Submitted:
29 July 2025
Posted:
30 July 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Vaccine Technologies and Platforms
2.1. Anti-Viral Vaccines
- i.
- Protein Platform
- 1.
- Whole Inactivated (killed) vaccines
- 2.
- Live Attenuated Vaccines
- 3.
- Virus-like Particles (VLP) Vaccines
- 4.
- Synthetic Peptide Vaccines (SPV)
- ii.
- Nucleic Acid Platforms (NAP)
- Bacterial and Viral Vectored Vaccines
- 2.
- Synthetic DNA Vaccines (SDNAV)
- 3.
- mRNA based Vaccines (mRNAV)
2.2. Anti-Cancer Vaccines (ACV)
ACV Technologies and Platforms
- Tumor Antigens
- (a)
- Neoantigens
- (b) Shared antigens
- 2.
- ACV Platforms
- 3.
- In-situ Vaccines (ISV)
- 4.
- Combined Vaccination
2.3. Adjuvants
3. The Oncogenic Virus Orchestra, Tumors and Vaccines
3.1. Human Papilloma Virus (HPV)
3.1.1. Virology and Oncogenesis
3.1.2. HPV-Associated Cancers
3.1.3. Vaccines
- Prophylactic Vaccines (PV)
- 2.
- Therapeutic Vaccines (TV)
3.1.4. The Viral Orchestra
- The duplet HPV and HIV
- 2.
- The duplet HPV and EBV
- 3.
- The trios HPV, HIV and EBV
3.2. Epstein-Barr Virus (EBV)
3.2.1. Virology and Oncogenesis
3.2.2. EBV-Associated Cancers
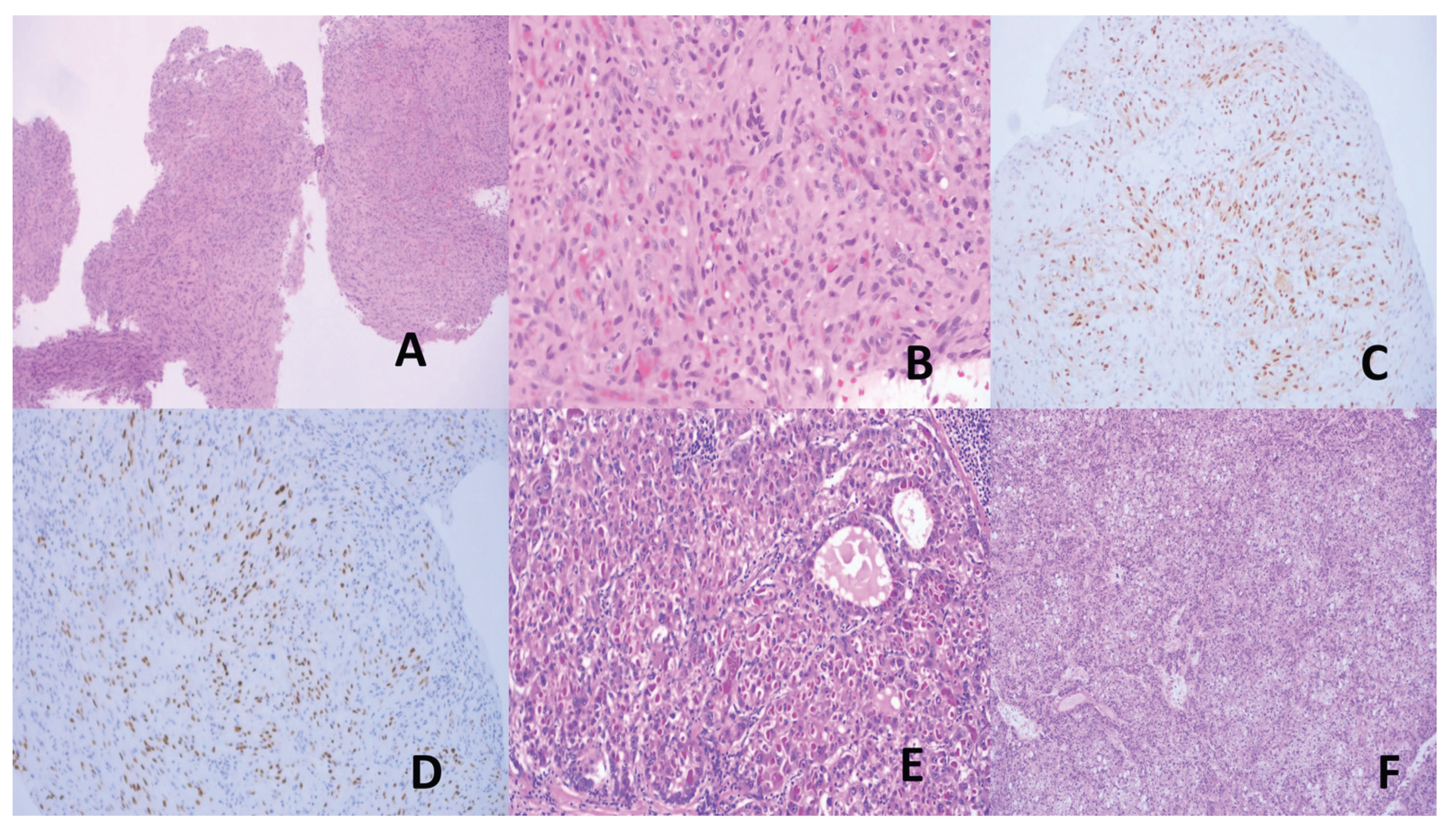
- Nasopharyngeal Carcinoma (NPC)
- 2.
- Gastric Cancer (GC)
- 3.
- Lymphoepithelial carcinoma of the lung (LEC)
- 4.
- Thymus Epithelial Tumors (TET)
- 5.
- Burkitt Lymphoma (BL)
- 6.
- Hodgkin Lymphoma (HL)
- 7.
- Extranodal NK/T –cell lymphoma (ENNKTL)
- 8.
- EBV+ Nodal T- and NK-cell Lymphoma (NTNKL)
- 9.
- Systemic EBV+ T-cell Lymphoma of Childhood (SEBVTCLC)
- 10.
- Aggressive NK-cell Leukemia (ANKL)
- 11.
- EBV+ Inflammatory Folicular Dendritic Cell Sarcoma (IFDCS)
- 12.
- Other EBV-associated lymphoproliferative diseases (LPD)
3.2.3. EBV Vaccines
- Prophylactic Vaccine
- 2.
- Therapeutic Vaccine
3.2.4. The Viral Orchestra
- The duplet EBV and HPV
- 2.
- The duplet EBV and Kaposi sarcoma-associated herpes virus (KSHV)
- 3.
- The duplet EBV and HIV
3.3. Kaposi Sarcoma-Associated Herpes Virus (KSHV)
3.3.1. Virology and Oncogenesis
3.3.2. KSHV-Associated Cancers
- Kaposi Sarcoma (KS)
- 2.
- Primary Effusion Lymphoma (PEL)
- 3.
- KSHV-positive Diffuse Large B-cell Lymphoma (DLBL)
3.3.3. KSHV Vaccines
3.3.4. The Viral Orchestra
- The duplet KSHV and HIV
- 2.
- The duplet HSHV and EBV
- 3.
- The trios KSHV, HIV and EBV
3.4. Human Immunodeficiency Virus (HIV)
3.4.1. Virology and Role in Oncogenesis
3.4.2. HIV Vaccines
3.4.3. The Viral Orchestra
3.5. Hepatitis Viruses
3.5.1. Virology and Oncogenesis
- Hepatitis B (HBV)
- 2.
- Hepatitis C (HCV)
3.5.2. Hepatitis Virus – Associated Cancers
3.5.3. Hepatitis Virus Vaccines
- HBV
- 2.
- HCV
3.5.4. The Viral Orchestra
- The Duplet HBV and HIV or HCV and HIV
- 2.
- The Duplet HBV and HCV
- 3.
- The trios
3.6. The Merkel Cell Polyoma Virus (McPyV)
3.6.1. Virology and Oncogenesis
3.6.2. The McPyV-Associated Cancers
3.6.3. McPyV Vaccines
3.6.4. The Viral Orchestra
3.7. The Human T-Cell Leukemia Virus Type-1 (HTLV-1)/Human Tlymphotropic Virus
3.7.1. Virology and Oncogenesis
3.7.2. HTLV-1-Associated Cancer
3.7.3. HTLV-1 Vaccine
3.8. SARS-Cov-2 Virus
3.8.1. Virology and Possible Oncogenetic Mechanisms
3.8.2. Vaccines
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Viruses | Oncogenicity | Cancers Caused | Cancer promoting products | Vaccines |
|---|---|---|---|---|
| HPV | Yes | ● Uterine cervical SCC ● Vaginal, vulval, penile, anal, oropharyngeal SCC ● Urinary bladder |
E2, E4, E5,E6, E7 | ● Gardasil (bi-,quadri-, nona-valent) ● Cervarix (bivalent) |
| EBV | Yes | ● Burkitt lymphoma ● Hodgkin lymphoma ● ENNKTL ● NPC ● GC ● LEC, lung and thymus ● IFDCS ● ANKL ● EBV+DLBL ● EBV+ nodal T and NK lymphoma ● SEBVTCLC |
● Latency products: LMP1, LMP2, EBNA1, EBNA2, EBER, microRNA ● Lytic products: BZL1, BHRF1, BALF1, vbcl2, BZLF1, BRLF1, BLLF3, microRNA (BART, BHRF1-2) |
No available vaccine |
| KSHV | Yes | ● KS ● PEL |
LANA, Kaposin, v-cyclin, vFLIP, vGPCR, K1, K15, vIL-6, microRNA | No available vaccine |
| HBV | Yes | Hepatocellular carcinoma (HCC) | HBx | ● Energix-B ● Recombivax HB ● HEPLISAV-B# ● PREHEVBRIO ● Twinrix# ● Pediarix# ● Vaxelis# |
| HCV | Yes | HCC | Core, NS3, NS4, NS5 | No available vaccine |
| McPyV | Yes | Merkel cell carcinoma | Small T, truncated large T | No available vaccine |
| HTLV-1 | Yes | Adult T-cell leukemia/lymphoma | Tax, HBZ | No available vaccine |
References
- Gaglia, M.M.; Munger, K. More than just oncogenes: mechanisms of tumorigenesis by human viruses. Curr. Opin. Virol. 2018, 32, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.S. From the midfacial destructive drama to the unfolding EBV story: a short history of EBV-positive NK-cell and T-cell lymphoproliferative diseases. Pathology 2024, 56, 773–785. [Google Scholar] [CrossRef] [PubMed]
- Mahdavi, P.; Javaherdehi, A.P.; Khanjanpoor, P.; Aminian, H.; Zakeri, M.; Zafarani, A.; Razizadeh, M.H. The role of c-Myc in Epstein-Barr virus-associated cancers: Mechanistic insights and therapeutic implications. Microb. Pathog. 2024, 197, 107025. [Google Scholar] [CrossRef]
- Allday, M.J. How does Epstein–Barr virus (EBV) complement the activation of Myc in the pathogenesis of Burkitt's lymphoma? Semin. Cancer Biol. 2009, 19, 366–376. [Google Scholar] [CrossRef]
- Moore, P.S.; Chang, Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat. Rev. Cancer 2010, 10, 878–889. [Google Scholar] [CrossRef]
- Park, N.H.; Chung, Y.-H.; Lee, H.-S. Impacts of Vaccination on Hepatitis B Viral Infections in Korea over a 25-Year Period. Intervirology 2010, 53, 20–28. [Google Scholar] [CrossRef]
- Schiller, J.T.; Lowy, D.R. Understanding and learning from the success of prophylactic human papillomavirus vaccines. Nat. Rev. Microbiol. 2012, 10, 681–692. [Google Scholar] [CrossRef]
- Cohen, J.I.; Fauci, A.S.; Varmus, H.; Nabel, G.J. Epstein-Barr Virus: An Important Vaccine Target for Cancer Prevention. Sci. Transl. Med. 2011, 3, 107fs7–107fs7. [Google Scholar] [CrossRef]
- Casper, C.; Corey, L.; Cohen, J.I.; Damania, B.; Gershon, A.A.; Kaslow, D.C.; Krug, L.T.; Martin, J.; Mbulaiteye, S.M.; Mocarski, E.S.; et al. KSHV (HHV8) vaccine: promises and potential pitfalls for a new anti-cancer vaccine. npj Vaccines 2022, 7, 1–10. [Google Scholar] [CrossRef]
- Ghattas, M.; Dwivedi, G.; Lavertu, M.; Alameh, M.-G. Vaccine Technologies and Platforms for Infectious Diseases: Current Progress, Challenges, and Opportunities. Vaccines 2021, 9, 1490. [Google Scholar] [CrossRef]
- Saxena, M.; van der Burg, S.H.; Melief, C.J.M.; Bhardwaj, N. Therapeutic cancer vaccines. Nat. Rev. Cancer 2021, 21, 360–378. [Google Scholar] [CrossRef] [PubMed]
- Bayani, F.; Hashkavaei, N.S.; Arjmand, S.; Rezaei, S.; Uskoković, V.; Alijanianzadeh, M.; Uversky, V.N.; Siadat, S.O.R.; Mozaffari-Jovin, S.; Sefidbakht, Y. An overview of the vaccine platforms to combat COVID-19 with a focus on the subunit vaccines. Prog. Biophys. Mol. Biol. 2023, 178, 32–49. [Google Scholar] [CrossRef] [PubMed]
- Folorunso, OS; Sebolai, OM. Overview of the Development, Impacts and Challenges of Live-attenuated Oral Rotavirus Vaccines. Vaccines 2020, 8, 341.
- Vignuzzi, M.; Wendt, E.; Andino, R. Engineering attenuated virus vaccines by controlling replication fidelity. Nat. Med. 2008, 14, 154–161. [Google Scholar] [CrossRef]
- Groenke, N.; Trimpert, J.; Merz, S.; Conradie, A.M.; Wyler, E.; Zhang, H.; Hazapis, O.-G.; Rausch, S.; Landthaler, M.; Osterrieder, N.; et al. Mechanism of Virus Attenuation by Codon Pair Deoptimization. Cell Rep. 2020, 31, 107586. [Google Scholar] [CrossRef]
- Mak, TW; Saunders, ME. Vaccines and Clinical Immunization. In: The Immune Response. Mak, TW; Saunders, ME (Eds). Academic Press, Burlington, MA, USA. 2006, 695-749.
- Fehlner-Gardiner, C.; Nadin-Davis, S.; Armstrong, J.; Muldoon, F.; Bachmann, P.; Wandeler, A. ERA VACCINE-DERIVED CASES OF RABIES IN WILDLIFE AND DOMESTIC ANIMALS IN ONTARIO, CANADA, 1989–2004. J. Wildl. Dis. 2008, 44, 71–85. [Google Scholar] [CrossRef]
- Frederiksen, L.S.F.; Zhang, Y.; Foged, C.; Thakur, A. The Long Road Toward COVID-19 Herd Immunity: Vaccine Platform Technologies and Mass Immunization Strategies. Front. Immunol. 2020, 11, 1817. [Google Scholar] [CrossRef]
- Syomin, B.V.; Ilyin, Y.V. Virus-Like Particles as an Instrument of Vaccine Production. Mol. Biol. 2019, 53, 323–334. [Google Scholar] [CrossRef]
- Cimica, V.; Galarza, J.M. Adjuvant formulations for virus-like particle (VLP) based vaccines. Clin. Immunol. 2017, 183, 99–108. [Google Scholar] [CrossRef]
- Lua, L.H.L.; Connors, N.K.; Sainsbury, F.; Chuan, Y.P.; Wibowo, N.; Middelberg, A.P.J. Bioengineering virus-like particles as vaccines. Biotechnol. Bioeng. 2014, 111, 425–440. [Google Scholar] [CrossRef]
- da Silva, A.J.; Zangirolami, T.C.; Novo-Mansur, M.T.M.; Giordano, R.d.C.; Martins, E.A.L. Live bacterial vaccine vectors: An overview. Braz. J. Microbiol. 2014, 45, 1117–1129. [Google Scholar] [CrossRef]
- Humphreys, I.R.; Sebastian, S. Novel viral vectors in infectious diseases. Immunology 2018, 153, 1–9. [Google Scholar] [CrossRef]
- Ertl, H.C. Viral vectors as vaccine carriers. Curr. Opin. Virol. 2016, 21, 1–8. [Google Scholar] [CrossRef]
- Lambricht, L.; Lopes, A.; Kos, S.; Sersa, G.; Préat, V.; Vandermeulen, G. Clinical potential of electroporation for gene therapy and DNA vaccine delivery. Expert Opin. Drug Deliv. 2015, 13, 295–310. [Google Scholar] [CrossRef]
- Wang, Z.; Troilo, P.J.; Wang, X.; Griffiths, T.G.; Pacchione, S.J.; Barnum, A.B.; Harper, L.B.; Pauley, C.J.; Niu, Z.; Denisova, L.; et al. Detection of integration of plasmid DNA into host genomic DNA following intramuscular injection and electroporation. Gene Ther. 2004, 11, 711–721. [Google Scholar] [CrossRef]
- Alameh, M-G; Weissman, D; Pardi, N. Messenger RNA-based Vaccines against Infectious Diseases. Springer, Berlin/Heidelberg, Germany, 2020,1-35.
- Andries, O.; Mc Cafferty, S.; De Smedt, S.C.; Weiss, R.; Sanders, N.N.; Kitada, T. N1-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release 2015, 217, 337–344. [Google Scholar] [CrossRef]
- Chaudhary, N; Weissman, D; Whitehead, KA. mRNA Vaccines for Infectious Diseases. Principles, Delivery and Clinical Translation. Nat Rev Drug Discov 2021, 20, 817–838.
- Magini, D.; Giovani, C.; Mangiavacchi, S.; Maccari, S.; Cecchi, R.; Ulmer, J.B.; De Gregorio, E.; Geall, A.J.; Brazzoli, M.; Bertholet, S.; et al. Self-Amplifying mRNA Vaccines Expressing Multiple Conserved Influenza Antigens Confer Protection against Homologous and Heterosubtypic Viral Challenge. PLOS ONE 2016, 11, e0161193. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Alameh, M-G; Tombacz, I; Bettini, E; Lederer, K; Sittplangkoon, C; Wilmore, JR; Gaudette, BT; Soliman, OY; Pine, M; Hicks, P, et al. Lipid Nanoparticles Enhance the Efficacy of mRNA and Protein Subunit Vaccines by Inducing Robust T Follicular Helper Cell and Humoral Responses. Immunity 2021, 54, 2877–2892.
- Verdegaal, E.M.E.; de Miranda, N.F.C.C.; Visser, M.; Harryvan, T.; van Buuren, M.M.; Andersen, R.S.; Hadrup, S.R.; van der Minne, C.E.; Schotte, R.; Spits, H.; et al. Neoantigen landscape dynamics during human melanoma–T cell interactions. Nature 2016, 536, 91–95. [Google Scholar] [CrossRef]
- Sade-Feldman, M.; Jiao, Y.J.; Chen, J.H.; Rooney, M.S.; Barzily-Rokni, M.; Eliane, J.-P.; Bjorgaard, S.L.; Hammond, M.R.; Vitzthum, H.; Blackmon, S.M.; et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat. Commun. 2017, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hammerich, L; Binder, A; Brody, JD. In situ Vaccination: Cancer Immunotherapy both Personalized and Off-the-shelf. Mol Oncol 2015, 9, 1966–1981. [CrossRef]
- Mariathasan, S.; Turley, S.J.; Nickles, D.; Castiglioni, A.; Yuen, K.; Wang, Y.; Kadel, E.E., III; Koeppen, H.; Astarita, J.L.; Cubas, R.; et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018, 554, 544–548. [Google Scholar] [CrossRef] [PubMed]
- Oweida, A.; Hararah, M.K.; Phan, A.; Binder, D.; Bhatia, S.; Lennon, S.; Bukkapatnam, S.; Van Court, B.; Uyanga, N.; Darragh, L.; et al. Resistance to Radiotherapy and PD-L1 Blockade Is Mediated by TIM-3 Upregulation and Regulatory T-Cell Infiltration. Clin. Cancer Res. 2018, 24, 5368–5380. [Google Scholar] [CrossRef]
- Smith, C.C.; Selitsky, S.R.; Chai, S.; Armistead, P.M.; Vincent, B.G.; Serody, J.S. Alternative tumour-specific antigens. Nat. Rev. Cancer 2019, 19, 465–478. [Google Scholar] [CrossRef]
- Roudko, V.; Bozkus, C.C.; Orfanelli, T.; McClain, C.B.; Carr, C.; O’dOnnell, T.; Chakraborty, L.; Samstein, R.; Huang, K.-L.; Blank, S.V.; et al. Shared Immunogenic Poly-Epitope Frameshift Mutations in Microsatellite Unstable Tumors. Cell 2020, 183, 1634–1649.e17. [Google Scholar] [CrossRef]
- Engelhard, V.H.; Obeng, R.C.; Cummings, K.L.; Petroni, G.R.; Ambakhutwala, A.L.; A Chianese-Bullock, K.; Smith, K.T.; Lulu, A.; Varhegyi, N.; E Smolkin, M.; et al. MHC-restricted phosphopeptide antigens: preclinical validation and first-in-humans clinical trial in participants with high-risk melanoma. J. Immunother. Cancer 2020, 8, e000262. [Google Scholar] [CrossRef]
- Brentville, V.; Vankemmelbeke, M.; Metheringham, R.; Durrant, L. Post-translational modifications such as citrullination are excellent targets for cancer therapy. Semin. Immunol. 2020, 47, 101393. [Google Scholar] [CrossRef]
- Yarchoan, M.; Albacker, L.A.; Hopkins, A.C.; Montesion, M.; Murugesan, K.; Vithayathil, T.T.; Zaidi, N.; Azad, N.S.; Laheru, D.A.; Frampton, G.M.; et al. PD-L1 expression and tumor mutational burden are independent biomarkers in most cancers. J. Clin. Investig. 2019, 4. [Google Scholar] [CrossRef]
- Bulk, J.v.D.; Verdegaal, E.M.E.; Ruano, D.; Ijsselsteijn, M.E.; Visser, M.; van der Breggen, R.; Duhen, T.; van der Ploeg, M.; de Vries, N.L.; Oosting, J.; et al. Neoantigen-specific immunity in low mutation burden colorectal cancers of the consensus molecular subtype 4. Genome Med. 2019, 11, 1–15. [Google Scholar] [CrossRef]
- Sahin, U.; Oehm, P.; Derhovanessian, E.; Jabulowsky, R.A.; Vormehr, M.; Gold, M.; Maurus, D.; Schwarck-Kokarakis, D.; Kuhn, A.N.; Omokoko, T.; et al. An RNA vaccine drives immunity in checkpoint-inhibitor-treated melanoma. Nature 2020, 585, 107–112. [Google Scholar] [CrossRef]
- Van der Burg, SH; Arens, R; Ossendorp, F; van Hall, T; Mielief, CJ. Vaccines foe Established Cancer: Overcomin the Challenges Posed by Immune Evasion. Nat Rev Cancer 2016, 16, 219–233.
- Melief, CJ, van Hall, T; Arens, R; Ossendorp, F; van der Burg, SH. Therapeutic Cancer Vaccines. J Clin Invest 2015, 125, 3401–3412.
- Baharom, F.; Ramirez-Valdez, R.A.; Tobin, K.K.S.; Yamane, H.; Dutertre, C.-A.; Khalilnezhad, A.; Reynoso, G.V.; Coble, V.L.; Lynn, G.M.; Mulè, M.P.; et al. Intravenous nanoparticle vaccination generates stem-like TCF1+ neoantigen-specific CD8+ T cells. Nat. Immunol. 2020, 22, 41–52. [Google Scholar] [CrossRef] [PubMed]
- van der Maaden, K.; Heuts, J.; Camps, M.; Pontier, M.; van Scheltinga, A.T.; Jiskoot, W.; Ossendorp, F.; Bouwstra, J. Hollow microneedle-mediated micro-injections of a liposomal HPV E743–63 synthetic long peptide vaccine for efficient induction of cytotoxic and T-helper responses. J. Control. Release 2018, 269, 347–354. [Google Scholar] [CrossRef]
- Li, A.W.; Sobral, M.C.; Badrinath, S.; Choi, Y.; Graveline, A.; Stafford, A.G.; Weaver, J.C.; Dellacherie, M.O.; Shih, T.-Y.; Ali, O.A.; et al. A facile approach to enhance antigen response for personalized cancer vaccination. Nat. Mater. 2018, 17, 528–534. [Google Scholar] [CrossRef]
- Meshii, N.; Takahashi, G.; Okunaga, S.; Hamada, M.; Iwai, S.; Takasu, A.; Ogawa, Y.; Yura, Y. Enhancement of systemic tumor immunity for squamous cell carcinoma cells by an oncolytic herpes simplex virus. Cancer Gene Ther. 2013, 20, 493–498. [Google Scholar] [CrossRef]
- Cerullo, V; Diaconu, J; Romano, V; Hirvinen, M; Ugolini, M; Escutenaire, S; Holm, S_L; Kipar, A; Kanerva, A; Hemminki, A. An Oncolytic Adenovirus Enhanced for Toll-like Receptor 9 Stimulation Increases Antitumor Clearance. Mol Ther 2012, 20, 2076–2086. [CrossRef]
- Motoyoshi, Y.; Kaminoda, K.; Saitoh, O.; Hamasaki, K.; Nakao, K.; Ishii, N.; Nagayama, Y.; Eguchi, K. Different mechanisms for anti-tumor effects of low- and high-dose cyclophosphamide. Oncol. Rep. 2006, 16, 141–146. [Google Scholar] [CrossRef]
- Cerullo, V.; Diaconu, I.; Kangasniemi, L.; Rajecki, M.; Escutenaire, S.; Koski, A.; Romano, V.; Rouvinen, N.; Tuuminen, T.; Laasonen, L.; et al. Immunological Effects of Low-dose Cyclophosphamide in Cancer Patients Treated With Oncolytic Adenovirus. Mol. Ther. 2011, 19, 1737–1746. [Google Scholar] [CrossRef] [PubMed]
- Banissi, C.; Ghiringhelli, F.; Chen, L.; Carpentier, A.F. Treg depletion with a low-dose metronomic temozolomide regimen in a rat glioma model. Cancer Immunol. Immunother. 2009, 58, 1627–1634. [Google Scholar] [CrossRef] [PubMed]
- Zamarin, D.; Holmgaard, R.B.; Subudhi, S.K.; Park, J.S.; Mansour, M.; Palese, P.; Merghoub, T.; Wolchok, J.D.; Allison, J.P. Localized Oncolytic Virotherapy Overcomes Systemic Tumor Resistance to Immune Checkpoint Blockade Immunotherapy. Sci. Transl. Med. 2014, 6, 226ra32–226ra32. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Cai, Y.; Jiang, Y.; He, X.; Wei, Y.; Yu, Y.; Tian, X. Vaccine adjuvants: mechanisms and platforms. Signal Transduct. Target. Ther. 2023, 8, 1–24. [Google Scholar] [CrossRef]
- Coffman, R.L.; Sher, A.; Seder, R.A. Vaccine Adjuvants: Putting Innate Immunity to Work. Immunity 2010, 33, 492–503. [Google Scholar] [CrossRef]
- Xie, C.; Yao, R.; Xia, X. The advances of adjuvants in mRNA vaccines. npj Vaccines 2023, 8, 1–6. [Google Scholar] [CrossRef]
- Parkin, D.M. The global health burden of infection-associated cancers in the year 2002. Int. J. Cancer 2006, 118, 3030–3044. [Google Scholar] [CrossRef]
- Morales-Sánchez, A.; Fuentes-Pananá, E.M. Human Viruses and Cancer. Viruses 2014, 6, 4047–4079. [Google Scholar] [CrossRef]
- De Martel, C; Georges, D; Bray, F’ Ferlay, J; Clifford, GM. Global Burden of Cancer Attributable to Infections in 2018: A Worldwide Incidence Analysis. Lancet Glob Health 2020, 8, e180–e190.
- Molina, M.A.; Steenbergen, R.D.; Pumpe, A.; Kenyon, A.N.; Melchers, W.J. HPV integration and cervical cancer: a failed evolutionary viral trait. Trends Mol. Med. 2024, 30, 890–902. [Google Scholar] [CrossRef]
- WHO Vaccine Position Papers. World Health Prganization. Geneva (www.who.int/teams/immunization-vaccines-and biologicals/policies/position papers, accessed August 2022).
- Cancer Genome Atlas Research Network, Albert Einstein College of Medicine; Analytic Biological services, Barretos Cancer Hospital, Baylor College of Medicine, City of Hope; Buck Institute of research on Aging; Canada’s Michael Smith Genome Science Centre; Harvard Medical School; Helen F Graham Cancer & Research Institute at Christiana Care Health Services; Hudson Alpha Institute for Biotechnology, IL Sbio, LLC, Indiana University School of Medicine; Institute of Human Virology, Institute for System Biology, International Genomics Consortium, Leidos Biomedical, Massachusetts General Hospital; McDonnell Denome Institute at Washington University, Medical College of Wisconsin, et al. Integrated Genomic and Molecular Characterization of Cervical Cancer. Nature 2017,543,378-384. Doi:10.1038/nature21386.
- Ren, S.; Gaykalova, D.A.; Guo, T.; Favorov, A.V.; Fertig, E.J.; Tamayo, P.; Callejas-Valera, J.L.; Allevato, M.; Gilardi, M.; Santos, J.; et al. HPV E2, E4, E5 drive alternative carcinogenic pathways in HPV positive cancers. Oncogene 2020, 39, 6327–6339. [Google Scholar] [CrossRef]
- Maehama, T.; Nishio, M.; Otani, J.; Mak, T.W.; Suzuki, A. The role of Hippo-YAP signaling in squamous cell carcinomas. Cancer Sci. 2020, 112, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Bossler, F.; Hoppe-Seyler, K.; Hoppe-Seyler, F. PI3K/AKT/mTOR Signaling Regulates the Virus/Host Cell Crosstalk in HPV-Positive Cervical Cancer Cells. Int. J. Mol. Sci. 2019, 20, 2188. [Google Scholar] [CrossRef] [PubMed]
- von Knebel Doeberitz, M.; Prigge, E.-S. Role of DNA methylation in HPV associated lesions. Papillomavirus Res. 2019, 7, 180–183. [Google Scholar] [CrossRef] [PubMed]
- Saco, A; Mills, AM; Park, KJ; Focchi, GRA; Carrhilho, C; Regauer, S; Kong, CS. Squamous Cell Carcinoma. HPV-associated, of the Uterine Cervix. In: WHO Classification of Female Genital Tumours, 5th edition, 2020, pp.347-349; Herrington, CS; Ordi, J (eds). Lyons, France, IARC.
- Saco, A; Mills, AM; Park, KJ; Focchi, GRA; Carrhilho, C; Regauer, S; Kong, CS. Squamous Cell Carcinoma, HPV-independent, of the Uterine Cervix. In: WHO Classification of Female Genital Tumours, 5th edition, 2020, p.350. Herrington, CS; Ordi, J (eds). Lyons, France, IARC.
- Lam, E.W.; Chan, J.Y.-W.; Chan, A.B.; Ng, C.S.; Lo, S.T.; Lam, V.S.; Chan, M.M.; Ngai, C.M.; Vlantis, A.; Ma, R.K.; et al. Prevalence, Clinicopathological Characteristics, and Outcome of Human Papillomavirus–Associated Oropharyngeal Cancer in Southern Chinese Patients. Cancer Epidemiol. Biomark. Prev. 2016, 25, 165–173. [Google Scholar] [CrossRef]
- Lei, J; Ploner, A; Elfstrom, M; Wang, J; Roth, A; Fang, F’ Sundstrom, K; Dillner, J. HPV Vaccination and the Risk of Invasive Cervical Cancer. N Eng J Med 2020, 383, 1340–1348. [CrossRef]
- Falcaro, M.; Soldan, K.; Ndlela, B.; Sasieni, P. Effect of the HPV vaccination programme on incidence of cervical cancer and grade 3 cervical intraepithelial neoplasia by socioeconomic deprivation in England: population based observational study. BMJ 2024, 385, e077341. [Google Scholar] [CrossRef]
- Suksanpaisan, L; Xu, T; Tesfay, MZ; Bomidi, C; Hamm, S; Vandergaast, R; Jenks, N; Steele, MB; Ota-Setlik, A; Akhtar, H, et al. Preclinical Development of Oncolytic Immunotherapy for Treatment of HPV-positive Cancers. Mol Ther Oncolytics 2018, 10, 1–13.
- Bushara, O.; Krogh, K.; Weinberg, S.E.; Finkelman, B.S.; Sun, L.; Liao, J.; Yang, G.-Y. Human Immunodeficiency Virus Infection Promotes Human Papillomavirus-Mediated Anal Squamous Carcinogenesis: An Immunologic and Pathobiologic Review. Pathobiology 2021, 89, 1–12. [Google Scholar] [CrossRef]
- Liu, G.; Sharma, M.; Tan, N.; Barnabas, R.V. HIV-positive women have higher risk of human papilloma virus infection, precancerous lesions, and cervical cancer. AIDS 2018, 32, 795–808. [Google Scholar] [CrossRef]
- Palefsky, J.M. Human papillomavirus-associated anal and cervical cancers in HIV-infected individuals. Curr. Opin. HIV AIDS 2017, 12, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Lien, K.; Mayer, W.; Herrera, R.; Padilla, N.T.; Cai, X.; Lin, V.; Pholcharoenchit, R.; Palefsky, J.; Tugizov, S.M.; Sinclair, A. HIV-1 Proteins gp120 and Tat Promote Epithelial-Mesenchymal Transition and Invasiveness of HPV-Positive and HPV-Negative Neoplastic Genital and Oral Epithelial Cells. Microbiol. Spectr. 2022, 10, e0362222. [Google Scholar] [CrossRef] [PubMed]
- Isaguliants, M.; Bayurova, E.; Avdoshina, D.; Kondrashova, A.; Chiodi, F.; Palefsky, J.M. Oncogenic Effects of HIV-1 Proteins, Mechanisms Behind. Cancers 2021, 13, 305. [Google Scholar] [CrossRef] [PubMed]
- Gordon, K.J.; Blobe, G.C. Role of transforming growth factor-β superfamily signaling pathways in human disease. Biochim. et Biophys. Acta (BBA) - Mol. Basis Dis. 2008, 1782, 197–228. [Google Scholar] [CrossRef]
- Glauser, D.A.; Schlegel, W. Sequential actions of ERK1/2 on the AP-1 transcription factor allow temporal integration of metabolic signals in pancreatic β cells. FASEB J. 2007, 21, 3240–3249. [Google Scholar] [CrossRef]
- Di Mascio, M.; Srinivasula, S.; Bhattacharjee, A.; Cheng, L.; Martiniova, L.; Herscovitch, P.; Lertora, J.; Kiesewetter, D. Antiretroviral Tissue Kinetics: In Vivo Imaging Using Positron Emission Tomography. Antimicrob. Agents Chemother. 2009, 53, 4086–4095. [Google Scholar] [CrossRef]
- Boomgarden, A.C.; Upadhyay, C. Progress and Challenges in HIV-1 Vaccine Research: A Comprehensive Overview. Vaccines 2025, 13, 148. [Google Scholar] [CrossRef]
- Silver, M.I.; Paul, P.; Sowjanya, P.; Ramakrishna, G.; Vedantham, H.; Kalpana, B.; Shah, K.V.; Gravitt, P.E. Shedding of Epstein-Barr Virus and Cytomegalovirus from the Genital Tract of Women in a Periurban Community in Andhra Pradesh, India. J. Clin. Microbiol. 2011, 49, 2435–2439. [Google Scholar] [CrossRef]
- Kahla, S.; Oueslati, S.; Achour, M.; Kochbati, L.; Chanoufi, M.B.; Maalej, M.; Oueslati, R. Correlation between ebv co-infection and HPV16 genome integrity in Tunisian cervical cancer patients. Braz. J. Microbiol. 2012, 43, 744–753. [Google Scholar] [CrossRef]
- de Lima, M.A.P.; Neto, P.J.N.; Lima, L.P.M.; Júnior, J.G.; Junior, A.G.T.; Teodoro, I.P.P.; Facundo, H.T.; da Silva, C.G.L.; Lima, M.V.A. Association between Epstein-Barr virus (EBV) and cervical carcinoma: A meta-analysis. Gynecol. Oncol. 2018, 148, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Blanco, R.; Carrillo-Beltrán, D.; Osorio, J.C.; Calaf, G.M.; Aguayo, F. Role of Epstein-Barr Virus and Human Papillomavirus Coinfection in Cervical Cancer: Epidemiology, Mechanisms and Perspectives. Pathogens 2020, 9, 685. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, F.; Rasizadeh, R.; Sharaflou, S.; Aghbash, P.S.; Shamekh, A.; Jafari-Sales, A.; Baghi, H.B. Coinfection of EBV with other pathogens: a narrative review. Front. A J. Women Stud. 2024, 4, 1482329. [Google Scholar] [CrossRef]
- Dillner, J; Lenner, P; Lehtinen, M; Eklundl, C; Heino, P; Wiklund, F; Hallmans, G; Stendahl, U. A Population-based Seroepidemiological Study of Cervical Cancer. Cancer Res 1994,54,134-141.
- Rodrigues, FR; Miranda, NL; Fonseca, EC; Pires, ARC; Dias, EP. Investigation of the LMPEBV and Co-infection by HPV in Genital Lesions of Patients Infected or Not by HIV. J Bras Patol Med Lab 2010, 46, 415–420.
- Kaiser, C.; Laux, G.; Eick, D.; Jochner, N.; Bornkamm, G.W.; Kempkes, B. The Proto-Oncogene c- myc Is a Direct Target Gene of Epstein-Barr Virus Nuclear Antigen 2. J. Virol. 1999, 73, 4481–4484. [Google Scholar] [CrossRef]
- Sausen, D.G.; Basith, A.; Muqeemuddin, S. EBV and Lymphomagenesis. Cancers 2023, 15, 2133. [Google Scholar] [CrossRef]
- Cui, X.; Snapper, C.M. Epstein Barr Virus: Development of Vaccines and Immune Cell Therapy for EBV-Associated Diseases. Front. Immunol. 2021, 12, 734471. [Google Scholar] [CrossRef]
- Glover, A.; Shannon-Lowe, C. From pathobiology to targeted treatment in Epstein Barr virus related T cell and Natural Killer cell lymphoproliferative diseases. Ann. Lymphoma 2021, 5, 31–31. [Google Scholar] [CrossRef]
- Yin, H.; Qu, J.; Peng, Q.; Gan, R. Molecular mechanisms of EBV-driven cell cycle progression and oncogenesis. Med Microbiol. Immunol. 2018, 208, 573–583. [Google Scholar] [CrossRef]
- Lurain, K.A.; Ramaswami, R.; Krug, L.T.; Whitby, D.; Ziegelbauer, J.M.; Wang, H.-W.; Yarchoan, R.; Forrest, G.N. HIV-associated cancers and lymphoproliferative disorders caused by Kaposi sarcoma herpesvirus and Epstein-Barr virus. Clin. Microbiol. Rev. 2024, 37, e0002223. [Google Scholar] [CrossRef]
- Nash, A.; Ryan, E.J. The oncogenic gamma herpesviruses Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV) hijack retinoic acid-inducible gene I (RIG-I) facilitating both viral and tumour immune evasion. Tumour Virus Res. 2022, 14, 200246. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Liu, Y.; Wang, C.; Gan, R. Signaling pathways of EBV-induced oncogenesis. Cancer Cell Int. 2021, 21, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Rosemarie, Q.; Sugden, B. Epstein–Barr Virus: How Its Lytic Phase Contributes to Oncogenesis. Microorganisms 2020, 8, 1824. [Google Scholar] [CrossRef] [PubMed]
- Cross, JR; Postigo, A; Blight, K; Downward, J. Viral Pro-survival Proteins Block Separate Stages in Bax Inactivation but Changes in Mitochondrial Ultrastructure Still Occur. Cell death Differ 2008, 15, 997.
- Fanidi, A.; Hancock, D.C.; Littlewood, T.D. Suppression of c-Myc-Induced Apoptosis by the Epstein-Barr Virus Gene Product BHRF1. J. Virol. 1998, 72, 8392–8395. [Google Scholar] [CrossRef]
- Chan, JKC; Peterson, BF; Bray, F; Lee, AWM; Rajadurai, P; Lo, KW. Nasopharyngeal Carcinoma. In: WHO Classification of Head and Neck Tumours 5th edition, 2023.
- Wei, K.-R.; Zheng, R.-S.; Zhang, S.-W.; Liang, Z.-H.; Li, Z.-M.; Chen, W.-Q. Nasopharyngeal carcinoma incidence and mortality in China, 2013. Chin. J. Cancer 2017, 36, 1–8. [Google Scholar] [CrossRef]
- Lin, J.-C.; Wang, W.-Y.; Chen, K.Y.; Wei, Y.-H.; Liang, W.-M.; Jan, J.-S.; Jiang, R.-S. Quantification of Plasma Epstein–Barr Virus DNA in Patients with Advanced Nasopharyngeal Carcinoma. New Engl. J. Med. 2004, 350, 2461–2470. [Google Scholar] [CrossRef]
- Su, W.-H.; Hildesheim, A.; Chang, Y.-S. Human Leukocyte Antigens and Epstein–Barr Virus-Associated Nasopharyngeal Carcinoma: Old Associations Offer New Clues into the Role of Immunity in Infection-Associated Cancers. Front. Oncol. 2013, 3, 299. [Google Scholar] [CrossRef]
- Bei, J.-X.; Li, Y.; Jia, W.-H.; Feng, B.-J.; Zhou, G.; Chen, L.-Z.; Feng, Q.-S.; Low, H.-Q.; Zhang, H.; He, F.; et al. A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nat. Genet. 2010, 42, 599–603. [Google Scholar] [CrossRef]
- Bruce, JP; To, KF; Lui, VEY; Chung, GTY; Chan, Y; Tsang, CM; Yip, KY; Ma, BBY; Woo, JKS; Hui, EP, et al. Whole-Genome Profiling of Nasopharyngeal Carcinoma Reveals Viral-Host Co-operation in Inflammatory NF-KB Activation and Immune Escape. Nat Commun 2021, 12, 4193. [CrossRef]
- Carneiro, F; Fukayama, M; Grabsch, HI; Yasui, W. Gastric Adenocarcinoma. In: WHO Classification of Digestive system Tumours, 5th edition, 2019, p.91. Lyons, France, IARC.
- Nishikawa, J.; Iizasa, H.; Yoshiyama, H.; Shimokuri, K.; Kobayashi, Y.; Sasaki, S.; Nakamura, M.; Yanai, H.; Sakai, K.; Suehiro, Y.; et al. Clinical Importance of Epstein–Barr Virus-Associated Gastric Cancer. Cancers 2018, 10, 167. [Google Scholar] [CrossRef]
- Saito, M.; Kono, K. Landscape of EBV-positive gastric cancer. Gastric Cancer 2021, 24, 983–989. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Cho, H.J.; Seo, J.; Park, K.B.; Kwon, Y.H.; Bae, H.I.; Na Seo, A.; Kim, M. Genetic landscape and PD-L1 expression in Epstein–Barr virus-associated gastric cancer according to the histological pattern. Sci. Rep. 2023, 13, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Salnikov, M.Y.; MacNeil, K.M.; Mymryk, J.S. The viral etiology of EBV-associated gastric cancers contributes to their unique pathology, clinical outcomes, treatment responses and immune landscape. Front. Immunol. 2024, 15, 1358511. [Google Scholar] [CrossRef] [PubMed]
- Saito, R; Abe, H; Kunita, A; Yamashita, H; Setoo, Y; Fukayama, M. Overexpression of PD-L1 in Cancer Cells and PD-L1+ Immune Cells in Epstein-Barr Virus-associated Gastric Cancer, the Prognostic Implications. Mod Pathol 2017, 30, 427–439. [CrossRef]
- Chou, T; Wong, P; Chang, Y. Lymphoepithelial Carcinoma of the Lung.In: WHO Classification of Thoracic Tumours, 5th edition, 2021, pp.94-96. Travis, WD; Chan, JKC (eds). Lyons, France, IARC.
- The, Y; Kao, H; Lee,K; Wu, M; Ho, H; Chou, T. Epstein-Barr Virus-associated pulmonary Carcinoma: Proposing an Alternative Term and Expanding the Histologic Spectrum of Lymphoepithelioma-like Carcinoma of the Lung. Am J Surg Pathol 2019, 43,211-219. [CrossRef]
- Chang, Y.-L.; Wu, C.-T.; Shih, J.-Y.; Lee, Y.-C. New Aspects in Clinicopathologic and Oncogene Studies of 23 Pulmonary Lymphoepithelioma-Like Carcinomas. Am. J. Surg. Pathol. 2002, 26, 715–723. [Google Scholar] [CrossRef]
- Zhao, L.; Ding, J.-Y.; Tao, Y.-L.; Zhu, K.; Chen, G. Detection of Epstein–Barr virus infection in thymic epithelial tumors by nested PCR and Epstein–Barr-encoded RNA ISH. Infect. Agents Cancer 2023, 18, 1–8. [Google Scholar] [CrossRef]
- Wu, T.-C.; Kuo, T.-T. Study of Epstein-Barr virus early RNA1 (EBER1) expression by in situ hybridization in thymic epithelial tumors of Chinese patients in Taiwan. Hum. Pathol. 1993, 24, 235–238. [Google Scholar] [CrossRef]
- Zhang, G.; Yu, Z.; Shen, G.; Chai, Y.; Liang, C. Association between Epstein-Barr virus and Thymic epithelial tumors: a systematic review. Infect. Agents Cancer 2019, 14, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chan, JKC, Chalabreysse, L; Mukai, K, Tateyama, H. Lymphoepithelial Carcinoma of the Thymus. In: WHO Classification of Thoracic Tumours, 5th edition, 2021, pp. 361-363. Lyons, France, IARC.
- Huang, J.; Ahmad, U.; Antonicelli, A.; Catlin, A.C.; Fang, W.; Gomez, D.; Loehrer, P.; Lucchi, M.; Marom, E.; Nicholson, A.; et al. Development of the International Thymic Malignancy Interest Group International Database: An Unprecedented Resource for the Study of a Rare Group of Tumors. J. Thorac. Oncol. 2014, 9, 1573–1578. [Google Scholar] [CrossRef] [PubMed]
- Sayed, S; Leoncini, L; Siebert, R; Ferry, JA; Meideiros, LJ; Cheuk, W, Klapper, W; d’Amore, ESG; Naresh, KN; Dave, SS, et al. Burkitt Lymphoma. In: WHO Classification of Hematolymphoid Tumours, 5th edition, 2024.
- Johnston, W.; Erdmann, F.; Newton, R.; Steliarova-Foucher, E.; Schüz, J.; Roman, E. Childhood cancer: Estimating regional and global incidence. Cancer Epidemiology 2021, 71, 101662. [Google Scholar] [CrossRef] [PubMed]
- Leoncini, L. Epstein–Barr virus positivity as a defining pathogenetic feature of Burkitt lymphoma subtypes. Br. J. Haematol. 2021, 196, 468–470. [Google Scholar] [CrossRef]
- Grande, B.M.; Gerhard, D.S.; Jiang, A.; Griner, N.B.; Abramson, J.S.; Alexander, T.B.; Allen, H.; Ayers, L.W.; Bethony, J.M.; Bhatia, K.; et al. Genome-wide discovery of somatic coding and noncoding mutations in pediatric endemic and sporadic Burkitt lymphoma. Blood 2019, 133, 1313–1324. [Google Scholar] [CrossRef]
- Robbiani, D.F.; Deroubaix, S.; Feldhahn, N.; Oliveira, T.Y.; Callen, E.; Wang, Q.; Jankovic, M.; Silva, I.T.; Rommel, P.C.; Bosque, D.; et al. Plasmodium Infection Promotes Genomic Instability and AID-Dependent B Cell Lymphoma. Cell 2015, 162, 727–737. [Google Scholar] [CrossRef]
- Ramaswami, R; Chia, G; Pria, AD; Pinato, DJ; Parker, K; Nelson, M; Bower, M. Evolution of HIV-associated Lymphoma over 3 Decades. J Acquir Immune Defic Synd 2016,72,177-183. [CrossRef]
- Mundo, L.; Del Porro, L.; Granai, M.; Siciliano, M.C.; Mancini, V.; Santi, R.; Marcar, L.; Vrzalikova, K.; Vergoni, F.; Di Stefano, G.; et al. Correction: Frequent traces of EBV infection in Hodgkin and non-Hodgkin lymphomas classified as EBV-negative by routine methods: expanding the landscape of EBV-related lymphomas. Mod. Pathol. 2020, 33, 2637–2637. [Google Scholar] [CrossRef]
- Ng, C.S.; Qin, J. New Facets of Hematolymphoid Eponymic Diseases. Lymphatics 2025, 3, 9. [Google Scholar] [CrossRef]
- Alsharif, R.; Dunleavy, K. Burkitt Lymphoma and Other High-Grade B-Cell Lymphomas with or without MYC, BCL2, and/or BCL6 Rearrangements. Hematol. Clin. North Am. 2019, 33, 587–596. [Google Scholar] [CrossRef]
- Borges, AM; Delabie, J; Vielh, P; d’Amore, ESG; Hebeda, KM; Diepstra, A; Naresh, KM; Garcia, JF; Tamarn, J; Laskar, S, et al. Classic Hodgkin Lymphoma. In: WHO Classification of Haematolymphoid Tumours, 5th edition, 2024.
- Kanzler, H.; Küppers, R.; Hansmann, M.L.; Rajewsky, K. Hodgkin and Reed-Sternberg cells in Hodgkin's disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J. Exp. Med. 1996, 184, 1495–1505. [Google Scholar] [CrossRef] [PubMed]
- Harabuchi, Y; Imai, S; Wakashima, J; Hirao, M; Katanra, A; Osato, T; Kon, S. Nasal T-cell Lymphoma Causally Associated with Epstein-Barr Virus: Clinicopathologic, Phenotypic and Genotypic Studies. Cancer 1996, 77, 2137–2149.
- Wang, H.; Fu, B.-B.; Gale, R.P.; Liang, Y. NK-/T-cell lymphomas. Leukemia 2021, 35, 2460–2468. [Google Scholar] [CrossRef] [PubMed]
- Au, W.-Y.; Pang, A.; Choy, C.; Chim, C.-S.; Kwong, Y.-L. Quantification of circulating Epstein-Barr virus (EBV) DNA in the diagnosis and monitoring of natural killer cell and EBV-positive lymphomas in immunocompetent patients. Blood 2004, 104, 243–249. [Google Scholar] [CrossRef]
- Wai, C.M.M.; Chen, S.; Phyu, T.; Fan, S.; Leong, S.M.; Zheng, W.; Low, L.C.Y.; Choo, S.-N.; Lee, C.-K.; Chung, T.-H.; et al. Immune pathway upregulation and lower genomic instability distinguish EBV-positive nodal T/NK-cell lymphoma from ENKTL and PTCL-NOS. Haematologica 2022, 107, 1864–1879. [Google Scholar] [CrossRef]
- Cohen, J.I.; Kimura, H.; Nakamura, S.; Ko, Y.-H.; Jaffe, E.S. Epstein–Barr virus-associated lymphoproliferative disease in non-immunocompromised hosts: a status report and summary of an international meeting, 8–9 September 2008. Ann. Oncol. 2009, 20, 1472–1482. [Google Scholar] [CrossRef]
- Tang, Y.-T.; Wang, D.; Luo, H.; Xiao, M.; Zhou, H.-S.; Liu, D.; Ling, S.-P.; Wang, N.; Hu, X.-L.; Luo, Y.; et al. Aggressive NK-cell leukemia: clinical subtypes, molecular features, and treatment outcomes. Blood Cancer J. 2017, 7, 1–5. [Google Scholar] [CrossRef]
- Jiang, X.-N.; Zhang, Y.; Xue, T.; Chen, J.-Y.; Chan, A.C.F.; Cheuk, W.F.; Chan, J.K.F.; Li, X.-Q. New Clinicopathologic Scenarios of EBV+ Inflammatory Follicular Dendritic Cell Sarcoma. Am. J. Surg. Pathol. 2020, 45, 765–772. [Google Scholar] [CrossRef]
- Cheuk, W.M.; Chan, J.K.C.M.; Shek, T.W.H.M.; Chang, J.H.; Tsou, M.-H.; Yuen, N.W.F.M.; Ng, W.-F.M.; Chan, A.C.L.M.; Prat, J. Inflammatory Pseudotumor-Like Follicular Dendritic Cell Tumor. Am. J. Surg. Pathol. 2001, 25, 721–731. [Google Scholar] [CrossRef]
- Li, Y.; Yang, X.; Tao, L.; Zeng, W.B.; Zuo, M.; Li, S.; Wu, L.B.; Lin, Y.; Zhang, Z.; Yun, J.; et al. Challenges in the Diagnosis of Epstein-Barr Virus-positive Inflammatory Follicular Dendritic Cell Sarcoma. Am. J. Surg. Pathol. 2022, 47, 476–489. [Google Scholar] [CrossRef]
- Natkunam, Y; Bhagat, G; Chadbum, A; Naresh, KN; Chan, J; Michelow, P; Satou, A; Satom Y; Bower, M; Gratzinger, D, et al. EBV-mucocutaneous Ulcer. In: WHO Classification of Haematolymphoid Tumours, 5th edition, 2024. deJong, D; Siebert, R; Alaggio, R; Coupland, SE (eds). Lyoms, France, IARC (available online from https://tumourclassification.iarc.who.int/chapters/63).
- Anagnostopoulos, I; Siebert, R; Nicholson, AG; Deckert, M. Lymphomatoid Granulomatosis. In: WHO Classification of Haematolymphoid Tumours, 5th edition, 2024. Oh, G; de jong, D (eds). Lyons, France, IARC (available online from https://tumourclassification.iarc.who.int/chapters/63).
- Anagnostopoulis, I; Medeiros, J; Klapper, W; de Jong, D; Miles, RR; Lenz, G; Asano, N; Chapman, JR; Steidi, C. EBV-positive Diffuse Large B-cell Lymphoma. In: WHO Classification of Haematolymphoid Tumours, 5th edition, 2024. Oh, G; Chan, J (eds). Lyons, france, IARC (available online from https://tumourclassification.iarc.who.int/chapters/63.).
- Montes-Morento, S; Leoncini, L; Miranda, R; Louissaint, A, Jr; sengar, M. Plasmablastic Lymphoma. In: WHO Classification of Haematolymphoid Tumours, 5th edition, 2024. Oh, G; de Jong, D; Dave, SS (eds).Lyons, France, IARC (available online from https://tumour classification.iarc.who.int/chapters/63).
- Cui, X.; Cao, Z.; Ishikawa, Y.; Cui, S.; Imadome, K.-I.; Snapper, C.M. Immunization with Epstein–Barr Virus Core Fusion Machinery Envelope Proteins Elicit High Titers of Neutralizing Activities and Protect Humanized Mice from Lethal Dose EBV Challenge. Vaccines 2021, 9, 285. [Google Scholar] [CrossRef] [PubMed]
- Moutschen, M.; Léonard, P.; Sokal, E.M.; Smets, F.; Haumont, M.; Mazzu, P.; Bollen, A.; Denamur, F.; Peeters, P.; Dubin, G.; et al. Phase I/II studies to evaluate safety and immunogenicity of a recombinant gp350 Epstein–Barr virus vaccine in healthy adults. Vaccine 2007, 25, 4697–4705. [Google Scholar] [CrossRef] [PubMed]
- Draper, S.J.; Angov, E.; Horii, T.; Miller, L.H.; Srinivasan, P.; Theisen, M.; Biswas, S. Recent advances in recombinant protein-based malaria vaccines. Vaccine 2015, 33, 7433–7443. [Google Scholar] [CrossRef]
- Zhong, L.; Zhao, Q.; Zeng, M.-S.; Zhang, X. Prophylactic vaccines against Epstein–Barr virus. Lancet 2024, 404, 845–845. [Google Scholar] [CrossRef]
- Li, P.; Meng, Z.; Zhou, Z.; Zhong, Z.; Kang, M. Therapeutic vaccines for Epstein–Barr virus: a way forward. Lancet 2024, 403, 2779–2780. [Google Scholar] [CrossRef]
- Chang Y; Cesarman, E; Pessin, MS; Lee,F; Culpepper, J; Knowles, DM; Moore, PS. Identification of Herpes-like DNA Sequences in AIDS-associated Kaposi’s Sarcoma. Science 1994, 266, 1865–1869.
- Ablashi, D.V.; Chatlynne, L.G.; Whitman, J.J.E.; Cesarman, E. Spectrum of Kaposi's Sarcoma-Associated Herpesvirus, or Human Herpesvirus 8, Diseases. Clin. Microbiol. Rev. 2002, 15, 439–464. [Google Scholar] [CrossRef]
- Cesarman, E.; Damania, B.; Krown, S.E.; Martin, J.; Bower, M.; Whitby, D. Kaposi sarcoma. Nat. Rev. Dis. Prim. 2019, 5, 1–21. [Google Scholar] [CrossRef]
- Gantts,S; i Casper, C. Human Herpesvirus 8-associated Neoplasms: The Roles of Viral Replication and Antiviral Treatment. Curr Opin. Infect. Dis. 2011, 24, 295–301. [Google Scholar] [CrossRef]
- Suda, T; Katano, H; Delso, G; Kakiuchi, C; Nakamura, T; Shiota, T; Higashihara, M; Mori, S. HHV-8 Infection Status of AIDS-unrelated and AIDS-associated Multicentric Castleman Disease. Pathol. Int. 2001, 51, 671–679. [Google Scholar] [CrossRef]
- Chadbum, A; Said, JW; Du, M; Vega, F. KSHV/HHV8-positive Germinotropic Lymohoproliferative Disorder. In: WHO Classification of Haematolymphoid Tumours, 5th Edition, 2024.
- Douglas, Jl; Gustin, JK; Moses, AV; Dezube, BJ; Pantanowitz, L. Kaposi Sarcoma Pathogenesis: A Triad of Viral Infection, Oncogenesis and Chronic Inflammation. Transl Biomed 2010,1,172.
- Radu, O; Pantanowitz, L. Kpaosi Sarcoma. Arch. Pathol. Lab. Med. 2013, 137, 289–294. [Google Scholar] [CrossRef]
- Guillet, S.; Gérard, L.; Meignin, V.; Agbalika, F.; Cuccini, W.; Denis, B.; Katlama, C.; Galicier, L.; Oksenhendler, E. Classic and extracavitary primary effusion lymphoma in 51 HIV-infected patients from a single institution. Am. J. Hematol. 2015, 91, 233–237. [Google Scholar] [CrossRef]
- Bruce-Brand, C.; Rigby, J. Kaposi Sarcoma With Intravascular Primary Effusion Lymphoma in the Skin: A Potential Pitfall in HHV8 Immunohistochemistry Interpretation. Int. J. Surg. Pathol. 2020, 28, 868–871. [Google Scholar] [CrossRef]
- Said, JW; Chadbum, A; Medeiros, LJ; Bacon, CM; Cesarman, E; Michelow, P; Du, M; Bower, M. Primary Effusion Lymphoma. In: WHO Classification of Haematolymphoid Tumours, 5th Edition, 2024.
- Rossi, G.; Cozzi, I.; Della Starza, I.; De Novi, L.A.; De Propris, M.S.; Gaeta, A.; Petrucci, L.; Pulsoni, A.; Pulvirenti, F.; Ascoli, V. Human herpesvirus-8–positive primary effusion lymphoma in HIV-negative patients: Single institution case series with a multidisciplinary characterization. Cancer Cytopathol. 2020, 129, 62–74. [Google Scholar] [CrossRef] [PubMed]
- Das, D.K. Serous effusions in malignant lymphomas: A review. Diagn. Cytopathol. 2006, 34, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Lurain, K.; Polizzotto, M.N.; Aleman, K.; Bhutani, M.; Wyvill, K.M.; Gonçalves, P.H.; Ramaswami, R.; Marshall, V.A.; Miley, W.; Steinberg, S.M.; et al. Viral, immunologic, and clinical features of primary effusion lymphoma. Blood 2019, 133, 1753–1761. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.-K.; Foreman, K.; Shin, J.W.; Hirakawa, S.; Curry, C.L.; Sage, D.R.; Libermann, T.; Dezube, B.J.; Fingeroth, J.D.; Detmar, M. Lymphatic reprogramming of blood vascular endothelium by Kaposi sarcoma–associated herpesvirus. Nat. Genet. 2004, 36, 683–685. [Google Scholar] [CrossRef]
- Dittmer, D.P.; Damania, B. Kaposi sarcoma associated herpesvirus pathogenesis (KSHV)—an update. Curr. Opin. Virol. 2013, 3, 238–244. [Google Scholar] [CrossRef]
- Mariggiò, G.; Koch, S.; Schulz, T.F. Kaposi sarcoma herpesvirus pathogenesis. Philos. Trans. R. Soc. B: Biol. Sci. 2017, 372, 20160275. [Google Scholar] [CrossRef]
- Pantanowitz, L.; Moses, A.V.; Dezube, B.J. The inflammatory component of Kaposi sarcoma. Exp. Mol. Pathol. 2009, 87, 163–165. [Google Scholar] [CrossRef]
- Kaposi Idiopathisches multiples Pigmentsarkom der Haut. Arch. Dermatol. Res. 1872, 4, 265–273. [CrossRef]
- Bigi, R.; Landis, J.T.; An, H.; Caro-Vegas, C.; Raab-Traub, N.; Dittmer, D.P. Epstein–Barr virus enhances genome maintenance of Kaposi sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. 2018, 115, 201810128–E11387. [Google Scholar] [CrossRef]
- Cesarmann, E; Chang, Y; Moore, Y; Said, JW; Knowles, DM. Kaposi’s Sarcoma-associated Herpesvirus-like DNA Sequences in AIDS-related Body Cavity-based Lymphoma. N Eng J Med 1995,4,1186-1191. Doi:10.1056/NEJM199505043321802.
- Vega, F; Chadbum, A; Said, JW; Cesarmann, E; Du, M; Bower, M. KSH/HHV8-positive Diffuse Large B-cell Lymphoma. In; WHO Classification of Haematolymphoid Tumours, 5th Edition, 2024. De Jong, D; Naresh, KN; Chan, J (eds). Lyons, France, IARC (available online from https://tumourclassification.iarc.who.int/chapters/63).
- Casper, C.; Corey, L.; Cohen, J.I.; Damania, B.; Gershon, A.A.; Kaslow, D.C.; Krug, L.T.; Martin, J.; Mbulaiteye, S.M.; Mocarski, E.S.; et al. KSHV (HHV8) vaccine: promises and potential pitfalls for a new anti-cancer vaccine. npj Vaccines 2022, 7, 1–10. [Google Scholar] [CrossRef]
- Chandran, B. Early Events in Kaposi's Sarcoma-Associated Herpesvirus Infection of Target Cells. J. Virol. 2010, 84, 2188–2199. [Google Scholar] [CrossRef] [PubMed]
- Mulama, D.H.; Mutsvunguma, L.Z.; Totonchy, J.; Ye, P.; Foley, J.; Escalante, G.M.; Rodriguez, E.; Nabiee, R.; Muniraju, M.; Wussow, F.; et al. A multivalent Kaposi sarcoma-associated herpesvirus-like particle vaccine capable of eliciting high titers of neutralizing antibodies in immunized rabbits. Vaccine 2019, 37, 4184–4194. [Google Scholar] [CrossRef] [PubMed]
- Muniraju, M.; Mutsvunguma, L.Z.; Foley, J.; Escalante, G.M.; Rodriguez, E.; Nabiee, R.; Totonchy, J.; Mulama, D.H.; Nyagol, J.; Wussow, F.; et al. Kaposi Sarcoma-Associated Herpesvirus Glycoprotein H Is Indispensable for Infection of Epithelial, Endothelial, and Fibroblast Cell Types. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [PubMed]
- Kwun, H.J.; da Silva, S.R.; Qin, H.; Ferris, R.L.; Tan, R.; Chang, Y.; Moore, P.S. The central repeat domain 1 of Kaposi's sarcoma-associated herpesvirus (KSHV) latency associated-nuclear antigen 1 (LANA1) prevents cis MHC class I peptide presentation. Virology 2011, 412, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Sugden, A.U.; Hayes, M.; Sugden, B. How Epstein–Barr Virus and Kaposi’s Sarcoma-Associated Herpesvirus Are Maintained Together to Transform the Same B-Cell. Viruses 2021, 13, 1478. [Google Scholar] [CrossRef]
- Böni, M.; Rieble, L.; Münz, C. Co-Infection of the Epstein–Barr Virus and the Kaposi Sarcoma-Associated Herpesvirus. Viruses 2022, 14, 2709. [Google Scholar] [CrossRef]
- Julius, P.; Kang, G.; Siyumbwa, S.; Musumali, J.; Tso, F.Y.; Ngalamika, O.; Kaile, T.; Maate, F.; Moonga, P.; West, J.T.; et al. Co-infection and co-localization of Kaposi sarcoma-associated herpesvirus and Epstein-Barr virus in HIV-associated Kaposi sarcoma: a case report. Front. Cell. Infect. Microbiol. 2023, 13, 1270935. [Google Scholar] [CrossRef]
- King, SR. HIV: Virology and Mechanisms of Disease. Ann. Emerg. Med. 1994, 24, 3. [Google Scholar] [CrossRef]
- Chinen, J.; Shearer, W.T. Molecular virology and immunology of HIV infection. J. Allergy Clin. Immunol. 2002, 110, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Kaposi’s Sarcoma and Pneucystitis Pneumonia Among Homosexual Men – New York City and California. MMWR Morb Mortal Wkly Rep 1981, 30, 305–308.
- Yarchoan, R.; Uldrick, T.S. HIV-Associated Cancers and Related Diseases. N. Engl. J. Med. 2018, 378, 1029–1041. [Google Scholar] [CrossRef]
- Isaguliants, M.; Bayurova, E.; Avdoshina, D.; Kondrashova, A.; Chiodi, F.; Palefsky, J.M. Oncogenic Effects of HIV-1 Proteins, Mechanisms Behind. Cancers 2021, 13, 305. [Google Scholar] [CrossRef]
- Omar, A.; Marques, N.; Crawford, N. Cancer and HIV: The Molecular Mechanisms of the Deadly Duo. Cancers 2024, 16, 546. [Google Scholar] [CrossRef]
- Lurain, K.A.; Ramaswami, R.; Krug, L.T.; Whitby, D.; Ziegelbauer, J.M.; Wang, H.-W.; Yarchoan, R.; Forrest, G.N. HIV-associated cancers and lymphoproliferative disorders caused by Kaposi sarcoma herpesvirus and Epstein-Barr virus. Clin. Microbiol. Rev. 2024, 37, e0002223. [Google Scholar] [CrossRef]
- Zhao, L.-H.; Liu, X.; Yan, H.-X.; Li, W.-Y.; Zeng, X.; Yang, Y.; Zhao, J.; Liu, S.P.; Zhuang, X.-H.; Lin, C.; et al. Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat. Commun. 2016, 7, 12992. [Google Scholar] [CrossRef]
- Nevola, R.; Beccia, D.; Rosato, V.; Ruocco, R.; Mastrocinque, D.; Villani, A.; Perillo, P.; Imbriani, S.; Femine, A.D.; Criscuolo, L.; et al. HBV Infection and Host Interactions: The Role in Viral Persistence and Oncogenesis. Int. J. Mol. Sci. 2023, 24, 7651. [Google Scholar] [CrossRef]
- Zhang, M; Chen, H; Liu, H; Tang, H. The Impact of Integrated Hepatitis B Virus DNA on Oncogenesis and Anti-viral Therapy. Biomarkers Res. 2024, 12, 84. [Google Scholar] [CrossRef]
- Flores, J.E.; Thompson, A.J.; Ryan, M.; Howell, J. The Global Impact of Hepatitis B Vaccination on Hepatocellular Carcinoma. Vaccines 2022, 10, 793. [Google Scholar] [CrossRef] [PubMed]
- Forner, A; Llovet, JM; Brnix, J. Hepatocellular Carcinoma. Lancet 2012, 379, 1245–1255. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.R.; Barnes, E.; Cox, A.L. Approaches, Progress, and Challenges to Hepatitis C Vaccine Development. Gastroenterology 2019, 156, 418–430. [Google Scholar] [CrossRef] [PubMed]
- Virzì, A.; Suarez, A.A.R.; Baumert, T.F.; Lupberger, J. Oncogenic Signaling Induced by HCV Infection. Viruses 2018, 10, 538. [Google Scholar] [CrossRef]
- Fiehn, F.; Beisel, C.; Binder, M. Hepatitis C virus and hepatocellular carcinoma: carcinogenesis in the era of direct-acting antivirals. Curr. Opin. Virol. 2024, 67, 101423. [Google Scholar] [CrossRef]
- Martineau, C.-A.; Rivard, N.; Bisaillon, M. From viruses to cancer: exploring the role of the hepatitis C virus NS3 protein in carcinogenesis. Infect. Agents Cancer 2024, 19, 1–14. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Sanyal, A.J.; Yoon, S.K.; Lencioni, R. The Etiology of Hepatocellular Carcinoma and Consequences for Treatment. Oncol. 2010, 15, 14–22. [Google Scholar] [CrossRef]
- Torbenson, M; Ng, I; Park, YN; Roncalli, M; Sakamato, M. Hepatocellular Carcinoma. In: WHO Classification of Digestive System Tumours, 5th Edition, 2019, p.229-239. Shirmacher, P (eds). Lyons, France, IARC.
- Burt, A.D.; Alves, V.; Bedossa, P.; Clouston, A.; Guido, M.; Hübscher, S.; Kakar, S.; Ng, I.; Park, Y.N.; Reeves, H.; et al. Data set for the reporting of intrahepatic cholangiocarcinoma, perihilar cholangiocarcinoma and hepatocellular carcinoma: recommendations from the International Collaboration on Cancer Reporting (ICCR). Histopathology 2018, 73, 369–385. [Google Scholar] [CrossRef]
- Romano, L.; Paladini, S.; Galli, C.; Raimondo, G.; Pollicino, T.; Zanetti, A.R. Hepatitis B Vaccination. Hum. Vaccin. Immunother. 2015, 11, 53–57. [Google Scholar] [CrossRef]
- Vesikari, T.; Finn, A.; van Damme, P.; Leroux-Roels, I.; Leroux-Roels, G.; Segall, N.; Toma, A.; Vallieres, G.; Aronson, R.; Reich, D.; et al. Immunogenicity and Safety of a 3-Antigen Hepatitis B Vaccine vs a Single-Antigen Hepatitis B Vaccine. JAMA Netw. Open 2021, 4, e2128652–e2128652. [Google Scholar] [CrossRef]
- Chang, M.-H.; You, S.-L.; Chen, C.-J.; Liu, C.-J.; Lee, C.-M.; Lin, S.-M.; Chu, H.-C.; Wu, T.-C.; Yang, S.-S.; Kuo, H.-S.; et al. Decreased Incidence of Hepatocellular Carcinoma in Hepatitis B Vaccinees: A 20-Year Follow-up Study. JNCI J. Natl. Cancer Inst. 2009, 101, 1348–1355. [Google Scholar] [CrossRef]
- Chen, T.-W. Paths toward hepatitis B immunization in South Korea and Taiwan. Clin. Exp. Vaccine Res. 2013, 2, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, Y.; Allain, J.-P.; Li, C. Chronic and occult hepatitis B virus infections in the vaccinated Chinese population. Ann. Blood 2016, 2, 4–4. [Google Scholar] [CrossRef]
- Cao, M.; Fan, J.; Lu, L.; Fan, C.; Wang, Y.; Chen, T.; Zhang, S.; Yu, Y.; Xia, C.; Lu, J.; et al. Long term outcome of prevention of liver cancer by hepatitis B vaccine: Results from an RCT with 37 years. Cancer Lett. 2022, 536, 215652. [Google Scholar] [CrossRef]
- Wong, NS; Chan, DPC, Poon, CM; Chan, CP; Lau, LHW; Yeoh, EK; Lee, SS. Hepatitis B Burden and Population Immunity in a High Endemicity City – a Geographically Random Household Epidemiology Study for Evaluating Achievability of Elimination. Epidemiol Infect 2022, 151, e22, 1–11.
- Shoukry, N.H. Hepatitis C vaccines, antibodies, and T cells. Front. Immunol. 2018, 9, 1480. [Google Scholar] [CrossRef]
- Czarnota, A.; Raszplewicz, A.; Sławińska, A.; Bieńkowska-Szewczyk, K.; Grzyb, K. Minicircle-based vaccine induces potent T-cell and antibody responses against hepatitis C virus. Sci. Rep. 2024, 14, 1–11. [Google Scholar] [CrossRef]
- Garcia-Samaniego, J.; Rodriguez, M.; Berenguer, J.; Rodriguez-Rosado, R.; Carbo, J.; Asensi, V.; Soriano, V. Hepatocellular carcinoma in HIV-infected patients with chronic hepatitis C. Am. J. Gastroenterol. 2001, 96, 179–183. [Google Scholar] [CrossRef]
- Gjærde, L.I.; Shepherd, L.; Jablonowska, E.; Lazzarin, A.; Rougemont, M.; Darling, K.; Battegay, M.; Braun, D.; Martel-Laferriere, V.; Lundgren, J.D.; et al. Trends in Incidences and Risk Factors for Hepatocellular Carcinoma and Other Liver Events in HIV and Hepatitis C Virus–coinfected Individuals From 2001 to 2014: A Multicohort Study. Clin. Infect. Dis. 2016, 63, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Awadh, A.A.; Alharthi, A.A.; Alghamdi, B.A.; Alghamdi, S.T.; Baqays, M.K.; Binrabaa, I.S.; Malli, I.A. Coinfection of Hepatitis B and C Viruses and Risk of Hepatocellular Carcinoma: Systematic Review and Meta-analysis. J. Glob. Infect. Dis. 2024, 16, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Zampino, R.; Pisaturo, M.A.; Cirillo, G.; Marrone, A.; Macera, M.; Rinaldi, L.; Stanzione, M.; Durante-Mangoni, E.; Gentile, I.; Sagnelli, E.; et al. Hepatocellular carcinoma in chronic HBV-HCV co-infection is correlated to fibrosis and disease duration. Ann. Hepatol. 2015, 14, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Maqsood, Q.; Sumrin, A.; Iqbal, M.; Younas, S.; Hussain, N.; Mahnoor, M.; Wajid, A. Hepatitis C virus/Hepatitis B virus coinfection: Current prospectives. Antivir. Ther. 2023, 28. [Google Scholar] [CrossRef]
- Mehershanhi, S.; Haider, A.; Kandhi, S.; Sun, H.; Patel, H. Prevalence of Hepatocellular Carcinoma in HIV Patients Co-infected or Triple Infected With Hepatitis B and Hepatitis C in a Community Hospital in South Bronx. Cureus 2022, 14, e26089. [Google Scholar] [CrossRef]
- Feng, H.; Shuda, M.; Chang, Y.; Moore, P.S. Clonal Integration of a Polyomavirus in Human Merkel Cell Carcinoma. Science 2008, 319, 1096–1100. [Google Scholar] [CrossRef]
- Spurgeon, M.E.; Lambert, P.F. Merkel cell polyomavirus: A newly discovered human virus with oncogenic potential. Virology 2013, 435, 118–130. [Google Scholar] [CrossRef]
- Silling, S.; Kreuter, A.; Gambichler, T.; Meyer, T.; Stockfleth, E.; Wieland, U. Epidemiology of Merkel Cell Polyomavirus Infection and Merkel Cell Carcinoma. Cancers 2022, 14, 6176. [Google Scholar] [CrossRef]
- Yang, J.F.; You, J. Merkel cell polyomavirus and associated Merkel cell carcinoma. Tumour Virus Res. 2021, 13, 200232. [Google Scholar] [CrossRef]
- DeCaprio, J.A. Merkel cell polyomavirus and Merkel cell carcinoma. Philos. Trans. R. Soc. B: Biol. Sci. 2017, 372, 20160276. [Google Scholar] [CrossRef]
- Zhou, X.; Yin, C.; Lin, Z.; Yan, Z.; Wang, J. Merkel Cell Polyomavirus Co-Infection in HIV/AIDS Individuals: Clinical Diagnosis, Consequences and Treatments. Pathogens 2025, 14, 134. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-Y.; Lai, Y.-R.; Cheng, P.-S.; Yu, W.-W.; Wang, R.C.; Shen, W.-L.; Chuang, S.-S. Merkel cell carcinoma in Taiwan: a subset is chronic arsenicism-related, and the Merkel cell polyomavirus-negative cases are pathologically distinct from virus-related cases with a poorer outcome. Pathology 2024, 57, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Jiang, S.; He, Y.; Jin, X.; Zhao, G.; Wang, B. Development of a therapeutic vaccine targeting Merkel cell polyomavirus capsid protein VP1 against Merkel cell carcinoma. npj Vaccines 2021, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Frey, A.; Pevzy, C.; olino, K.; Ishizwka, J. Development of an mRNA Therapeutic Vaccine for Virally Driven Merkel Cell Carcinoma. J. Clin. Oncol. 2024, 42, s2637. [Google Scholar] [CrossRef]
- Gambichler, T.; Schrama, D.; Käpynen, R.; Weyer-Fahlbusch, S.S.; Becker, J.C.; Susok, L.; Kreppel, F.; Abu Rached, N. Current Progress in Vaccines against Merkel Cell Carcinoma: A Narrative Review and Update. Vaccines 2024, 12, 533. [Google Scholar] [CrossRef]
- Rosean, C.B.; Leyder, E.C.; Hamilton, J.; Carter, J.J.; Galloway, D.A.; Koelle, D.M.; Nghiem, P.; Heiland, T. LAMP1 targeting of the large T antigen of Merkel cell polyomavirus results in potent CD4 T cell responses and tumor inhibition. Front. Immunol. 2023, 14, 1253568. [Google Scholar] [CrossRef]
- Zeng, Q.; Gomez, B.P.; Viscidi, R.P.; Peng, S.; He, L.; Ma, B.; Wu, T.-C.; Hung, C.-F. Development of a DNA vaccine targeting Merkel cell polyomavirus. Vaccine 2011, 30, 1322–1329. [Google Scholar] [CrossRef]
- A Engels, E.; Frisch, M.; Goedert, J.J.; Biggar, R.J.; Miller, R.W. Merkel cell carcinoma and HIV infection. Lancet 2002, 359, 497–498. [Google Scholar] [CrossRef]
- Izikson, L.; Nornhold, E.; Iyer, J.G.; Nghiem, P.; Zeitouni, N.C. Merkel cell carcinoma associated with HIV: review of 14 patients. AIDS 2011, 25, 119–121. [Google Scholar] [CrossRef]
- Brites, C.; Grassi, M.F.; Quaresma, J.A.S.; Ishak, R.; Vallinoto, A.C.R. Pathogenesis of HTLV-1 infection and progression biomarkers: An overview. Braz. J. Infect. Dis. 2021, 25, 101594. [Google Scholar] [CrossRef]
- Forlani, G.; Shallak, M.; Accolla, R.S.; Romanelli, M.G. HTLV-1 Infection and Pathogenesis: New Insights from Cellular and Animal Models. Int. J. Mol. Sci. 2021, 22, 8001. [Google Scholar] [CrossRef]
- Hirons, A.; Khoury, G.; Purcell, D.F.J. Human T-cell lymphotropic virus type-1: a lifelong persistent infection, yet never truly silent. Lancet Infect. Dis. 2021, 21, e2–e10. [Google Scholar] [CrossRef] [PubMed]
- GonçaLves, D.U.; Proietti, F.A.; Ribas, J.G.R.; AraújO, M.G.; Pinheiro, S.R.; Guedes, A.C.; Carneiro-Proietti, A.B.F. Epidemiology, Treatment, and Prevention of Human T-Cell Leukemia Virus Type 1-Associated Diseases. Clin. Microbiol. Rev. 2010, 23, 577–589. [Google Scholar] [CrossRef]
- Bangham, C.R.M. HTLV-1 persistence and the oncogenesis of adult T cell leukemia/lymphoma. Blood 2023, 141, 2299–2306. [Google Scholar] [CrossRef] [PubMed]
- Letafati, A.; Bahari, M.; Ardekani, O.S.; Jazi, N.N.; Nikzad, A.; Norouzi, F.; Mahdavi, B.; Aboofazeli, A.; Mozhgani, S.-H. HTLV-1 vaccination Landscape: Current developments and challenges. Vaccine: X 2024, 19, 100525. [Google Scholar] [CrossRef] [PubMed]
- Attygalle, A.D.; Karube, K.; Jeon, Y.K.; Cheuk, W.; Bhagat, G.; Chan, J.K.C.; Naresh, K.N. The fifth edition of the WHO classification of mature T cell, NK cell and stroma-derived neoplasms. J. Clin. Pathol. 2025, 78, 217–232. [Google Scholar] [CrossRef]
- Suehiro, Y.; Hasegawa, A.; Iino, T.; Sasada, A.; Watanabe, N.; Matsuoka, M.; Takamori, A.; Tanosaki, R.; Utsunomiya, A.; Choi, I.; et al. Clinical outcomes of a novel therapeutic vaccine with Tax peptide-pulsed dendritic cells for adult T cell leukaemia/lymphoma in a pilot study. Br. J. Haematol. 2015, 169, 356–367. [Google Scholar] [CrossRef]
- McFee, R.B. SARS2 Human Coronavirus (COVID-19, SARSCoV2). Dis. Mon. 2020, 66, 10163. [Google Scholar] [CrossRef]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef]
- Policard, M.; Jain, S.; Rego, S.; Dakshanamurthy, S. Immune characterization and profiles of SARS-CoV-2 infected patients reveals potential host therapeutic targets and SARS-CoV-2 oncogenesis mechanism. Virus Res. 2021, 301, 198464–198464. [Google Scholar] [CrossRef]
- Jahankhani, K.; Ahangari, F.; Adcock, I.M.; Mortaz, E. Possible cancer-causing capacity of COVID-19: Is SARS-CoV-2 an oncogenic agent? Biochimie 2023, 213, 130–138. [Google Scholar] [CrossRef]
- Stingi, A.; Crillo, L. SARS-Cov-2 Infection and Cancer. Bio. Essays. 2021, 43, 2000289. [Google Scholar] [CrossRef] [PubMed]
- López-Romero, R.; Nambo-Lucio, M.d.J.; Salcedo-Carrillo, E.; Hernández-Cueto, M.d.L.Á.; Salcedo, M. El gran desafío de la latencia de SARS-CoV-2: el testículo como reservorio. Gac. medica de Mex. 2020, 156, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Goubran, H.; Stakiw, J.; Seghatchian, J.; Ragab, G.; Burnouf, T. SARS-CoV-2 and cancer: the intriguing and informative cross-talk. Transfus. Apher. Sci. 2022, 61, 103488–103488. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, M.; De Giglio, M.A.R.; Roviello, G.N. Deciphering the Relationship between SARS-CoV-2 and Cancer. Int. J. Mol. Sci. 2023, 24, 7803. [Google Scholar] [CrossRef]
- De Souza, L.M.; Ismail, M.; Elaskandrany, M.-A.; Fratella-Calabrese, A.; Grossman, I.R. Primary Hepatic EBV-DLBCL Lymphoma in the Setting of COVID-19 Infection. ACG Case Rep. J. 2024, 11, e01276. [Google Scholar] [CrossRef]
- Pietroluongo, E.; Luciano, A.; Peddio, A.; Buonaiuto, R.; Caltavituro, A.; Servetto, A.; De Angelis, C.; Arpino, G.; Palmieri, G.; Veneziani, B.M.; et al. Exploring the interplay between Kaposi's sarcoma and SARS-CoV-2 infection: A case series and systematic review. J. Med Virol. 2024, 96, e29849. [Google Scholar] [CrossRef]
- Pasi, F; Calveri, MM; Pizzarelli, G; Calabresse, A; Andreoli, M; Bongiovanni, I; Cattaneo, C; Rignanese, G. Oncolytic Effect of SARS-Cov-2 in a Patient with NK Lymphoma. Acta Biomed 2020,91,e2020047.
- Challenor, S; Tucker, D. SARS-Cov-2 Induced Remission of Hodgkin Lymphoma. Br J Haematol 2021, 192, 415.
- Ohadi, L.; Hosseinzadeh, F.; Dadkhahfar, S.; Nasiri, S. Oncolytic effect of SARS-CoV-2 in a patient with mycosis fungoides: A case report. Clin. Case Rep. 2022, 10, e05682. [Google Scholar] [CrossRef]
- Han, X.; Xu, P.; Ye, Q. Analysis of COVID-19 vaccines: Types, thoughts, and application. J. Clin. Lab. Anal. 2021, 35, e23937. [Google Scholar] [CrossRef]
- Panagiotakopoulos, L. Use of 2025-2026 Covid-19 Vaccines-Work Group Considerations. CDC Advisory Committee on Immunization Practices, April 15, 2025.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




