1. Introduction
The question on the origin of life, “how did the first life emerge on the primitive Earth” has been asked by many philosophers and scientists for many years. However, correct answer to the question has not been obtained still now, although some ideas for explaining the origin of life have been proposed, such as RNA world hypothesis [
1], hydrothermal vent hypothesis [
2,
3,
4] and [GADV]-protein world hypothesis or GADV hypothesis [
5,
6,
7] and so on. In addition, it is also important to answer to the question, “how did the first life or the first universal common ancestor (FUCA) [
8] evolved to so-called the last universal common ancestor (LUCA) [
9]”, because LUCA is literally only the last common ancestor connecting with all modern organisms living on the present Earth and, therefore, LUCA is unrelated to the origin of life.
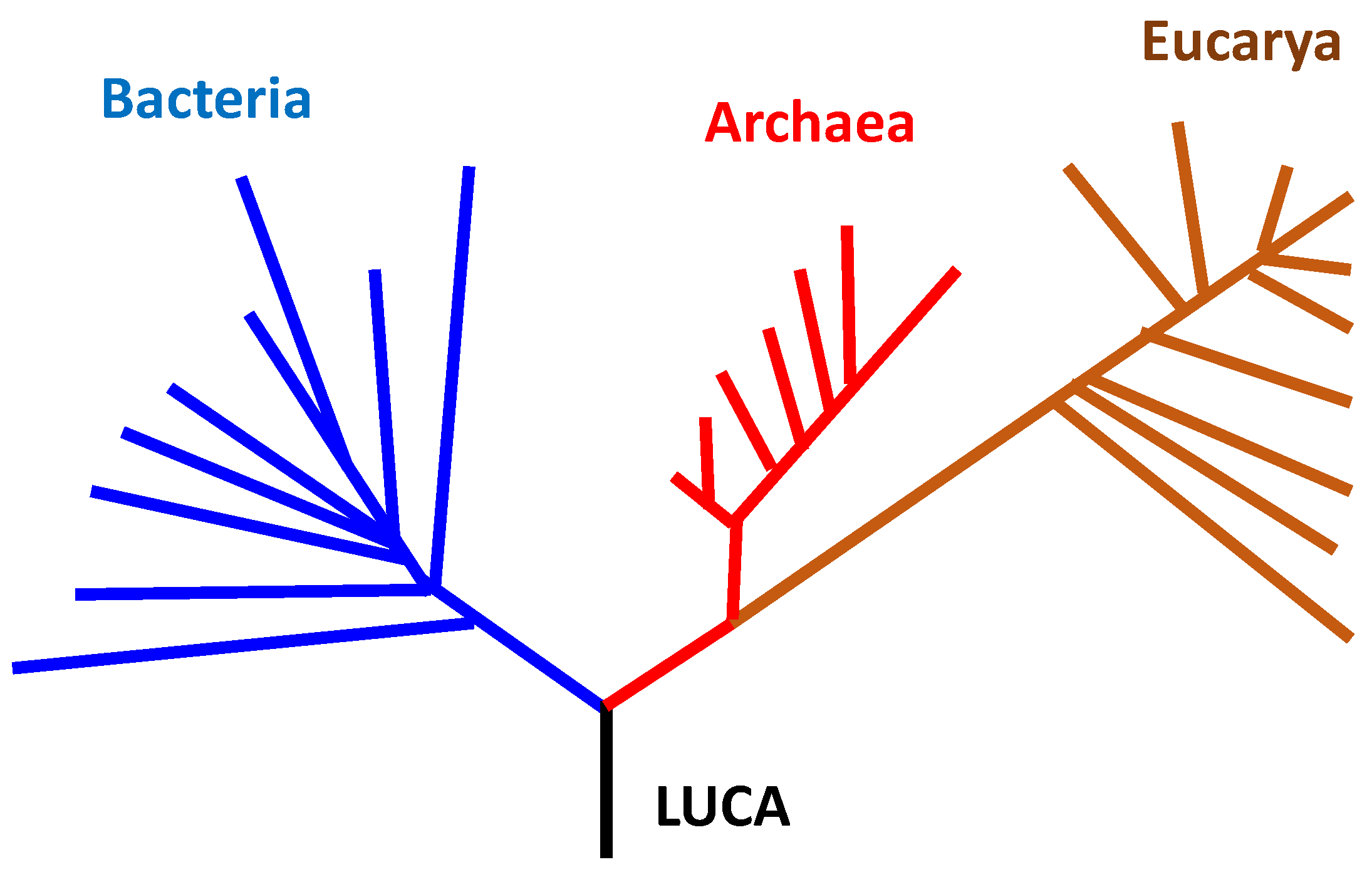
On the other hand, many amino acid sequences of proteins and many base sequences of genomes of modern organisms have been made clear and stored in some databanks, such as Genbank. Various phylogenetic trees are drawn accompanied by the rapid development of genome analyses [
10] and of mathematical techniques exploring close relationship among amino acid sequences and base sequences [
11,
12,
13,
14]. One representative phylogenetic tree, which was proposed by Woese et al. [
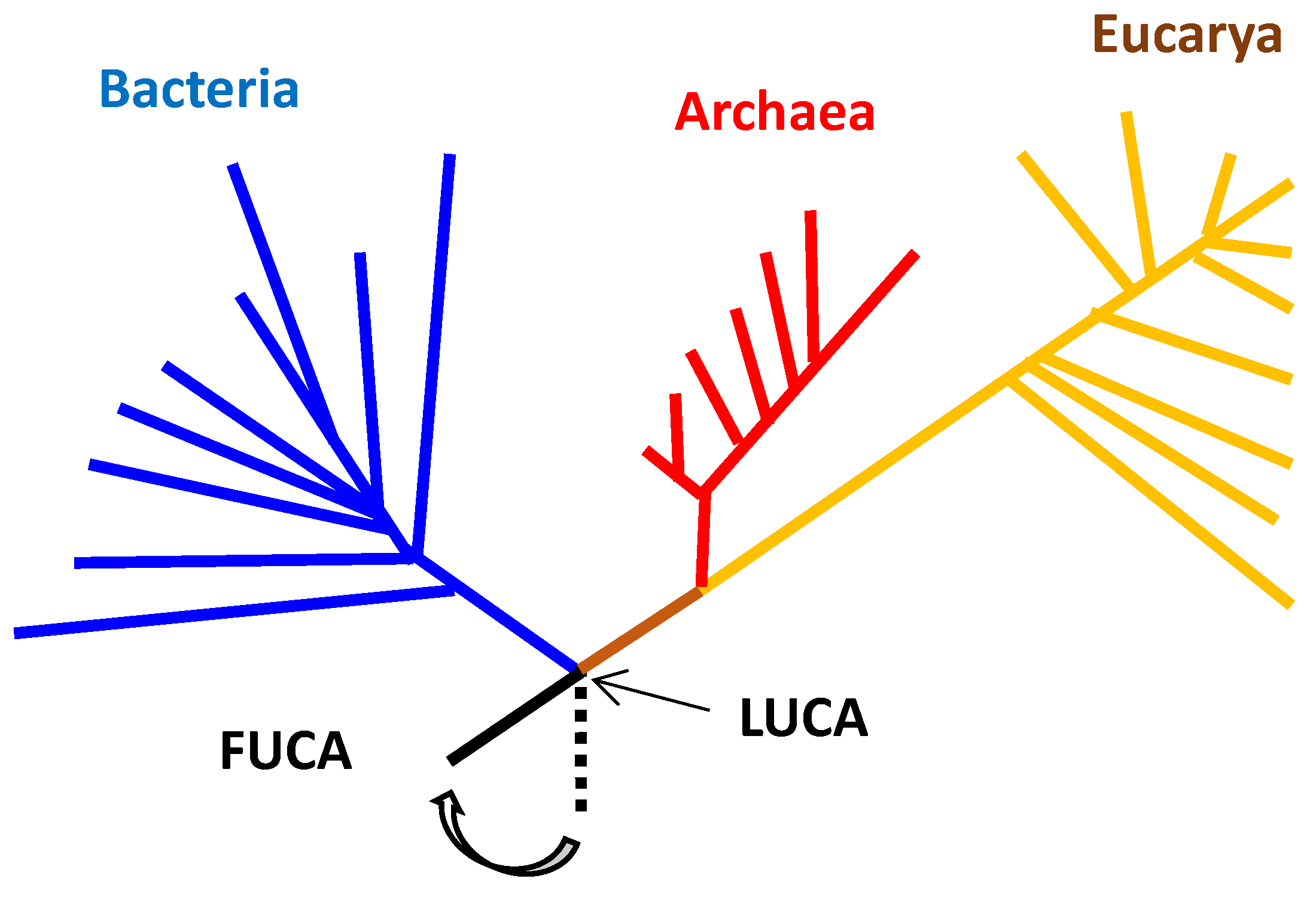
15], is shown in
Figure 1. The figure is frequently shown when the origin of life is discussed based on hydrothermal vent hypothesis [
2,
3,
4]. The reason is, because many archaea living under high temperature are seen at the position close to the root of the phylogenetic tree (
Figure 1).
2. Properties of LUCA, Which Viewed from Phylogenetic Tree
As described above, the origin of life is frequently discussed as connecting with LUCA. The reason would be because it is currently impossible to know entities before LUCA. Then, it is first discussed here what properties had LUCA. The number of genes used in LUCA is estimated as 355 based on the idea that LUCA should use genes, which are commonly used in genomes of modern organisms [
10]. However, it would be impossible to answer to the question, “how many number of genes LUCA were actually used, because the phylogenetic tree is drawn with properties of modern organisms. However, there are debates against the idea described above, because LUCA carrying 355 genes is too refined. Another property of LUCA can be also supposed that LUCA would have lived under the universal genetic code, based on the fact that almost all organisms living on the present Earth use the twenty natural amino acids [
10].
It would be impossible to reveal the origin of life by exploring the properties of LUCA, because as a matter of course, it is principally unknown what happened around the root of the phylogenetic tree, which has been drawn with the genetic properties of modern organisms. Thus, there are some fatal flaws in exploring the origin of life from the phylogenetic tree and also in revealing evolutionary process from the first life to LUCA, because the studies on the origin of life used phylogenetic trees are carried out by top-down approach, under which properties of modern organisms affects those of LUCA too strongly. In addition, it must be emphasized here that, as a matter of course, LUCA is quite different from FUCA, which was a key entity we must understand to reveal the origin of life.
3. Relationship Between LUCA and FUCA Viewed from GADV Hypothesis
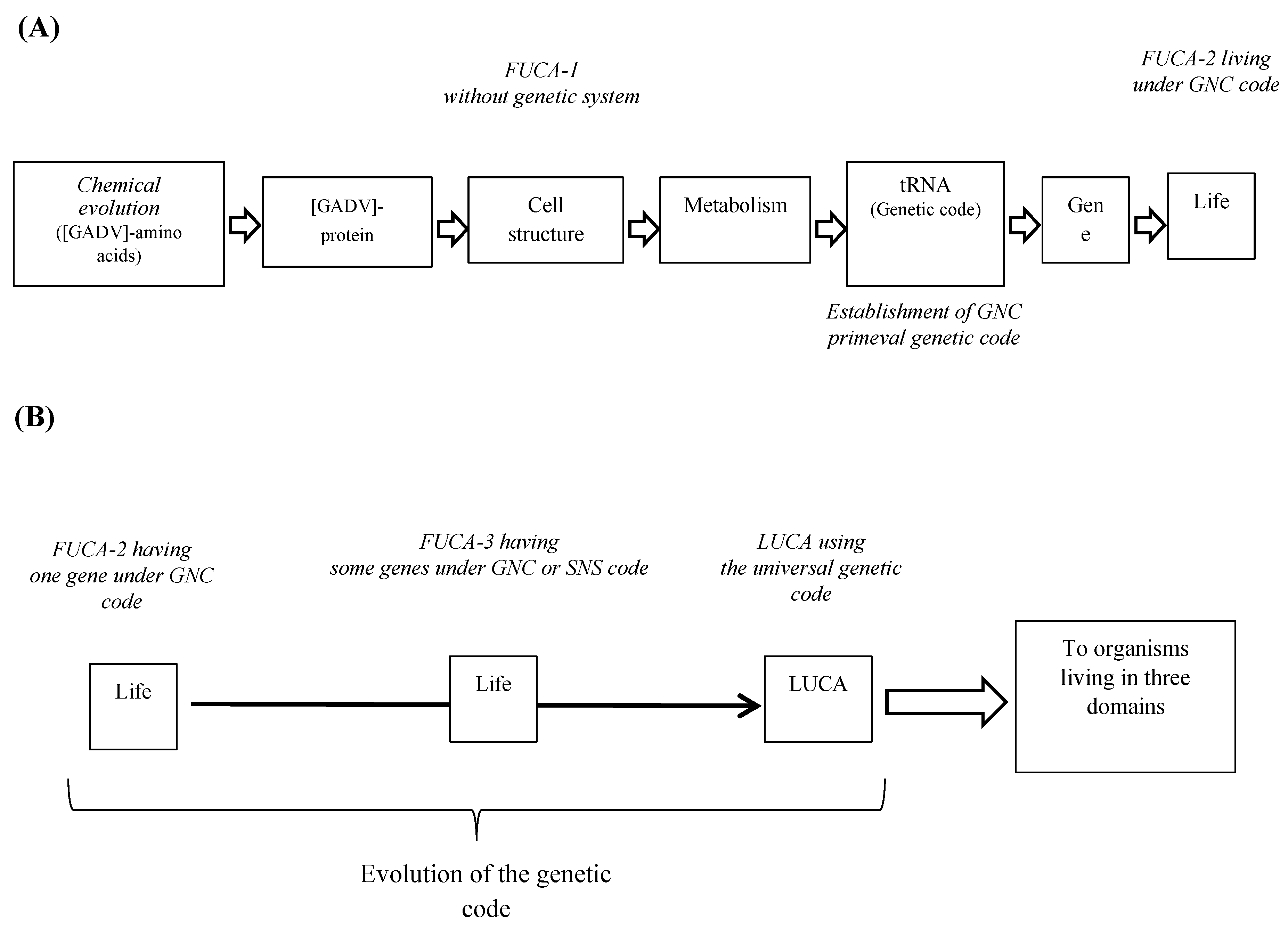
The problem, “what is LUCA?”, is discussed afresh from the standpoint of GADV hypothesis on the origin of life, which I have proposed [
6,
7]. Viewed from the standpoint, it is naturally supposed that a kind of evolution proceeded even during chemical evolution, when organic compounds were produced from inorganic compounds, just as the name implies (
Figure 2 (A)). As a result of further evolution, which could be achieved through selection of [GADV]-microspheres having larger proliferation ability than others, it is assumed that the first life or FUCA emerged at some point in time. However, it would be impossible to understand evolution process from LUCA to organisms belonging to three domains under GADV hypothesis (
Figure 2 (B)), because the study, which is carried out by the bottom-up approach, is strongly forced to understanding the emergence of life or FUCA.
Of course, it cannot be considered that FUCA had equipped 355 genes, because it is natural to consider that everything begins from one and the number increases gradually. It is considered from the standpoint of GADV hypothesis that the first genetic system composed of gene, genetic code (tRNA) and protein was completed, when the first (GNC)
n gene was formed as seen in
Figure 2 (
A) [
16]. Thereafter, the number of genes increased as homologous genes were formed from sense strand of the first (GNC)
n gene after gene duplication as expected by S. Ohno [
17] (
Figure 2 (B)). On the other hand, entirely new or non-homologous genes were generated from antisense strand of the first (GNC)
n gene after gene duplication as deduced by GC-NSF(a) hypothesis on entirely new gene formation (
Figure 2) [
18].
Accompanied by evolution of FUCA, the GNC primeval genetic code evolved to the universal genetic code through SNS code [
19]. It can be deduced that LUCA carrying 355 genes emerged after FUCA further evolved and accumulated a number of genes (
Figure 1 (B)). Refer my book [
7]) about these discussions, because formation process of new genes cannot be discussed here in detail due to paper limitations.
4. How Can the Whole Evolutionary Process from Chemical Evolution to Modern Organisms Be Grasped?
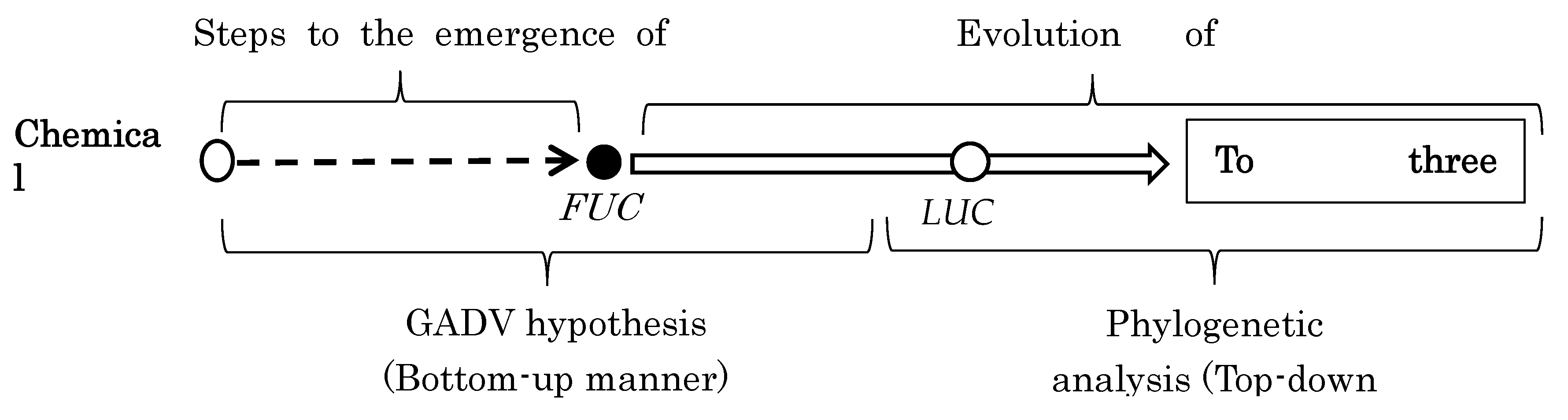
Then, how can we know the whole process of evolution from chemical evolution to prosperity of diverse modern organisms? It can be viewed for the first time by connecting LUCA, which is obtained from analysis of phylogenetic tree (top-down approach), with FUCA, which is deduced from GADV hypothesis (bottom-up manner) (
Figure 3).
5. Which Traits Did LUCA Have, Bacteria or Archaea?
As a matter of course, it is considered that FUCA carrying one or several genes evolved to the organisms belonging three domains, bacteria, archaea and eukarya, through the LUCA (
Figure 3). It would be important to reconsider from the standpoint of GADV hypothesis what happened during from FUCA to the formation of three domains. For the purpose, similarities or differences among bacteria, archaea and eukerya are written out from “Archaea” of Wikipedia, as shown in
Table 1 and
Table 2. As can be seen in
Table 1, cell size, genome size, properties of cytoplasm, mRNA and ribosome size are similar between bacteria and archaea, although those properties are quite different from eukarya. From the five properties shown in
Table 1, it is reasonably supposed that the five properties were established during evolution from FUCA to proto-bacteria, which is located at branch point to bacteria domain, and those were succeeded to archaea.
On the contrary, properties of archaea, such as promoter, transcription mechanism, RNA polymerase, tRNA and rRNA are similar to eukarya, although those are different from bacteria (
Table 2). On the other hand, life-forms separated from archaea further evolved to eukarya. Life-forms heading to eukarya developed further the five properties shown in
Table 1, to adapt to the respective environments.
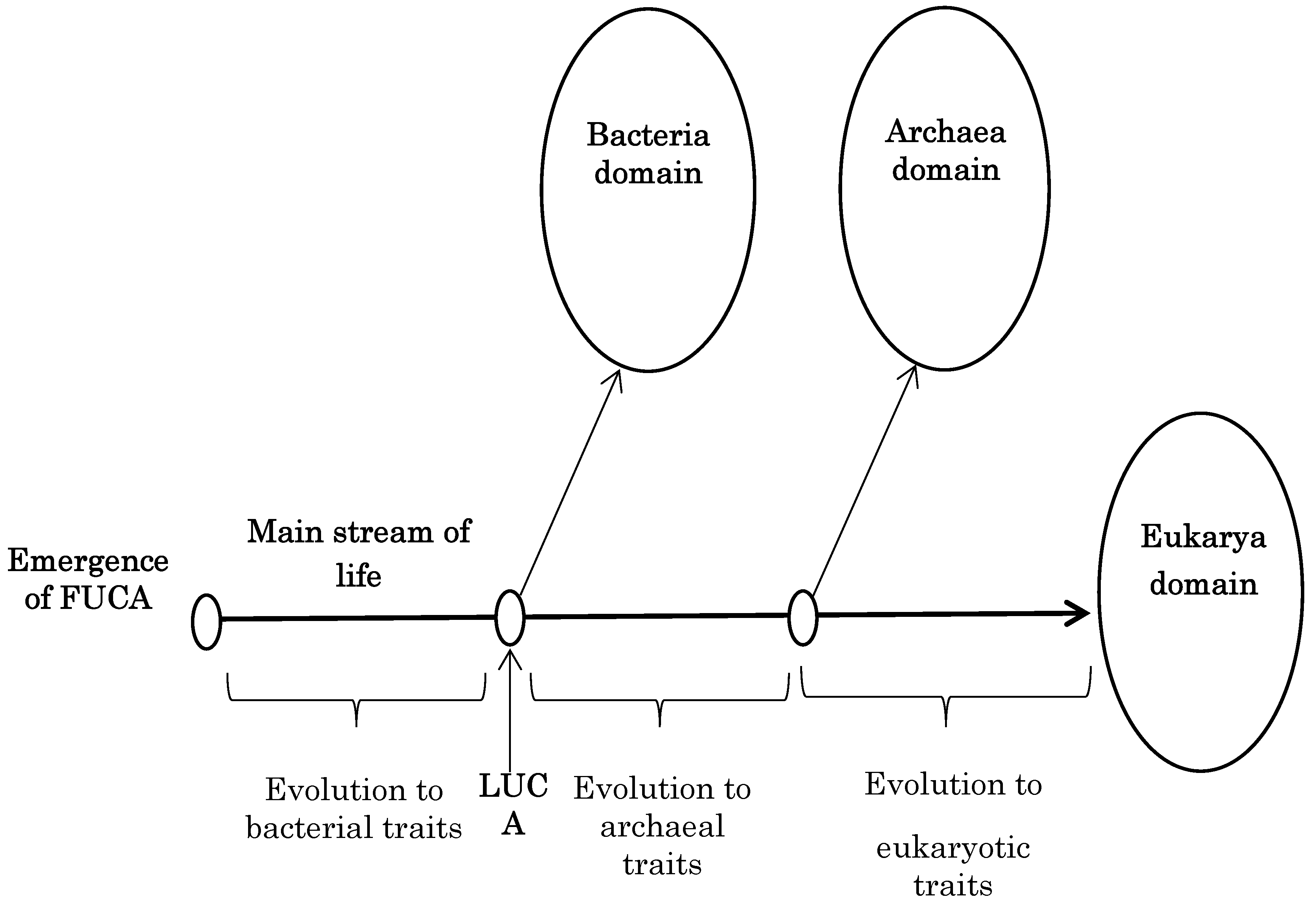
6. Proposition of a New Phylogenetic Tree
As can be seen in the two
Table 1 and
Table 2, it is understood that the five properties of archaea, which are shown in
Table 1, are similar to those of bacteria having a simpler structure. On the contrary, the other five properties of archaea shown in
Table 2 are similar to those of eukarya having more complex structures. These clearly indicate that archaea have intermediately complex structures or are using more complex ones than bacteria (
Table 2) and simpler structures than eukarya (
Table 1). From the five properties shown in
Figure 4, it can be supposed that the five properties were established during evolution from LUCA to archaea and were succeeded to eukarya. Therefore, it can be concluded that modern organisms emerged in order of bacteria, archaea and eukarya (
Figure 4).
Nevertheless, it is sometimes insisted that LUCA, which is a quite different entity from FUCA, emerged around hydrothermal vents on the basis of that thermophilic microorganisms belonging to many archaea and a few bacteria, can be seen around the root of the phylogenetic tree (
Figure 1). In other words, it is considered in the hydrothermal vent hypothesis that archaea-like thermophilic LUCA first emerged among organisms belonging to the three domains. One of the reasons, why such hypothesis has been proposed, would be because the phylogenetic tree shown in
Figure 1 was misinterpreted under the condition that the origin of life cannot be well understood.
In
Figure 5, a phylogenetic tree newly redrawn is given. In the phylogenetic tree, only one short line shown in
Figure 1 is moved as can be seen in the
Figure 5. That is, one short line showing evolutionary pathway from LUCA to the branch point entering bacterial domain and heading further to archaeal domain is different between
Figure 5 (blue bold line) and
Figure 1 (blue broken line). Note that topology of the phylogenetic tree has unchanged at all between
Figure 1 and
Figure 5.
7. Interpretation of the New Philogenetic Tree
Two large differences arise in interpretations as described below in spite of the small change between the two phylogenetic trees (
Figure 1 and
Figure 5).
- (1)
The formation order of three domains changes. In
Figure 1, the order is assumed as archaea, bacteria and eukarya. On the contrary, in
Figure 5, the order is definitely interpreted as bacteria, archaea and eukarya.
- (2)
It can be concluded that LUCA emerged as a proto-microorganism evolving to bacteria, successively heading to archaea and eukarya.
It has been insisted in hydrothermal vent hypothesis insisting that the first life might emerge based on existence of energy and metal catalysts, which are necessary to synthesize organic compounds from inorganic compounds. However new large changes in interpretation inevitably further occur by introduction of the small angular change at the root around LUCA (
Figure 5).
That is, the hydrothermal vent hypothesis on the origin of life must loose the basis insisting that the first life emerged around hydrothermal vents. The two new interpretations described above are inconsistent with the idea that life arose around hydrothermal vents.
8. Discussion
In this article, only one short line around the root of the phylogenetic tree, based on which hydrothermal vent hypothesis was proposed, is redrawn in
Figure 5, as considering steps to the emergence of life deduced from GADV hypothesis (
Figure 2) and properties of organisms belonging to three biological domains (
Table 1 and
Table 2). The small change caused some large differences in interpretation of the phylogenetic tree. The differences are enumerated below.
- (1)
It is deduced that LUCA emerged as preproto-bacteria but not preproto-archaea.
- (2)
LUCA was not thermophilic but mesophilic entity.
- (3)
Therefore, it cannot be always stated that life arose around hydrothermal vents.
Furthermore, some new interpretations of the phylogenetic tree given in
Figure 5 can be added as follows.
- (1)
Evolutionary pathway from FUCA to LUCA or entrance of three biological domains is drawn with one line. This means that only one species sharing one gene pool survived and reached to the entrance.
- (2)
Evolutionary processes from LUCA to the respective branch points and the processes from the branch points heading to the respective species in the domains are also drawn by one line without crossing. These mean that only one descendant survived among diverse life-forms and evolved further to the respective modern organisms.
- (3)
The reason why the phylogenetic tree is drawn with many lines in the respective domains, is because the respective species had differentiated to adapt to the respective environments or their niches.
- (4)
Similarly, the reason, why evolutionary processes reaching all modern organisms are drawn with the respective straight lines, is because only modern organisms are living. In other words, all organisms, which had lived on the evolutionary pathways, disappeared. That is, only organisms, which are situated at the tips on branches of the phylogenetic tree, are living.
- (5)
In bacteria and eukarya domains, the line length from branch point to the tip is the closer to the point, at which was branched off from evolutionary main stream, is generally the longer. The reason is because more mutations accumulated due to longer time passed.
- (6)
However, line lengths in archaea domain are generally shorter than lines in the other two domains. This means that evolution velocities or mutation accumulation rates of species in archaea domain were slower or smaller than those in the other two domains. The reason must be because extremophiles in archaea domain could not proliferate at a fast speed due to the respective extreme environments.
Next, consider the reason why diverse organisms are living on the present Earth?
- (1)
The reason would be probably because FUCA, which was composed of organic compounds with a simple structure, had pluripotency in the meaning of that FUCA could evolve any kind of organisms.
- (2)
Life fundamentally proceeded in subdivision processes to adapt to various environments.
- (3)
Once an organism arose into a biological domain, the organism never transfer to any organism living in other domain. The reason is because every evolutionary process is irreversible. In other words, evolution is always carried out by addition of a new ability. Therefore, retrogression, which looses an ability and goes against the flow of evolution, never be realized. This means that prosperity of diverse organisms never continue forever and any organisms must become extinct someday.
Informed Consent Statement
Not applicable.
Acknowledgements
I am very grateful to Dr. Tadashi Oishi (G&L Kyosei Institute, Emeritus professor of Nara Women’s University) for encouragement throughout my research on the origin and evolution of the fundamental life system.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gilbert, W. The origin of life: RNA world. Nature. 1986, 319, 618. [Google Scholar] [CrossRef]
- Corliss, J.B.; Dymond, J.; Gordon, L.I.; Edmond, J.M.; von Herzen, R.P.; Ballard, R.D.; Green, K.; Williams, D.; Bainbridge, A.; Crane, K.; van Andel, T.H. Submarine thermal springs on the galapagos rift. Science. 1979, 203, 1073–1083. [Google Scholar] [CrossRef] [PubMed]
- Holm, N.G.; Andersson, E. , Hydrothermal simulation experiments as a tool for studies of the origin of life on Earth and other terrestrial planets. A review. Astrobiology, 2005, 5, 444–460. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tian, G.; Gao, J.; Han, M.; Su, R.; Wang, Y.; Feng, S. Prebiotic synthesis of glycine from ethanolamine in simulated Archean alkaline hydrothermal vents. Orig. Life Evol. Biosph. 2017, 47, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Ikehara, K. Origins of gene, genetic code, protein and life: Comprehensive view of life system from a GNC-SNS primitive genetic code hypothesis. J. Biosci. 2002, 27, 165–186. [Google Scholar] [CrossRef] [PubMed]
- Ikehara, K. Possible steps to the emergence of life: The [GADV]-protein world hypothesis. Chem. Rec. 2005, 5, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Ikehara, K. Towards Revealing the Origin of life.—Presenting the GADV Hypothesis; Springer Nature: Gewerbestrasse Cham, Switzerland, 2021. [Google Scholar]
- Prosdocimi, F.; Jheeta, S.; Torres de Farias, S. Conceptual challenges for the emergence of the biological system: Cell theory and self-replication. Med Hypotheses. 2018, 119, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Archaea in Wikipedia; https://en.wikipedia.org/wiki/Archaea.
- Weiss, M.C.; Sousa, F. L.; Mrnjavac, N.; Neukirchen, S.; Roettger, M.; Nelson-Sathi, S.; Martin, W. F. The physiology and habitat of the last universal common ancestor. Nat. Microbiol., 2016, 1, 16116. [Google Scholar] [CrossRef] [PubMed]
- Pal, J.; Saha, S.; Maji, B.; Bhattacharya, D.K. PTGAC Model: A machine learning approach for constructing phylogenetic tree to compare protein sequences. J Bioinform Comput Biol. 2023, 21, 2250028. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Wu, S.; Tsang, A. Phylogenetic Analysis of Protein Family. Methods Mol Biol. 2018, 1775, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Kapli, P.; Yang, Z.; Telford, M.J. Phylogenetic tree building in the genomic age. Nat. Rev. Genet. 2020, 21, 428–444. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Lou, F.; Yin, X.; Cong, B.; Liu, S.; Zhao, L.; Zheng, L. Whole-genome survey and phylogenetic analysis of Gadus macrocephalus. Biosci. Rep. 2022, 42, BSR20221037. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R.; Kandler, O.; Wheelis, M.L. “Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya”. Proc. Nat. Acad. Sci., 1990, 87, 4576–4579. [Google Scholar] [CrossRef] [PubMed]
- Ikehara, K. The first genetic system was established not in the top-down manner (RNA world hypothesis) but in the bottom-up manner (GADV hypothesis). Europ. Soc. Med., 2024, 12. [Google Scholar] [CrossRef]
- Ohno, S. Evolution by gene duplication theory; Springer: Heiderberg, 1970. [Google Scholar]
- Ikehara, K.; Amada, F.; Yoshida, S.; Mikata, Y.; Tanaka, A. A possible origin of newly-born bacterial genes: Significance of GC-rich nonstop frame on antisense strand. Nucl. Acids Res. 1996, 24, 4249–4255. [Google Scholar] [CrossRef] [PubMed]
- Ikehara, K.; Omori, Y.; Arai, R.; Hirose, A. A Novel Theory on the Origin of the Genetic Code: A GNC-SNS Hypothesis. J. Mol. Evol. 2002, 54, 530–538. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).