Submitted:
23 July 2025
Posted:
24 July 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Patients and Tissue Material
2.2. Tissue Microarrays and Immunohistochemical Staining
2.3. Evaluation of Immunohistochemical Staining
2.4. Statistical Analysis
3. Results
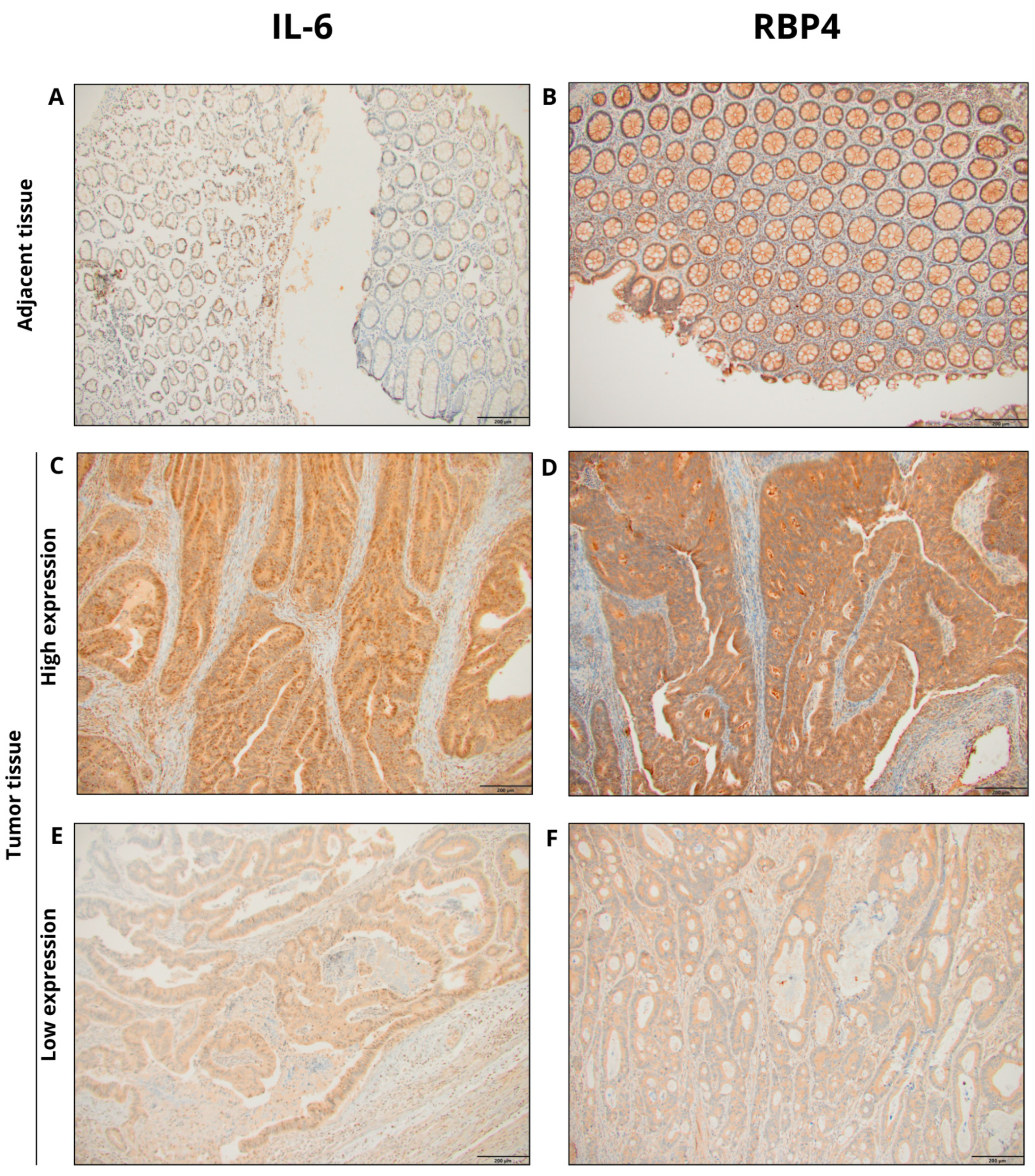
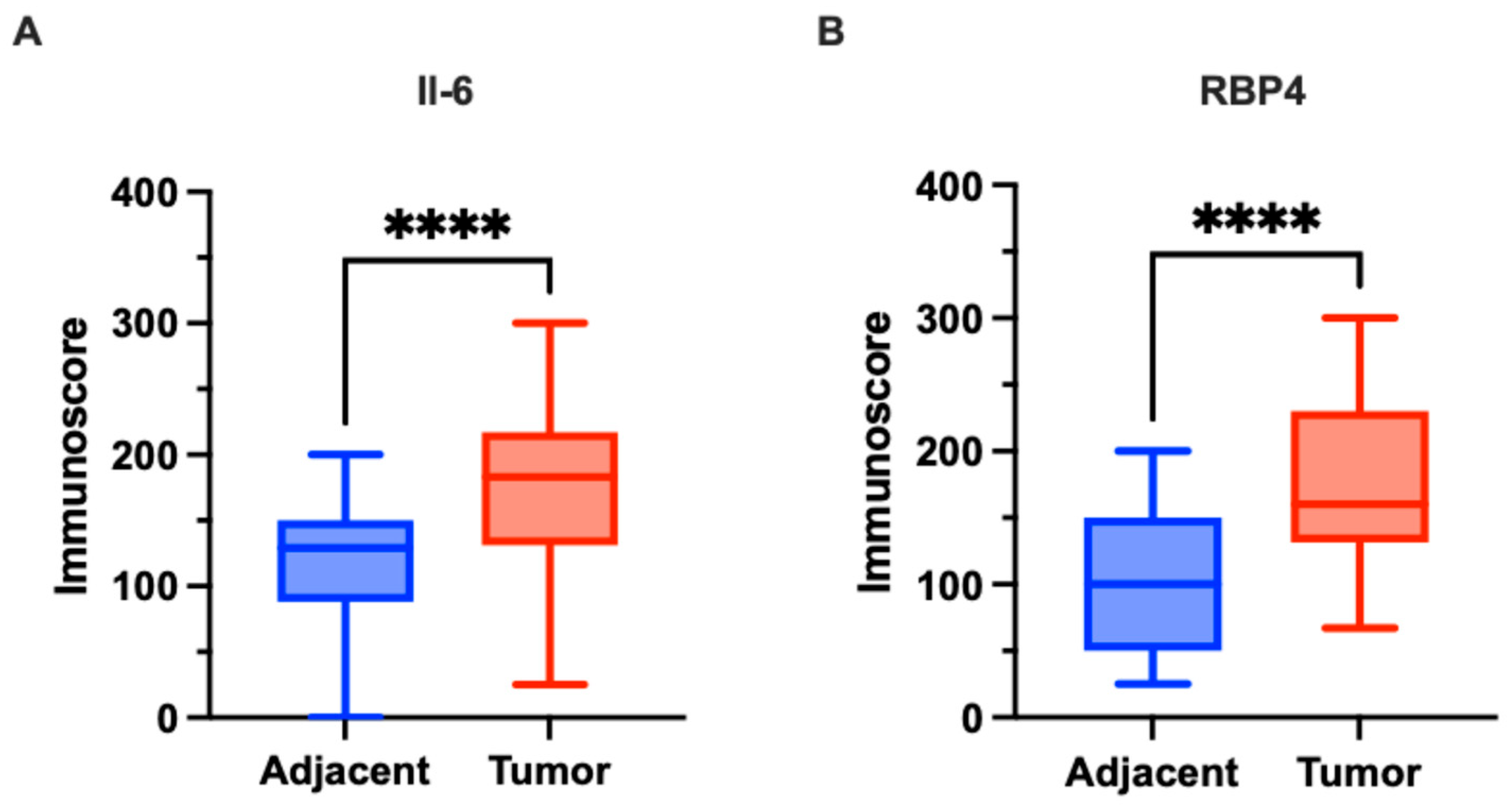
3.1. Immunoexpression of Il-6 and RBP4 in Tumor and Normal Adjacent Tissue and Its Clinicopathological Associations of Our Cohort
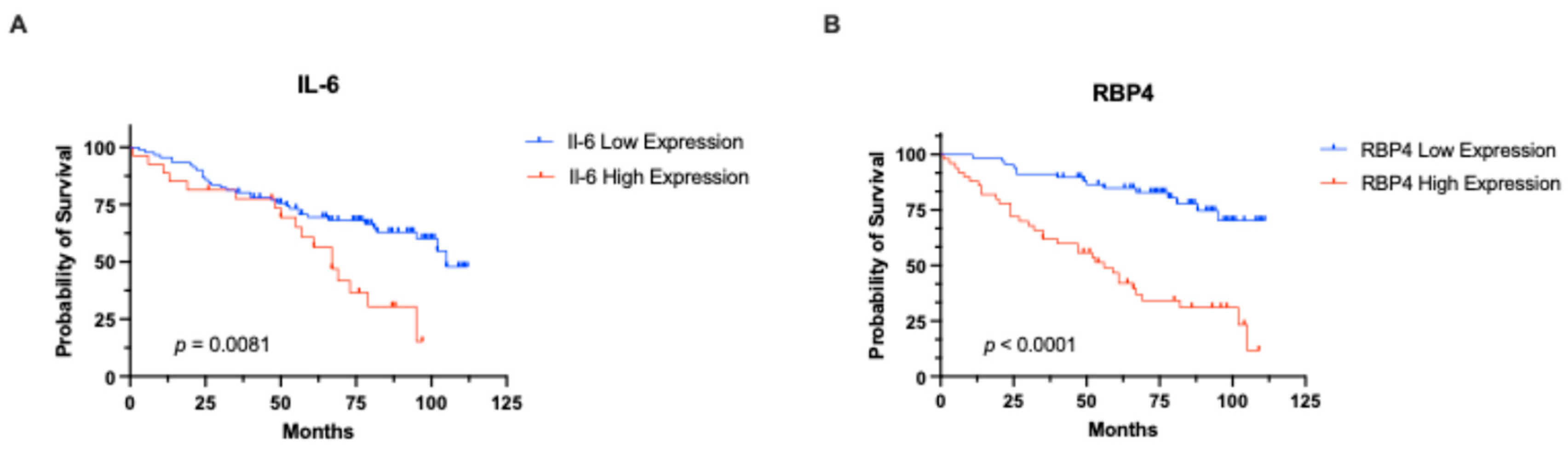
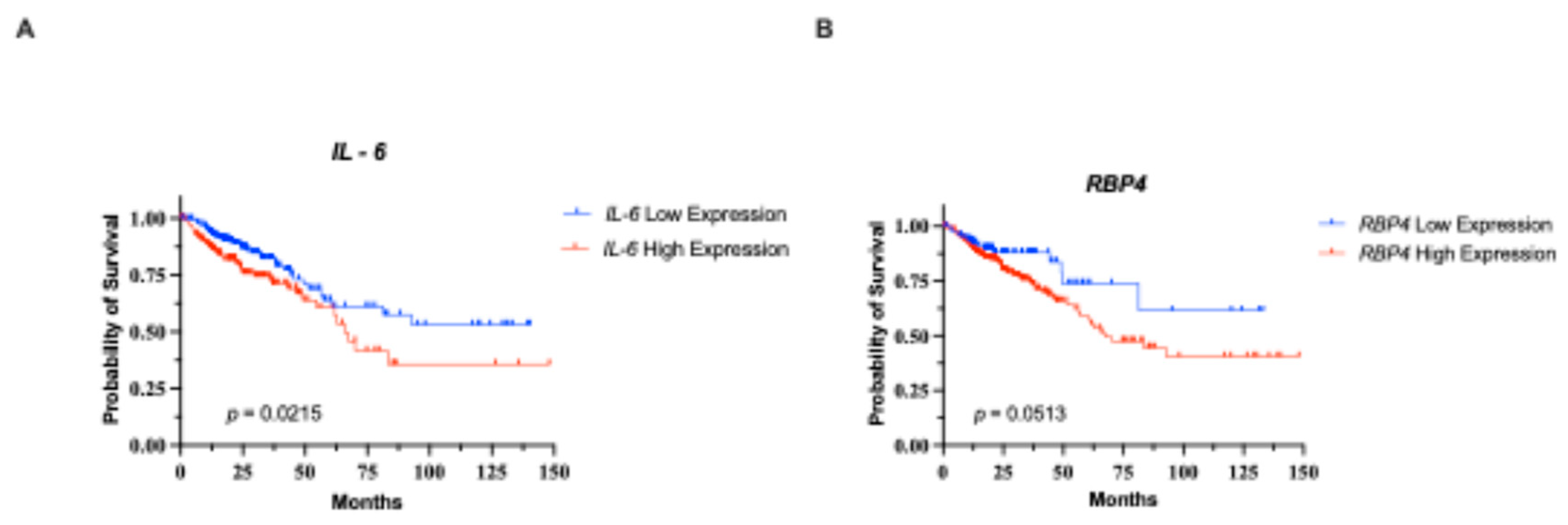
3.2. Association with the Clinical Outcome of Our Cohort
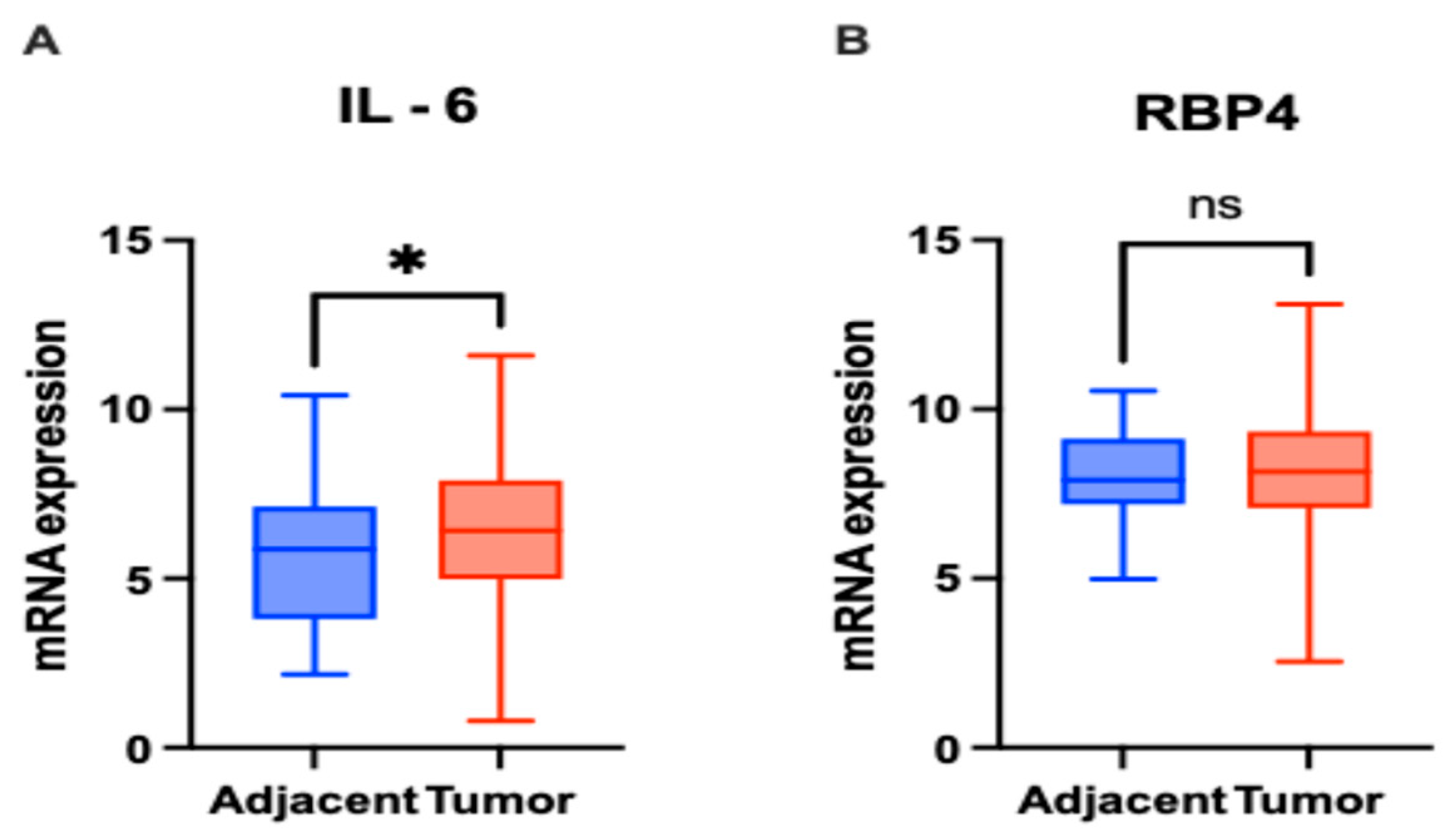
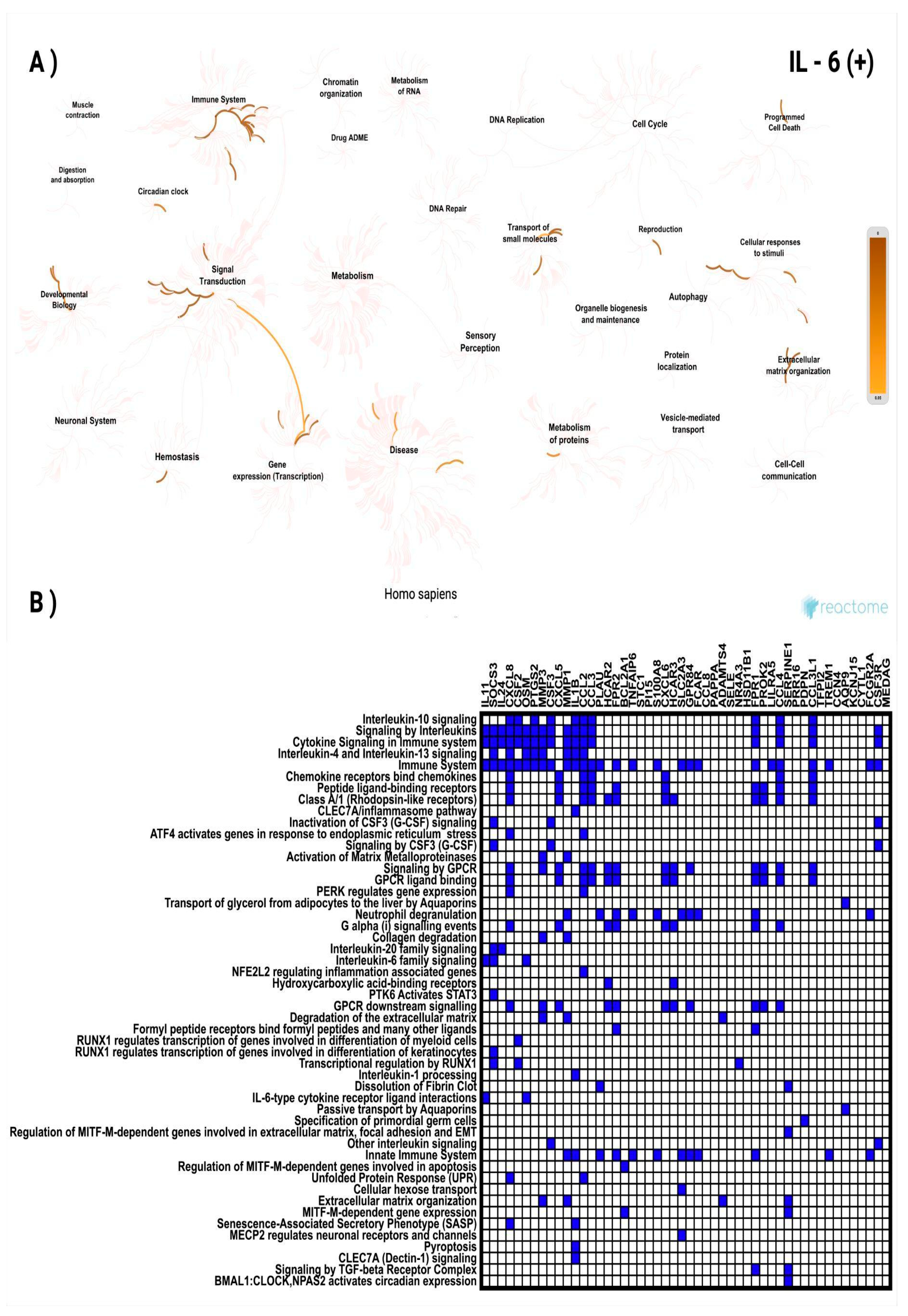
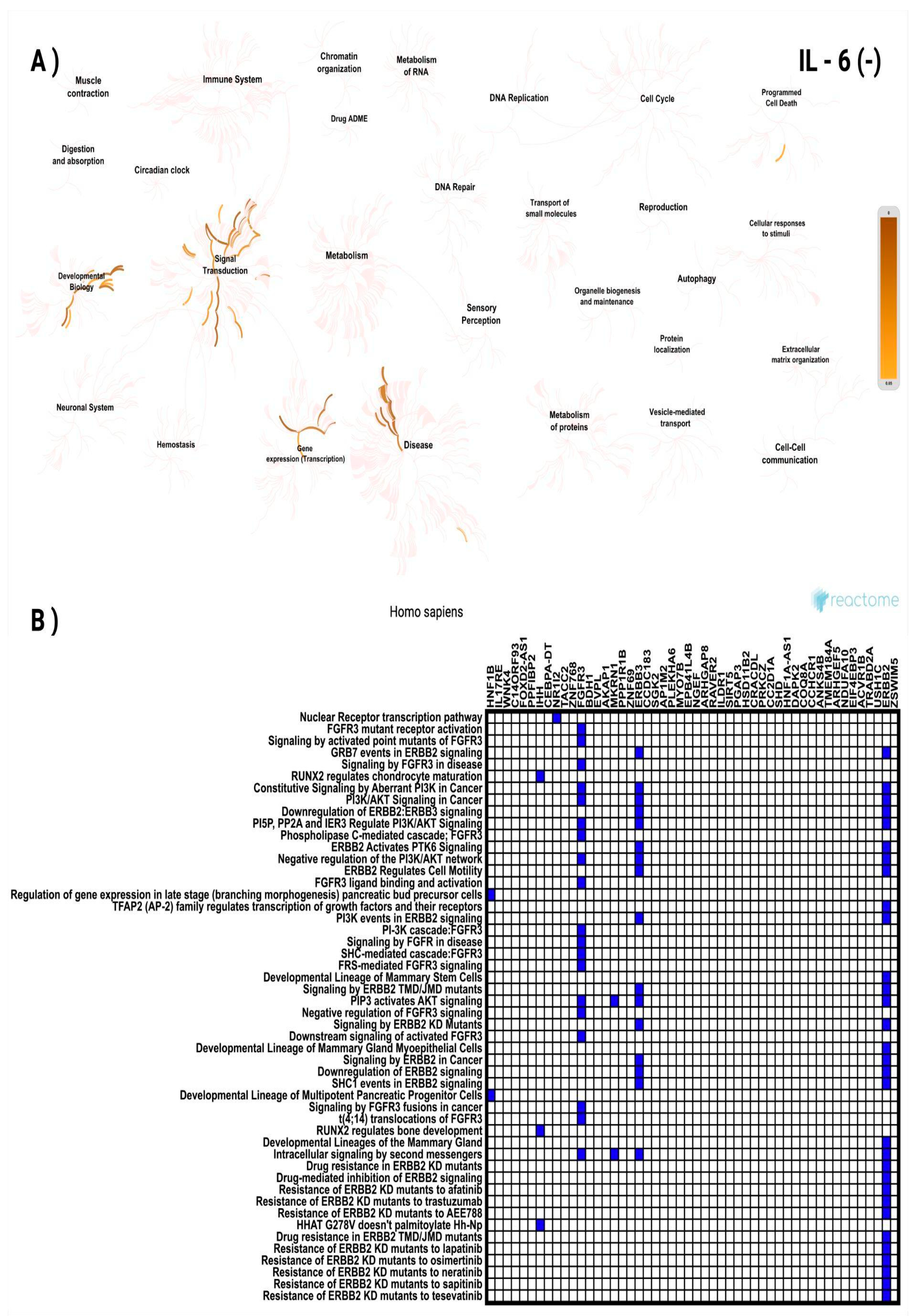
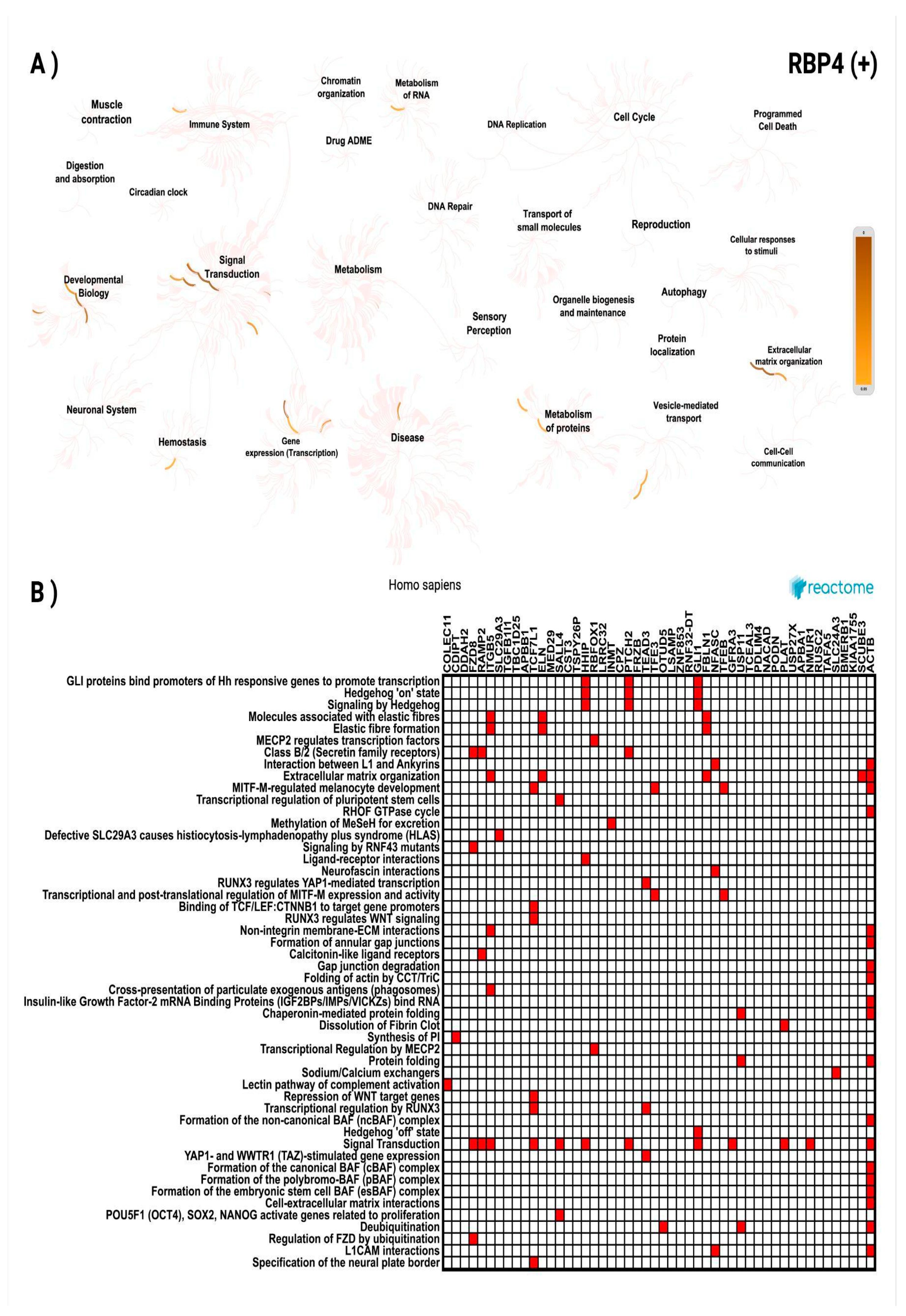
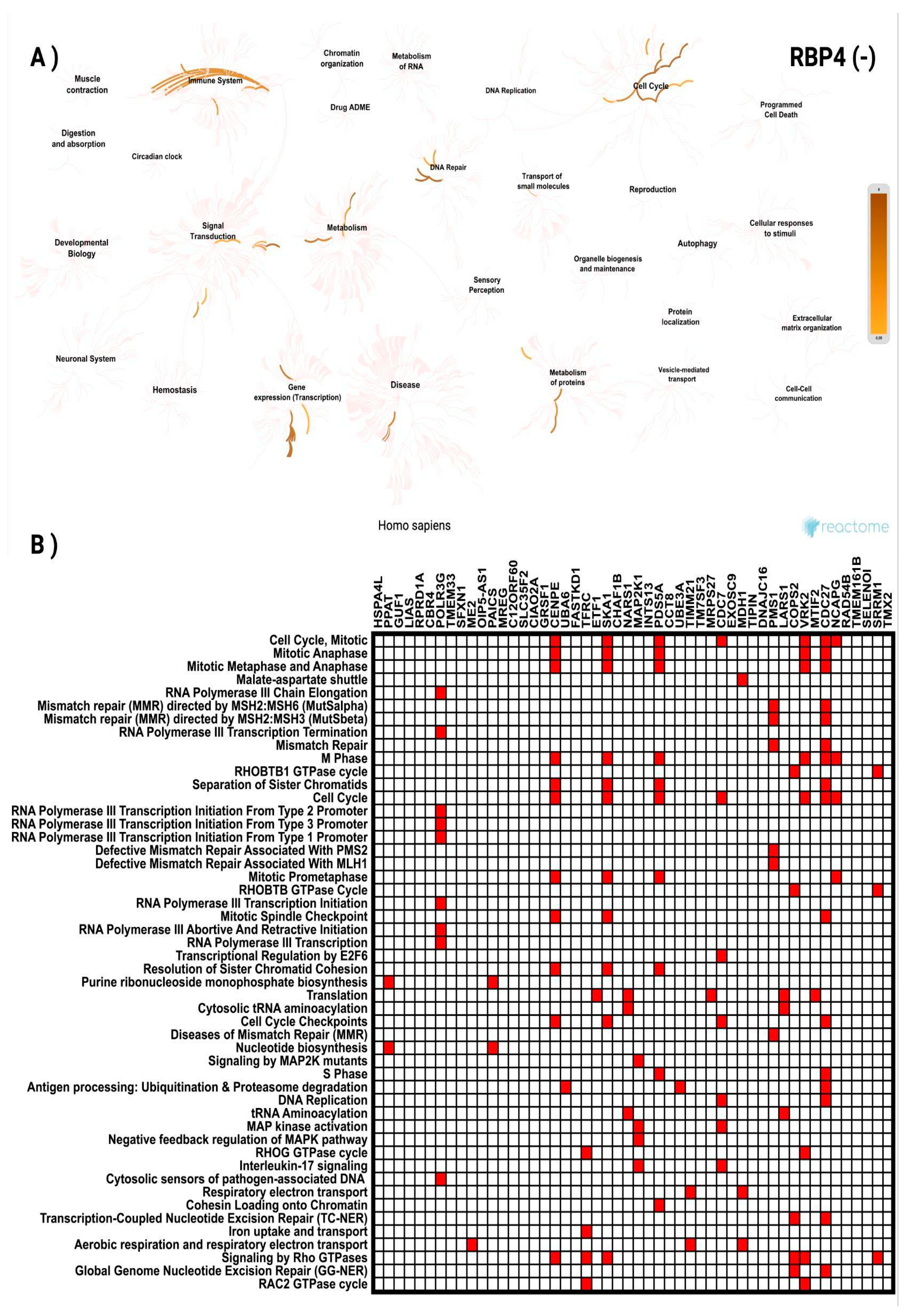
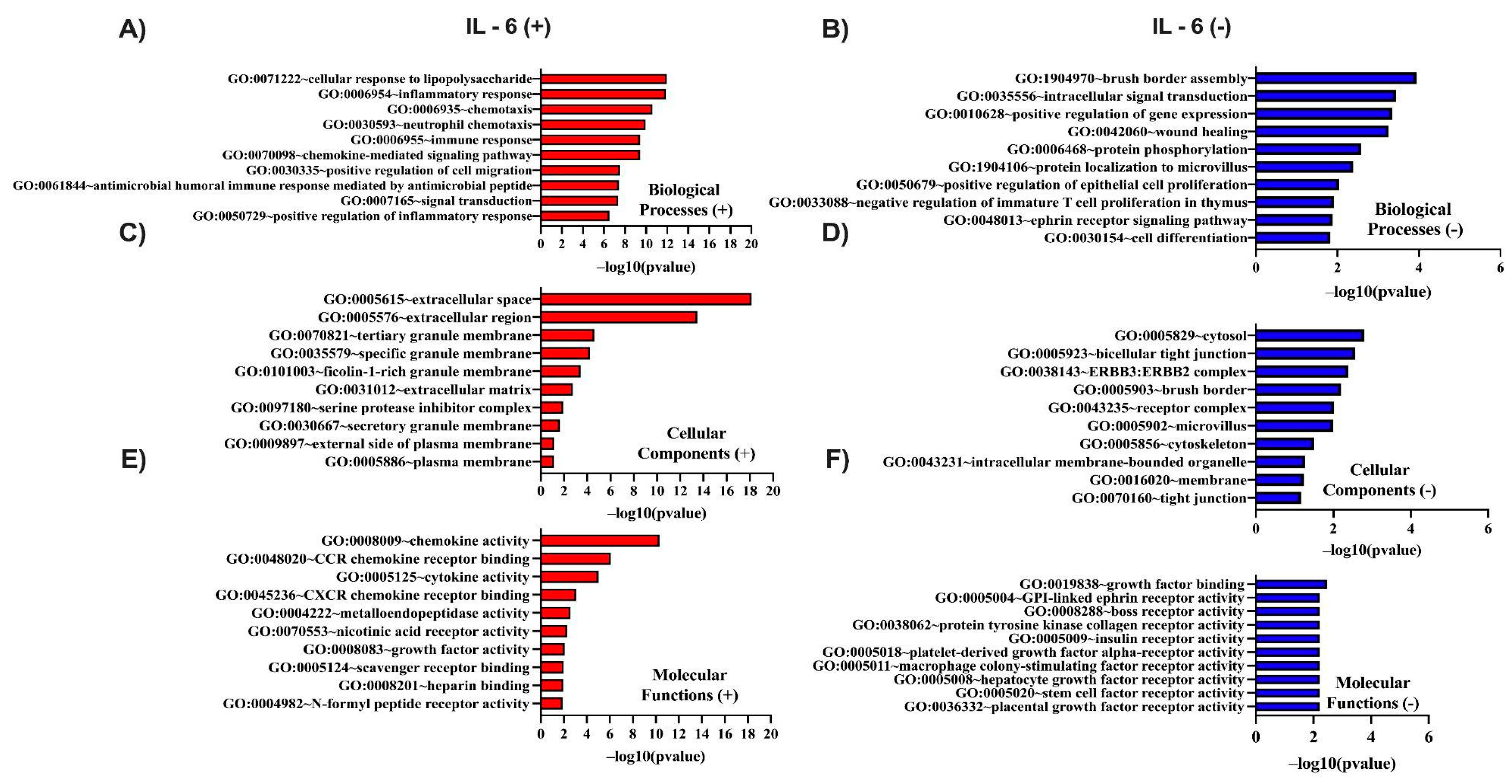
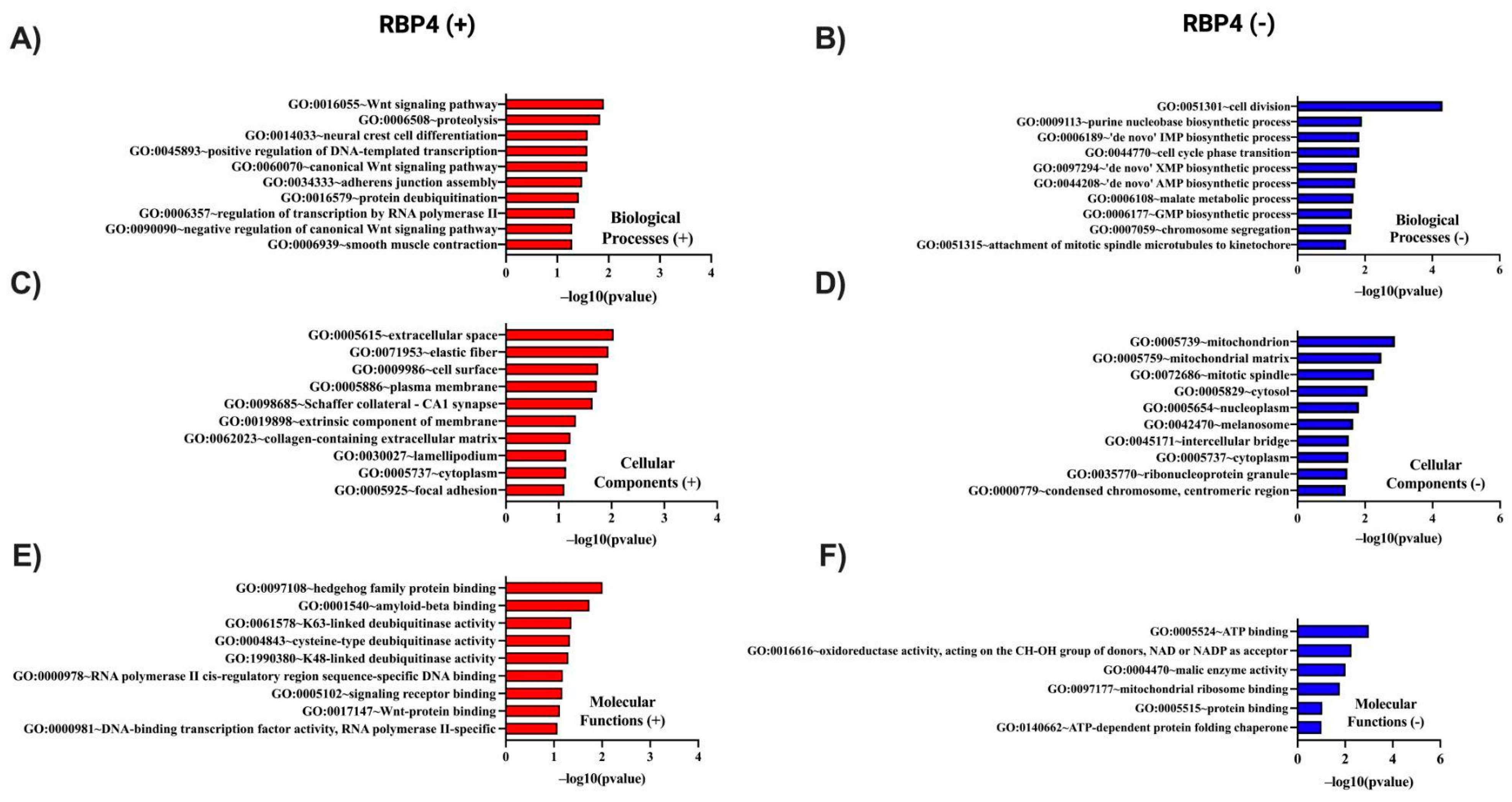
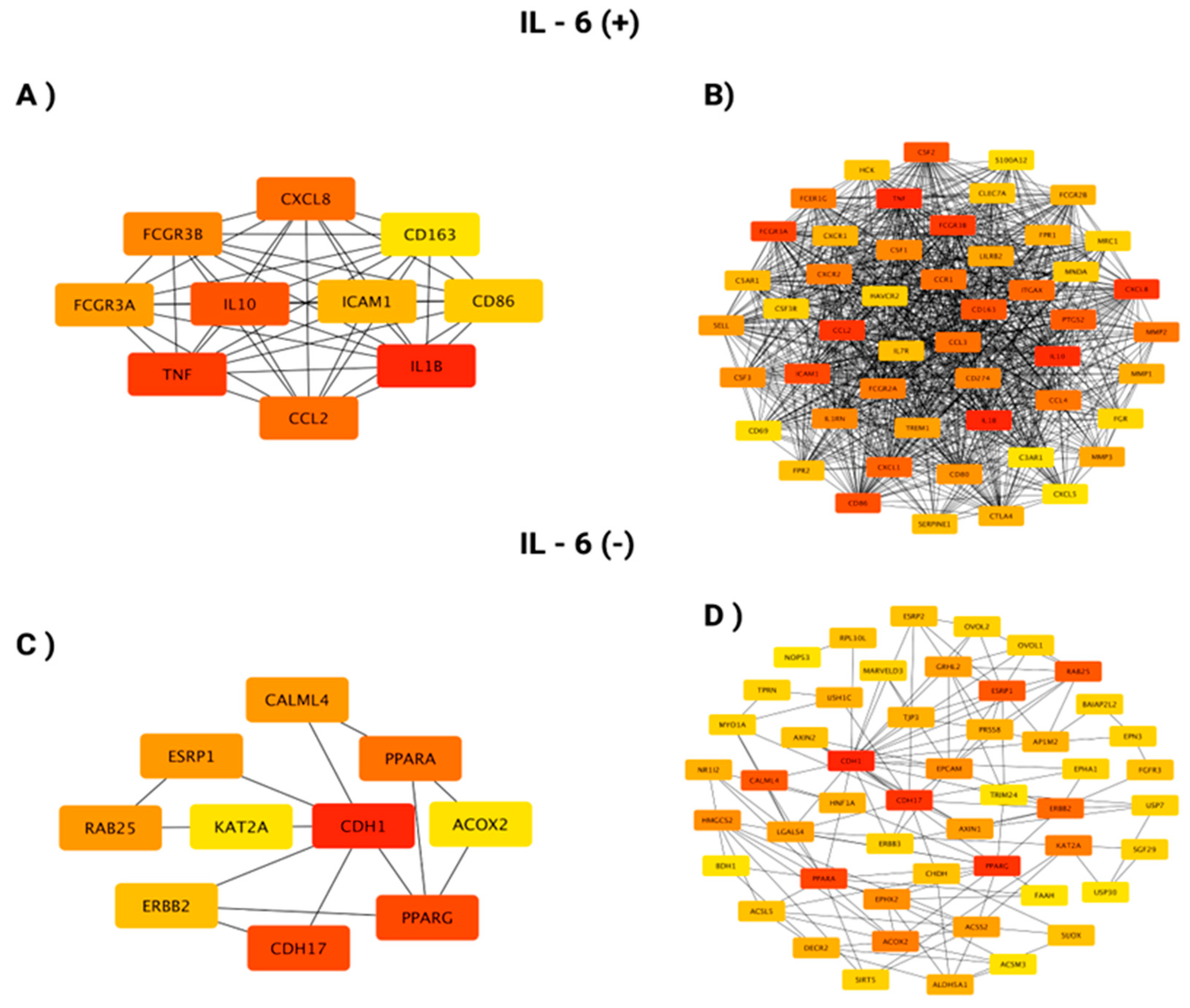
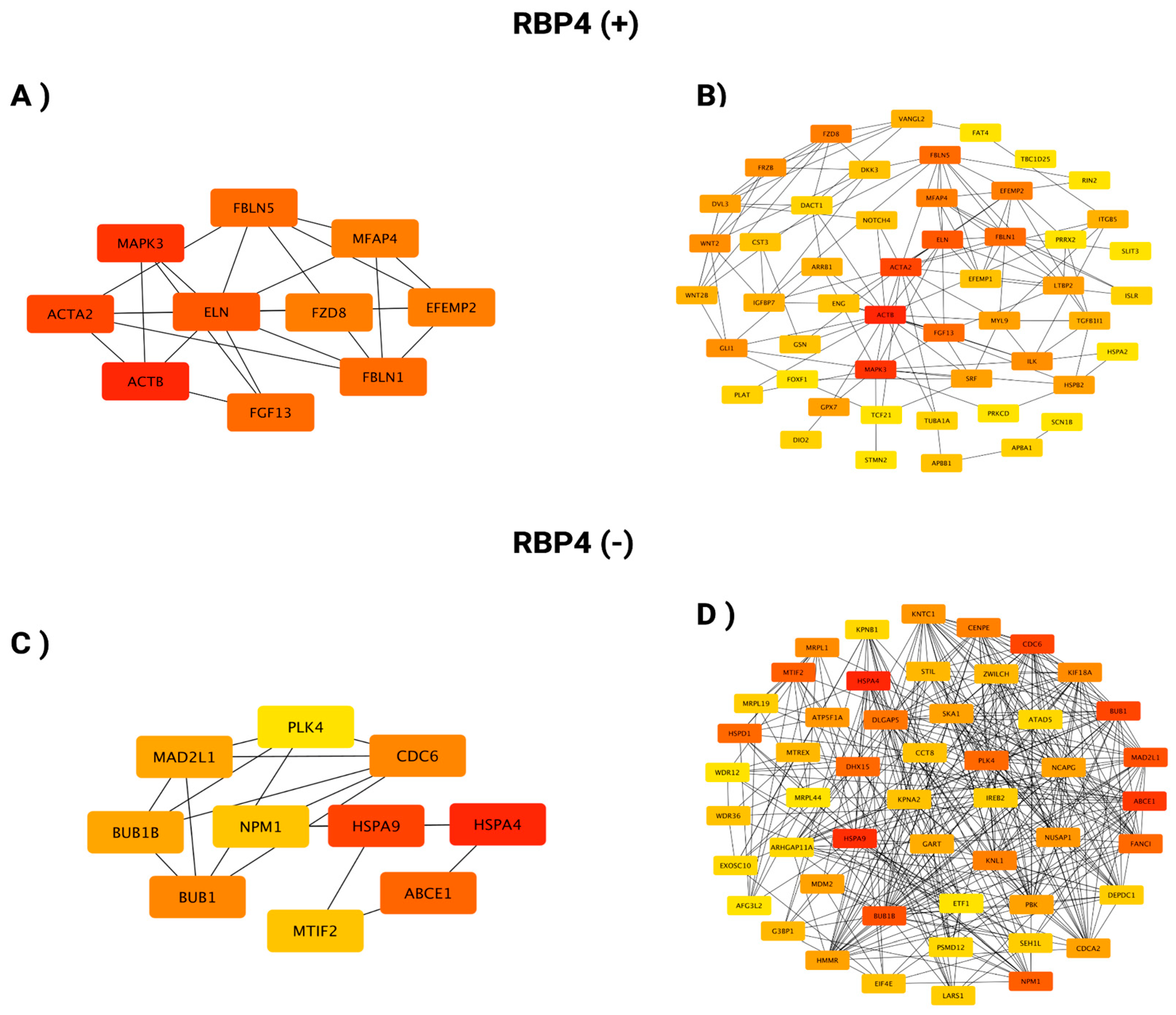
3.3. In Silico Investigation of Functional Pathways Connected to RBP4 and IL-6-Related Genes of the TCGA Cohort
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2024, 74, 229–263. [CrossRef]
- Fabregas, J.C.; Ramnaraign, B.; George, T.J. Clinical Updates for Colon Cancer Care in 2022. Clin Colorectal Cancer 2022, 21, 198–203. [CrossRef]
- Dekker, E.; Tanis, P.J.; Vleugels, J.L.A.; Kasi, P.M.; Wallace, M.B. Colorectal Cancer. The Lancet 2019, 394, 1467–1480. [CrossRef]
- Hanna, T.P.; King, W.D.; Thibodeau, S.; Jalink, M.; Paulin, G.A.; Harvey-Jones, E.; O’Sullivan, D.E.; Booth, C.M.; Sullivan, R.; Aggarwal, A. Mortality Due to Cancer Treatment Delay: Systematic Review and Meta-Analysis. The BMJ 2020, 371, m4087. [CrossRef]
- Hugo, H.J.; Lebret, S.; Tomaskovic-Crook, E.; Ahmed, N.; Blick, T.; Newgreen, D.F.; Thompson, E.W.; Ackland, M.L. Contribution of Fibroblast and Mast Cell (Afferent) and Tumor (Efferent) IL-6 Effects within the Tumor Microenvironment. Cancer Microenvironment 2012, 5, 83. [CrossRef]
- Guo, Y.; Wang, B.; Wang, T.; Gao, L.; Yang, Z.J.; Wang, F.F.; Shang, H.W.; Hua, R.; Xu, J.D. Biological Characteristics of IL-6 and Related Intestinal Diseases. Int J Biol Sci 2021, 17, 204. [CrossRef]
- Kaur, S.; Bansal, Y.; Kumar, R.; Bansal, G. A Panoramic Review of IL-6: Structure, Pathophysiological Roles and Inhibitors. Bioorg Med Chem 2020, 28. [CrossRef]
- Schneider, M.R.; Hoeflich, A.; Fischer, J.R.; Wolf, E.; Sordat, B.; Lahm, H. Interleukin-6 Stimulates Clonogenic Growth of Primary and Metastatic Human Colon Carcinoma Cells. Cancer Lett 2000, 151, 31–38. [CrossRef]
- Nikiteas, N.I.; Tzanakis, N.; Gazouli, M.; Rallis, G.; Daniilidis, K.; Theodoropoulos, G. Serum IL-6, TNFalpha and CRP Levels in Greek Colorectal Cancer Patients: Prognostic Implications. World J Gastroenterol 2005, 11, 1639–1643. [CrossRef]
- Lee, W.S.; Baek, J.H.; You, D.H.; Nam, M.J. Prognostic Value of Circulating Cytokines for Stage III Colon Cancer. J Surg Res 2013, 182, 49–54. [CrossRef]
- Okugawa, Y.; Miki, C.; Toiyama, Y.; Yasuda, H.; Yokoe, T.; Saigusa, S.; Hiro, J.; Tanaka, K.; Inoue, Y.; Kusunoki, M. Loss of Tumoral Expression of Soluble IL-6 Receptor Is Associated with Disease Progression in Colorectal Cancer. Br J Cancer 2010, 103, 787. [CrossRef]
- Belluco, C.; Nitti, D.; Frantz, M.; Toppan, P.; Basso, D.; Plebani, M.; Lise, M.; Jessup, J.M. Interleukin-6 Blood Level Is Associated with Circulating Carcinoembryonic Antigen and Prognosis in Patients with Colorectal Cancer. Ann Surg Oncol 2000, 7, 133–138. [CrossRef]
- Tanner, J.; Tosato, G. Impairment of Natural Killer Functions by Interleukin 6 Increases Lymphoblastoid Cell Tumorigenicity in Athymic Mice. J Clin Invest 1991, 88, 239–247. [CrossRef]
- Matsumoto, S.; Hara, T.; Mitsuyama, K.; Yamamoto, M.; Tsuruta, O.; Sata, M.; Scheller, J.; Rose-John, S.; Kado, S.; Takada, T. Essential Roles of IL-6 Trans -Signaling in Colonic Epithelial Cells, Induced by the IL-6/Soluble–IL-6 Receptor Derived from Lamina Propria Macrophages, on the Development of Colitis-Associated Premalignant Cancer in a Murine Model . The Journal of Immunology 2010, 184, 1543–1551. [CrossRef]
- Karunanithi, S.; Levi, L.; DeVecchio, J.; Karagkounis, G.; Reizes, O.; Lathia, J.D.; Kalady, M.F.; Noy, N. RBP4-STRA6 Pathway Drives Cancer Stem Cell Maintenance and Mediates High-Fat Diet-Induced Colon Carcinogenesis. Stem Cell Reports 2017, 9, 438–450. [CrossRef]
- Noy, N.; Li, L.; Abola, M. V.; Berger, N.A. Is Retinol Binding Protein 4 a Link between Adiposity and Cancer? Horm Mol Biol Clin Investig 2015, 23, 39–46. [CrossRef]
- Wang, Y.; Wang, Y.; Zhang, Z. Adipokine RBP4 Drives Ovarian Cancer Cell Migration. J Ovarian Res 2018, 11. [CrossRef]
- Zhang, H.; Zuo, L.; Li, J.; Geng, Z.; Ge, S.; Song, X.; Wang, Y.; Zhang, X.; Wang, L.; Zhao, T.; et al. Construction of a Fecal Immune-Related Protein-Based Biomarker Panel for Colorectal Cancer Diagnosis: A Multicenter Study. Front Immunol 2023, 14. [CrossRef]
- Abola, M. V.; Thompson, C.L.; Chen, Z.; Chak, A.; Berger, N.A.; Kirwan, J.P.; Li, L. Serum Levels of Retinol-Binding Protein 4 and Risk of Colon Adenoma. Endocr Relat Cancer 2015, 22, L1–L4. [CrossRef]
- Yu, Y.; Zhang, C.; Sun, Q.; Baral, S.; Ding, J.; Zhao, F.; Yao, Q.; Gao, S.; Liu, B.; Wang, D. Retinol Binding Protein 4 Serves as a Potential Tumor Biomarker and Promotes Malignant Behavior in Gastric Cancer. Cancer Management and Research 2024, 16, 891–908. [CrossRef]
- Wlodarczyk, B.; Gasiorowska, A.; Borkowska, A.; Malecka-Panas, E. Evaluation of Insulin-like Growth Factor (IGF-1) and Retinol Binding Protein (RBP-4) Levels in Patients with Newly Diagnosed Pancreatic Adenocarcinoma (PDAC). Pancreatology 2017, 17, 623–628. [CrossRef]
- Sayagués, J.M.; Corchete, L.A.; Gutiérrez, M.L.; Sarasquete, M.E.; Abad, M. del M.; Bengoechea, O.; Fermiñán, E.; Anduaga, M.F.; del Carmen, S.; Iglesias, M.; et al. Genomic Characterization of Liver Metastases from Colorectal Cancer Patients. Oncotarget 2016, 7, 72908–72922. [CrossRef]
- El-Mesallamy, H.O.; Hamdy, N.M.; Zaghloul, A.S.; Sallam, A.M. Serum Retinol Binding Protein-4 and Neutrophil Gelatinase-Associated Lipocalin Are Interrelated in Pancreatic Cancer Patients. Scand J Clin Lab Invest 2012, 72, 602–607. [CrossRef]
- Sobotka, R.; Apoun, O.; Kalousová, M.; Hanuŝ, T.; Zima, T.; Koŝtíová, M.; Soukup, V. Prognostic Importance of Vitamins A, E and Retinol-Binding Protein 4 in Renal Cell Carcinoma Patients. Anticancer Res 2017, 37, 3801–3806. [CrossRef]
- Nono Nankam, P.A.; Blüher, M. Retinol-Binding Protein 4 in Obesity and Metabolic Dysfunctions. Mol Cell Endocrinol 2021, 531. [CrossRef]
- Antosik, P.; Durślewicz, J.; Smolińska-Świtała, M.; Podemski, J.; Podemska, E.; Neska-Długosz, I.; Jóźwicki, J.; Grzanka, D. KIF11 and KIF14 Are a Novel Potential Prognostic Biomarker in Patients with Endometrioid Carcinoma. Cancers (Basel) 2025, 17, 804. [CrossRef]
- Ogłuszka, M.; Orzechowska, M.; Jędroszka, D.; Witas, P.; Bednarek, A.K. Evaluate Cutpoints: Adaptable Continuous Data Distribution System for Determining Survival in Kaplan-Meier Estimator. Comput Methods Programs Biomed 2019, 177, 133–139. [CrossRef]
- Fabregas, J.C.; Ramnaraign, B.; George, T.J. Clinical Updates for Colon Cancer Care in 2022. Clin Colorectal Cancer 2022, 21, 198–203. [CrossRef]
- Dekker, E.; Tanis, P.J.; Vleugels, J.L.A.; Kasi, P.M.; Wallace, M.B. Colorectal Cancer. The Lancet 2019, 394, 1467–1480. [CrossRef]
- Hanna, T.P.; King, W.D.; Thibodeau, S.; Jalink, M.; Paulin, G.A.; Harvey-Jones, E.; O’Sullivan, D.E.; Booth, C.M.; Sullivan, R.; Aggarwal, A. Mortality Due to Cancer Treatment Delay: Systematic Review and Meta-Analysis. The BMJ 2020, 371, m4087. [CrossRef]
- Wan, F.; Zhu, Y.; Wu, F.; Huang, X.; Chen, Y.; Zhou, Y.; Li, H.; Liang, L.; Qin, L.; Wang, Q.; et al. Retinol-Binding Protein 4 as a Promising Serum Biomarker for the Diagnosis and Prognosis of Hepatocellular Carcinoma. Transl Oncol 2024, 45. [CrossRef]
- Wang, D.D.; Zhao, Y.M.; Wang, L.; Ren, G.; Wang, F.; Xia, Z.G.; Wang, X.L.; Zhang, T.; Pan, Q.; Dai, Z.; et al. Preoperative Serum Retinol-Binding Protein 4 Is Associated with the Prognosis of Patients with Hepatocellular Carcinoma after Curative Resection. J Cancer Res Clin Oncol 2011, 137, 651–658. [CrossRef]
- Komoda, H.; Tanaka, Y.; Honda, M.; Matsuo, Y.; Hazama, K.; Takao, T. Interleukin-6 Levels in Colorectal Cancer Tissues. World J Surg 1998, 22, 895–898. [CrossRef]
- Lu, C.C.; Kuo, H.C.; Wang, F.S.; Jou, M.H.; Lee, K.C.; Chuang, J.H. Upregulation of TLRs and IL-6 as a Marker in Human Colorectal Cancer. Int J Mol Sci 2014, 16, 159–177. [CrossRef]
- Nagasaki, T.; Hara, M.; Nakanishi, H.; Takahashi, H.; Sato, M.; Takeyama, H. Interleukin-6 Released by Colon Cancer-Associated Fibroblasts Is Critical for Tumour Angiogenesis: Anti-Interleukin-6 Receptor Antibody Suppressed Angiogenesis and Inhibited Tumour-Stroma Interaction. Br J Cancer 2014, 110, 469–478. [CrossRef]
- Chung, Y.C.; Chang, Y.F. Serum Interleukin-6 Levels Reflect the Disease Status of Colorectal Cancer. J Surg Oncol 2003, 83, 222–226. [CrossRef]
- Galizia, G.; Orditura, M.; Romano, C.; Lieto, E.; Castellano, P.; Pelosio, L.; Imperatore, V.; Catalano, G.; Pignatelli, C.; De Vita, F. Prognostic Significance of Circulating IL-10 and IL-6 Serum Levels in Colon Cancer Patients Undergoing Surgery. Clinical Immunology 2002, 102, 169–178. [CrossRef]
- Milan, J.C. Effects of Natural Aging and Gender on Pro-Inflammatory Markers. [CrossRef]
- Brozek, W.; Bises, G.; Fabjani, G.; Cross, H.S.; Peterlik, M. Clone-Specific Expression, Transcriptional Regulation, and Action of Interleukin-6 in Human Colon Carcinoma Cells. BMC Cancer 2008, 8. [CrossRef]
- Legrand-Poels, S.; Schoonbroodt, S.; Piette, J. Regulation of Interleukin-6 Gene Expression by pro-Inflammatory Cytokines in a Colon Cancer Cell Line. Biochemical Journal 2000, 349, 765–773. [CrossRef]
- OLSEN, J.; KIRKEBY, L.T.; OLSEN, J.; EIHOLM, S.; JESS, P.; GÖGENUR, I.; TROELSEN, J.T. High Interleukin-6 MRNA Expression Is a Predictor of Relapse in Colon Cancer. Anticancer Res 2015, 35.
- Tsakogiannis, D.; Kalogera, E.; Zagouri, F.; Zografos, E.; Balalis, D.; Bletsa, G. Determination of FABP4, RBP4 and the MMP-9/NGAL Complex in the Serum of Women with Breast Cancer. Oncol Lett 2021, 21. [CrossRef]
- Lorkova, L.; Pospisilova, J.; Lacheta, J.; Leahomschi, S.; Zivny, J.; Cibula, D.; Zivny, J.; Petrak, J. Decreased Concentrations of Retinol-Binding Protein 4 in Sera of Epithelial Ovarian Cancer Patients: A Potential Biomarker Identified by Proteomics. Oncol Rep 2012, 27, 318–324. [CrossRef]
- Jiao, C.; Cui, L.; Ma, A.; Li, N.; Si, H. Elevated Serum Levels of Retinol-Binding Protein 4 Are Associated with Breast Cancer Risk: A Case-Control Study. PLoS One 2016, 11. [CrossRef]
- Tsunoda, S.; Smith, E.; De Young, N.J.; Wang, X.; Tian, Z.Q.; Liu, J.F.; Jamieson, G.G.; Drew, P.A. Methylation of CLDN6, FBN2, RBP1, RBP4, TFPI2, and TMEFF2 in Esophageal Squamous Cell Carcinoma. Oncol Rep 2009, 21, 1067–1073. [CrossRef]
- Zhao, J.; Liu, Y.; Zhou, L.; Liu, Y. Retinol-Binding Protein 4 as a Biomarker in Cancer: Insights from a Pan-Cancer Analysis of Expression, Immune Infiltration, and Methylation. Genes (Basel) 2025, 16, 150. [CrossRef]
- Wan, F.; Zhu, Y.; Wu, F.; Huang, X.; Chen, Y.; Zhou, Y.; Li, H.; Liang, L.; Qin, L.; Wang, Q.; et al. Retinol-Binding Protein 4 as a Promising Serum Biomarker for the Diagnosis and Prognosis of Hepatocellular Carcinoma. Transl Oncol 2024, 45, 101979. [CrossRef]
- Jiao, C.; Cui, L.; Ma, A.; Li, N.; Si, H. Elevated Serum Levels of Retinol-Binding Protein 4 Are Associated with Breast Cancer Risk: A Case-Control Study. PLoS One 2016, 11. [CrossRef]
- Papiernik, D.; Urbaniak, A.; Kłopotowska, D.; Nasulewicz-Goldeman, A.; Ekiert, M.; Nowak, M.; Jarosz, J.; Cuprych, M.; Strzykalska, A.; Ugorski, M.; et al. Retinol-Binding Protein 4 Accelerates Metastatic Spread and Increases Impairment of Blood Flow in Mouse Mammary Gland Tumors. Cancers (Basel) 2020, 12, 623. [CrossRef]
- Deng, X.; Ren, J.; Bi, Z.; Fu, Z. Positive Expression of Retinol-Binding Protein 4 Is Related to the Malignant Clinical Features Leading to Poor Prognosis of Glioblastoma. Genet Res (Camb) 2022, 2022. [CrossRef]
- Chu, Q.; Shen, D.; He, L.; Wang, H.; Liu, C.; Zhang, W. Prognostic Significance of SOCS3 and Its Biological Function in Colorectal Cancer. Gene 2017, 627, 114–122. [CrossRef]
- Li, Y.; De Haar, C.; Chen, M.; Deuring, J.; Gerrits, M.M.; Smits, R.; Xia, B.; Kuipers, E.J.; Van Janneke Der Woude, C. Disease-Related Expression of the IL6/STAT3/SOCS3 Signalling Pathway in Ulcerative Colitis and Ulcerative Colitis-Related Carcinogenesis. Gut 2010, 59, 227–235. [CrossRef]
- Bártů, M.; Hojný, J.; Hájková, N.; Michálková, R.; Krkavcová, E.; Simon, K.; Frýba, V.; Stružinská, I.; Němejcová, K.; Dundr, P. Expression, Epigenetic, and Genetic Changes of HNF1B in Colorectal Lesions: An Analysis of 145 Cases. Pathol Oncol Res 2020, 26, 2337–2350. [CrossRef]
- Razi, S.; Baradaran Noveiry, B.; Keshavarz-Fathi, M.; Rezaei, N. IL-17 and Colorectal Cancer: From Carcinogenesis to Treatment. Cytokine 2019, 116, 7–12. [CrossRef]
- Zhou, Y.; Song, X.; Yuan, M.; Li, Y. The Diverse Function of IL-6 in Biological Processes and the Advancement of Cancer. Immune Netw 2025, 25. [CrossRef]
- Du, W.; Xia, X.; Gou, Q.; Qiu, Y. Mendelian Randomization and Transcriptomic Analysis Reveal a Positive Cause-and-Effect Relationship between Alzheimer’s Disease and Colorectal Cancer. Transl Oncol 2024, 51, 102169. [CrossRef]
- Wang, J.X.; Cao, B.; Ma, N.; Wu, K.Y.; Chen, W.B.; Wu, W.; Dong, X.; Liu, C.F.; Gao, Y.F.; Diao, T.Y.; et al. Collectin-11 Promotes Cancer Cell Proliferation and Tumor Growth. JCI Insight 2023, 8, e159452. [CrossRef]
| Clinicopathological parameters | n (%) | Il-6 Expression | p Value | RBP4 Expression | p Value | ||
| n = 118 | |||||||
| Low | High | Low | High | ||||
| n = 91 | n = 27 | n = 68 | n = 50 | ||||
| Age (years) | |||||||
| ≤65 | 50 (42.37) | 44 (88.00) | 6 (12.00) | 0.0254 | 34 (68.00) | 16 (32.00) | 0.0607 |
| >65 | 68 (57.63) | 47 (69.12) | 21 (30.88) | 34 (50.00) | 34 (50.00) | ||
| Gender | |||||||
| Male | 71 | 54 | 17 | 0.8248 | 36 | 35 | 0.0864 |
| Female | 47 | 37 | 10 | 32 | 15 | ||
| Grading | |||||||
| G1-G2 | 113 | 86 | 27 | 0.5876 | 63 | 50 | 0.0717 |
| G3 | 5 | 5 | 0 | 5 | 0 | ||
| pT status | |||||||
| T1-T2 | 61 | 49 | 12 | 40 | 21 | ||
| T3 | 54 | 39 | 15 | 0.2757 | 26 | 28 | 0.0398 |
| T4 | 3 | 2 | 1 | 1 | 2 | ||
| pN status | |||||||
| N0 | 80 | 59 | 21 | 0.8005 | 53 | 27 | 0.0044 |
| N1–N2 | 35 | 21 | 6 | 13 | 22 | ||
| pM status | |||||||
| M0 | 115 | 88 | 27 | 0.5599 | 67 | 48 | 0.5733 |
| M1 | 3 | 2 | 1 | 1 | 2 | ||
| TNM stage | |||||||
| I | 51 | 40 | 11 | 0.9851 | 34 | 17 | 0.0119 |
| II | 28 | 18 | 10 | 18 | 10 | ||
| III | 33 | 27 | 6 | 13 | 20 | ||
| IV | 3 | 2 | 1 | 1 | 2 | ||
| VI | |||||||
| Absent | 105 | 80 | 25 | 0.7298 | 64 | 41 | 0.0708 |
| Present | 13 | 11 | 2 | 4 | 9 | ||
| PNI | |||||||
| Absent | 111 | 87 | 24 | 0.1955 | 66 | 45 | 0.1323 |
| Present | 7 | 4 | 3 | 2 | 5 | ||
| Resection margin | |||||||
| R0 | 114 | 89 | 25 | 0.2243 | 66 | 48 | >0.9999 |
| R1−R2 | 4 | 2 | 2 | 2 | 2 | ||
| Primary tumor location | |||||||
| Right colon | 40 | 30 | 10 | 0.9018 | 26 | 14 | 0.3779 |
| Left colon | 40 | 33 | 7 | 21 | 19 | ||
| Rectum | 38 | 28 | 10 | 21 | 17 | ||
| Obesity | |||||||
| BMI <30 kg/m2 | 83 | 74 | 9 | < 0.0001 | 48 | 35 | 0.0264 |
| BMI ≥30 kg/m2 | 35 | 17 | 18 | 12 | 23 | ||
| Waist circumference | |||||||
| <88 cm (female) / <102 cm (male) | 48 | 40 | 8 | 0.0127 | 32 | 16 | 0.0396 |
| ≥88 cm (female) / ≥102 cm (male) | 47 | 28 | 19 | 21 | 26 | ||
| Clinicopathological parameters | n (%) | Il-6 Expression | p Value | RBP4 Expression | p Value | ||
| n = 350 | |||||||
| Low | High | Low | High | ||||
| n = 191 | n = 159 | n = 95 | n = 255 | ||||
| Age (years) | |||||||
| ≤65 | 169 (48.29) | 88 (52.07) | 81 (47.93) | 0.364 | 48 (28.4) | 121 (71.6) | 0.6086 |
| >65 | 181 (51.71) | 103 (56.91) | 78 (43.09) | 47 (25.97) | 134 (74.03) | ||
| Gender | |||||||
| Male | 189 (54) | 110 (58.2) | 79 (41.8) | 0.1395 | 60 (31.75) | 129 (68.25) | 0.0359 |
| Female | 161 (46) | 81 (50.31) | 80 (49.69) | 35 (21.74) | 126 (78.26) | ||
| pT status | |||||||
| T1-T2 | 64 (18.29) | 35 (54.69) | 29 (45.31) | 0.3761 | 17 (26.56) | 47 (73.44) | 0.9912 |
| T3 | 239 (68.29) | 126 (52.72) | 113 (47.28) | 65 (27.2) | 174 (72.8) | ||
| T4 | 47 (13.43) | 30 (63.83) | 17 (36.17) | 13 (27.66) | 34 (72.34) | ||
| pN status | |||||||
| N0 | 195 (55.71) | 101 (51.79) | 94 (48.21) | 0.2386 | 40 (20.51) | 155 (79.49) | 0.0018 |
| N1–N2 | 155 (44.29) | 90 (58.06) | 65 (41.94) | 55 (35.48) | 100 (54.52) | ||
| pM status | |||||||
| M0 | 247 (70.57) | 137 (55.47) | 110 (44.53) | 0.1364 | 62 (25.1) | 185 (74.9) | 0.0086 |
| M1 | 48 (13.71) | 21 (43.75) | 27 (56.25) | 21 (43.75) | 27 (56.25) | ||
| MX | 55 (15.71) | 33 (60) | 22 (40) | 12 (21.82) | 43 (78.18) | ||
| TNM stage | |||||||
| I | 56 (16) | 31 (55.36) | 25 (44.64) | 0.9259 | 15 (26.79) | 41 (73.21) | 0.0017 |
| II | 132 (37.71) | 69 (52.27) | 63 (47.73) | 22 (16.67) | 110 (83.33) | ||
| III | 110 (31.43) | 62 (56.36) | 48 (43.64) | 36 (32.72) | 74 (67.27) | ||
| IV | 52 (14.86) | 29 (55.77) | 23 (44.23) | 22 (42.31) | 30 (57.69) | ||
|
IL-6 (+) Correlated Gene |
Cytoband | Spearman's Correlation | p Value |
IL-6 (+) Correlated Gene |
Cytoband | Spearman's Correlation | p Value |
| IL11 | 19q13.42 | 0.765 | 5.83 × 10⁻¹⁰² | GPR84 | 12q13.13 | 0.608 | 2.37 × 10⁻⁵⁴ |
| SOCS3 | 17q25.3 | 0.753 | 9.05 × 10⁻⁹⁷ | FCAR | 19q13.42 | 0.603 | 3.64 × 10⁻⁵³ |
| IL24 | 1q32.1 | 0.742 | 1.34 × 10⁻⁹² | CCL8 | 17q12 | 0.601 | 7.69 × 10⁻⁵³ |
| CXCL8 | 4q13.3 | 0.738 | 3.66 × 10⁻⁹¹ | PAPPA | 9q33.1 | 0.601 | 9.60 × 10⁻⁵³ |
| CSF2 | 5q31.1 | 0.724 | 2.24 × 10⁻⁸⁶ | ADAMTS4 | 1q23.3 | 0.600 | 1.48 × 10⁻⁵² |
| OSM | 22q12.2 | 0.721 | 3.34 × 10⁻⁸⁵ | SELE | 1q24.2 | 0.598 | 4.18 × 10⁻⁵² |
| PTGS2 | 1q31.1 | 0.710 | 1.07 × 10⁻⁸¹ | NR4A3 | 9q31.1 | 0.596 | 8.60 × 10⁻⁵² |
| MMP3 | 11q22.2 | 0.708 | 6.63 × 10⁻⁸¹ | HSD11B1 | 1q32.2 | 0.595 | 1.37 × 10⁻⁵¹ |
| CSF3 | 17q21.1 | 0.706 | 3.81 × 10⁻⁸⁰ | FPR1 | 19q13.41 | 0.595 | 1.85 × 10⁻⁵¹ |
| CXCL5 | 4q13.3 | 0.685 | 6.51 × 10⁻⁷⁴ | PROK2 | 3p13 | 0.589 | 3.74 × 10⁻⁵⁰ |
| MMP1 | 11q22.2 | 0.682 | 8.15 × 10⁻⁷³ | LILRA5 | 19q13.42 | 0.585 | 2.35 × 10⁻⁴⁹ |
| IL1B | 2q14.1 | 0.673 | 2.25 × 10⁻⁷⁰ | CCL4 | 17q12 | 0.585 | 2.43 × 10⁻⁴⁹ |
| CCL2 | 17q12 | 0.655 | 1.42 × 10⁻⁶⁵ | SERPINE1 | 7q22.1 | 0.583 | 4.88 × 10⁻⁴⁹ |
| CCL3 | 17q12 | 0.650 | 3.82 × 10⁻⁶⁴ | PRR16 | 5q23.1 | 0.580 | 2.21 × 10⁻⁴⁸ |
| PLAU | 10q22.2 | 0.644 | 1.07 × 10⁻⁶² | PDPN | 1p36.21 | 0.578 | 4.03 × 10⁻⁴⁸ |
| HCAR2 | 12q24.31 | 0.641 | 4.90 × 10⁻⁶² | CCL3L1 | 17q12 | 0.578 | 4.27 × 10⁻⁴⁸ |
| FPR2 | 19q13.41 | 0.626 | 2.02 × 10⁻⁵⁸ | TFPI2 | 7q21.3 | 0.576 | 1.21 × 10⁻⁴⁷ |
| BCL2A1 | 15q25.1 | 0.620 | 4.85 × 10⁻⁵⁷ | TREM1 | 6p21.1 | 0.574 | 3.02 × 10⁻⁴⁷ |
| TNFAIP6 | 2q23.3 | 0.620 | 5.66 × 10⁻⁵⁷ | CCN4 | 8q24.22 | 0.571 | 1.07 × 10⁻⁴⁶ |
| STC1 | 8p21.2 | 0.619 | 7.72 × 10⁻⁵⁷ | AQP9 | 15q21.3 | 0.566 | 9.93 × 10⁻⁴⁶ |
| PI15 | 8q21.13 | 0.616 | 4.78 × 10⁻⁵⁶ | KCNJ15 | 21q22.13-q22.2 | 0.564 | 2.75 × 10⁻⁴⁵ |
| S100A8 | 1q21.3 | 0.614 | 1.15 × 10⁻⁵⁵ | CYTL1 | 4p16.2 | 0.562 | 7.00 × 10⁻⁴⁵ |
| CXCL6 | 4q13.3 | 0.614 | 1.51 × 10⁻⁵⁵ | FCGR2A | 1q23.3 | 0.554 | 1.87 × 10⁻⁴³ |
| HCAR3 | 12q24.31 | 0.610 | 8.46 × 10⁻⁵⁵ | CSF3R | 1p34.3 | 0.554 | 2.02 × 10⁻⁴³ |
| SLC2A3 | 12p13.31 | 0.610 | 1.20 × 10⁻⁵⁴ | MEDAG | 13q12.3 | 0.552 | 3.54 × 10⁻⁴³ |
|
IL-6 (-) Correlated Gene |
Cytoband | Spearman's Correlation | p Value |
IL-6 (-) Correlated Gene |
Cytoband | Spearman's Correlation | p Value |
| HNF1B | 17q12 | -0.356 | 4.10 × 10⁻¹⁷ | NGEF | 2q37.1 | -0.292 | 9.47 × 10⁻¹² |
| IL17RE | 3p25.3 | -0.356 | 4.39 × 10⁻¹⁷ | ARHGAP8 | 22q13.31 | -0.292 | 9.73 × 10⁻¹² |
| WNK4 | 17q21.2 | -0.350 | 1.49 × 10⁻¹⁶ | RAVER2 | 1p31.3 | -0.291 | 1.04 × 10⁻¹¹ |
| C14ORF93 | 14q11.2 | -0.328 | 1.36 × 10⁻¹⁴ | ILDR1 | 3q13.33 | -0.291 | 1.06 × 10⁻¹¹ |
| FOXD2-AS1 | 1p33 | -0.326 | 1.88 × 10⁻¹⁴ | SIRT5 | 6p23 | -0.291 | 1.07 × 10⁻¹¹ |
| PPFIBP2 | 11p15.4 | -0.323 | 3.67 × 10⁻¹⁴ | PGAP3 | 17q12 | -0.289 | 1.44 × 10⁻¹¹ |
| IHH | 2q35 | -0.322 | 4.12 × 10⁻¹⁴ | HSD11B2 | 16q22.1 | -0.287 | 2.00 × 10⁻¹¹ |
| CEBPA-DT | 19q13.11 | -0.314 | 1.92 × 10⁻¹³ | CRACDL | 2q11.2 | -0.287 | 2.15 × 10⁻¹¹ |
| NR1I2 | 3q13.33 | -0.313 | 2.08 × 10⁻¹³ | PRKCZ | 1p36.33 | -0.287 | 2.31 × 10⁻¹¹ |
| TACC2 | 10q26.13 | -0.310 | 3.63 × 10⁻¹³ | CC2D1A | 19p13.12 | -0.286 | 2.42 × 10⁻¹¹ |
| ZNF768 | 16p11.2 | -0.310 | 4.03 × 10⁻¹³ | SHD | 19p13.3 | -0.286 | 2.63 × 10⁻¹¹ |
| FGFR3 | 4p16.3 | -0.308 | 6.06 × 10⁻¹³ | HNF1A-AS1 | 12q24.31 | -0.284 | 3.72 × 10⁻¹¹ |
| BDH1 | 3q29 | -0.307 | 6.83 × 10⁻¹³ | DAPK2 | 15q22.31 | -0.282 | 5.03 × 10⁻¹¹ |
| EVPL | 17q25.1 | -0.306 | 7.81 × 10⁻¹³ | COQ8A | 1q42.13 | -0.280 | 6.64 × 10⁻¹¹ |
| AKAP1 | 17q22 | -0.304 | 1.08 × 10⁻¹² | CCHCR1 | 6p21.33 | -0.280 | 6.80 × 10⁻¹¹ |
| MKRN1 | 7q34 | -0.304 | 1.18 × 10⁻¹² | ANKS4B | 16p12.2 | -0.280 | 7.05 × 10⁻¹¹ |
| PPP1R1B | 17q12 | -0.303 | 1.28 × 10⁻¹² | TMEM184A | 7p22.3 | -0.279 | 7.41 × 10⁻¹¹ |
| ZNF69 | 19p13.2 | -0.302 | 1.76 × 10⁻¹² | ARHGEF5 | 7q35 | -0.278 | 8.93 × 10⁻¹¹ |
| ERBB3 | 12q13.2 | -0.300 | 2.26 × 10⁻¹² | NDUFA10 | 2q37.3 | -0.278 | 9.17 × 10⁻¹¹ |
| CCDC183 | 9q34.3 | -0.297 | 3.63 × 10⁻¹² | EIF4EBP3 | 5q31.3 | -0.278 | 9.23 × 10⁻¹¹ |
| SGK2 | 20q13.12 | -0.294 | 6.30 × 10⁻¹² | ACVR1B | 12q13.13 | -0.277 | 1.06 × 10⁻¹⁰ |
| AP1M2 | 19p13.2 | -0.294 | 6.53 × 10⁻¹² | TRABD2A | 2p11.2 | -0.276 | 1.30 × 10⁻¹⁰ |
| PLEKHA6 | 1q32.1 | -0.292 | 8.76 × 10⁻¹² | USH1C | 11p15.1 | -0.275 | 1.49 × 10⁻¹⁰ |
| MYO7B | 2q14.3 | -0.292 | 9.00 × 10⁻¹² | ERBB2 | 17q12 | -0.275 | 1.54 × 10⁻¹⁰ |
| EPB41L4B | 9q31.3 | -0.292 | 9.03 × 10⁻¹² | ZSWIM5 | 1p34.1 | -0.274 | 1.88 × 10⁻¹⁰ |
|
RBP4 (+) Correlated Gene |
Cytoband |
Spearman's Correlation |
p Value | RBP4 (+) Correlated Gene | Cytoband | Spearman's Correlation | p Value |
| COLEC11 | 2p25.3 | 0.369 | 2.19 × 10⁻¹⁸ | OTUD5 | Xp11.23 | 0.319 | 7.83 × 10⁻¹⁴ |
| CDIPT | 16p11.2 | 0.368 | 2.95 × 10⁻¹⁸ | LSAMP | 3q13.31 | 0.319 | 8.05 × 10⁻¹⁴ |
| DDAH2 | 6p21.33 | 0.367 | 3.61 × 10⁻¹⁸ | ZNF853 | 7p22.1 | 0.318 | 8.74 × 10⁻¹⁴ |
| FZD8 | 10p11.21 | 0.360 | 1.61 × 10⁻¹⁷ | RNF32-DT | 7q36.3 | 0.317 | 1.06 × 10⁻¹³ |
| RAMP2 | 17q21.2 | 0.358 | 2.61 × 10⁻¹⁷ | GLI1 | 12q13.3 | 0.317 | 1.08 × 10⁻¹³ |
| ITGB5 | 3q21.2 | 0.355 | 5.04 × 10⁻¹⁷ | FBLN1 | 22q13.31 | 0.316 | 1.24 × 10⁻¹³ |
| SLC29A3 | 10q22.1 | 0.353 | 8.12 × 10⁻¹⁷ | NFASC | 1q32.1 | 0.316 | 1.30 × 10⁻¹³ |
| TGFB1I1 | 16p11.2 | 0.350 | 1.67 × 10⁻¹⁶ | TFEB | 6p21.1 | 0.315 | 1.54 × 10⁻¹³ |
| TBC1D25 | Xp11.23 | 0.346 | 3.57 × 10⁻¹⁶ | GFRA3 | 5q31.2 | 0.314 | 1.78 × 10⁻¹³ |
| APBB1 | 11p15.4 | 0.346 | 3.59 × 10⁻¹⁶ | USP11 | Xp11.3 | 0.314 | 1.81 × 10⁻¹³ |
| TCF7L1 | 2p11.2 | 0.342 | 8.46 × 10⁻¹⁶ | TCEAL3 | Xq22.2 | 0.314 | 1.83 × 10⁻¹³ |
| ELN | 7q11.23 | 0.339 | 1.45 × 10⁻¹⁵ | PDLIM4 | 5q31.1 | 0.314 | 1.95 × 10⁻¹³ |
| MED29 | 19q13.2 | 0.339 | 1.50 × 10⁻¹⁵ | NACAD | 7p13 | 0.312 | 2.83 × 10⁻¹³ |
| SALL4 | 20q13.2 | 0.333 | 5.08 × 10⁻¹⁵ | PODN | 1p32.3 | 0.312 | 2.89 × 10⁻¹³ |
| CST3 | 20p11.21 | 0.333 | 5.10 × 10⁻¹⁵ | PLAT | 8p11.21 | 0.311 | 3.00 × 10⁻¹³ |
| TSPY26P | 20q11.21 | 0.327 | 1.67 × 10⁻¹⁴ | USP27X | Xp11.23 | 0.311 | 3.23 × 10⁻¹³ |
| HHIP | 4q31.21 | 0.326 | 2.08 × 10⁻¹⁴ | APBA1 | 9q21.12 | 0.310 | 3.87 × 10⁻¹³ |
| RBFOX1 | 16p13.3 | 0.326 | 2.09 × 10⁻¹⁴ | NMUR1 | 2q37.1 | 0.309 | 4.76 × 10⁻¹³ |
| LRRC32 | 11q13.5 | 0.325 | 2.38 × 10⁻¹⁴ | RUSC2 | 9p13.3 | 0.308 | 5.70 × 10⁻¹³ |
| INMT | 7p14.3 | 0.323 | 3.18 × 10⁻¹⁴ | TAFA5 | 22q13.32 | 0.308 | 5.79 × 10⁻¹³ |
| CPZ | 4p16.1 | 0.323 | 3.55 × 10⁻¹⁴ | SLC24A3 | 20p11.23 | 0.307 | 7.31 × 10⁻¹³ |
| PTCH2 | 1p34.1 | 0.322 | 3.98 × 10⁻¹⁴ | BMERB1 | 16p13.11 | 0.306 | 7.47 × 10⁻¹³ |
| FRZB | 2q32.1 | 0.322 | 4.22 × 10⁻¹⁴ | KIAA1755 | 20q11.23 | 0.306 | 8.41 × 10⁻¹³ |
| TEAD3 | 6p21.31 | 0.319 | 6.87 × 10⁻¹⁴ | SCUBE3 | 6p21.3 | 0.306 | 8.70 × 10⁻¹³ |
| TFE3 | Xp11.23 | 0.319 | 7.78 × 10⁻¹⁴ | ACTB | 7p22.1 | 0.305 | 8.97 × 10⁻¹³ |
| RBP4 (-) Correlated Gene | Cytoband | Spearman's Correlation | p Value | RBP4 (-) Correlated Gene | Cytoband | Spearman's Correlation | p Value |
| HSPA4L | 4q28.1 | -0.398 | 2.71 × 10⁻²¹ | MAP2K1 | 15q22.31 | -0.294 | 6.63 × 10⁻¹² |
| PPAT | 4q12 | -0.365 | 6.35 × 10⁻¹⁸ | INTS13 | 12p11.23 | -0.294 | 6.65 × 10⁻¹² |
| GUF1 | 4p12 | -0.346 | 3.50 × 10⁻¹⁶ | PDS5A | 4p14 | -0.293 | 7.66 × 10⁻¹² |
| LIAS | 4p14 | -0.336 | 2.47 × 10⁻¹⁵ | CCT8 | 21q21.3 | -0.293 | 7.69 × 10⁻¹² |
| RPRD1A | 18q12.2 | -0.325 | 2.40 × 10⁻¹⁴ | UBE3A | 15q11.2 | -0.290 | 1.33 × 10⁻¹¹ |
| CBR4 | 4q32.3 | -0.322 | 4.06 × 10⁻¹⁴ | TIMM21 | 18q22.3 | -0.289 | 1.46 × 10⁻¹¹ |
| POLR3G | 5q14.3 | -0.321 | 5.49 × 10⁻¹⁴ | TM7SF3 | 12p11.23 | -0.289 | 1.58 × 10⁻¹¹ |
| TMEM33 | 4p13 | -0.320 | 5.83 × 10⁻¹⁴ | MRPS27 | 5q13.2 | -0.289 | 1.59 × 10⁻¹¹ |
| SFXN1 | 5q35.2 | -0.316 | 1.21 × 10⁻¹³ | CDC7 | 1p22.1 | -0.287 | 2.17 × 10⁻¹¹ |
| ME2 | 18q21.2 | -0.316 | 1.38 × 10⁻¹³ | EXOSC9 | 4q27 | -0.286 | 2.38 × 10⁻¹¹ |
| OIP5-AS1 | 15q15.1 | -0.315 | 1.46 × 10⁻¹³ | MDH1 | 2p15 | -0.286 | 2.46 × 10⁻¹¹ |
| PAICS | 4q12 | -0.312 | 2.77 × 10⁻¹³ | TIPIN | 15q22.31 | -0.286 | 2.59 × 10⁻¹¹ |
| MREG | 2q35 | -0.311 | 3.18 × 10⁻¹³ | DNAJC16 | 1p36.21 | -0.285 | 2.86 × 10⁻¹¹ |
| C12ORF60 | 12p12.3 | -0.310 | 3.75 × 10⁻¹³ | PMS1 | 2q32.2 | -0.285 | 3.10 × 10⁻¹¹ |
| SLC35F2 | 11q22.3 | -0.307 | 7.12 × 10⁻¹³ | LARS1 | 5q32 | -0.283 | 3.89 × 10⁻¹¹ |
| CIAO2A | 15q22.31 | -0.306 | 8.36 × 10⁻¹³ | COPS2 | 15q21.1 | -0.283 | 4.23 × 10⁻¹¹ |
| GRSF1 | 4q13.3 | -0.305 | 9.43 × 10⁻¹³ | VRK2 | 2p16.1 | -0.282 | 4.68 × 10⁻¹¹ |
| CENPE | 4q24 | -0.303 | 1.35 × 10⁻¹² | MTIF2 | 2p16.1 | -0.282 | 4.69 × 10⁻¹¹ |
| UBA6 | 4q13.2 | -0.302 | 1.54 × 10⁻¹² | CDC27 | 17q21.32 | -0.282 | 4.70 × 10⁻¹¹ |
| FASTKD1 | 2q31.1 | -0.302 | 1.75 × 10⁻¹² | NCAPG | 4p15.31 | -0.282 | 4.90 × 10⁻¹¹ |
| TFRC | 3q29 | -0.301 | 1.86 × 10⁻¹² | RAD54B | 8q22.1 | -0.282 | 5.16 × 10⁻¹¹ |
| ETF1 | 5q31.2 | -0.296 | 4.50 × 10⁻¹² | TMEM161B | 5q14.3 | -0.281 | 5.46 × 10⁻¹¹ |
| SKA1 | 18q21.1 | -0.296 | 4.70 × 10⁻¹² | SELENOI | 2p23.3 | -0.281 | 5.91 × 10⁻¹¹ |
| CHAF1B | 21q22.12-q22.13 | -0.295 | 5.30 × 10⁻¹² | SRRM1 | 1p36.11 | -0.280 | 6.30 × 10⁻¹¹ |
| NARS1 | 18q21.31 | -0.295 | 5.83 × 10⁻¹² | TMX2 | 11q12.1 | -0.279 | 7.61 × 10⁻¹¹ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





