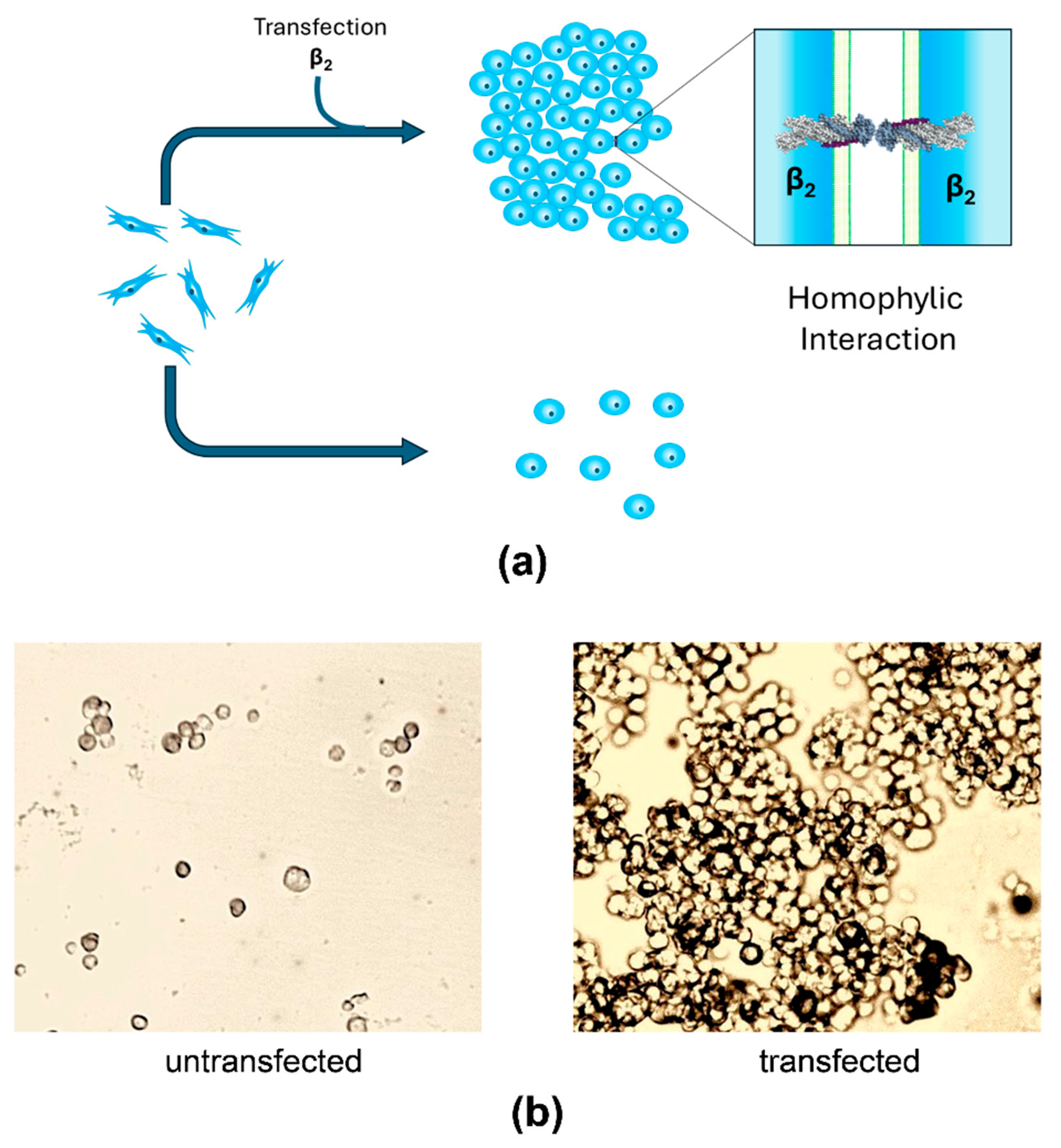
4. Recent Findings
5. Looking for the Partner
5.1. Experimental Strategies
5.2. Candidates for AMOG/β2 Receptor
- TSPAN31: As a member of the Tetraspanin family, TSPAN31 is notable for its role in organizing membrane microdomains and mediating lateral interactions between cell surface proteins. Tetraspanins act as molecular scaffolds, clustering adhesion molecules, integrins, and signaling receptors into functional complexes [99,100,101]. TSPAN31 has been implicated in cell adhesion, migration, and membrane signaling functions aligning closely with AMOG/β2 activities. TSPAN31 might associate in cis with a neuronal adhesion receptor, creating a complex that interacts in trans with AMOG/β2 on astrocytes. Alternatively, TSPAN31 might directly stabilize or present the neuronal partner required for AMOG/β2 recognition. Its potential involvement raises intriguing questions about how these microdomain organizations contribute to AMOG/β2 function.
- RTN4 (Nogo-A): Known for inhibiting neurite outgrowth, RTN4 has a complex topology and is present not only in the endoplasmic reticulum but also on the plasma membrane of axons and dendrites [102]. Its interactions with membrane proteins suggest it could serve as a scaffold or modulator for a receptor complex capable of interacting with AMOG/β2. If RTN4 is enriched in specific neuronal compartments, such as dendritic spines or axon terminals, its spatial distribution could explain the specificity and context-dependence of AMOG/β2-mediated adhesion during synaptogenesis or glial ensheathment. RTN4’s role in membrane dynamics and its interaction network make it a compelling candidate for further investigation.
6. Functional Implications in the CNS
7. Future Directions
7.1. Research Avenues and Implications
7.2. Therapeutic Targeting Potential
8. Summary
9. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AMOG/β2 | Adhesion molecule on Glia |
| CNS | Central nervous system |
| TSPAN31 | Tetraspanin 31 |
| RTN4 | Reticulon 4 |
| CAMs | Cell adhesion molecules |
| IgCAMs | Inmunoglobulin superfamily cell adhesion molecules |
| CTLDs | C-type lectin-like domain proteins |
| Pcdh | Protocadherin |
| PAJs | Puncta adherentia junctions |
| Necls | Nectin-like molecules |
| NCAM | Neural Cell Adhesion molecule |
| FGFR | Fibroblast growth factor receptor |
| ECM | Extracellular matrix |
| NgCAM | Neuron-glia cell adhesion molecule |
| CNTNs | Contactins |
| TAG-1 | Transient Axonal Glycoprotein-1 |
| GABA | Gamma-aminobutyric acid |
| ATP | Adenosine triphosphate |
| BBB | Blood-brain barrier |
| EMT | Epithelial-mesenchymal transition |
| U87-MG | Uppsala 87 malignant glioma |
| YFP | Yellow fluorescence protein |
| CHO | Chinese hamster ovary cells |
| MDCK | Madin-Darby Canine Kidney |
| MD | Molecular dynamic |
| HEK | Human embryonic kidney |
| CRISPR | Clustered Regularly Interspaced short palindromic repeats |
| FRET | Förster resonance energy transfer |
| BiFC | Biomolecular Fluorescence Complementation |
References
- Kennedy, M.B. Synaptic Signaling in Learning and Memory. Cold Spring Harb. Perspect. Biol. 2016, 8, a016824. [CrossRef]
- Noriega-Prieto, J.A.; Araque, A. Sensing and Regulating Synaptic Activity by Astrocytes at Tripartite Synapse. Neurochem. Res. 2021, 46, 2580–2585. [CrossRef]
- Accogli, A.; Addour-Boudrahem, N.; Srour, M. Chapter 4 - Neurogenesis, Neuronal Migration, and Axon Guidance. In Handbook of Clinical Neurology; Gallagher, A., Bulteau, C., Cohen, D., Michaud, J.L., Eds.; Neurocognitive Development: Normative Development; Elsevier, 2020; Vol. 173, pp. 25–42.
- Affrald R, J.; Narayan, S. A Review: Oligodendrocytes in Neuronal Axonal Conduction and Methods for Enhancing Their Performance. Int. J. Neurosci. 0, 1–22. [CrossRef]
- Norris, G.T.; Kipnis, J. Immune Cells and CNS Physiology: Microglia and Beyond. J. Exp. Med. 2018, 216, 60–70. [CrossRef]
- Thalhammer, A.; Cingolani, L.A. Cell Adhesion and Homeostatic Synaptic Plasticity. Neuropharmacology 2014, 78, 23–30. [CrossRef]
- Ben Achour, S.; Pascual, O. Glia: The Many Ways to Modulate Synaptic Plasticity. Neurochem. Int. 2010, 57, 440–445. [CrossRef]
- Durkee, C.A.; Araque, A. Diversity and Specificity of Astrocyte–Neuron Communication. Neuroscience 2019, 396, 73–78. [CrossRef]
- Benarroch, E.E. Neuron-Astrocyte Interactions: Partnership for Normal Function and Disease in the Central Nervous System. Mayo Clin. Proc. 2005, 80, 1326–1338. [CrossRef]
- Antonicek, H.; Persohn, E.; Schachner, M. Biochemical and Functional Characterization of a Novel Neuron-Glia Adhesion Molecule That Is Involved in Neuronal Migration. J. Cell Biol. 1987, 104, 1587–1595. [CrossRef]
- Gloor, S.; Antonicek, H.; Sweadner, K.J.; Pagliusi, S.; Frank, R.; Moos, M.; Schachner, M. The Adhesion Molecule on Glia (AMOG) Is a Homologue of the Beta Subunit of the Na,K-ATPase. J. Cell Biol. 1990, 110, 165–174. [CrossRef]
- Martin-Vasallo, P.; Dackowski, W.; Emanuel, J.R.; Levenson, R. Identification of a Putative Isoform of the Na,K-ATPase β Subunit: Primary Structure and Tissue-Specific Expression. J. Biol. Chem. 1989, 264, 4613–4618. [CrossRef]
- Contreras, R.G.; Torres-Carrillo, A.; Flores-Maldonado, C.; Shoshani, L.; Ponce, A. Na+/K+-ATPase: More than an Electrogenic Pump. Int. J. Mol. Sci. 2024, 25, 6122. [CrossRef]
- Lutsenko, S.; Kaplan, J.H. An Essential Role for the Extracellular Domain of the Sodium-Potassium-ATPase .Beta.-Subunit in Cation Occlusion. Biochemistry 1993, 32, 6737–6743. [CrossRef]
- Rajasekaran, S.A.; Palmer, L.G.; Moon, S.Y.; Peralta Soler, A.; Apodaca, G.L.; Harper, J.F.; Zheng, Y.; Rajasekaran, A.K. Na,K-ATPase Activity Is Required for Formation of Tight Junctions, Desmosomes, and Induction of Polarity in Epithelial Cells. Mol. Biol. Cell 2001, 12, 3717–3732. [CrossRef]
- Shoshani, L.; Contreras, R.G.; Roldán, M.L.; Moreno, J.; Lázaro, A.; Balda, M.S.; Matter, K.; Cereijido, M. The Polarized Expression of Na+,K+-ATPase in Epithelia Depends on the Association between β-Subunits Located in Neighboring Cells. Mol. Biol. Cell 2005, 16, 1071–1081. [CrossRef]
- Padilla-Benavides, T.; Roldán, M.L.; Larre, I.; Flores-Benitez, D.; Villegas-Sepúlveda, N.; Contreras, R.G.; Cereijido, M.; Shoshani, L. The Polarized Distribution of Na+,K+-ATPase: Role of the Interaction between β Subunits. Mol. Biol. Cell 2010, 21, 2217–2225. [CrossRef]
- Vagin, O.; Dada, L.A.; Tokhtaeva, E.; Sachs, G. The Na-K-ATPase A1β1 Heterodimer as a Cell Adhesion Molecule in Epithelia. Am. J. Physiol.-Cell Physiol. 2012, 302, C1271–C1281. [CrossRef]
- Vagin, O.; Tokhtaeva, E.; Sachs, G. The Role of the Β1 Subunit of the Na,K-ATPase and Its Glycosylation in Cell-Cell Adhesion *. J. Biol. Chem. 2006, 281, 39573–39587. [CrossRef]
- Tokhtaeva, E.; Sachs, G.; Sun, H.; Dada, L.A.; Sznajder, J.I.; Vagin, O. Identification of the Amino Acid Region Involved in the Intercellular Interaction between the Β1 Subunits of Na+/K+-ATPase. J. Cell Sci. 2012, 125, 1605–1616. [CrossRef]
- Tokhtaeva, E.; Sachs, G.; Souda, P.; Bassilian, S.; Whitelegge, J.P.; Shoshani, L.; Vagin, O. Epithelial Junctions Depend on Intercellular Trans-Interactions between the Na,K-ATPase Β1 Subunits*. J. Biol. Chem. 2011, 286, 25801–25812. [CrossRef]
- Antonicek, H.; Schachner, M. The Adhesion Molecule on Glia (AMOG) Incorporated into Lipid Vesicles Binds to Subpopulations of Neurons. J. Neurosci. 1988, 8, 2961–2966. [CrossRef]
- Togashi, H.; Sakisaka, T.; Takai, Y. Cell Adhesion Molecules in the Central Nervous System. Cell Adhes. Migr. 2009, 3, 29–35. [CrossRef]
- Saint-Martin, M.; Goda, Y. Astrocyte–Synapse Interactions and Cell Adhesion Molecules. FEBS J. 2023, 290, 3512–3526. [CrossRef]
- Shapiro, L.; Love, J.; Colman, D.R. Adhesion Molecules in the Nervous System: Structural Insights into Function and Diversity. Annu. Rev. Neurosci. 2007, 30, 451–474. [CrossRef]
- Uchida, N.; Honjo, Y.; Johnson, K.R.; Wheelock, M.J.; Takeichi, M. The Catenin/Cadherin Adhesion System Is Localized in Synaptic Junctions Bordering Transmitter Release Zones. J. Cell Biol. 1996, 135, 767–779. [CrossRef]
- Hirano, S.; Takeichi, M. Cadherins in Brain Morphogenesis and Wiring. Physiol. Rev. 2012, 92, 597–634. [CrossRef]
- de Agustín-Durán, D.; Mateos-White, I.; Fabra-Beser, J.; Gil-Sanz, C. Stick around: Cell–Cell Adhesion Molecules during Neocortical Development. Cells 2021, 10, 118. [CrossRef]
- Wu, Q.; Jia, Z. Wiring the Brain by Clustered Protocadherin Neural Codes. Neurosci. Bull. 2021, 37, 117–131. [CrossRef]
- Mizoguchi, A.; Nakanishi, H.; Kimura, K.; Matsubara, K.; Ozaki-Kuroda, K.; Katata, T.; Honda, T.; Kiyohara, Y.; Heo, K.; Higashi, M.; et al. Nectin : An Adhesion Molecule Involved in Formation of Synapses. J. Cell Biol. 2002, 156, 555–565. [CrossRef]
- Mizutani, K.; Miyata, M.; Shiotani, H.; Kameyama, T.; Takai, Y. Nectins and Nectin-like Molecules in Synapse Formation and Involvement in Neurological Diseases. Mol. Cell. Neurosci. 2021, 115, 103653. [CrossRef]
- Jozsef Zoltan Kiss; Dominique Müller Contribution of the Neural Cell Adhesion Molecule to Neuronal and Synaptic Plasticity. Rev. Neurosci. 2001, 12, 297–310. [CrossRef]
- Walsh, F.S.; Doherty, P. NEURAL CELL ADHESION MOLECULES OF THE IMMUNOGLOBULIN SUPERFAMILY: Role in Axon Growth and Guidance. Annu. Rev. Cell Dev. Biol. 1997, 13, 425–456. [CrossRef]
- Kerrisk, M.E.; Cingolani, L.A.; Koleske, A.J. Chapter 5 - ECM Receptors in Neuronal Structure, Synaptic Plasticity, and Behavior. In Progress in Brain Research; Dityatev, A., Wehrle-Haller, B., Pitkänen, A., Eds.; Brain Extracellular Matrix in Health and Disease; Elsevier, 2014; Vol. 214, pp. 101–131.
- Zuko, A.; Kleijer, K.T.E.; Oguro-Ando, A.; Kas, M.J.H.; van Daalen, E.; van der Zwaag, B.; Burbach, J.P.H. Contactins in the Neurobiology of Autism. Eur. J. Pharmacol. 2013, 719, 63–74. [CrossRef]
- Masuda, T. Contactin-2/TAG-1, Active on the Front Line for Three Decades. Cell Adhes. Migr. 2017, 11, 524–531. [CrossRef]
- Shen, K.; Fetter, R.D.; Bargmann, C.I. Synaptic Specificity Is Generated by the Synaptic Guidepost Protein SYG-2 and Its Receptor, SYG-1. Cell 2004, 116, 869–881. [CrossRef]
- Shen, K.; Bargmann, C.I. The Immunoglobulin Superfamily Protein SYG-1 Determines the Location of Specific Synapses in C. Elegans. Cell 2003, 112, 619–630. [CrossRef]
- Goodman, K.M.; Yamagata, M.; Jin, X.; Mannepalli, S.; Katsamba, P.S.; Ahlsén, G.; Sergeeva, A.P.; Honig, B.; Sanes, J.R.; Shapiro, L. Molecular Basis of Sidekick-Mediated Cell-Cell Adhesion and Specificity. eLife 2016, 5, e19058. [CrossRef]
- Craig, A.M.; Kang, Y. Neurexin–Neuroligin Signaling in Synapse Development. Curr. Opin. Neurobiol. 2007, 17, 43–52. [CrossRef]
- Bang, M.L.; Owczarek, S. A Matter of Balance: Role of Neurexin and Neuroligin at the Synapse. Neurochem. Res. 2013, 38, 1174–1189. [CrossRef]
- Clausen, M.V.; Hilbers, F.; Poulsen, H. The Structure and Function of the Na,K-ATPase Isoforms in Health and Disease. Front. Physiol. 2017, 8. [CrossRef]
- Álvarez, J.A.L.; Murillo, T. del C.L.; Nestor, C.A.V.; Gutierrez, M.L.R.; Gómez, O.P.; Shoshani, L.; Álvarez, J.A.L.; Murillo, T. del C.L.; Nestor, C.A.V.; Gutierrez, M.L.R.; et al. Epithelial Na+,K+-ATPase — A Sticky Pump. In Cell Biology - New Insights; IntechOpen, 2016 ISBN 978-953-51-2242-5.
- Cereijido, M.; Contreras, R.G.; Shoshani, L.; Larre, I. The Na+-K+-ATPase as Self-Adhesion Molecule and Hormone Receptor. Am. J. Physiol.-Cell Physiol. 2012, 302, C473–C481. [CrossRef]
- Blanco, G. Na,K-ATPase Subunit Heterogeneity as a Mechanism for Tissue-Specific Ion Regulation. Semin. Nephrol. 2005, 25, 292–303. [CrossRef]
- Peng, L.; Martin-Vasallo, P.; Sweadner, K.J. Isoforms of Na,K-ATPase α and β Subunits in the Rat Cerebellum and in Granule Cell Cultures. J. Neurosci. 1997, 17, 3488–3502. [CrossRef]
- Levenson, R. Isoforms of the Na,K-ATPase: Family Members in Search of Function. In Reviews of Physiology, Biochemistry and Pharmacology, Volume 123: Volume: 123; Springer: Berlin, Heidelberg, 1994; pp. 1–45 ISBN 978-3-540-48217-8.
- Lingrel, J.B. Na,K-ATPase: Isoform Structure, Function, and Expression. J. Bioenerg. Biomembr. 1992, 24, 263–270. [CrossRef]
- Lingrel, J.B.; Orlowski, J.; Shull, M.M.; Price, E.M. Molecular Genetics of Na,K-ATPase. In Progress in Nucleic Acid Research and Molecular Biology; Cohn, W.E., Moldave, K., Eds.; Academic Press, 1990; Vol. 38, pp. 37–89.
- Sweadner, K.J. Overlapping and Diverse Distribution of Na–K ATPase Isozymes in Neurons and Glia. Can. J. Physiol. Pharmacol. 1992, 70, S255–S259. [CrossRef]
- Brines, M.L.; Robbins, R.J. Cell-Type Specific Expression of Na+,K+-ATPase Catalytic Subunits in Cultured Neurons and Glia: Evidence for Polarized Distribution in Neurons. Brain Res. 1993, 631, 1–11. [CrossRef]
- Cameron, R.; Klein, L.; Shyjan, A.W.; Rakic, P.; Levenson, R. Neurons and Astroglia Express Distinct Subsets of Na,K-ATPase α and β Subunits. Mol. Brain Res. 1994, 21, 333–343. [CrossRef]
- Fink, D.; Knapp, P.E.; Mata, M. Differential Expression of Na,K-ATPase Isoforms in Oligodendrocytes and Astrocytes. Dev. Neurosci. 1996, 18, 319–326. [CrossRef]
- Wetzel, R.K.; Arystarkhova, E.; Sweadner, K.J. Cellular and Subcellular Specification of Na,K-ATPase α and β Isoforms in the Postnatal Development of Mouse Retina. J. Neurosci. 1999, 19, 9878–9889. [CrossRef]
- Lavoie, L.; Levenson, R.; Martin-Vasallo, P.; Klip, A. The Molar Ratios of α and β Subunits of the Na+−K+-ATPase Differ in Distinct Subcellular Membranes from Rat Skeletal Muscle. Biochemistry 1997, 36, 7726–7732. [CrossRef]
- Shyjan, A.W.; Ceña, V.; Klein, D.C.; Levenson, R. Differential Expression and Enzymatic Properties of the Na+,K(+)-ATPase Alpha 3 Isoenzyme in Rat Pineal Glands. Proc. Natl. Acad. Sci. 1990, 87, 1178–1182. [CrossRef]
- Pagliusi, S.R.; Schachner, M.; Seeburg, P.H.; Shivers, B.D. The Adhesion Molecule on Glia (AMOG) Is Widely Expressed by Astrocytes in Developing and Adult Mouse Brain. Eur. J. Neurosci. 1990, 2, 471–480. [CrossRef]
- Malik, N.; Canfield, V.A.; Beckers, M.-C.; Gros, P.; Levenson, R. Identification of the Mammalian Na,K-ATPase Β3 Subunit *. J. Biol. Chem. 1996, 271, 22754–22758. [CrossRef]
- Arystarkhova, E.; Sweadner, K.J. Tissue-Specific Expression of the Na,K-ATPase Β3 Subunit: THE PRESENCE OF Β3 IN LUNG AND LIVER ADDRESSES THE PROBLEM OF THE MISSING SUBUNIT *. J. Biol. Chem. 1997, 272, 22405–22408. [CrossRef]
- Book, C.B.; Wilson, R.P.; Ng, Y.C. Cardiac Hypertrophy in the Ferret Increases Expression of the Na(+)-K(+)-ATPase Alpha 1- but Not Alpha 3-Isoform. Am. J. Physiol.-Heart Circ. Physiol. 1994, 266, H1221–H1227. [CrossRef]
- Charlemagne, D.; Swynghedauw, B. Myocardial Phenotypic Changes in Na+, K+ATPase in Left Ventricular Hypertrophy: Pharmacological Consequences. Eur. Heart J. 1995, 16, 20–23. [CrossRef]
- Charlemagne, D.; Orlowski, J.; Oliviero, P.; Rannou, F.; Sainte Beuve, C.; Swynghedauw, B.; Lane, L.K. Alteration of Na,K-ATPase Subunit mRNA and Protein Levels in Hypertrophied Rat Heart. J. Biol. Chem. 1994, 269, 1541–1547. [CrossRef]
- Ewart, H.S.; Klip, A. Hormonal Regulation of the Na(+)-K(+)-ATPase: Mechanisms Underlying Rapid and Sustained Changes in Pump Activity. Am. J. Physiol.-Cell Physiol. 1995, 269, C295–C311. [CrossRef]
- Zahler, R.; Gilmore-Hebert, M.; Sun, W.; Benz, E.J. Na, K-ATPase Isoform Gene Expression in Normal and Hypertrophied Dog Heart. Basic Res. Cardiol. 1996, 91, 256–266. [CrossRef]
- Araque, A.; Parpura, V.; Sanzgiri, R.P.; Haydon, P.G.; Araque, A.; Parpura, V.; Sanzgiri, R.P.; Haydon, P.G. Tripartite Synapses: Glia, the Unacknowledged Partner. Trends Neurosci. 1999, 22, 208–215. [CrossRef]
- Chung, W.-S.; Allen, N.J.; Eroglu, C. Astrocytes Control Synapse Formation, Function, and Elimination. Cold Spring Harb. Perspect. Biol. 2015, 7, a020370. [CrossRef]
- Allen, N.J.; Eroglu, C. Cell Biology of Astrocyte-Synapse Interactions. Neuron 2017, 96, 697–708. [CrossRef]
- Baldwin, K.T.; Eroglu, C. Molecular Mechanisms of Astrocyte-Induced Synaptogenesis. Curr. Opin. Neurobiol. 2017, 45, 113–120. [CrossRef]
- Kilb, W.; Kirischuk, S. GABA Release from Astrocytes in Health and Disease. Int. J. Mol. Sci. 2022, 23, 15859. [CrossRef]
- Harada, K.; Kamiya, T.; Tsuboi, T. Gliotransmitter Release from Astrocytes: Functional, Developmental and Pathological Implications in the Brain. Front. Neurosci. 2016, 9. [CrossRef]
- Abbott, N.J.; Rönnbäck, L.; Hansson, E. Astrocyte–Endothelial Interactions at the Blood–Brain Barrier. Nat. Rev. Neurosci. 2006, 7, 41–53. [CrossRef]
- Pietrobon, D.; Conti, F. Astrocytic Na+, K+ ATPases in Physiology and Pathophysiology. Cell Calcium 2024, 118, 102851. [CrossRef]
- Pagliusi, S.; Antonicek, H.; Gloor, S.; Frank, R.; Moos, M.; Schachner, M. Identification of a cDNA Clone Specific for the Neural Cell Adhesion Molecule AMOG. J. Neurosci. Res. 1989, 22, 113–119. [CrossRef]
- Müller-Husmann, G.; Gloor, S.; Schachner, M. Functional Characterization of Beta Isoforms of Murine Na,K-ATPase. The Adhesion Molecule on Glia (AMOG/Beta 2), but Not Beta 1, Promotes Neurite Outgrowth. J. Biol. Chem. 1993, 268, 26260–26267. [CrossRef]
- Magyar, J.P.; Bartsch, U.; Wang, Z.Q.; Howells, N.; Aguzzi, A.; Wagner, E.F.; Schachner, M. Degeneration of Neural Cells in the Central Nervous System of Mice Deficient in the Gene for the Adhesion Molecule on Glia, the Beta 2 Subunit of Murine Na,K-ATPase. J. Cell Biol. 1994, 127, 835–845. [CrossRef]
- Weber, P.; Bartsch, U.; Schachner, M.; Montag, D. Na,K-ATPase Subunit Β1 Knock-in Prevents Lethality of Β2 Deficiency in Mice. J. Neurosci. 1998, 18, 9192–9203. [CrossRef]
- Isenmann, S.; Molthagen, M.; Brandner, S.; Bartsch, U.; Kühne, G.; Magyar, J.P.; Sure, U.; Schachner, M.; Aguzzi, A. The AMOG/Β2 Subunit of Na, K-ATPase Is Not Necessary for Long-Term Survival of Telencephalic Grafts. Glia 1995, 15, 377–388. [CrossRef]
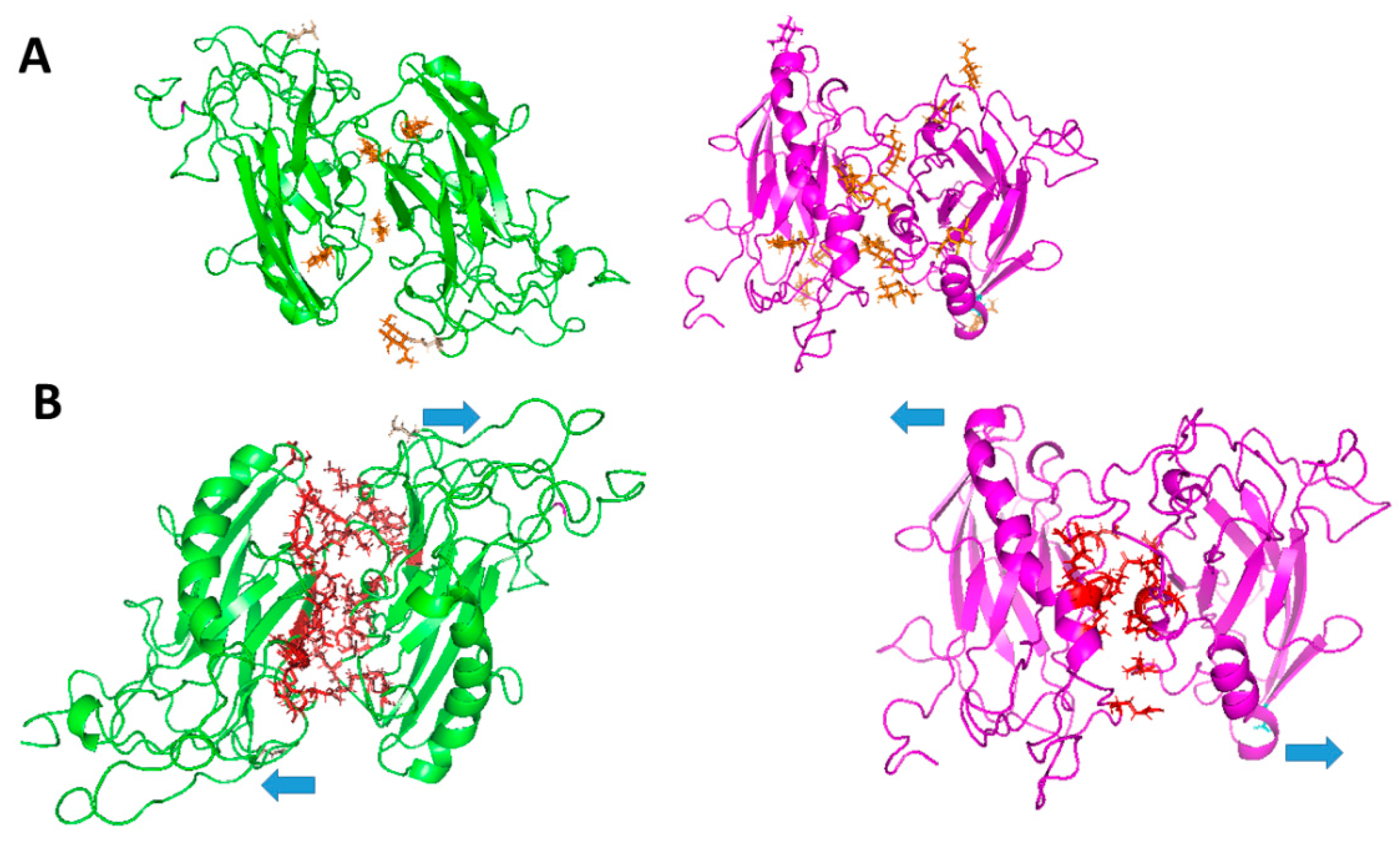
- Páez, O.; Martínez-Archundia, M.; Villegas-Sepúlveda, N.; Roldan, M.L.; Correa-Basurto, J.; Shoshani, L. A Model for the Homotypic Interaction between Na+,K+-ATPase Β1 Subunits Reveals the Role of Extracellular Residues 221–229 in Its Ig-Like Domain. Int. J. Mol. Sci. 2019, 20, 4538. [CrossRef]
- Scheidenhelm, D.K.; Cresswell, J.; Haipek, C.A.; Fleming, T.P.; Mercer, R.W.; Gutmann, D.H. Akt-Dependent Cell Size Regulation by the Adhesion Molecule on Glia Occurs Independently of Phosphatidylinositol 3-Kinase and Rheb Signaling. Mol. Cell. Biol. 2005, 25, 3151–3162. [CrossRef]
- Barreto, N.; Caballero, M.; Bonfanti, A.P.; de Mato, F.C.P.; Munhoz, J.; da Rocha--e--Silva, T.A.A.; Sutti, R.; Vitorino-Araujo, J.L.; Verinaud, L.; Rapôso, C. Spider Venom Components Decrease Glioblastoma Cell Migration and Invasion through RhoA-ROCK and Na+/K+-ATPase Β2: Potential Molecular Entities to Treat Invasive Brain Cancer. Cancer Cell Int. 2020, 20, 576. [CrossRef]
- Litan, A.; Li, Z.; Tokhtaeva, E.; Kelly, P.; Vagin, O.; Langhans, S.A. A Functional Interaction Between Na,K-ATPase Β2-Subunit/AMOG and NF2/Merlin Regulates Growth Factor Signaling in Cerebellar Granule Cells. Mol. Neurobiol. 2019, 56, 7557–7571. [CrossRef]
- Kanai, R.; Ogawa, H.; Vilsen, B.; Cornelius, F.; Toyoshima, C. Crystal Structure of a Na+-Bound Na+,K+-ATPase Preceding the E1P State. Nature 2013, 502, 201–206. [CrossRef]
- Zlokovic, B.V.; Mackic, J.B.; Wang, L.; McComb, J.G.; McDonough, A. Differential Expression of Na,K-ATPase Alpha and Beta Subunit Isoforms at the Blood-Brain Barrier and the Choroid Plexus. J. Biol. Chem. 1993, 268, 8019–8025. [CrossRef]
- Boer, K.; Spliet, W.G.M.; van Rijen, P.C.; Jansen, F.E.; Aronica, E. Expression Patterns of AMOG in Developing Human Cortex and Malformations of Cortical Development. Epilepsy Res. 2010, 91, 84–93. [CrossRef]
- McGrail, K.M.; Phillips, J.M.; Sweadner, K.J. Immunofluorescent Localization of Three Na,K-ATPase Isozymes in the Rat Central Nervous System: Both Neurons and Glia Can Express More than One Na,K-ATPase. J. Neurosci. 1991, 11, 381–391. [CrossRef]
- Watts, A.G.; Sanchez-Watts, G.; Emanuel, J.R.; Levenson, R. Cell-Specific Expression of mRNAs Encoding Na+,K(+)-ATPase Alpha- and Beta-Subunit Isoforms within the Rat Central Nervous System. Proc. Natl. Acad. Sci. 1991, 88, 7425–7429. [CrossRef]
- Roldán, M.L.; Ramírez-Salinas, G.L.; Martinez-Archundia, M.; Cuellar-Perez, F.; Vilchis-Nestor, C.A.; Cancino-Diaz, J.C.; Shoshani, L. The Β2-Subunit (AMOG) of Human Na+, K+-ATPase Is a Homophilic Adhesion Molecule. Int. J. Mol. Sci. 2022, 23, 7753. [CrossRef]
- Ramírez-Salinas, G.; Shoshani, L.; Rosas-Trigueros, J.L.; Huerta, C.S.; Martínez-Archundia, M. In Silico Studies Provide New Structural Insights into Trans-Dimerization of Β1 and Β2 Subunits of the Na+, K+-ATPase. PLOS ONE 2025, 20, e0321064. [CrossRef]
- Goulding, S.P.; Szumlinski, K.K.; Contet, C.; MacCoss, M.J.; Wu, C.C. A Mass Spectrometry-Based Proteomic Analysis of Homer2-Interacting Proteins in the Mouse Brain. J. Proteomics 2017, 166, 127–137. [CrossRef]
- Roux, K.J.; Kim, D.I.; Burke, B.; May, D.G. BioID: A Screen for Protein-Protein Interactions. Curr. Protoc. Protein Sci. 2018, 91, 19.23.1-19.23.15. [CrossRef]
- Mannix, K.M.; Starble, R.M.; Kaufman, R.S.; Cooley, L. Proximity Labeling Reveals Novel Interactomes in Live Drosophila Tissue. Development 2019, 146, dev176644. [CrossRef]
- Tsiami, F.; Lago, C.; Pozza, N.; Piccioni, F.; Zhao, X.; Lülsberg, F.; Root, D.E.; Tiberi, L.; Kool, M.; Schittenhelm, J.; et al. Genome-Wide CRISPR-Cas9 Knockout Screens Identify DNMT1 as a Druggable Dependency in Sonic Hedgehog Medulloblastoma. Acta Neuropathol. Commun. 2024, 12, 125. [CrossRef]
- Charrin, S.; Naour, F.L.; Oualid, M.; Billard, M.; Faure, G.; Hanash, S.M.; Boucheix, C.; Rubinstein, E. The Major CD9 and CD81 Molecular Partner: IDENTIFICATION AND CHARACTERIZATION OF THE COMPLEXES *. J. Biol. Chem. 2001, 276, 14329–14337. [CrossRef]
- Hegazy, M.; Cohen-Barak, E.; Koetsier, J.L.; Najor, N.A.; Arvanitis, C.; Sprecher, E.; Green, K.J.; Godsel, L.M. Proximity Ligation Assay for Detecting Protein-Protein Interactions and Protein Modifications in Cells and Tissues in Situ. Curr. Protoc. Cell Biol. 2020, 89, e115. [CrossRef]
- Mateos-Martínez, P.; Coronel, R.; Sachse, M.; González-Sastre, R.; Maeso, L.; Rodriguez, M.J.; Terrón, M.C.; López-Alonso, V.; Liste, I. Human Cerebral Organoids: Cellular Composition and Subcellular Morphological Features. Front. Cell. Neurosci. 2024, 18. [CrossRef]
- Rakotomamonjy, J.; Rylaarsdam, L.; Fares-Taie, L.; McDermott, S.; Davies, D.; Yang, G.; Fagbemi, F.; Epstein, M.; Fairbanks-Santana, M.; Rozet, J.-M.; et al. PCDH12 Loss Results in Premature Neuronal Differentiation and Impeded Migration in a Cortical Organoid Model. Cell Rep. 2023, 42. [CrossRef]
- Huttlin, E.L.; Bruckner, R.J.; Paulo, J.A.; Cannon, J.R.; Ting, L.; Baltier, K.; Colby, G.; Gebreab, F.; Gygi, M.P.; Parzen, H.; et al. Architecture of the Human Interactome Defines Protein Communities and Disease Networks. Nature 2017, 545, 505–509. [CrossRef]
- Huttlin, E.L.; Bruckner, R.J.; Navarrete-Perea, J.; Cannon, J.R.; Baltier, K.; Gebreab, F.; Gygi, M.P.; Thornock, A.; Zarraga, G.; Tam, S.; et al. Dual Proteome-Scale Networks Reveal Cell-Specific Remodeling of the Human Interactome. Cell 2021, 184, 3022-3040.e28. [CrossRef]
- Charrin, S.; Jouannet, S.; Boucheix, C.; Rubinstein, E. Tetraspanins at a Glance. J. Cell Sci. 2014, 127, 3641–3648. [CrossRef]
- Ovalle, S.; Gutiérrez-López, M.D.; Olmo, N.; Turnay, J.; Lizarbe, M.A.; Majano, P.; Molina-Jiménez, F.; López-Cabrera, M.; Yáñez-Mó, M.; Sánchez-Madrid, F.; et al. The Tetraspanin CD9 Inhibits the Proliferation and Tumorigenicity of Human Colon Carcinoma Cells. Int. J. Cancer 2007, 121, 2140–2152. [CrossRef]
- Wang, J.; Zhou, Y.; Li, D.; Sun, X.; Deng, Y.; Zhao, Q. TSPAN31 Is a Critical Regulator on Transduction of Survival and Apoptotic Signals in Hepatocellular Carcinoma Cells. FEBS Lett. 2017, 591, 2905–2918. [CrossRef]
- Chen, M.S.; Huber, A.B.; van der Haar, M.E.; Frank, M.; Schnell, L.; Spillmann, A.A.; Christ, F.; Schwab, M.E. Nogo-A Is a Myelin-Associated Neurite Outgrowth Inhibitor and an Antigen for Monoclonal Antibody IN-1. Nature 2000, 403, 434–439. [CrossRef]
- Kleene, R.; Loers, G.; Langer, J.; Frobert, Y.; Buck, F.; Schachner, M. Prion Protein Regulates Glutamate-Dependent Lactate Transport of Astrocytes. J. Neurosci. 2007, 27, 12331–12340. [CrossRef]
- Rotoli, D.; Cejas, M.-M.; Maeso, M.-C.; Pérez-Rodríguez, N.-D.; Morales, M.; Ávila, J.; Mobasheri, A.; Martín-Vasallo, P. The Na, K-ATPase β-Subunit Isoforms Expression in Glioblastoma Multiforme: Moonlighting Roles. Int. J. Mol. Sci. 2017, 18, 2369. [CrossRef]
- Sun, M.Z.; Kim, J.M.; Oh, M.C.; Safaee, M.; Kaur, G.; Clark, A.J.; Bloch, O.; Ivan, M.E.; Kaur, R.; Oh, T.; et al. Na+/K+-ATPase Β2-Subunit (AMOG) Expression Abrogates Invasion of Glioblastoma-Derived Brain Tumor-Initiating Cells. Neuro-Oncol. 2013, 15, 1518–1531. [CrossRef]
- Lefranc, F.; Mijatovic, T.; Kiss, R. The Sodium Pump Could Constitute a New Target to Combat Glioblastomas. Bull. Cancer (Paris) 2008, 95, 271–281. [CrossRef]
- Senner, V.; Schmidtpeter, S.; Braune, S.; Püttmann, S.; Thanos, S.; Bartsch, U.; Schachner, M.; Paulus, W. AMOG/Β2 and Glioma Invasion: Does Loss of AMOG Make Tumour Cells Run Amok? Neuropathol. Appl. Neurobiol. 2003, 29, 370–377. [CrossRef]
- Kim, Y.S.; Choi, J.; Yoon, B.-E. Neuron-Glia Interactions in Neurodevelopmental Disorders. Cells 2020, 9, 2176. [CrossRef]
- Phatnani, H.; Maniatis, T. Astrocytes in Neurodegenerative Disease. Cold Spring Harb. Perspect. Biol. 2015, 7, a020628. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




