Submitted:
15 July 2025
Posted:
17 July 2025
You are already at the latest version
Abstract

Keywords:
1. Introduction
2. Materials and Methods
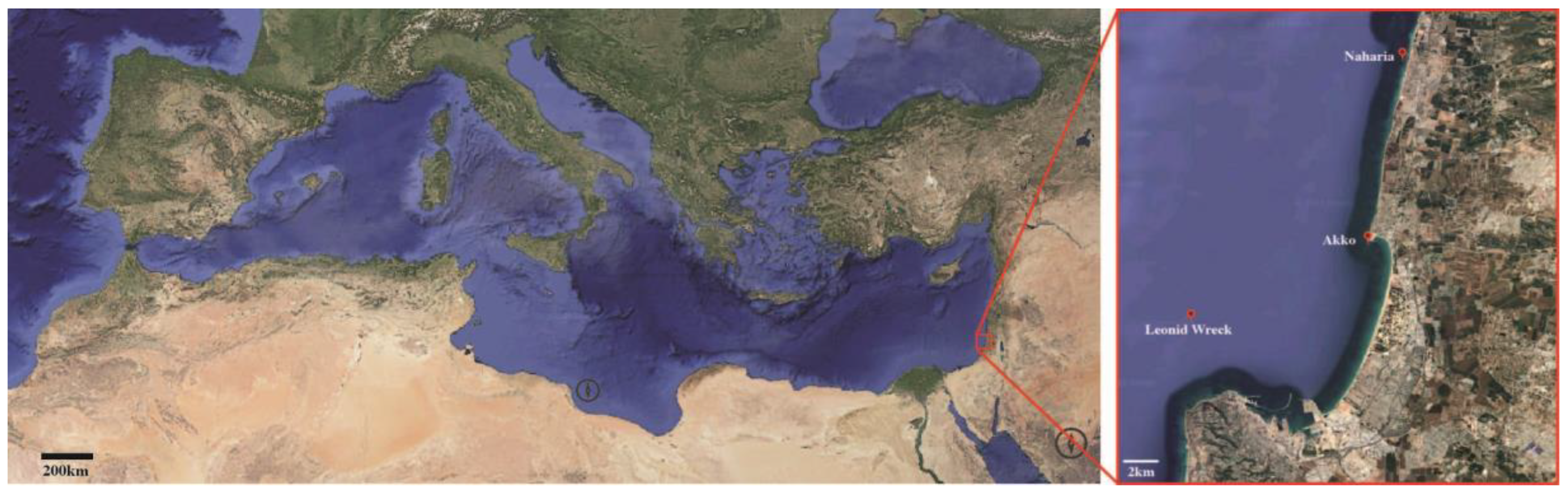
2.1. Collection
2.2. Photosynthetic Parameters Assessment
2.3. Skeletal Morphology
2.3.1. Scanning electron Microscopy
2.3.2. Computed Tomography Scanning
2.4. DNA Extraction
2.4.1. Modern Corals
2.4.2. Sub-Fossil Corals
2.4.3. Species Relationships
2.5. Compound Specific Stable Isotope Analysis of Amino Acids (CSIA-AA):
2.5.1. Sample Preparation
2.5.2. CSIA-AA
3. Results
3.1. Morphology:
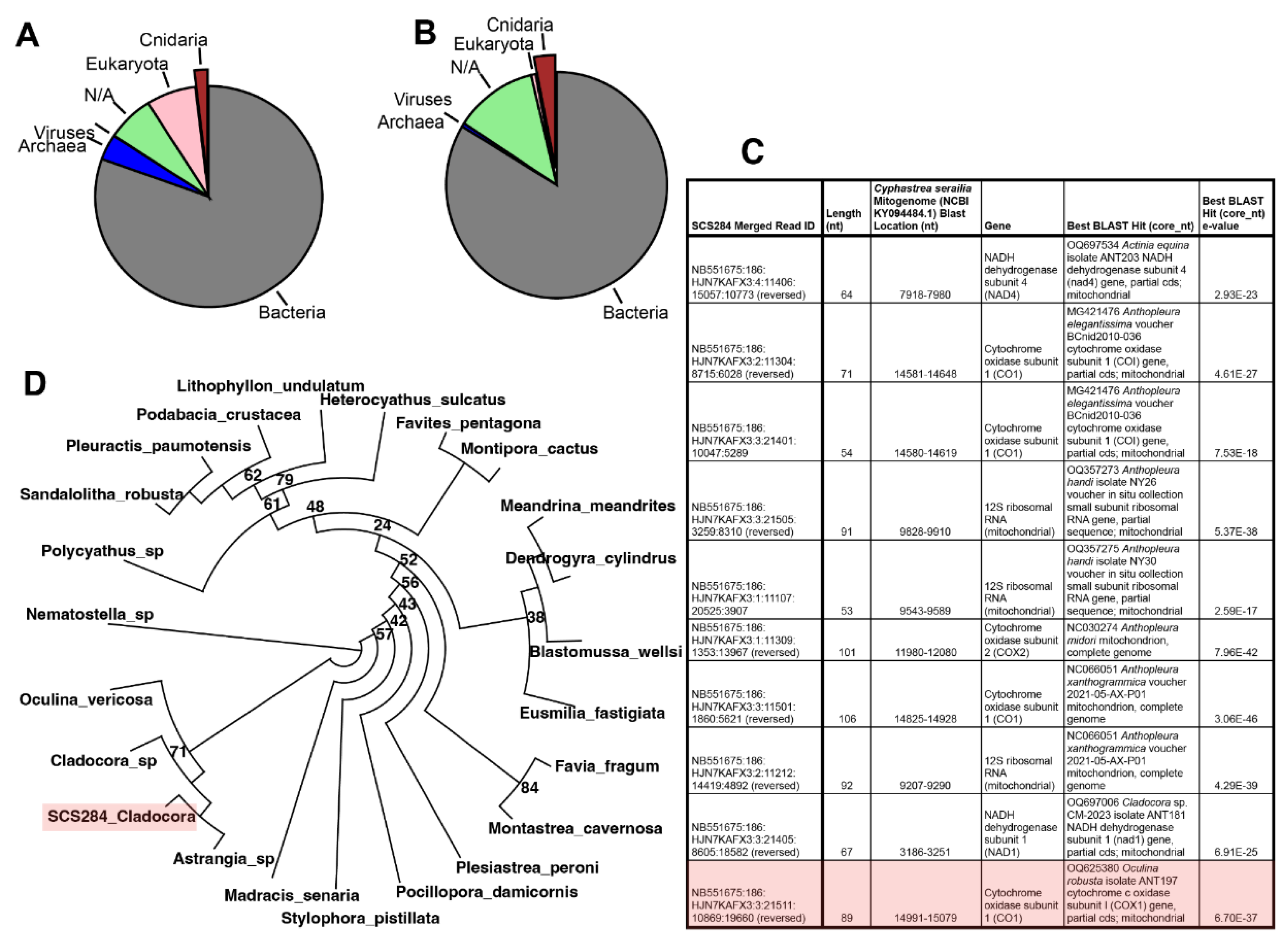
3.2. DNA Results:
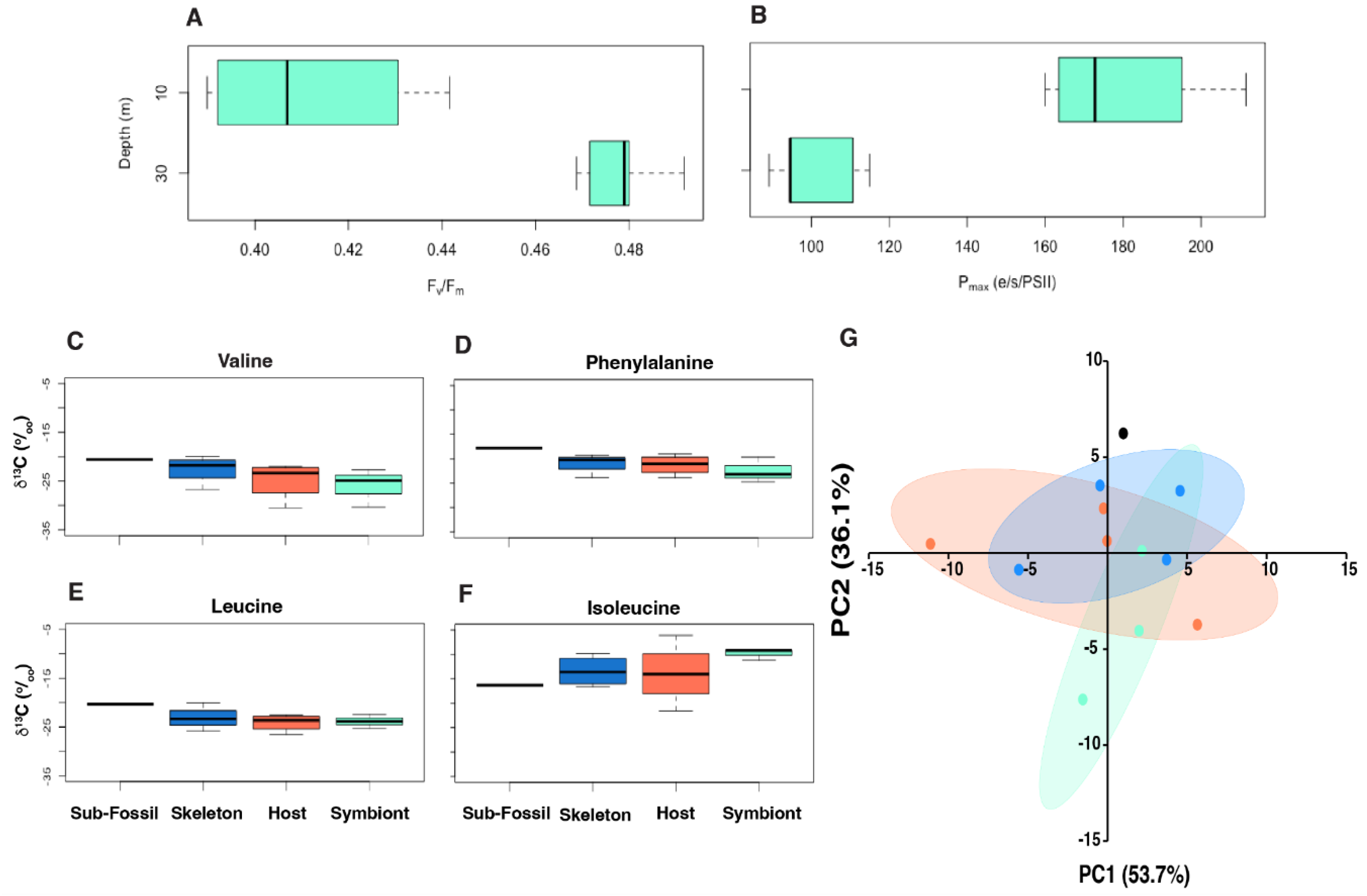
3.3. Photophysiology
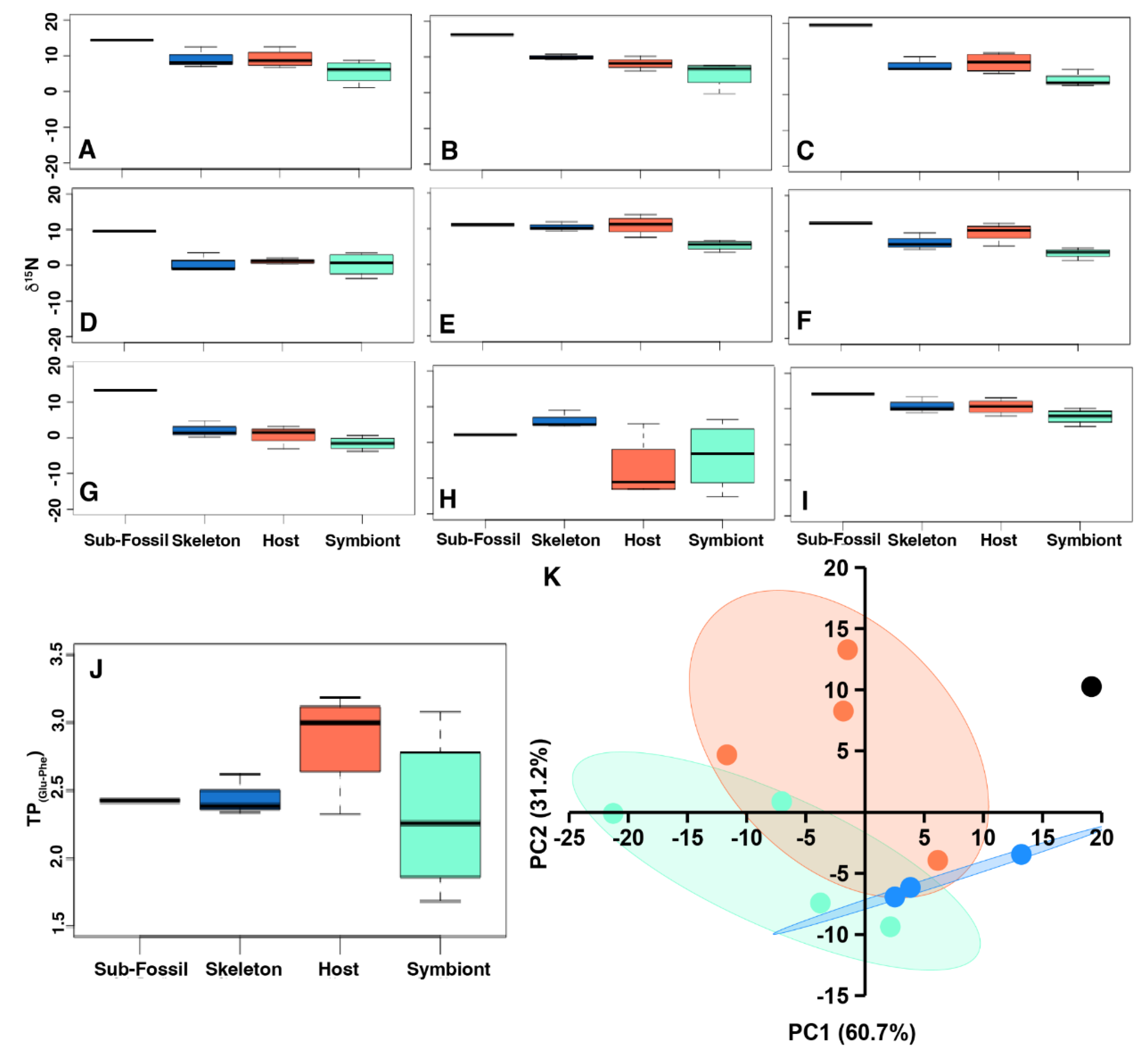
3.4. CSIA-AA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Drake, J.L.; Mass, T.; Stolarski, J.; Von Euw, S.; van de Schootbrugge, B.; Falkowski, P.G. How Corals Made Rocks through the Ages. Glob. Chang. Biol. 2020, 26, 31–53. [Google Scholar] [CrossRef]
- Knowlton, N.; Brainard, R.E.; Fisher, R.; Moews, M.; Plaisance, L.; Caley, M.J. Coral Reef Biodiversity. In Life in the World’s Oceans; Wiley-Blackwell: Oxford, UK, 2010; ISBN 9781444325508. [Google Scholar]
- Rocha, L.A.; Bowen, B.W. Speciation in Coral-reef Fishes. J. Fish Biol. 2008, 72, 1101–1121. [Google Scholar] [CrossRef]
- Mumby, P.J.; Broad, K.; Brumbaugh, D.R.; Dahlgren, C.P.; Harborne, A.R.; Hastings, A.; Holmes, K.E.; Kappel, C.V.; Micheli, F.; Sanchirico, J.N. Coral Reef Habitats as Surrogates of Species, Ecological Functions, and Ecosystem Services: Coral Reef Habitats as Surrogates. Conserv. Biol. 2008, 22, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Veron, J.E.N. Corals of the World; Australian Institute of Marine Sciences: Townsville, 2000. [Google Scholar]
- Malik, A.; Einbinder, S.; Martinez, S.; Tchernov, D.; Haviv, S.; Almuly, R.; Zaslansky, P.; Polishchuk, I.; Pokroy, B.; Stolarski, J.; et al. Molecular and Skeletal Fingerprints of Scleractinian Coral Biomineralization: From the Sea Surface to Mesophotic Depths. Acta Biomater. 2021, 120, 263–276. [Google Scholar] [CrossRef]
- Bellworthy, J.; Pardo, R.; Scucchia, F.; Zaslansky, P.; Goodbody-Gringley, G.; Mass, T. Physiological and Morphological Plasticity in Stylophora Pistillata Larvae from Eilat, Israel, to Shallow and Mesophotic Light Conditions. iScience 2023, 26, 106969. [Google Scholar] [CrossRef]
- Scucchia, F.; Wong, K.; Zaslansky, P.; Putnam, H.M.; Goodbody-Gringley, G.; Mass, T. Morphological and Genetic Mechanisms Underlying the Plasticity of the Coral Porites Astreoides across Depths in Bermuda. J. Struct. Biol. 2023, 215, 108036. [Google Scholar] [CrossRef]
- Goodbody-Gringley, G.; Waletich, J. Morphological Plasticity of the Depth Generalist Coral, Montastraea Cavernosa, on Mesophotic Reefs in Bermuda. Ecology 2018, 99, 1688–1690. [Google Scholar] [CrossRef]
- Mass, T.; Brickner, I.; Hendy, E.; Genin, A. Enduring Physiological and Reproductive Benefits of Enhanced Flow for a Stony Coral. Limnol. Oceanogr. 2011, 56, 2176–2188. [Google Scholar] [CrossRef]
- Mass, T.; Genin, A. Environmental versus Intrinsic Determination of Colony Symmetry in the Coral Pocillopora Verrucosa. Mar. Ecol. Prog. Ser. 2008, 369, 131–137. [Google Scholar] [CrossRef]
- Helmuth, B.; Sebens, K. The Influence of Colony Morphology and Orientation to Flow on Particle Capture by the Scleractinian Coral Agaricia Agaricites (Linnaeus). J. Exp. Mar. Bio. Ecol. 1993, 165, 251. [Google Scholar] [CrossRef]
- Patterson, M.R. A Mass-Transfer Explanation of Metabolic Scaling Relations in Some Aquatic Invertebrates and Algae. Science 1992, 255, 1421–1423. [Google Scholar] [CrossRef]
- Stanley, G.D. , Jr The Evolution of Modern Corals and Their Early History. Earth Sci. Rev. 2003, 60, 195–225. [Google Scholar] [CrossRef]
- Thompson, D.M. Environmental Records from Coral Skeletons: A Decade of Novel Insights and Innovation. Wiley Interdiscip. Rev. Clim. Change 2022, 13. [Google Scholar] [CrossRef]
- DeLong, K.L.; Quinn, T.M.; Taylor, F.W.; Shen, C.-C.; Lin, K. Improving Coral-Base Paleoclimate Reconstructions by Replicating 350years of Coral Sr/Ca Variations. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2013, 373, 6–24. [Google Scholar] [CrossRef]
- Trotter, J.; Montagna, P.; McCulloch, M.; Silenzi, S.; Reynaud, S.; Mortimer, G.; Martin, S.; Ferrier-Pagès, C.; Gattuso, J.-P.; Rodolfo-Metalpa, R. Quantifying the pH “vital Effect” in the Temperate Zooxanthellate Coral Cladocora Caespitosa: Validation of the Boron Seawater pH Proxy. Earth Planet. Sci. Lett. 2011, 303, 163–173. [Google Scholar] [CrossRef]
- Gothmann, A.M.; Stolarski, J.; Adkins, J.F.; Schoene, B.; Dennis, K.J.; Schrag, D.P.; Mazur, M.; Bender, M.L. Fossil Corals as an Archive of Secular Variations in Seawater Chemistry since the Mesozoic. Geochim. Cosmochim. Acta 2015, 160, 188–208. [Google Scholar] [CrossRef]
- Muscatine, L.; Goiran, C.; Land, L.; Jaubert, J.; Cuif, J.-P.; Allemand, D. Stable Isotopes (delta13C and delta15N) of Organic Matrix from Coral Skeleton. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 1525–1530. [Google Scholar] [CrossRef]
- Wang, X.T.; Sigman, D.M.; Cohen, A.L.; Sinclair, D.J.; Sherrell, R.M.; Weigand, M.A.; Erler, D.V.; Ren, H. Isotopic Composition of Skeleton-Bound Organic Nitrogen in Reef-Building Symbiotic Corals: A New Method and Proxy Evaluation at Bermuda. Geochim. Cosmochim. Acta 2015, 148, 179–190. [Google Scholar] [CrossRef]
- Jung, J.; Zoppe, S.F.; Söte, T.; Moretti, S.; Duprey, N.N.; Foreman, A.D.; Wald, T.; Vonhof, H.; Haug, G.H.; Sigman, D.M.; et al. Coral Photosymbiosis on Mid-Devonian Reefs. Nature 2024, 636, 647–653. [Google Scholar] [CrossRef]
- Frankowiak, K.; Wang, X.T.; Sigman, D.M.; Gothmann, A.M.; Kitahara, M.V.; Mazur, M.; Meibom, A.; Stolarski, J. Photosymbiosis and the Expansion of Shallow-Water Corals. Sci. Adv. 2016, 2, e1601122. [Google Scholar] [CrossRef]
- Wang, X.T.; Sigman, D.M.; Cohen, A.L.; Sinclair, D.J.; Sherrell, R.M.; Cobb, K.M.; Erler, D.V.; Stolarski, J.; Kitahara, M.V.; Ren, H. Influence of Open Ocean Nitrogen Supply on the Skeletal δ15N of Modern Shallow-Water Scleractinian Corals. Earth Planet. Sci. Lett. 2016, 441, 125–132. [Google Scholar] [CrossRef]
- Martinez, S.; Kolodny, Y.; Shemesh, E.; Scucchia, F.; Nevo, R.; Levin-Zaidman, S.; Paltiel, Y.; Keren, N.; Tchernov, D.; Mass, T. Energy Sources of the Depth-Generalist Mixotrophic Coral Stylophora Pistillata. Front Mar Sci 2020, 7, 988. [Google Scholar] [CrossRef] [PubMed]
- Kast, E.R.; Griffiths, M.L.; Kim, S.L.; Rao, Z.C.; Shimada, K.; Becker, M.A.; Maisch, H.M.; Eagle, R.A.; Clarke, C.A.; Neumann, A.N.; et al. Cenozoic Megatooth Sharks Occupied Extremely High Trophic Positions. Sci. Adv. 2022, 8, eabl6529. [Google Scholar] [CrossRef] [PubMed]
- Kast, E.R.; Stolper, D.A.; Auderset, A.; Higgins, J.A.; Ren, H.; Wang, X.T.; Martínez-García, A.; Haug, G.H.; Sigman, D.M. Nitrogen Isotope Evidence for Expanded Ocean Suboxia in the Early Cenozoic. Science 2019, 364, 386–389. [Google Scholar] [CrossRef]
- Tornabene, C.; Martindale, R.C.; Wang, X.T.; Schaller, M.F. Detecting Photosymbiosis in Fossil Scleractinian Corals. Sci. Rep. 2017, 7, 9465. [Google Scholar] [CrossRef]
- Wang, X.T.; Wang, Y.; Auderset, A.; Sigman, D.M.; Ren, H.; Martínez-García, A.; Haug, G.H.; Su, Z.; Zhang, Y.G.; Rasmussen, B.; et al. Oceanic Nutrient Rise and the Late Miocene Inception of Pacific Oxygen-Deficient Zones. Proc. Natl. Acad. Sci. USA 2022, 119, e2204986119. [Google Scholar] [CrossRef]
- Zibrowius, H. Les Scléractiniaires de La Méditerranée et de l’Atlantique Nord-Oriental. Mem. Inst. Oceanogr. Monaco 1980.
- Aguirre, J.; Jiménez, A.P. Fossil Analogues of Present-Day Cladocora Caespitosa Coral Banks: Sedimentary Setting, Dwelling Community, and Taphonomy (Late Pliocene, W Mediterranean). Coral Reefs 1998, 17, 203–213. [Google Scholar] [CrossRef]
- Kružić, P.; Žuljević, A.; Nikolić, V. Spawning of the Colonial Coral Cladocora Caespitosa (Anthozoa, Scleractinia) in the Southern Adriatic Sea. Coral Reefs 2008, 27, 337–341. [Google Scholar] [CrossRef]
- Kersting, D.-K.; Linares, C. Cladocora Caespitosa Bioconstructions in the Columbretes Islands Marine Reserve (Spain, NW Mediterranean): Distribution, Size Structure and Growth: Cladocora Caespitosa bioconstructions in the Columbretes Islands Marine Reserve. Mar. Ecol. 2012, 33, 427–436. [Google Scholar] [CrossRef]
- Peirano, A.; Kružić, P. Growth Comparison between Ligurian and Adriatic Samples of the Coral Cladocora Caespitosa: First Results. Biologia marina mediterranea 2004, 11, 166–168. [Google Scholar]
- Silenzi, S.; Bard, E.; Montagna, P.; Antonioli, F. Isotopic and Elemental Records in a Non-Tropical Coral (Cladocora Caespiosa): Discovery of a New High-Resolution Climate Archive for the Mediterranean Sea. Glob. Planet. Change 2005, 49, 94–120. [Google Scholar] [CrossRef]
- Nantet, E. The Rise of the Tonnage in the Hellenistic Period. In Sailing from polis to empire: Ships in the eastern Mediterranean during the Hellenistic period; Nantet, E., Ed.; OpenBook: Adelaide, SA, Australia, 2020; pp. 75–89. [Google Scholar]
- Sisma-Ventura, G.; Yam, R.; Shemesh, A. Recent Unprecedented Warming and Oligotrophy of the Eastern Mediterranean Sea within the Last Millennium. Geophys. Res. Lett. 2014, 41, 5158–5166. [Google Scholar] [CrossRef]
- Ozer, T.; Gertman, I.; Kress, N.; Silverman, J.; Herut, B. Interannual Thermohaline (1979–2014) and Nutrient (2002–2014) Dynamics in the Levantine Surface and Intermediate Water Masses, SE Mediterranean Sea. Glob. Planet. Change 2017, 151, 60–67. [Google Scholar] [CrossRef]
- Grainger, J.D. The Syrian Wars; Brill, 2010.
- Ben-Yosef, D. Akko Bay: Hinterland of Phoenician Commercial City during the Persian Period. In In the Hill-Country, and in the Shephelah, and in the Arabah, Studies and Researches Presented to Adam Zertal in the Thirtieth Anniversary of the Manasseh Hill-Country Survey; Bar, S., Ed.; Jerusalem, 2008; pp. 271–290.
- Sharvit, J.; Planer, D.; Buxton, B.; Hale, J.; Barkai, O. Akko, Underwater Excavation; Hadashot Arkeologiot, Excavation and Surveys in Israel, 2023;
- Makhouly, N.; Johns, C.N. Guide to Acre; Government of Palestine Department of Antiquities: Jerusalem, 1946. [Google Scholar]
- Linder, E.; Raban, A. From the Diary of the Acre Expedition. Bimtzuloth-Yam 1965, 7, 23–27. [Google Scholar]
- Sharvit, J.; Buxton, B.; Hale, J.R.; Ratzlaff, A. The Hellenistic-Early Roman Harbour of Akko: Preliminary Finds from Archaeological Excavations at the Foot of the Southeastern Seawall at Akko, 2008-2014. In Under the Mediterranean I: Studies in Maritime Archaeology; Demesticha, S., Blue, L., Eds.; Sidestone Press: Leiden, Netherlands, 2021; pp. 163–180. [Google Scholar]
- Pietraszek, A. The Submerged Hellenistic to Early Roman Harbor at Akko: A Geoarchaeological Approach, University of Haifa, 2018.
- Peirano, A.; Morri, C.; Bianchi, C.N. Skeleton Growth and Density Pattern of the Temperate, Zooxanthellate Scleractinian Cladocora Caespitosa from the Ligurian Sea (NW Mediterranean). Mar. Ecol. Prog. Ser. 1999, 185, 195–201. [Google Scholar] [CrossRef]
- Peirano, A.; Morri, C.; Bianchi, C.N.; Rodolfo-Metalpa, R. Biomass, Carbonate Standing Stock and Production of the Mediterranean coral Cladocora Caespitosa (L.). Facies 2001, 44, 75–80. [Google Scholar] [CrossRef]
- Gorbunov, M.Y.; Falkowski, P.G. Using Chlorophyll Fluorescence Kinetics to Determine Photosynthesis in Aquatic Ecosystems. Limnol. Oceanogr. 2021, 66, 1–13. [Google Scholar] [CrossRef]
- Carpenter, G.E.; Chequer, A.D.; Weber, S.; Mass, T.; Goodbody-Gringley, G. Light and Photoacclimatization Drive Distinct Differences between Shallow and Mesophotic Coral Communities. Ecosphere 2022, 13. [Google Scholar] [CrossRef]
- Schöne, B.R.; Dunca, E.; Fiebig, J.; Pfeiffer, M. Mutvei’s Solution: An Ideal Agent for Resolving Microgrowth Structures of Biogenic Carbonates. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2005, 228, 149–166. [Google Scholar] [CrossRef]
- Martinez, S.; Bellworthy, J.; Ferrier-Pagès, C.; Mass, T. Selection of Mesophotic Habitats by Oculina Patagonica in the Eastern Mediterranean Sea Following Global Warming. Sci. Rep. 2021, 11, 18134. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA Primers for Amplification of Mitochondrial Cytochrome c Oxidase Subunit I from Diverse Metazoan Invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Arif, C.; Daniels, C.; Bayer, T.; Banguera-Hinestroza, E.; Barbrook, A.; Howe, C.J.; LaJeunesse, T.C.; Voolstra, C.R. Assessing Symbiodinium Diversity in Scleractinian Corals via next-Generation Sequencing-Based Genotyping of the ITS2 rDNA Region. Mol. Ecol. 2014, 23, 4418–4433. [Google Scholar] [CrossRef] [PubMed]
- Hume, B.C.C.; Smith, E.G.; Ziegler, M.; Warrington, H.J.M.; Burt, J.A.; LaJeunesse, T.C.; Wiedenmann, J.; Voolstra, C.R. SymPortal: A Novel Analytical Framework and Platform for Coral Algal Symbiont next-Generation Sequencing ITS2 Profiling. Mol. Ecol. Resour. 2019, 19, 1063–1080. [Google Scholar] [CrossRef] [PubMed]
- Orlando, L.; Allaby, R.; Skoglund, P.; Der Sarkissian, C.; Stockhammer, P.W.; Ávila-Arcos, M.C.; Fu, Q.; Krause, J.; Willerslev, E.; Stone, A.C.; et al. Ancient DNA Analysis. Nat. Rev. Methods Primers 2021, 1. [Google Scholar] [CrossRef]
- Knapp, M.; Clarke, A.C.; Horsburgh, K.A.; Matisoo-Smith, E.A. Setting the Stage - Building and Working in an Ancient DNA Laboratory. Ann. Anat. 2012, 194, 3–6. [Google Scholar] [CrossRef]
- Kapp, J.D.; Green, R.E.; Shapiro, B. A Fast and Efficient Single-Stranded Genomic Library Preparation Method Optimized for Ancient DNA. Journal of Heredity 2021, 112, 241–249. [Google Scholar] [CrossRef]
- López-Márquez, V.; Lozano-Martín, C.; Hadjioannou, L.; Acevedo, I.; Templado, J.; Jimenez, C.; Taviani, M.; Machordom, A. Asexual Reproduction in Bad Times? The Case of Cladocora Caespitosa in the Eastern Mediterranean Sea. Coral Reefs 2021, 40, 663–677. [Google Scholar]
- Di Tommaso, P.; Moretti, S.; Xenarios, I.; Orobitg, M.; Montanyola, A.; Chang, J.-M.; Taly, J.-F.; Notredame, C. T-Coffee: A Web Server for the Multiple Sequence Alignment of Protein and RNA Sequences Using Structural Information and Homology Extension. Nucleic Acids Res. 2011, 39, W13–W17. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Docherty, G.; Jones, V.; Evershed, R.P. Practical and Theoretical Considerations in the Gas Chromatography/combustion/isotope Ratio Mass Spectrometry delta(13)C Analysis of Small Polyfunctional Compounds: Practical and Theoretical Considerations in GC/C/IRMS. Rapid Commun. Mass Spectrom. 2001, 15, 730–738. [Google Scholar] [CrossRef]
- Francey, R.J.; Allison, C.E.; Etheridge, D.M.; Trudinger, C.M.; Enting, I.G.; Leuenberger, M.; Langenfelds, R.L.; Michel, E.; Steele, L.P. A 1000-Year High Precision Record of delta13C in Atmospheric CO2. Tellus B Chem. Phys. Meteorol. 1999, 51, 170–193. [Google Scholar] [CrossRef]
- Chikaraishi, Y.; Ogawa, N.O.; Kashiyama, Y.; Takano, Y.; Suga, H.; Tomitani, A.; Miyashita, H.; Kitazato, H.; Ohkouchi, N. Determination of Aquatic Food-Web Structure Based on Compound-Specific Nitrogen Isotopic Composition of Amino Acids. Limnol. Oceanogr. Methods 2009, 7, 740–750. [Google Scholar] [CrossRef]
- Frankowiak, K.; Kret, S.; Mazur, M.; Meibom, A.; Kitahara, M.V.; Stolarski, J. Fine-Scale Skeletal Banding Can Distinguish Symbiotic from Asymbiotic Species among Modern and Fossil Scleractinian Corals. PLoS One 2016, 11, e0147066. [Google Scholar] [CrossRef] [PubMed]
- Boudouresque, C.-F. Marine Biodiversity in the Mediterranean: Status of Species, Populations and Communities. Travaux scientifiques du Parc national de Port-Cros 2004, 20, 97–146. [Google Scholar]
- Templado, J. Future Trends of Mediterranean Biodiversity. In The Mediterranean Sea; Springer Netherlands: Dordrecht, 2014; ISBN 9789400767034. [Google Scholar]
- Coll, M.; Piroddi, C.; Steenbeek, J.; Kaschner, K.; Ben Rais Lasram, F.; Aguzzi, J.; Ballesteros, E.; Bianchi, C.N.; Corbera, J.; Dailianis, T.; et al. The Biodiversity of the Mediterranean Sea: Estimates, Patterns, and Threats. PLoS One 2010, 5, e11842. [Google Scholar] [CrossRef]
- Cristino, J. Dabrio Mateu Esteban The Coral Reef of Nijar, Messinian (uppermost Miocene), Almeria Province, S. E. Spain. J. Sediment. Res. 1981, 51. [Google Scholar] [CrossRef]
- Pomar, L. Reef Geometries, Erosion Surfaces and High-frequency Sea-level Changes, Upper Miocene Reef Complex, Mallorca, Spain. Sedimentology 1991, 38, 243–269. [Google Scholar] [CrossRef]
- Martín, J.; Braga, J.C. Messinian Events in the Sorbas Basin in Southeastern Spain and Their Implications in the Recent History of the Mediterranean. Sediment. Geol. 1994, 90, 257–268. [Google Scholar] [CrossRef]
- Vertino, A.; Stolarski, J.; Bosellini, F.R.; Taviani, M. Mediterranean Corals through Time: From Miocene to Present. In The Mediterranean Sea; Springer Netherlands: Dordrecht, 2014; ISBN 9789400767034. [Google Scholar]
- Freiwald, A. 5 Messinian Salinity Crisis: What Happened to Cold-Water Corals? In Mediterranean Cold-Water Corals. In Mediterranean Cold-Water Corals: Past, Present and Future; Springer International Publishing: Cham, 2019; ISBN 9783319916071. [Google Scholar]
- Zabala, M.; Ballesteros, E. Surface-Dependent Strategies and Energy Flux in Benthic Marine Communities Or, Why Corals Do Not Exist in the Mediterranean. Sci. Mar. 1989, 53, 3–17. [Google Scholar]
- Stambler, N. Life in the Mediterranean Sea: A Look at Habitat Changes; Nova Science Publishers, Inc, 2012;
- Schuhmacher, H.; Zibrowius, H. What Is Hermatypic?: A Redefinition of Ecological Groups in Corals and Other Organisms. Coral Reefs 1985, 4, 1–9. [Google Scholar] [CrossRef]
- Laborel, J. Marine Biogenic Constructions in the Mediterranean, a Review. Sci. Rep. Port-Cros natl. Park 1987, 13, 97–126. [Google Scholar]
- Peirano, A.; Morri, C.; Mastronuzzi, G.A.; Bianchi, C.N. The Coral Cladocora Caespitosa (Anthozoa, Scleractinia) as a Biotherm Builder in the Mediterranean Sea: A Short Review. Memorie Descrittive della Carta geologica d’Italia 1994, 52, 59–74. [Google Scholar]
- Kersting, D.K.; Teixidó, N.; Linares, C. Recruitment and Mortality of the Temperate Coral Cladocora Caespitosa: Implications for the Recovery of Endangered Populations. Coral Reefs 2014, 33, 403–407. [Google Scholar] [CrossRef]
- Kružić, P.; Sršen, P.; Benković, L. The Impact of Seawater Temperature on Coral Growth Parameters of the Colonial Coral Cladocora Caespitosa (Anthozoa, Scleractinia) in the Eastern Adriatic Sea. Facies 2012, 58, 477–491. [Google Scholar] [CrossRef]
- Morri, C.; Peirano, A.; Bianchi, C.N.; Sassarini, M. Present Day Bioconstructions of the Hard Coral, Cladocora Caespitosa (L.)(Anthozoa, Scleractinia), in the Eastern Ligurian Sea (NW Mediterranean). Biol. Mar. Mediterr. 1994.
- Schiller, C. Ecology of the Symbiotic Coral Cladocora Caespitosa (L.) (faviidae, Scleractinia) in the Bay of Piran (Adriatic Sea): I. Distribution and Biometry. Mar. Ecol. 1993, 14, 205–219. [Google Scholar]
- Kružić, P.; Požar-Domac, A. Banks of the Coral Cladocora Caespitosa (Anthozoa, Scleractinia) in the Adriatic Sea. Coral Reefs 2003, 22, 536–536. [Google Scholar] [CrossRef]
- Sanna, G.; Büscher, J.V.; Freiwald, A. Cold-Water Coral Framework Architecture Is Selectively Shaped by Bottom Current Flow. Coral Reefs 2023, 42, 483–495. [Google Scholar] [CrossRef]
- Zunino, S.; Pitacco, V.; Mavrič, B.; Orlando-Bonaca, M.; Kružić, P.; Lipej, L. The Ecology of the Mediterranean Stony Coral Cladocora Caespitosa (Linnaeus, 1767) in the Gulf of Trieste (northern Adriatic Sea): A 30-Year Long Story. Mar. Biol. Res. 2018, 14, 307–320. [Google Scholar] [CrossRef]
- Baron-Szabo, R.C. Geographic and Stratigraphic Distributions of the Caribbean Species of Cladocora (Scleractinia, Faviidae). Facies 2005, 51, 185–196. [Google Scholar] [CrossRef]
- Tremblay, P.; Ferrier-Pagès, C.; Maguer, J.F.; Rottier, C.; Legendre, L.; Grover, R. Controlling Effects of Irradiance and Heterotrophy on Carbon Translocation in the Temperate Coral Cladocora Caespitosa. PLoS One 2012, 7, e44672. [Google Scholar] [CrossRef]
- Kersting, D.K.; Linares, C. Living Evidence of a Fossil Survival Strategy Raises Hope for Warming-Affected Corals. Sci. Adv. 2019, 5, eaax2950. [Google Scholar] [CrossRef]
- Del Carmen Gomez Cabrera, M.; Young, J.M.; Roff, G.; Staples, T.; Ortiz, J.C.; Pandolfi, J.M.; Cooper, A. Broadening the Taxonomic Scope of Coral Reef Palaeoecological Studies Using Ancient DNA. Mol. Ecol. 2019, 28, 2636–2652. [Google Scholar] [CrossRef] [PubMed]
- Martin-Roy, R.; Thyrring, J.; Mata, X.; Bangsgaard, P.; Bennike, O.; Christiansen, G.; Funder, S.; Gotfredsen, A.B.; Gregersen, K.M.; Hansen, C.H.; et al. Advancing Responsible Genomic Analyses of Ancient Mollusc Shells. PLoS One 2024, 19, e0302646. [Google Scholar] [CrossRef] [PubMed]
- Der Sarkissian, C.; Möller, P.; Hofman, C.A.; Ilsøe, P.; Rick, T.C.; Schiøtte, T.; Sørensen, M.V.; Dalén, L.; Orlando, L. Unveiling the Ecological Applications of Ancient DNA from Mollusk Shells. Front. Ecol. Evol. 2020, 8. [Google Scholar] [CrossRef]
- Harney, É.; Cheronet, O.; Fernandes, D.M.; Sirak, K.; Mah, M.; Bernardos, R. ; Pinhasi A Minimally Destructive Protocol for DNA Extraction from Ancient Teeth. Genome research 2021, 31, 472–483. [Google Scholar] [CrossRef]
- Straube, N.; Lyra, M.L.; Paijmans, J.L.A.; Preick, M.; Basler, N.; Penner, J.; Rödel, M.-O.; Westbury, M.V.; Haddad, C.F.B.; Barlow, A.; et al. Successful Application of Ancient DNA Extraction and Library Construction Protocols to Museum Wet Collection Specimens. Mol. Ecol. Resour. 2021, 21, 2299–2315. [Google Scholar] [CrossRef]
- Korlević, P.; Gerber, T.; Gansauge, M.-T.; Hajdinjak, M.; Nagel, S.; Aximu-Petri, A.; Meyer, M. Reducing Microbial and Human Contamination in DNA Extractions from Ancient Bones and Teeth. Biotechniques 2015, 59, 87–93. [Google Scholar] [CrossRef]
- Ferrier-Pagès, C.; Gevaert, F.; Reynaud, S.; Beraud, E.; Menu, D.; Janquin, M.-A.; Cocito, S.; Peirano, A. In Situ Assessment of the Daily Primary Production of the Temperate Symbiotic Coral Cladocora Caespitosa. Limnol. Oceanogr. 2013, 58, 1409–1418. [Google Scholar] [CrossRef]
- Hoogenboom, M.; Rodolfo-Metalpa, R.; Ferrier-Pagès, C. Co-Variation between Autotrophy and Heterotrophy in the Mediterranean Coral Cladocora Caespitosa. Journal of Experimental Biology 2010, 213, 2399–2409. [Google Scholar] [CrossRef]
- Huang, D.; Licuanan, W.Y.; Baird, A.H.; Fukami, H. Cleaning up the “Bigmessidae”: Molecular Phylogeny of Scleractinian Corals from Faviidae, Merulinidae, Pectiniidae and Trachyphylliidae. BMC Evol. Biol. 2011, 11, 37. [Google Scholar] [CrossRef] [PubMed]
- Addamo, A.M.; Modrell, M.S.; Taviani, M.; Machordom, A. Unravelling the Relationships among Madrepora Linnaeus, 1758, Oculina Lamark, 1816 and Cladocora Ehrenberg, 1834 (Cnidaria: Anthozoa: Scleractinia). Invertebr. Syst. 2024, 38. [Google Scholar] [CrossRef] [PubMed]
- Ferrier-Pagès, C.; Martinez, S.; Grover, R.; Cybulski, J.; Shemesh, E.; Tchernov, D. Tracing the Trophic Plasticity of the Coral-Dinoflagellate Symbiosis Using Amino Acid Compound-Specific Stable Isotope Analysis. Microorganisms 2021, 9, 182. [Google Scholar] [CrossRef] [PubMed]
- Omata, T.; Suzuki, A.; Sato, T.; Minoshima, K.; Nomaru, E.; Murakami, A.; Murayama, S.; Kawahata, H.; Maruyama, T. Effect of Photosynthetic Light Dosage on Carbon Isotope Composition in the Coral Skeleton: Long-term Culture of Porites Spp: EFFECT OF LIGHT ONδ13C IN THE CORAL SKELETON. J. Geophys. Res. 2008, 113. [Google Scholar] [CrossRef]
- Prada, F.; Yam, R.; Levy, O.; Caroselli, E.; Falini, G.; Dubinsky, Z.; Goffredo, S.; Shemesh, A. Kinetic and Metabolic Isotope Effects in Zooxanthellate and Non-Zooxanthellate Mediterranean Corals along a Wide Latitudinal Gradient. Front. Mar. Sci. 2019, 6. [Google Scholar] [CrossRef]
- Einbinder, S.; Mass, T.; Brokovich, E.; Dubinsky, Z.; Erez, J.; Tchernov, D. Changes in Morphology and Diet of the Coral Styophora Pistillata along a Depth Gradient. Mar. Ecol. Prog. Ser. 2009, 381, 167–174. [Google Scholar] [CrossRef]
- Heikoop, J.M.; Dunn, J.J.; Risk, M.J.; Tomascik, T.; Schwarcz, H.P.; Sandeman, I.M.; Sammarco, P.W. δ 15 N and δ 13 C of Coral Tissue Show Significant Inter-Reef Variation. Coral Reefs-Journal of the International Society for Reef Studies 2000, 19, 189–193. [Google Scholar] [CrossRef]
- Muscatine, L.; Porter, J.W.; Kaplan, I.R. Resource Partitioning by Reef Corals as Determined from Stable Isotope Composition: I. δ 13C of Zooxanthellae and Animal Tissue vs Depth. Mar. Biol. 1989, 100, 185–193. [Google Scholar] [CrossRef]
- Nahon, S.; Richoux, N.B.; Kolasinski, J.; Desmalades, M.; Ferrier Pages, C.; Lecellier, G.; Planes, S.; Berteaux Lecellier, V. Spatial and Temporal Variations in Stable Carbon (δ13C) and Nitrogen (δ15N) Isotopic Composition of Symbiotic Scleractinian Corals. PLoS One 2013, 8, e81247. [Google Scholar] [CrossRef]
- Yannopoulos, S.; Yapijakis, C.; Kaiafa-Saropoulou, A.; Antoniou, G.; Angelakis, A.N. History of Sanitation and Hygiene Technologies in the Hellenic World. J. Water Sanit. Hyg. Dev. 2017, 7, 163–180. [Google Scholar] [CrossRef]
- Angelakis, A.N.; Capodaglio, A.G.; Dialynas, E.G. Wastewater Management: From Ancient Greece to Modern Times and Future. Water (Basel) 2022, 15, 43. [Google Scholar] [CrossRef]
- Barkai, O.; Jaijel, R.; Sharvit, J.; Goodman-Tchernov, B. The Location of the Coastline During the Late Roman and Early Byzantine Periods. Skyllis 6–13.
- Duprey, N.N.; Wang, T.X.; Kim, T.; Cybulski, J.D.; Vonhof, H.B.; Crutzen, P.J.; Haug, G.H.; Sigman, D.M.; Martínez-García, A.; Baker, D.M. Megacity Development and the Demise of Coastal Coral Communities: Evidence from Coral Skeleton δ15 N Records in the Pearl River Estuary. Glob. Chang. Biol. 2020, 26, 1338–1353. [Google Scholar] [CrossRef] [PubMed]
- Rico-Esenaro, S.D.; de Jesús Adolfo Tortolero-Langarica, J.; Iglesias-Prieto, R.; Carricart-Ganivet, J.P. The δ15N in Orbicella Faveolata Organic Matter Reveals Anthropogenic Impact by Sewage Inputs in a Mexican Caribbean Coral Reef Lagoon. Environ. Sci. Pollut. Res. Int. 2023, 30, 118872–118880. [Google Scholar] [CrossRef] [PubMed]
- Marion, G.S.; Dunbar, R.B.; Mucciarone, D.A.; Kremer, J.N.; Lansing, J.S.; Arthawiguna, A. Coral Skeletal δ15N Reveals Isotopic Traces of an Agricultural Revolution. Mar. Pollut. Bull. 2005, 50, 931–944. [Google Scholar] [CrossRef]
- Nothdurft, L.D.; Webb, G.E. Earliest Diagenesis in Scleractinian Coral Skeletons: Implications for Palaeoclimate-Sensitive Geochemical Archives. Facies 2009, 55, 161–201. [Google Scholar] [CrossRef]
- Chefaoui, R.M.; Casado-Amezúa, P.; Templado, J. Environmental Drivers of Distribution and Reef Development of the Mediterranean Coral Cladocora Caespitosa. Coral Reefs 2017, 36, 1195–1209. [Google Scholar] [CrossRef]
- Cramer, K.L.; O’Dea, A.; Carpenter, C.; Norris, R.D. A 3000 Year Record of Caribbean Reef Urchin Communities Reveals Causes and Consequences of Long-Term Decline in Diadema Antillarum. Ecography (Cop.) 2018, 41, 164–173. [Google Scholar] [CrossRef]
- McClenachan, L.; Rick, T.; Thurstan, R.H.; Trant, A.; Alagona, P.S.; Alleway, H.K.; Armstrong, C.; Bliege Bird, R.; Rubio-Cisneros, N.T.; Clavero, M.; et al. Global Research Priorities for Historical Ecology to Inform Conservation. Endanger. Species Res. 2024, 54, 285–310. [Google Scholar] [CrossRef]
- Drake, J.L.; Whitelegge, J.P.; Jacobs, D.K. First Sequencing of Ancient Coral Skeletal Proteins. Sci. Rep. 2020, 10, 19407. [Google Scholar] [CrossRef]
- Flewellen, A.O. The Biophysical Afterlife of Slavery Signaled through Coral Architectural Stones at Heritage Sites on St. Croix. Am. Antiq. 2024, 89, 591–607. [Google Scholar] [CrossRef]
- Sherwood, O.A.; Lapointe, B.E.; Risk, M.J.; Jamieson, R.E. Nitrogen Isotopic Records of Terrestrial Pollution Encoded in Floridian and Bahamian Gorgonian Corals. Environ. Sci. Technol. 2010, 44, 874–880. [Google Scholar] [CrossRef]
- Luu, V.H.; Ryu, Y.; Darling, W.S.; Oleynik, S.; de Putron, S.J.; Cohen, A.L.; Wang, X.T.; Sigman, D.M. Nitrogen Isotope Ratios across the Bermuda Coral Reef: Implications for Coral Nitrogen Sources and the Coral-Bound Nitrogen Isotope Proxy. Front. Mar. Sci. 2025, 12. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




