1. Introduction
In the evolving landscape of healthcare, there is a growing demand for intelligent systems capable of supporting clinical decisions through the integration and analysis of vast biomedical data. Oncology, which involves the study and treatment of cancer, is a highly complex and data-intensive medical field. With the increasing availability of biomedical literature and clinical records, there is a pressing need to extract structured domain knowledge that can assist oncologists in the diagnosis, planning of treatment, and understanding of disease pathways. Knowledge graphs (KG) have emerged as a powerful representation to capture and organize semantic relationships between medical concepts, enabling more interpretable and actionable clinical decision support systems (CDSS) [
1].
Traditional methods for constructing KGs in specialized domains, such as cancer, often rely heavily on manual annotations and domain expertise. However, acquiring high-quality labeled data in oncology is particularly challenging due to the need for expert knowledge and the high costs associated with manual labeling. Furthermore, rule-based approaches like Stanford CoreNLP and OpenIE often lack the flexibility to adapt to fine-grained domain-specific relationships and entities, thereby limiting their applicability in dynamic and complex fields like oncology [
2].
Recent advancements in deep learning and pretrained language models (PLMs) have enabled automated extraction of entities and relationships from unstructured biomedical text. Despite their success, these methods frequently require large annotated datasets, which again poses a challenge in specialized fields. This has led to increasing interest in weakly-supervised and distantly-supervised learning techniques, which attempt to bypass the need for manually labeled data by leveraging existing knowledge bases to automatically generate training labels [
3].
In particular, distant supervision provides a promising alternative by using a general-domain KG (e.g., a biomedical KG) to infer labels for unstructured text in a more specific domain, such as oncology. However, direct application of distant supervision often fails to capture fine-domain knowledge due to label noise and incomplete coverage of domain-specific entities and relations in the source KG [
4]. This gap necessitates an adaptive approach to KG construction that can iteratively refine and expand the graph based on new information gleaned from the fine domain.
To address these challenges, this paper proposes a novel Knowledge Graph Domain Adaptation (KGDA) framework designed for automatic and iterative construction of oncology-specific KGs using distant supervision. By iteratively fine-tuning NER and RE models on unlabeled oncology literature and incorporating feedback from previously discovered high-confidence entities and triples, the framework significantly enhances the discovery of both overlapping and novel domain-specific knowledge. This method reduces the dependency on manual annotations while maintaining high accuracy in entity and relation extraction [
5].
Our approach introduces a coarse-to-fine learning paradigm where the general biomedical KG serves as a foundational knowledge base to bootstrap the extraction process in the oncology domain. Through iterative refinement, the system dynamically learns to recognize previously unseen entities and relationships, thereby constructing a more comprehensive and accurate oncology KG. This also facilitates the continuous update and evolution of KGs as new biomedical literature becomes available.
The contributions of this study are threefold: (1) We develop an end-to-end KGDA framework that supports dynamic and scalable KG construction from raw text; (2) we demonstrate the effectiveness of our approach through extensive experiments on real-world oncology literature using different PLMs; and (3) we provide both held-out and manual evaluations to validate the accuracy and relevance of the extracted knowledge. Our work lays the groundwork for scalable and automated biomedical knowledge discovery systems, with the potential to significantly enhance clinical decision-making in oncology [
6].
2. Related Work
The construction of Knowledge Graphs (KGs) from biomedical text requires structured extraction of semantic information. Conventional approaches, which depend heavily on handcrafted rules or manual annotations, often lack the scalability and domain adaptability required for specialized fields such as oncology.
Early KG construction methods were primarily rule-based, using handcrafted linguistic and syntactic rules. Pipelines like Stanford CoreNLP, NLTK, spaCy, and OpenIE are often employed for entity and relation extraction. However, these systems are typically designed for general-purpose texts and exhibit limited performance on domain-specific corpora like biomedical literature. The inability to capture nuanced medical terminology and complex sentence structures impairs their utility in oncology applications [
2].
To overcome these limitations, data-driven techniques using machine learning have gained traction. Fully supervised models, especially those using deep learning architectures like Bi-LSTM and transformers (e.g., BERT), have achieved state-of-the-art results in biomedical NLP tasks. These models can learn complex patterns from annotated data, but their effectiveness is constrained by the availability of large, high-quality labeled datasets, which are costly and labor-intensive to produce in specialized domains [
5].
Distant supervision offers a pragmatic alternative to manual labeling. By aligning known triples from existing KGs with text mentions, it automatically generates pseudo-labeled data for training NER and RE models. While this approach significantly reduces annotation costs, it often introduces noisy labels due to misalignment or ambiguity, especially when applied to fine-grained subdomains such as oncology [
4].
One major challenge in distant supervision is the domain mismatch between the source KG (e.g., a general biomedical KG) and the target application domain (e.g., oncology). Entities and relations specific to oncology are often missing or underrepresented in the source KG. Consequently, models trained with distant supervision on general-domain data may fail to generalize well to the fine domain, missing crucial insights or misclassifying entities [
7].
To address these issues, hybrid frameworks that combine distant supervision with domain adaptation strategies have been proposed. These systems iteratively retrain NER and RE models using feedback from earlier predictions to gradually improve the accuracy and specificity of the extracted knowledge. Filtering mechanisms based on prediction confidence or frequency are employed to mitigate the effects of label noise and refine the KG construction process.
In this paper, we present a novel KG Domain Adaptation (KGDA) framework that utilizes distant supervision enhanced by iterative training. The proposed method is designed to dynamically discover and integrate oncology-specific knowledge, offering a scalable and annotation-free solution to KG construction in healthcare. Recent work has demonstrated the effectiveness of domain-specific pretrained models for biomedical text mining tasks [
5,
8]. Additionally, lightweight and automated KG construction strategies have shown promise in addressing scalability and generalizability issues in fine domains like oncology [
9,
10].
3. Methodology
The goal of Knowledge Graph Domain Adaptation (KGDA) is to construct a domain-specific knowledge graph (KG), denoted , from unlabeled text D in the fine domain (e.g., oncology), using an existing KG from a coarse domain (e.g., general biomedical knowledge). The resulting KG should capture both shared and domain-specific entities and relations, thus supporting downstream clinical applications.
Formally, let represent a sentence with n tokens. The dataset is a corpus of such sentences. A KG is defined as a set of triples , where and are head and tail entities and is the relation between them. We classify the triples in into:
Overlapping triples (): Present in both and .
Triples with new relations but known entities ().
Triples with new entities and possibly new relations ().
The KGDA framework proceeds through an iterative training strategy, designed to incrementally improve the model’s ability to extract fine-domain knowledge. The steps include:
1. Corpus Partitioning
The dataset D is split into n non-overlapping subsets, . is used to initialize the models, while subsequent subsets are used for iterative refinement.
2. Distant Supervision
Using , we generate distant supervision labels for entities and relations in . These labels are created via dictionary matching and heuristic rules to construct pseudo-labeled corpora for NER and RE training.
3. Model Training
We use pretrained language models (PLMs), such as BioBERT or ClinicalBERT, and fine-tune them on the distant supervision corpus. The NER model is trained using the BIO tagging scheme, while the RE model is trained using a sentence-level classification template.
4. Iterative Knowledge Discovery
For each (), the trained models from the previous round are used to identify new entities and triples. These predictions are filtered using confidence thresholds based on prediction probability (, ) and frequency (, ). High-confidence items are added to the supervision set for retraining.
5. Negative Sampling
To train the RE model to differentiate valid and invalid triples, we generate negative samples using two strategies: (a) randomly pairing unrelated entities, and (b) mixing in non-entity words as false entities. These are labeled with a NULL relation and included in the training set.
6. Algorithm Implementation
Algorithm 1 manages the full KGDA process over all . Algorithm 2 builds the distant supervision corpus using both and discovered knowledge (, ). Algorithm 3 filters and selects confident predictions for iterative refinement.
Through these steps, the KGDA framework incrementally adapts a general biomedical KG into a highly specialized oncology KG, identifying both known and novel knowledge without requiring any manual annotations.
4. Experimental Setup
To evaluate the proposed KGDA framework, we conduct comprehensive experiments focusing on knowledge graph construction from biomedical text. This section outlines the datasets used, model configurations, training details, and evaluation methodologies.
4.1. Datasets
We utilize two primary sources of data. The coarse-domain knowledge graph () is derived from a biomedical KG that includes over 5 million English-language entities and 7.3 million relational triples, covering 18 entity types (e.g., anatomy, drug, disease) and 19 relationship types (e.g., may_treat, is_part_of).
For the fine domain, we collect oncology-related texts from 12 peer-reviewed international medical journals. After cleaning and sentence segmentation, approximately 240,000 paragraphs are used, forming the unlabeled corpus D for fine-domain KG construction.
4.2. Model Settings
We evaluate three pretrained language models (PLMs) for Named Entity Recognition and Relation Extraction tasks:
BERT (Base, uncased) – general-purpose language model.
BioClinicalBERT – pre-trained on clinical notes and biomedical articles.
BiomedRoBERTa – optimized for biomedical scientific literature.
Each model is fine-tuned for both NER and RE using our distant supervision strategy.
4.3. Training Configuration
The corpus is divided into 6 subsets of 40,000 sentences each. The first 5 subsets are used for iterative training, and the last subset serves as a held-out test set. The experiments are executed on an Ubuntu server with 4 NVIDIA A100 GPUs. Hyperparameters are as follows:
Learning rate:
Batch size: 20
Epochs: 4
Confidence thresholds: , , ,
Negative sampling ratios: ,
4.4. Evaluation Methods
Held-out Evaluation: We compute precision, recall, and F1 score using a test subset labeled via distant supervision. For NER, evaluation is done using the seqeval library. For RE, evaluation focuses on entity pairs in the KG.
Manual Evaluation: To assess the framework’s ability to discover domain-specific knowledge, we manually evaluate 50 instances of each: new entities (), new relations with existing entities (), and new triples with new entities ().
Table 1.
Summary of Datasets and Model Configurations.
Table 1.
Summary of Datasets and Model Configurations.
| Item |
Description |
| Coarse KG |
5.2M entities, 7.3M triples |
| Fine Corpus |
240K oncology paragraphs |
| PLMs Used |
BERT, BioClinicalBERT, BiomedRoBERTa |
| Test Set Size |
40K sentences |
| Hardware |
4 × NVIDIA A100 GPUs |
5. Results and Discussion
This section presents the results of the held-out and manual evaluations, highlights the effectiveness of the iterative strategy, and analyzes the impact of different model configurations.
5.1. Held-Out Evaluation
BiomedRoBERTa consistently outperforms the other models across all NER and RE tasks, attributed to its specialization in biomedical literature.
Table 2.
Held-Out Evaluation of NER and RE Models
Table 2.
Held-Out Evaluation of NER and RE Models
| Model |
Precision |
Recall |
F1 Score |
| BERT |
0.908 |
0.900 |
0.904 |
| BioClinicalBERT |
0.909 |
0.895 |
0.902 |
| BiomedRoBERTa |
0.908 |
0.901 |
0.905 |
5.2. Manual Evaluation
Table 3.
Manual Evaluation of Fine-Domain Discoveries
Table 3.
Manual Evaluation of Fine-Domain Discoveries
| Model |
|
|
|
| BERT |
0.90 |
0.58 |
0.70 |
| BioClinicalBERT |
0.90 |
0.66 |
0.62 |
| BiomedRoBERTa |
0.94 |
0.76 |
0.74 |
The high precision of BiomedRoBERTa in detecting both entities and new relations demonstrates the value of iterative knowledge refinement.
5.3. Ablation Study
We conducted an ablation study to evaluate the impact of removing iteration and cumulative corpus construction.
Table 4.
Ablation Study on RE Precision
Table 4.
Ablation Study on RE Precision
| Model Variant |
RE Precision |
| BiomedRoBERTa (Full) |
0.987 |
| w/o Cumulative |
0.983 |
| w/o Iteration |
0.986 |
The results confirm that iterative refinement and corpus accumulation both contribute meaningfully to the system’s generalization capability.
5.4. Case Study
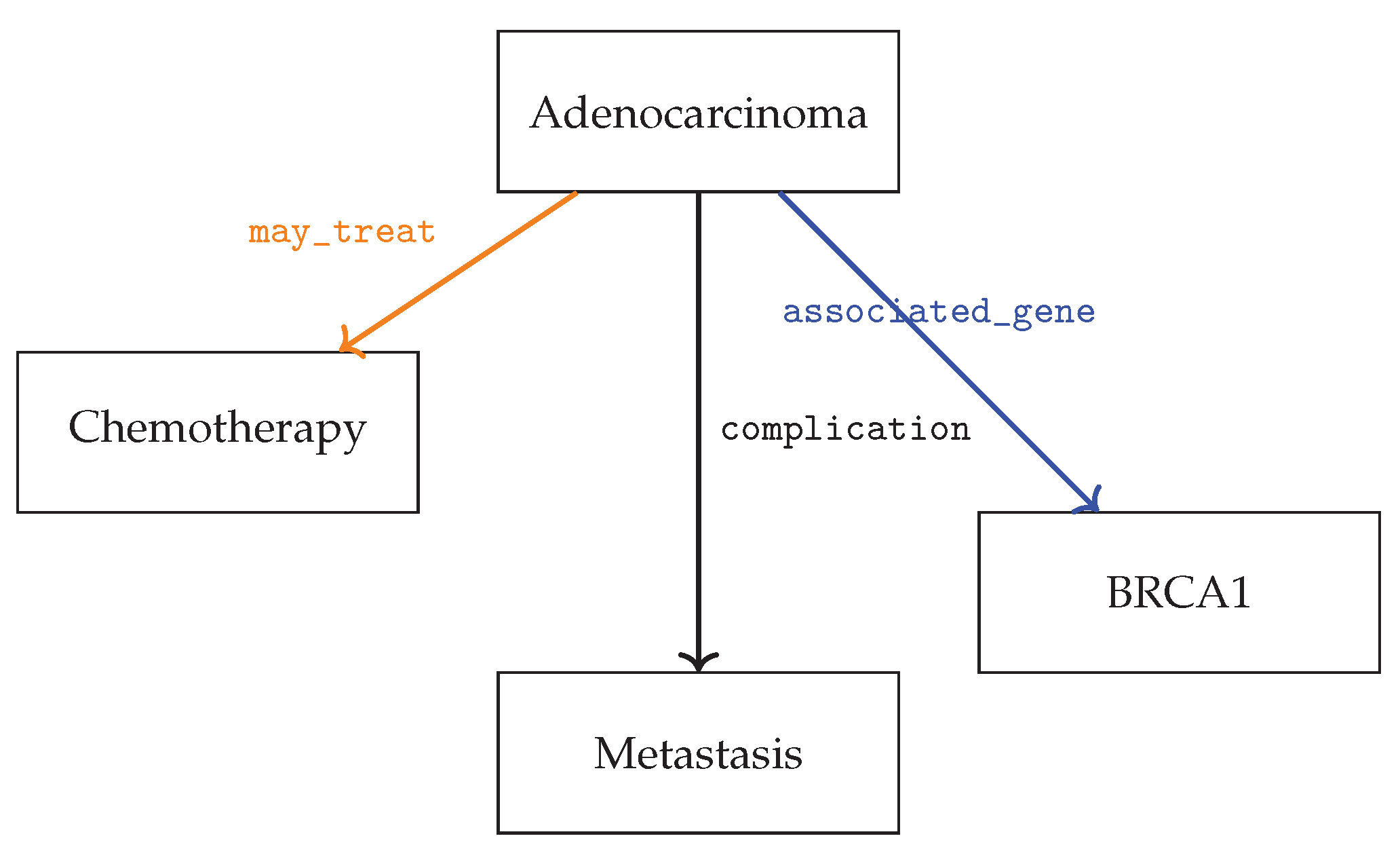
Figure 1 illustrates a portion of the oncology KG extracted by the model, focusing on adenocarcinoma and related entities.
6. Conclusion and Future Work
In this study, we proposed a novel KG Domain Adaptation (KGDA) framework for constructing oncology-specific knowledge graphs using distant supervision and iterative model refinement. The framework efficiently adapts coarse biomedical knowledge to fine domains by discovering domain-specific entities and relationships from unlabeled text.
Experimental results using various pretrained models confirm the robustness and scalability of the approach. Our ablation studies validate the importance of iterative training and cumulative corpus updates. Manual evaluations further demonstrate the framework’s ability to discover accurate and novel knowledge.
However, several limitations remain. The reliance on distant supervision introduces noisy labels, and the static definition of entity/relation types may restrict discovery in evolving domains. The current system also depends on a single source domain for supervision, which may limit knowledge coverage.
In future work, we plan to address these challenges by:
Incorporating human-in-the-loop verification for higher-quality extractions.
Leveraging clinical ontologies and multi-source KGs to enrich supervision.
Extending the framework to support dynamic entity/relation type discovery.
Adapting the KGDA approach to integrate with large generative language models like GPT-3.
Ultimately, our work aims to contribute to the development of scalable, accurate, and adaptive systems for biomedical knowledge discovery, with direct implications for clinical decision support in oncology and beyond.
References
- Bodenreider, O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Research 2004, 32, D267–D270. [Google Scholar] [CrossRef] [PubMed]
- Angeli, G.; Premkumar, M.J.; Manning, C.D. Leveraging linguistic structure for open domain information extraction. In Proceedings of the Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing (EMNLP), 2015, pp. 344–354.
- Mintz, M.; Bills, S.; Snow, R.; Jurafsky, D. Distant supervision for relation extraction without labeled data. In Proceedings of the Proceedings of the Joint Conference of the 47th Annual Meeting of the ACL and the 4th International Joint Conference on Natural Language Processing (ACL-IJCNLP), 2009, pp. 1003–1011.
- Zhang, D.; Wang, D. Relation classification via recurrent neural network. arXiv 2015. [Google Scholar] [CrossRef]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; So, C.H.; Kang, J. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020, 36, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- Verandani, A.; Smith, J.; Lee, K. Adaptive Knowledge Graph Construction for Oncology Literature. Journal of Biomedical Informatics In press. 2025. [Google Scholar]
- Wang, H.; Zhang, F.; Xie, X.; Guo, M. DKN: Deep knowledge-aware network for news recommendation. In Proceedings of the Proceedings of the 2018 World Wide Web Conference, 2018, pp. 1835–1844.
- Peng, Y.; Yan, S.; Lu, Z. Transfer learning in biomedical natural language processing: An evaluation of BERT and ELMo on ten benchmarking datasets. Bioinformatics 2019, 35, 3055–3062. [Google Scholar]
- Chen, L.; Zhang, Y.; Wang, H. DistilBERT-KG: Lightweight Knowledge Graph-Aware Language Model for Biomedical Relation Extraction. IEEE Transactions on Computational Biology and Bioinformatics 2023. [Google Scholar] [CrossRef]
- Yan, W.; Hu, J.; Sun, S.; Zhang, L. AutoKG: Towards Automated Construction and Refinement of Domain-Specific Biomedical Knowledge Graphs. In Proceedings of the Proceedings of the 2024 Annual Conference of the Association for Computational Linguistics (ACL); 2024. [Google Scholar]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).