Submitted:
27 July 2025
Posted:
29 July 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
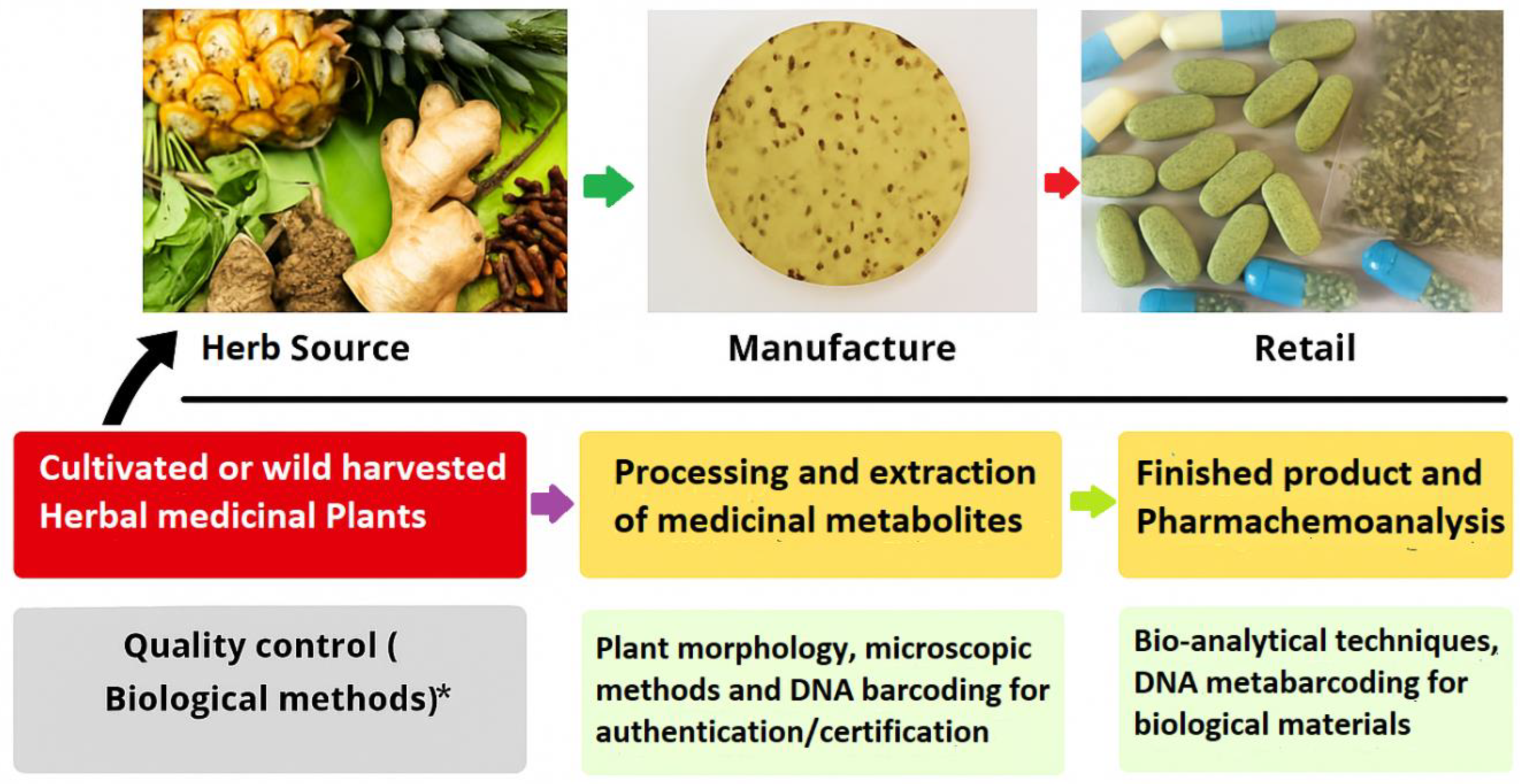
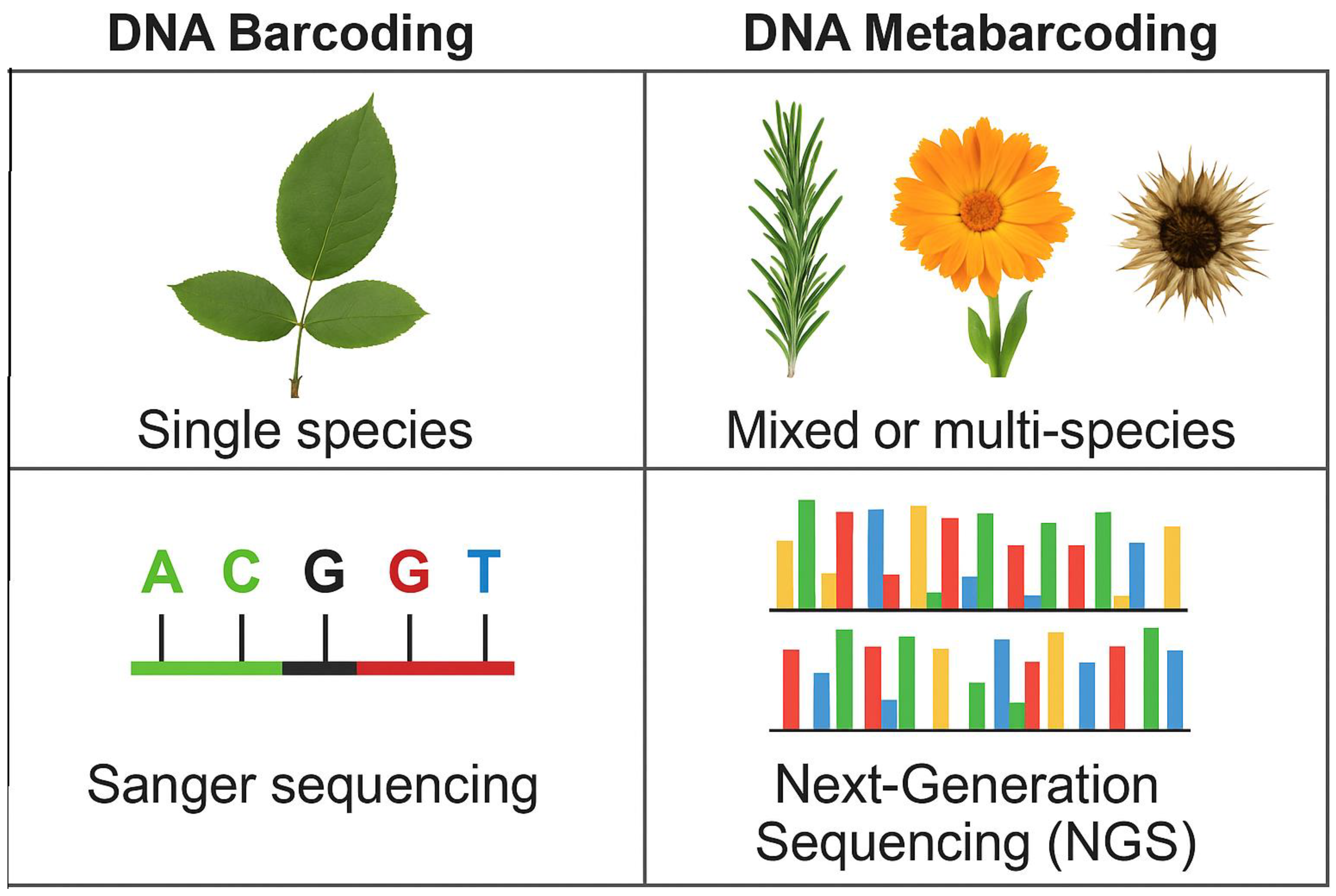
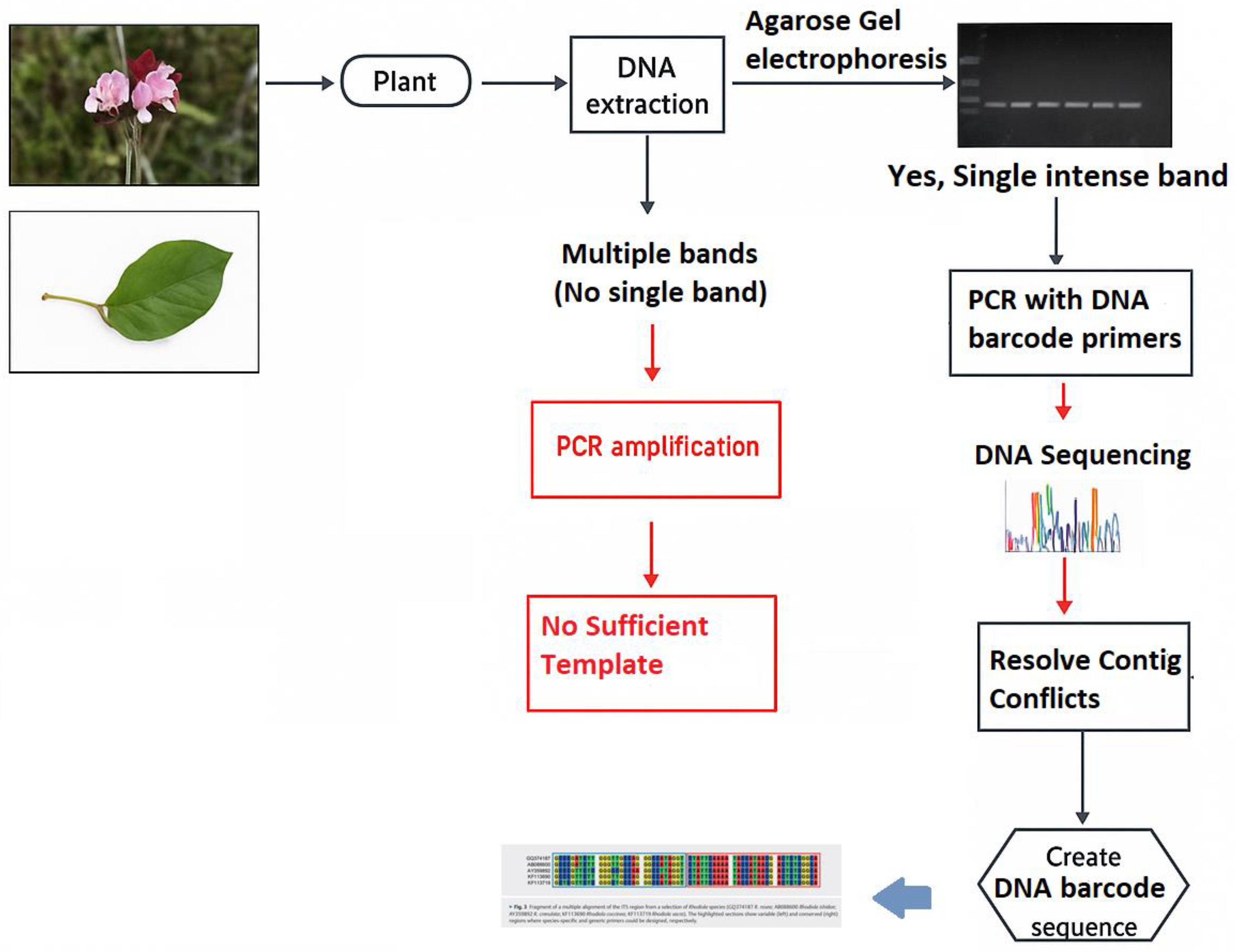
2. Methodology
- Used metabarcoding to examine commercial herbal preparations.
- Complicated metabarcoding with traditional identification.
- Provided definite rates of detection, mislabeling, or adulteration.
- Utilized DNA barcode verifications (e.g., ITS2, rbcL, matK, trnH-psbA).
- Addressed issues, i.e., DNA degradation, primer bias (Table 1), or database constraints.
3. Results
- High Adulteration Rates:
-
Commonly Detected Issues:
- ❖
- Presence of unlisted or banned species (e.g., Aristolochia spp.).
- ❖
- Mislabeling of traditional herbs (e.g., Panax ginseng replaced by Panax quinquefolius).
- ❖
- Use of low-cost fillers like Cuscuta or Amaranthus species.
-
Barcode Performance:
- ❖
- ITS2 was the most commonly used marker due to its high species resolution.
- ❖
- Dual-locus barcoding (ITS2 + rbcL) enhanced precision in complicated mixtures.
-
Technology and Workflow:
- ❖
- The majority of studies employed Illumina MiSeq platforms with in-house bioinformatics pipelines (e.g., QIIME, OBITools).
- ❖
- Sensitivity of detection was achieved for minor species at <1% relative abundance.
- Comparison with Traditional Methods:
4. Discussion
- ❖
- Primer bias can result in some taxa being under-sampled.
- ❖
- Damaged DNA can restrict detection in heat-treated or solvent-extracted samples.
- ❖
- Inadequate standard reference databases (e.g., errors in GenBank, lack of barcodes) impede taxonomic resolution.
5. Future Prospects
6. Conclusions
Acknowledgments
Conflicts of Interest
References
- Ahmed, S., Alawam, A. S., Rudayn, H. A., Allam, A. A., Mahmoud, R., Abd El-Raheem, H., & Alahmad, W. (2025). Advanced analytical and digital approaches for proactive detection of food fraud as an emerging contaminant threat. Talanta Open. Science Direct. Elsevier. 12, 100499.https://www.sciencedirect.com/science/article/pii/S2666831925001018?via%3Dihub. [CrossRef]
- Alamgir, A. N. M. (2017). Microscopy in pharmacognosy. In Therapeutic Use of Medicinal Plants and Their Extracts: Volume 1: Pharmacognosy. Cham: Springer International Publishing, 497-513. https://link.springer.com/chapter/10.1007/978-3-319-63862-1_11.
- Ali, M. U., Shaikh, E., Alishba, S., Rubeen, R., Ayesha, A., Akhlaq, M., Abbas, S.K., Kanwal, K., Rizwan, S., & Jahanzeb, M. (2025). ARTIFICIAL INTELLIGENCE-ASSISTED COMPARATIVE ANALYSIS OF DNA BARCODING SEQUENCES FOR HERBAL PLANT SPECIES IDENTIFICATION AND ADULTERATION DETECTION. Journal of Medical & Health Sciences Review, 2(3). https://jmhsr.com/index.php/jmhsr/article/view/484/606. [CrossRef]
- Crighton, E., Coghlan, M. L., Farrington, R., Hoban, C. L., Power, M. W., Nash, C., Mullaney, I., Byard, R.W., Trengove, R., Musgrave, I.F. and Bunce, M., & Maker, G. (2019). Toxicological screening and DNA sequencing detects contamination and adulteration in regulated herbal medicines and supplements for diet, weight loss and cardiovascular health. Journal of Pharmaceutical and Biomedical Analysis, 176, 112834. https://www.sciencedirect.com/science/article/abs/pii/S073170851931708X. [CrossRef]
- Heinrich, M., & Anagnostou, S. (2017). From pharmacognosia to DNA-based medicinal plant authentication–pharmacognosy through the centuries. Planta medica, 83(14/15), 1110-1116. https://www.thieme-connect.com/products/ejournals/html/10.1055/s-0043-108999. [CrossRef]
- Ichim, M. C. (2019). The DNA-based authentication of commercial herbal products reveals their globally widespread adulteration. Frontiers in pharmacology, 10, 1227. https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2019.01227/full. [CrossRef]
- Ichim, M. C., & Booker, A. (2021). Chemical authentication of botanical ingredients: a review of commercial herbal products. Frontiers in Pharmacology, 12, 666850. https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2021.666850/full. [CrossRef]
- Jiang, Y., Wei, S., Ge, H., Zhang, Y., Wang, H., Wen, X., Guo, C., Wang, S., Chen, Z. & Li, P. (2025). Advances in the Identification Methods of Food-Medicine Homologous Herbal Materials. Foods, 14(4), 608. https://www.mdpi.com/2304-8158/14/4/608. [CrossRef]
- Khade, H. D., & Variyar, P. S. (2025). Introduction to Spice Adulteration and Authenticity Testing. In Spices Production to Products (pp. 11-33). CRC Press. https://www.taylorfrancis.com/chapters/edit/10.1201/9781003470816-3/introduction-spice-adulteration-authenticity-testing-khade-prasad-variyar.
- Manisha, D. R. B., Begam, A. M., Chahal, K. S., & Ashok, M. A. (2025). Medicinal Plants and Traditional Uses and Modern Applications. Journal of Neonatal Surgery, 14(3). https://www.researchgate.net/profile/Akshay-Harale/publication/392629885_Akshay_Ashok_Harale_2025_Medicinal_Plants_and_Traditional_Uses_and_Modern_Applications/links/684b9b53e7be56124ab63b10/Akshay-Ashok-Harale-2025-Medicinal-Plants-and-Traditional-Uses-and-Modern-Applications.pdf.
- Naseem, I., Khan, M. A., Habib, U., Rana, R. M., Qasim, M., Alwahibi, M. S., M.S., Khanum, R., Shafiq, M., & Iqbal, R. (2025). Morphological profiling and DNA barcoding revealed genetic diversity and phylogeny of Mentha species cultivated in Pakistan. Genetic Resources and Crop Evolution, 72(3), 2977-2995. https://link.springer.com/article/10.1007/s10722-024-02140-x. [CrossRef]
- Nazar, N., Saxena, A., Sebastian, A., Slater, A., Sundaresan, V., & Sgamma, T. (2025). Integrating DNA barcoding within an orthogonal approach for herbal product authentication: A narrative review. Phytochemical Analysis, 36(1), 7-29. https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/full/10.1002/pca.3466. [CrossRef]
- Raclariu, A. C., Mocan, A., Popa, M. O., Vlase, L., Ichim, M. C., Crisan, G., Brysting, A.K., & de Boer, H. (2017). Veronica officinalis product authentication using DNA metabarcoding and HPLC-MS reveals widespread adulteration with Veronica chamaedrys. Frontiers in pharmacology, 8, 378. [CrossRef]
- https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2017.00378/full.
- Raclariu, A. C., Heinrich, M., Ichim, M. C., & de Boer, H. (2018). Benefits and limitations of DNA barcoding and metabarcoding in herbal product authentication. Phytochemical Analysis, 29(2), 123-128. [CrossRef]
- https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/full/10.1002/pca.2732.
- Raclariu-Manolică, A. C., & de Boer, H. J. (2022). DNA barcoding and metabarcoding for quality control of botanicals and derived herbal products. In Evidence-Based Validation of Herbal Medicine . Translational Research on Botanicals, 223-238. Elsevier. https://www.sciencedirect.com/science/article/abs/pii/B9780323855426000044. [CrossRef]
- Sgamma, T., Lockie-Williams, C., Kreuzer, M., Williams, S., Scheyhing, U., Koch, E., Slater, A., & Howard, C. (2017). DNA barcoding for industrial quality assurance. Planta Medica, 83(14/15), 1117-1129. https://www.thieme-connect.com/products/ejournals/html/10.1055/s-0043-113448. [CrossRef]
- Shaheen, S., Jaffer, M., Khalid, S., Khan, M. A., Hussain, K., Butt, M. M., Rauf Siddiqui, A., Ashfaq, M., Ahmad, M., Zafar, M., & Khan, F. (2019). Microscopic techniques used for the identification of medicinal plants: A case study of Senna. Microscopy Research and Technique, 82(10), 1660-1667. https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/abs/10.1002/jemt.23332. [CrossRef]
- Shi, Y., Zhao, M., Yao, H., Yang, P., Xin, T., Li, B., Sun, W., & Chen, S. (2017). Rapidly discriminate commercial medicinal Pulsatilla chinensis (Bge.) Regel from its adulterants using ITS2 barcoding and specific PCR-RFLP assay. Scientific reports, 7(1), 40000. https://www.nature.com/articles/srep40000. [CrossRef]
- Seethapathy, G. S., Raclariu-Manolica, A. C., Anmarkrud, J. A., Wangensteen, H., & de Boer, H. J. (2019). DNA metabarcoding authentication of ayurvedic herbal products on the European market raises concerns of quality and fidelity. Frontiers in plant science, 10, 68. https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2019.00068/full. [CrossRef]
- Staats, M., Arulandhu, A. J., Gravendeel, B., Holst-Jensen, A., Scholtens, I., Peelen, T., Prins, T.W., & Kok, E. (2016). Advances in DNA metabarcoding for food and wildlife forensic species identification. Analytical and bioanalytical chemistry, 408(17), 4615-4630. https://link.springer.com/article/10.1007/s00216-016-9595-8. [CrossRef]
- Travadi, T., Shah, A. P., Pandit, R., Sharma, S., Joshi, C., & Joshi, M. (2023). A combined approach of DNA metabarcoding collectively enhances the detection efficiency of medicinal plants in single and polyherbal formulations. Frontiers in Plant Science, 14, 1169984. https://www.frontiersin.org/journals/plant-science/articles/10.3389/fpls.2023.1169984/full. [CrossRef]
- Urumarudappa, S. K. J., Tungphatthong, C., Prombutara, P., & Sukrong, S. (2020). DNA metabarcoding to unravel plant species composition in selected herbal medicines on the National List of Essential Medicines (NLEM) of Thailand. Scientific Reports, 10(1), 18259. https://www.nature.com/articles/s41598-020-75305-0. [CrossRef]
- Vagare, R. D., Mane, S. R., & Bais, S. K. (2025). Review on Phytochemical Analysis of Finished Product by Chromatographic Techniques. Int. J. Pharm. Herbal. Technol, 3, 2583-8962.
- https://ijprdjournal.com/myapp/uploads/423-RITIKA%20VAGARE%20%203399-3418.pdf.
- Xiang, R., Wan, H., Sun, W., Duan, B., Chen, W., Cao, X., Wang, S., Song, C., Chen, S., Wang, Y. and Wahab, A.T., & Meng, X. (2025). TPMGD: A genomic database for the traditional medicines in Pakistan. Chinese Herbal Medicines, 17(1), 87-93. https://www.sciencedirect.com/science/article/pii/S167463842400042X. [CrossRef]
- Yameen, B., Abbas, W., Awan, M. U. F., Tahir, A., Mohammad, S., & Shoib, M. (2025). R bcLa marker based identification and phylogenetic analysis of Kasuri methi (Trigonella foenum-graecum L.): A native plant of Kasur district of Punjab (Pakistan). Genetic Resources and Crop Evolution, 72(2), 1663-1673. https://link.springer.com/article/10.1007/s10722-024-02044-w. [CrossRef]
| DNA Marker | Target Taxonomic Group | Primer Name | Primer Sequence (5'–3') | Amplicon Length (bp) | Remark |
| trnL (UAA) | Universal plant (minibarcode) | c | CGAAATCGGTAGACGCTACG | 250 | |
| h | CCATTGAGTCTCTGCACCTATC | ||||
| trnL (UAA) | Universal plant | c | CGAAATCGGTAGACGCTACG | 767 | |
| d | GGGGATAGAGGGACTTGAAC | ||||
| trnL (UAA) | Universal plant (minibarcode) | g | GGGCAATCCTGAGCCAA | 10–143 | p-loop region |
| h | CCATTGAGTCTCTGCACCTATC | ||||
| trnH-psbA | Universal plant | psbAF | GTTATGCATGAACGTAATGCTC | 264–792 | Forward |
| trnH2 | CGCGCATGGTGGATTCACAATCC | Reverse | |||
| nrITS | Universal angiosperm | 17SE | ACGAATTCATGGTCCGGTGAAGTGTTCG | 800 | Forward |
| 26SE | TAGAATTCCCCGGTTCGCTCGCCGTTAC | Reverse | |||
| nrITS2 | Universal plant | S2F | ATGCGATACTTGGTGTGAAT | 160–320 | Forward |
| S3R | GACGCTTCTCCAGACTACAAT | Reverse | |||
| nrITS2 | Universal plant | S2F | ATGCGATACTTGGTGTGAAT | 160–320 | Forward |
| ITS4 | TCCTCCGCTTATTGATATGC | Reverse | |||
| matK | Gymnosperms | matKpkF4 | CCCTATTCTATTCAYCCNGA | 656–889 | Forward |
| matKpkR1 | CGTATCGTGCTTTTRTGYTT | Reverse | |||
| matK | Gymnosperms | NY552F | CTGGATYCAAGATGCTCCTT | 656–889 | Forward |
| NY1150R | GGTCTTTGAGAAGAACGGAGA | Reverse | |||
| matK | Angiosperms & Gymnosperms | matK-390f | CGATCTATTCATTCAATATTTC | 656–889 | Forward |
| matK1326r | TCTAGCACACGAAAGTCGAAGT | Reverse | |||
| matK | Angiosperms & Gymnosperms | matKKIM1R | ACCCAGTCCATCTGGAAATCTTGGTTC | 656–889 | Forward |
| matKKIM3F | CGTACAGTACTTTTGTGTTTACGAG | Reverse | |||
| rbcL | Universal plant | rbcLa-F | ATGTCACCACAAACAGAGACTAAAGC | 654 | Forward |
| rbcLa-R | GTAAAATCAAGTCCACCRCG | Reverse |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





