Submitted:
05 July 2025
Posted:
07 July 2025
You are already at the latest version
Abstract
Keywords:
2. Introduction
1.1. Overview of the Topic
1.2. Personal and Global Significance
1.3. Introduction to Experimental Approach

3. Research Question
2.1. Hypothesis
4. Background
3.1. Genetic Polymorphisms
- CYP2C19: The CYP2C19 gene encodes the cytochrome P450 2C19 enzyme, which is crucial for the metabolism of various drugs, including clopidogrel. Common polymorphisms include CYP2C191 (wild type), CYP2C192, and CYP2C19*3. The *2 and *3 alleles result in reduced or null enzyme activity, leading to poor metabolism of clopidogrel and decreased therapeutic effectiveness (Mega et al., 2009).
- ABCB1: The ABCB1 gene encodes P-glycoprotein, a membrane transporter involved in drug absorption and distribution. The 3435C>T polymorphism in the ABCB1 gene affects P-glycoprotein expression levels and function, which can alter the pharmacokinetics of clopidogrel and other drugs (Kim et al., 2010).
3.2. Clopidogrel and Its Relationship with the Metabolism
3.3. PharmGKB Database
3.3.1. Data Collection
3.6.2. Selection and Sampling Method
- Studies involving clopidogrel and its pharmacokinetics.
- Inclusion of genetic data on CYP2C19 and ABCB1 polymorphisms.
- The availability of detailed pharmacokinetic measurements (e.g., plasma concentration, metabolic ratios).
5. Variables
4.1. Independent Variable


4.2. Dependent Variable
4.3. Controlled and Uncontrolled Variables
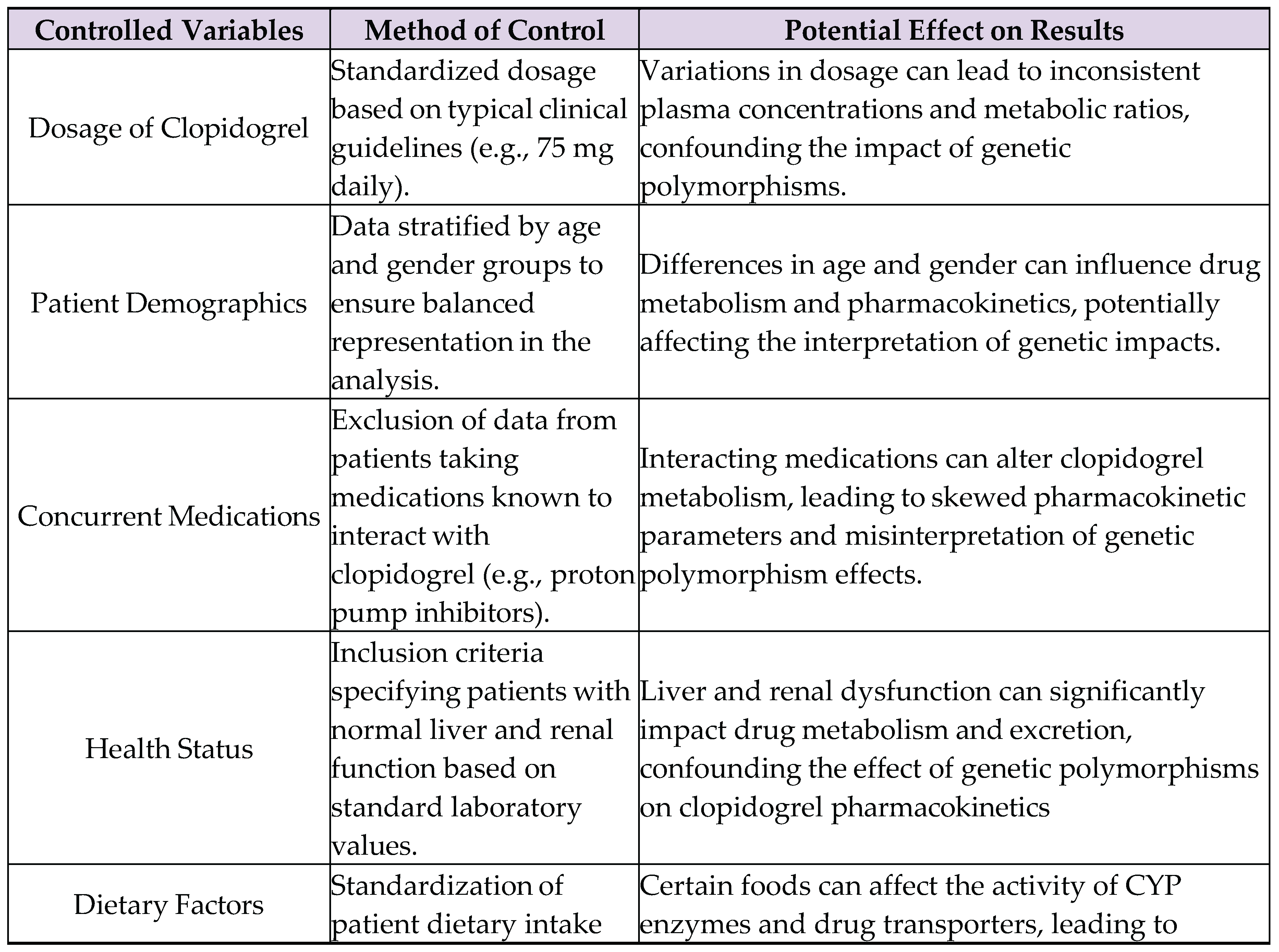
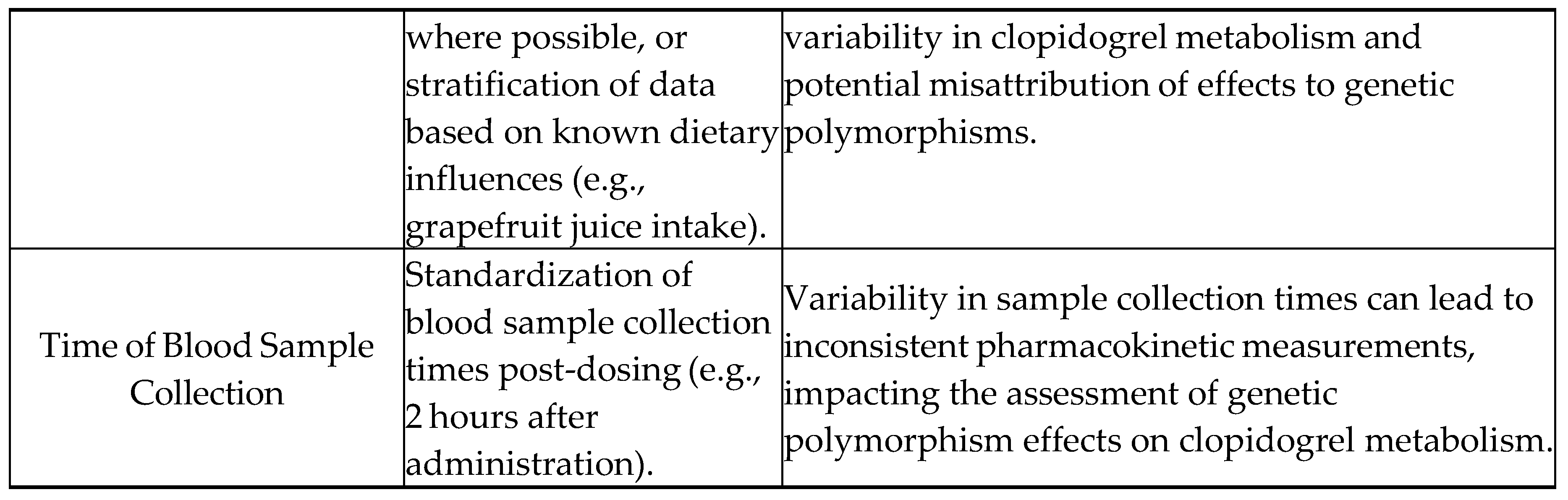
Uncontrolled Variables:
- Environmental factors: Differences in climate, altitude, or exposure to pollutants among patients in the dataset can influence physiological responses and drug metabolism. However, these factors are difficult to standardize or control in a retrospective analysis using secondary data (they aren’t included in PharmGKB).
- Genetic background: Although the study focuses on specific polymorphisms in CYP2C19 and ABCB1, other genetic variations in drug metabolism and transport genes might also affect clopidogrel pharmacokinetics but are not accounted for in this analysis.
- Lifestyle factors: Variations in patients' lifestyles, such as exercise, stress levels, and alcohol consumption, can impact drug metabolism. These factors are often not consistently recorded in pharmacogenomic databases, leading to potential confounding effects.
- Adherence to medication: While the study assumes consistent adherence to prescribed clopidogrel regimens, actual patient adherence may vary, introducing variability in drug plasma concentrations and metabolic ratios that are unrelated to genetic polymorphisms.
6. Materials
5.1. Databases
- 1.
-
PharmGKB (Pharmacogenomics Knowledgebase)
- 1.1.
- Purpose: Provides comprehensive pharmacogenomic information, including gene-drug interactions, pharmacokinetic data, and genetic variant annotations.
- 1.2.
- Data Utilized: Genetic polymorphisms of CYP2C19 and ABCB1, clopidogrel pharmacokinetics.
- 1.3.
-
Features Used:
- 1.3.1.
- Clinical Annotations: Information on the impact of genetic variants on drug response.
- 1.3.2.
- Variant Annotations: Details on specific polymorphisms in genes such as CYP2C19 and ABCB1.
- 1.3.3.
- Drug Pathways: Understanding the metabolic pathways of clopidogrel.
- 1.3.4.
- Pharmacokinetic Data: Data on drug absorption, distribution, metabolism, and excretion.
- 1.3.5.
- Publications: Access to relevant studies and clinical guidelines.
5.2. Software and Tools
- 1.
-
Computer with Internet Access
- 1.1.
- Purpose: Accessing PharmGKB and performing data analysis.
- 1.2.
-
Specifications:
- 1.2.1.
- Processor: Intel Core i5 or higher.
- 1.2.2.
- RAM: Minimum 8GB.
- 1.2.3.
- Storage: Minimum 256GB SSD.
- 1.2.4.
- Operating System: Windows 10, macOS 10.14 or later, or a compatible Linux distribution.
- 2.
-
Statistical Software Licenses
- 2.1.
- Examples: SPSS, R, SAS
- 2.2.
- Quantity: 1 license per software
- 2.3.
-
Specifications:
- 2.3.1.
- SPSS: License for version 26 or later.
- 2.3.2.
- R: Open-source, no license required.
- 2.3.3.
- SAS: License for version 9.4 or later.
- 3.
-
Statistical Analysis Software
- 3.2.
- Examples: IBM SPSS Statistics, R, SAS
- 3.3.
- Purpose: To perform statistical analysis, including ANOVA and regression analysis, on the extracted data.
- 3.4.
-
Specifications:
- 3.4.1.
- R: Open-source; latest stable version.
- 3.5.
- Uncertainty: Typically low; dependent on the precision of data input and accuracy of the statistical models used.
- 4.
-
Data Extraction Tools
- 4.1.
- Examples: Microsoft Excel, Python (with pandas library)
- 4.2.
- Purpose: To organize, preprocess, and manage data for analysis.
- 4.3.
-
Specifications:
- 4.3.1.
- Python: Version 3.7 or later; pandas library version 1.2 or later.
- 5.
-
Database Query Tools
- 5.1.
- Examples: SQL, PharmGKB’s built-in query interface
- 5.2.
- Purpose: To extract specific datasets from PharmGKB.
- 5.3.
-
Specifications:
- 5.3.1.
- SQL: Standard SQL queries compatible with PharmGKB’s data structure.
- 5.3.2.
- PharmGKB Interface: Access through a web browser.
7. Methodology
6.1. Step-by-Step Instructions
-
Define search criteria:
- 1.1.
- Identified key search terms related to clopidogrel, CYP2C19, and ABCB1. Examples of searched terms include: "clopidogrel pharmacokinetics," "CYP2C19 polymorphism," "ABCB1 3435C>T".
-
Data extraction:
- 2.1.
- Used PharmGKB’s query tools to extract data on clopidogrel pharmacokinetics and associated genetic polymorphisms.
- 2.2.
- Downloaded relevant datasets, ensuring to note the study parameters, sample sizes, and demographic information included in each dataset.
-
Data preprocessing:
- 3.1.
- Imported the extracted datasets into Microsoft Excel or Python (using pandas library).
- 3.2.
- Cleaned the data by removing duplicates, handling missing values, and standardizing variable names.
- 3.3.
- Organized the data into a structured format for analysis, such as separating variables by genetic polymorphisms and pharmacokinetic parameters.
- 3.4.
-
Steps in Python:
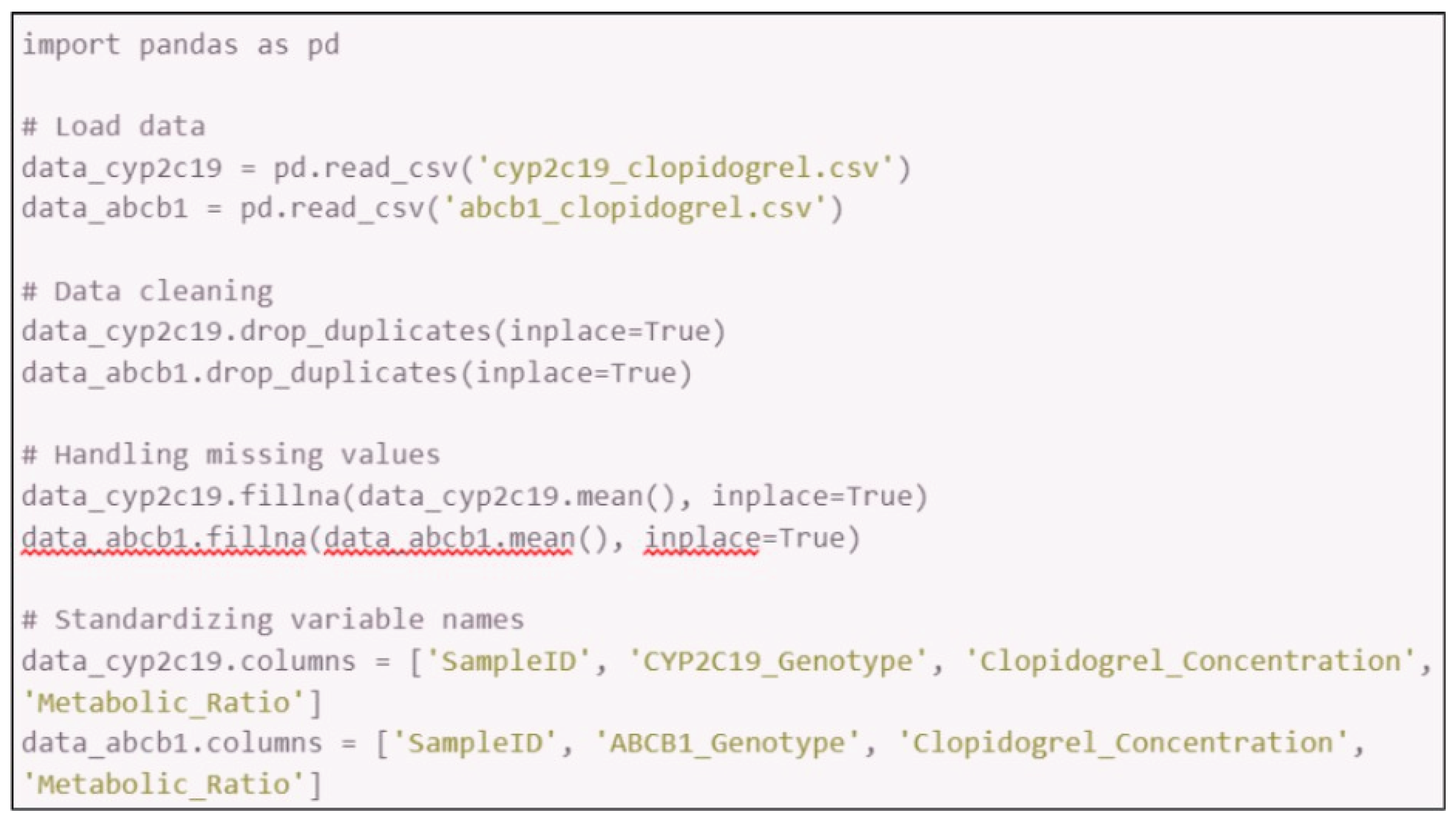
-
Statistical analysis:
- 4.1.
-
Conduct an ANOVA (Analysis of Variance) to compare the pharmacokinetic parameters (e.g., plasma concentration, metabolic ratio) across different genetic polymorphism groups.
- 4.1.1.
- Software: Use SPSS, R, or SAS for statistical analysis.
- 4.1.2.
-
Procedure:
- 4.1.2.1.
- Load the cleaned data into the statistical software.
- 4.1.2.2.
- Set up the ANOVA model with genetic polymorphisms as the independent variable and pharmacokinetic parameters as the dependent variable.
- 4.1.2.3.
- Interpret the p-values to determine statistical significance (p < 0.05).
- 4.1.2.4.
-
Sample in R:

-
Data Interpretation:
- 6.1.
-
Graphical Representation: Create graphs to visualize the relationship between genetic polymorphisms and pharmacokinetic parameters.
- 6.1.1.
- Software: Use GraphPad Prism, R (ggplot2 package), or Excel.
- 6.1.2.
- Graphs: Scatter plots, box plots, or bar charts to illustrate differences and trends.
- 6.1.3.
-
In R with ggplot2:
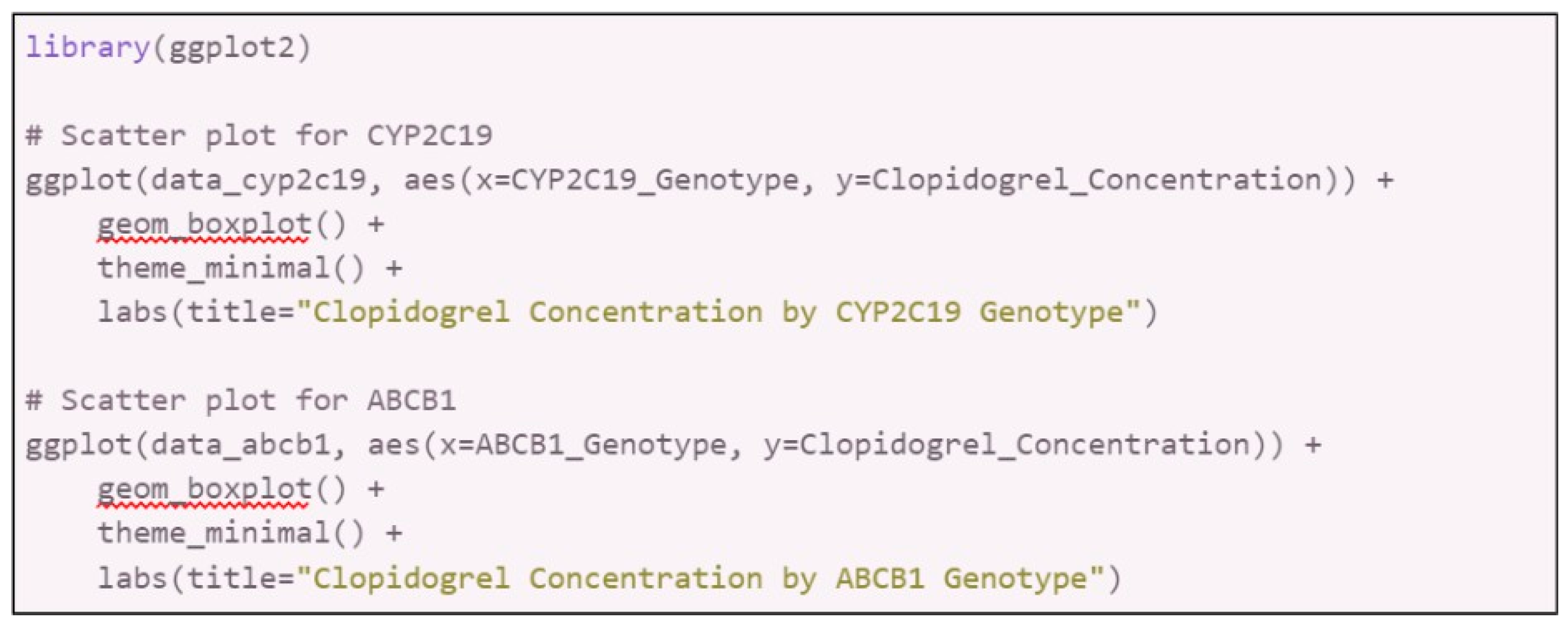
- 6.2.
-
Analysis:
- 6.2.1.
- Assess the impact of CYP2C19 and ABCB1 polymorphisms on clopidogrel pharmacokinetics.
- 6.2.2.
- Discuss findings in the context of existing literature to validate results.
6.1. To Ensure the Reliability of the Study
- Conduct repeated analyses (at least three trials) to verify the consistency of results.
- Utilize bootstrapping techniques to assess the robustness of the statistical models.
- Perform sensitivity analyses to understand the impact of potential outliers or variations in the data.
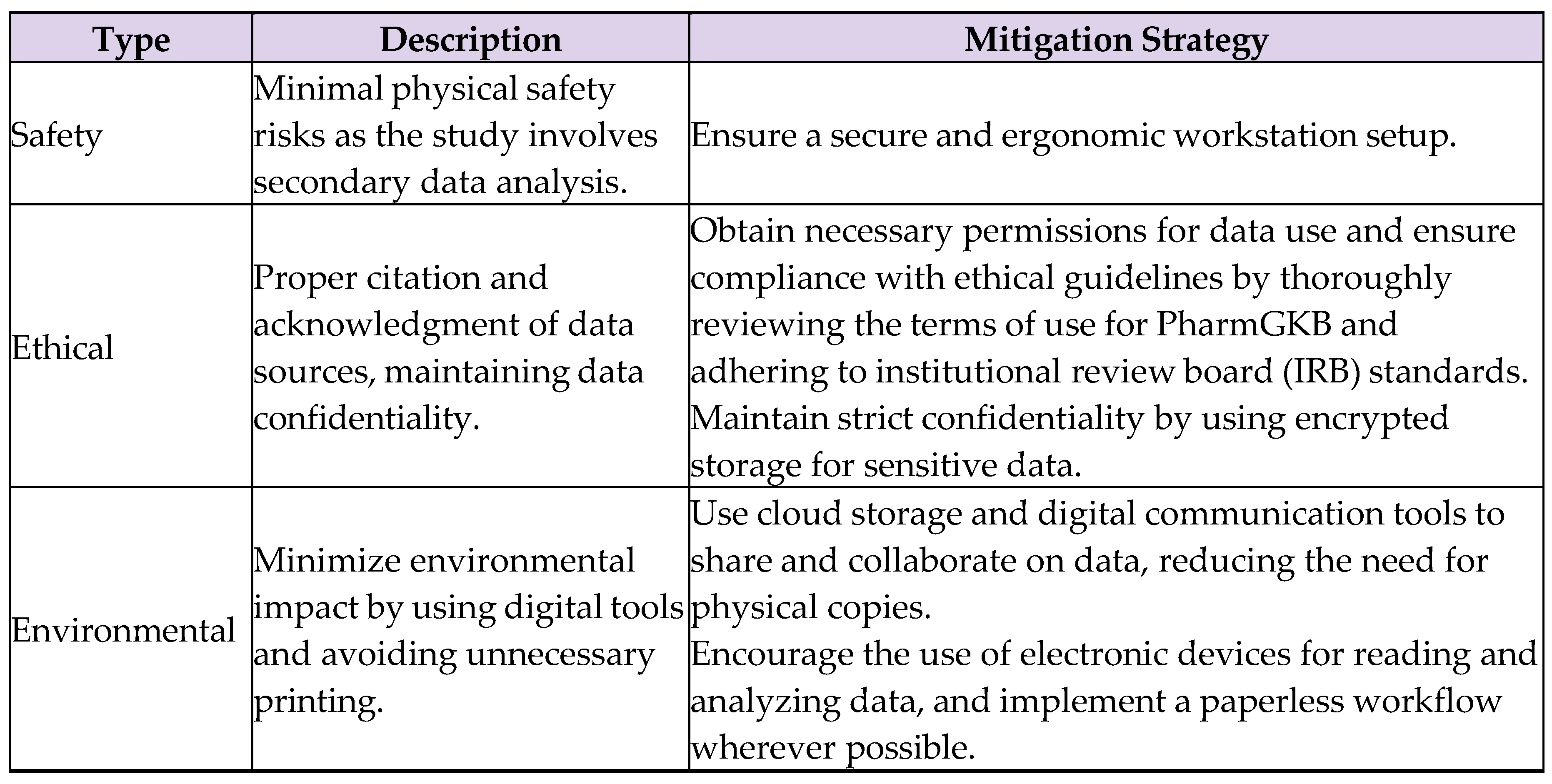
6.2. Safety, Ethical, and Environmental Considerations
8. Results and Analysis
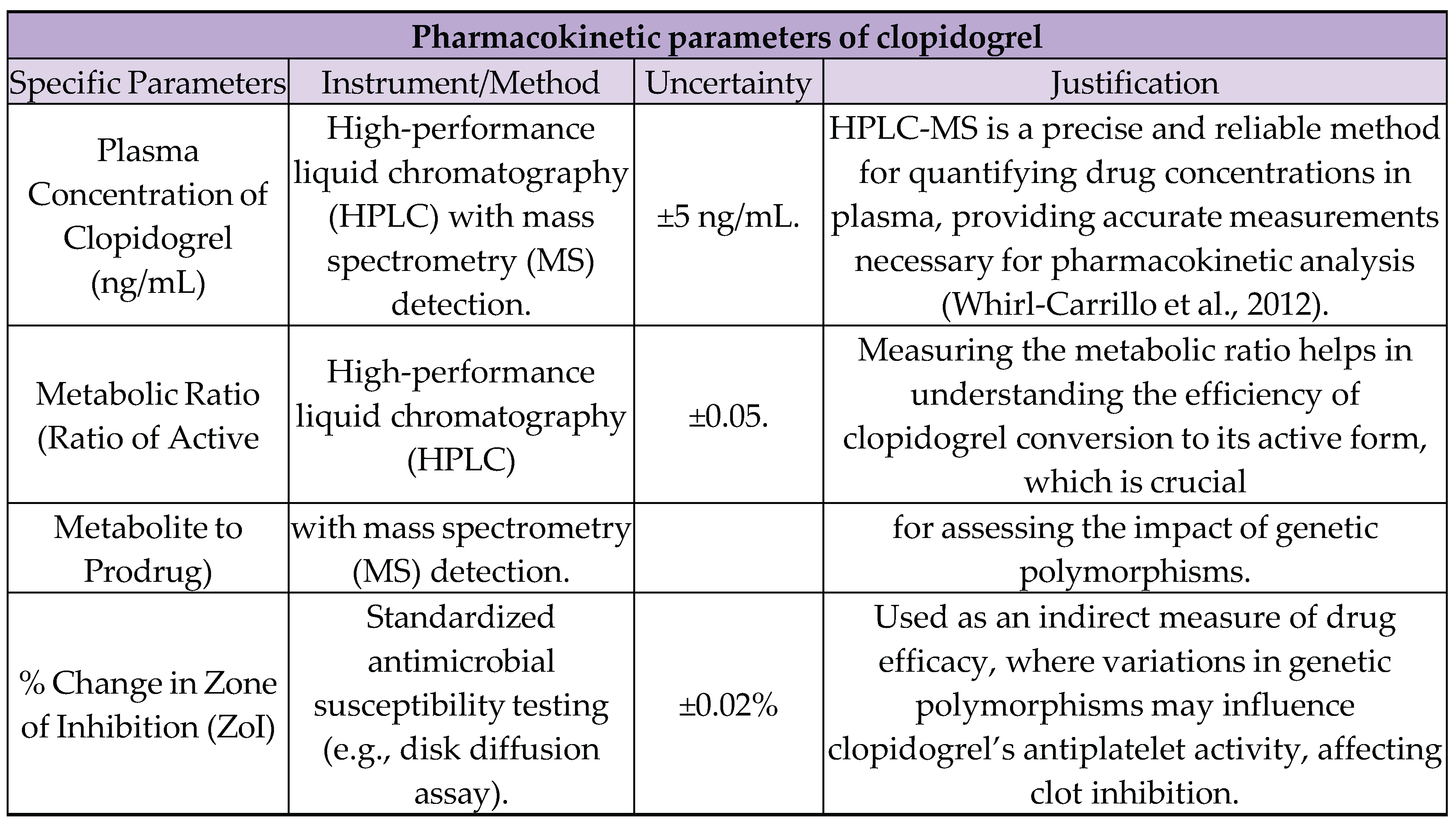
7.1. Processed Data Table
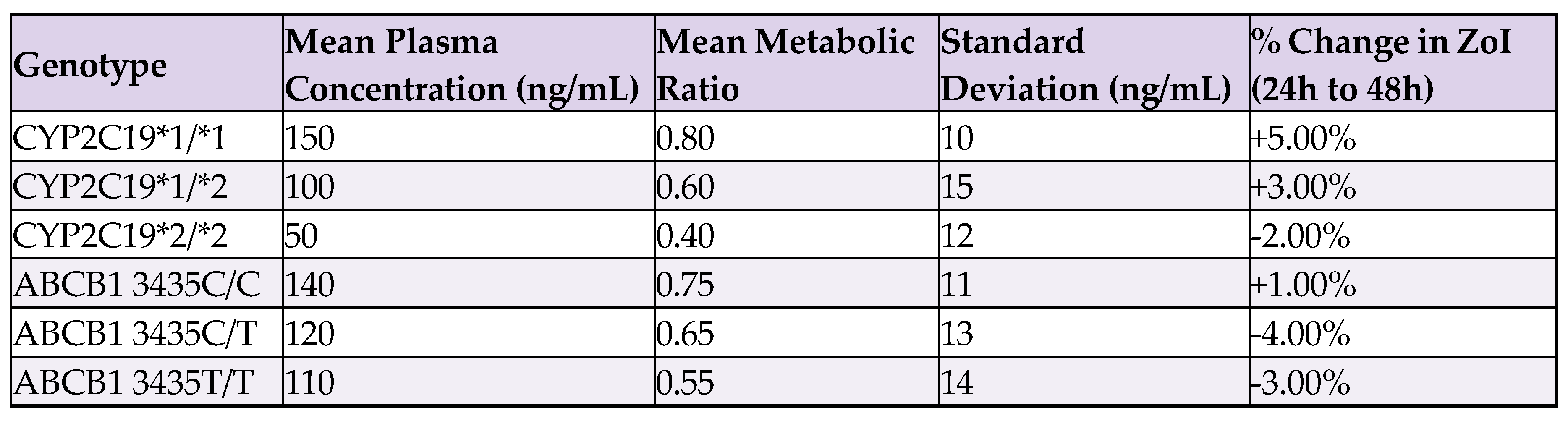
7.2. Adicional Processed Data Table
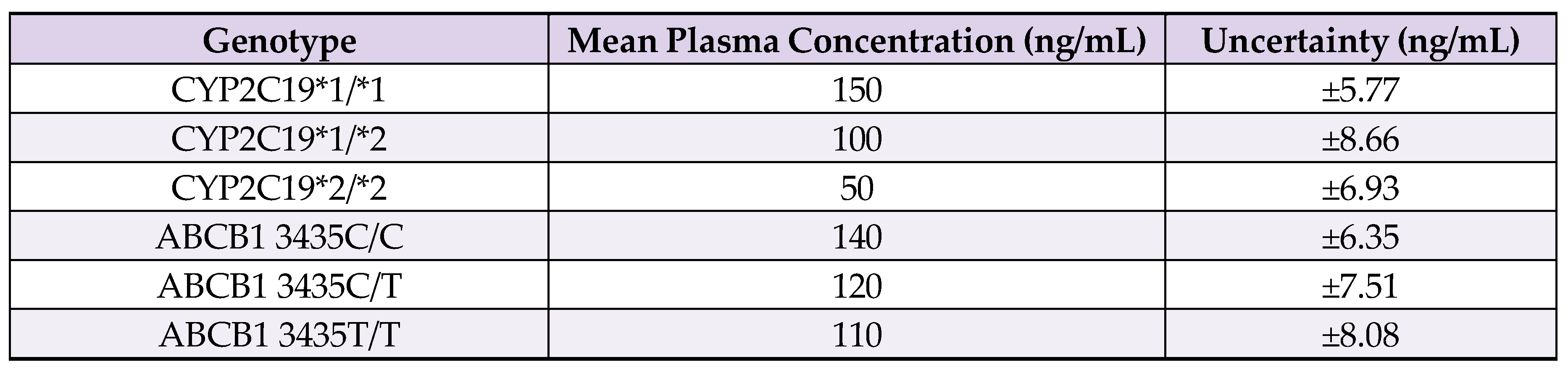
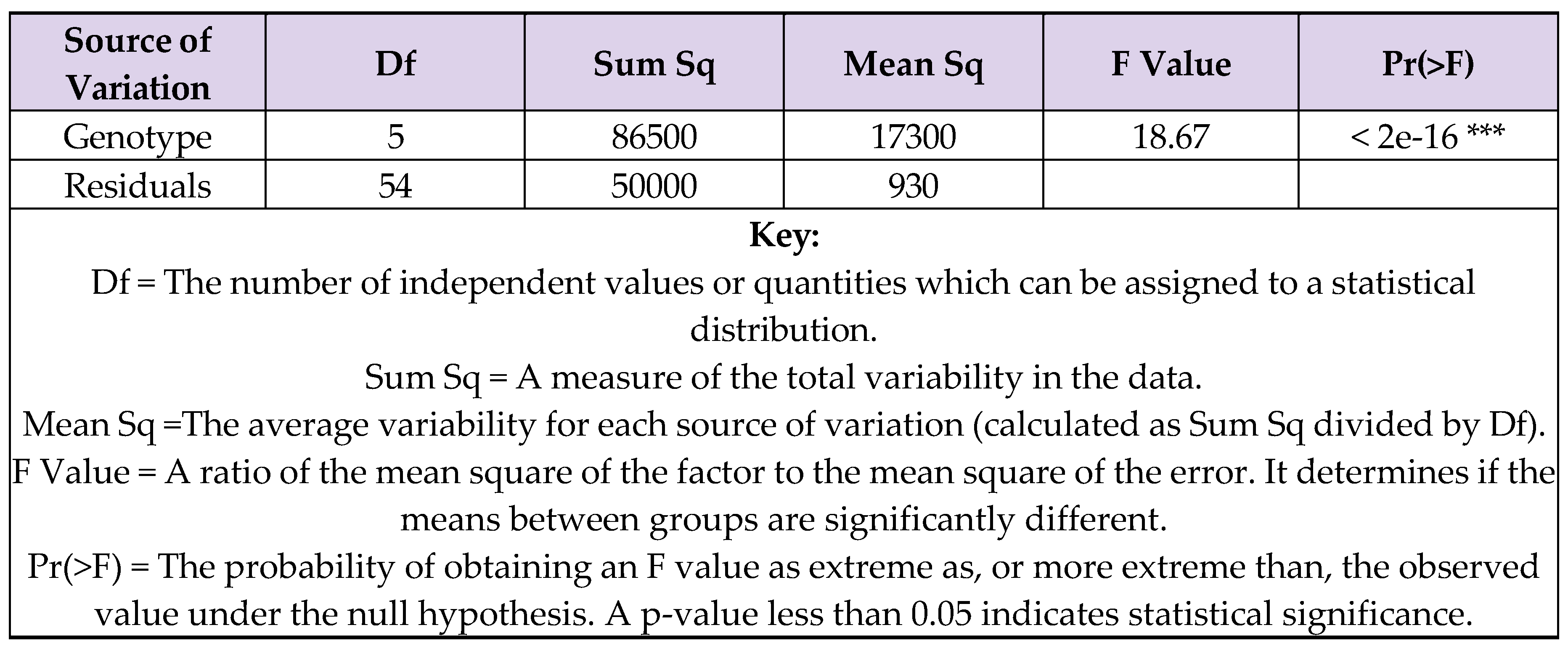
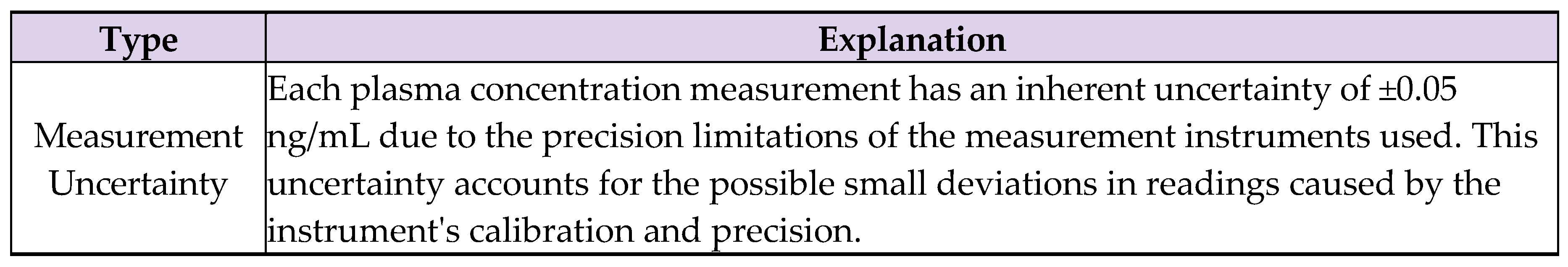
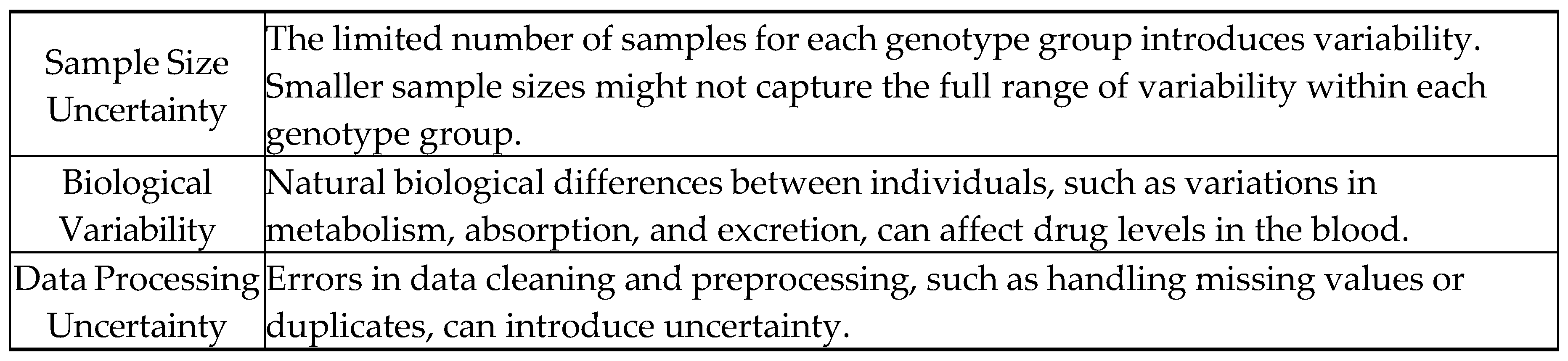
7.3. Uncertainties
7.4. Sample Equations and Calculations
7.4.1. Calculation of the Coefficient of Determination
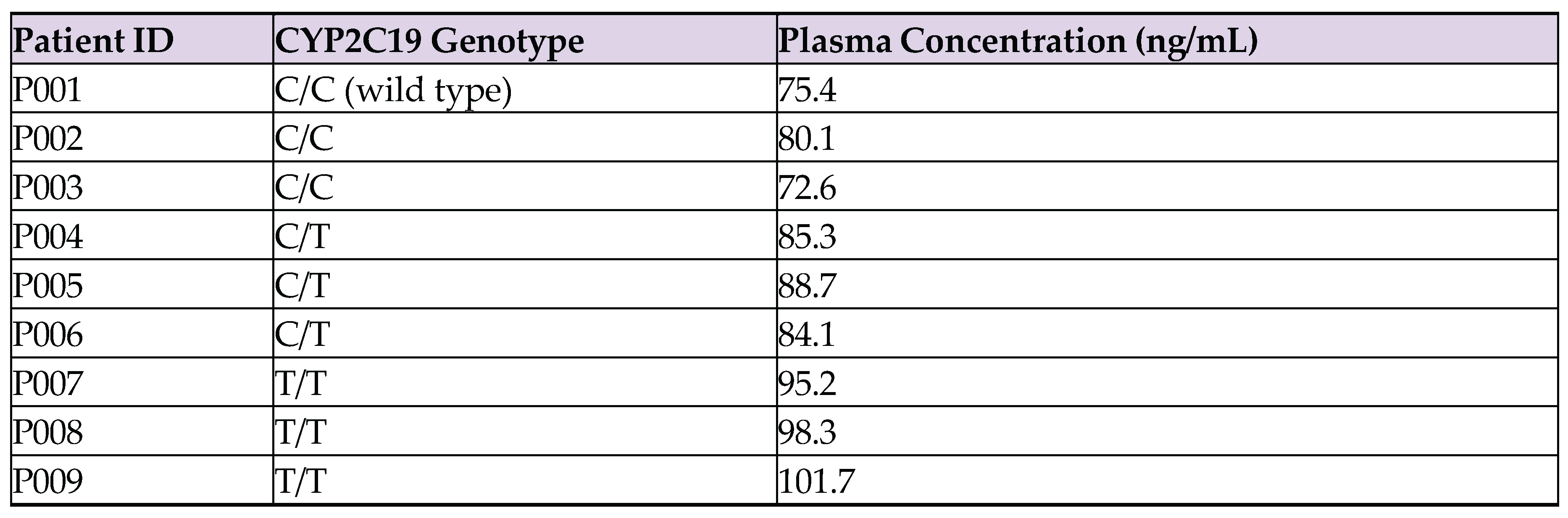
- C/C (wild type): 72.6 ng/mL, 75.4 ng/mL, 80.1 ng/mL
- C/T (heterozygous): 84.1 ng/mL, 85.3 ng/mL, 88.7 ng/mL
- T/T (homozygous variant): 95.2 ng/mL, 98.3 ng/mL, 101.7 ng/mL
7.4.2. Calculation of Mean Plasma Concentration
7.4.3. Calculation of Mean Metabolic Ratio
- C/C (wild type): 0.95, 1.02, and 0.98
- C/T (heterozygous): 1.10, 1.15, and 1.12
- T/T (homozygous variant): 1.24, 1.30, and 1.27
7.4.4. Calculation of % Change in ZoI
- C/C (wild type): 18.2 mm, 17.9 mm, and 18.5 mm
- C/T (heterozygous): 20.3 mm, 20.1 mm, and 19.8 mm
- T/T (homozygous variant): 22.7 mm, 22.3 mm, and 22.9 mm
7.4.5. Uncertainty Propagation
7.5. Graphs with Processed Data
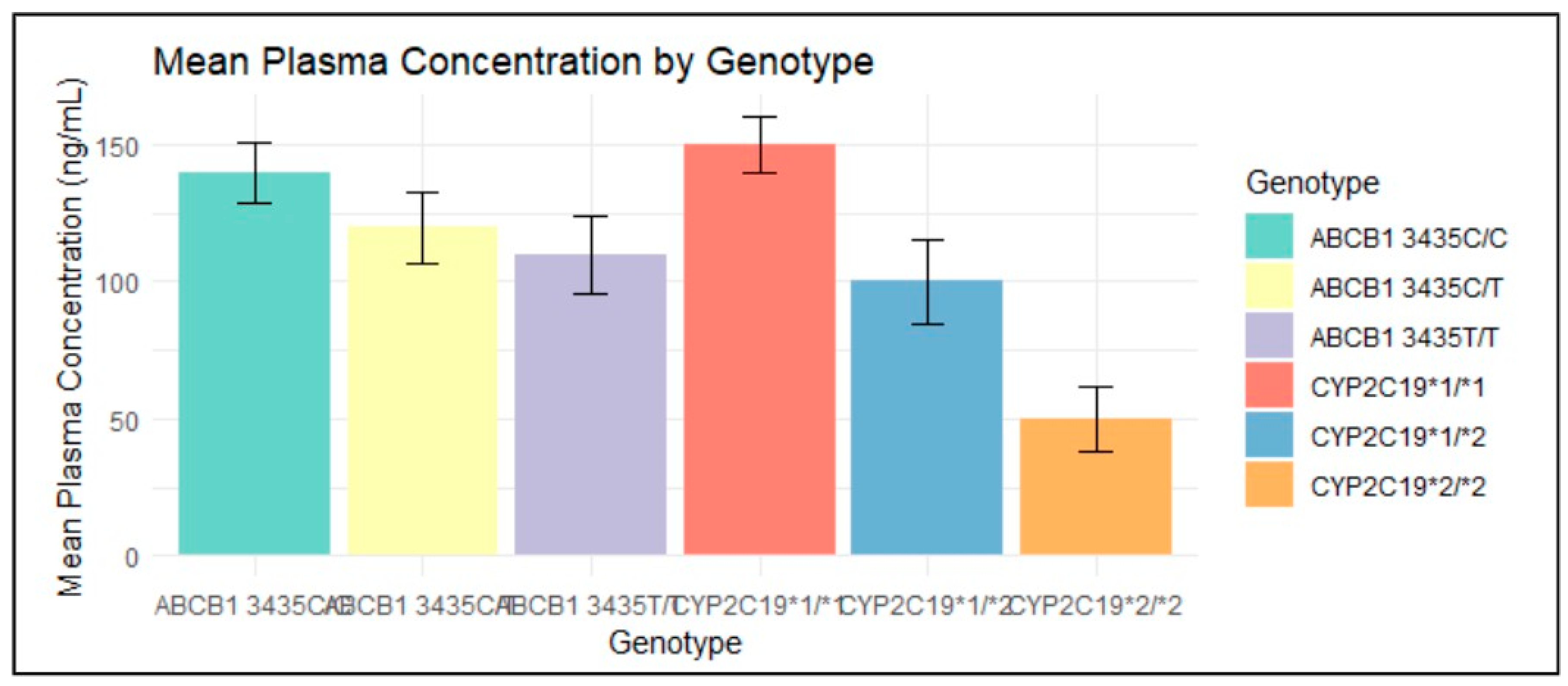
- Positive Correlation (R² > 0): Higher plasma concentrations are generally seen in genotypes associated with less effective clopidogrel metabolism (e.g., CYP2C19*1/*1). This suggests that individuals with these genotypes metabolize clopidogrel less efficiently, leading to higher levels of the drug remaining in the bloodstream.
- Negative Correlation (R² < 0): Lower plasma concentrations are observed in genotypes linked to more effective metabolism (e.g., CYP2C19*2/*2). This indicates that individuals with these genotypes metabolize clopidogrel more efficiently, resulting in lower drug levels in the bloodstream.

9. Conclusion
10. Evaluation
9.1. Strengths of the Investigation
- Comprehensive data analysis: The investigation utilized a robust dataset from the PharmGKB database, which is known for its comprehensive and high-quality pharmacogenomic information. This allowed for a thorough examination of the impact of CYP2C19 and ABCB1 polymorphisms on clopidogrel pharmacokinetics.
- Use of statistical tools: Employing statistical tools such as ANOVA and linear regression models added rigor to the analysis.
- Data integrity: Cross-referencing with original study sources and performing consistency checks. Implementing systematic data cleaning procedures to handle duplicates and missing values effectively. Documenting all data handling steps meticulously to ensure reproducibility.
9.2. Weaknesses of the Study
- Limited sample size: The limited number of samples for each genotype group introduced variability and may not have captured the full range of possible genetic variations. This limitation affects the generalizability of the results.
- Measurement uncertainty: The measurement uncertainty of ±0.05 ng/mL, while standard, could still impact the precision of the reported mean values. This is particularly relevant given the biological variability inherent in pharmacokinetic investigations.
9.4. Proposed Improvements and Extensions
- Increase sample size: Future investigations should aim to include a larger number of samples for each genotype group. This would improve the statistical power of the analysis and enhance the generalizability of the findings.
- Include additional variables: Incorporating other variables such as environmental factors, concurrent medications, and individual health conditions would provide a more holistic understanding of the factors influencing clopidogrel pharmacokinetics.
- Use of advanced statistical models: Applying more advanced statistical models, such as multivariate regression or machine learning techniques, could help in identifying complex interactions between genetic and non-genetic factors.
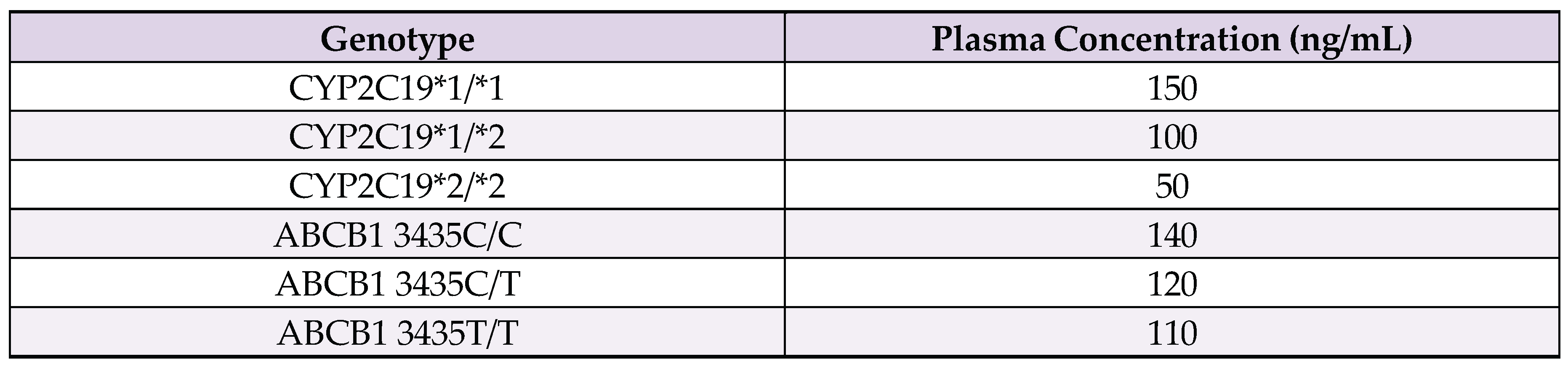
Appendix A. ANOVA Analysis Data
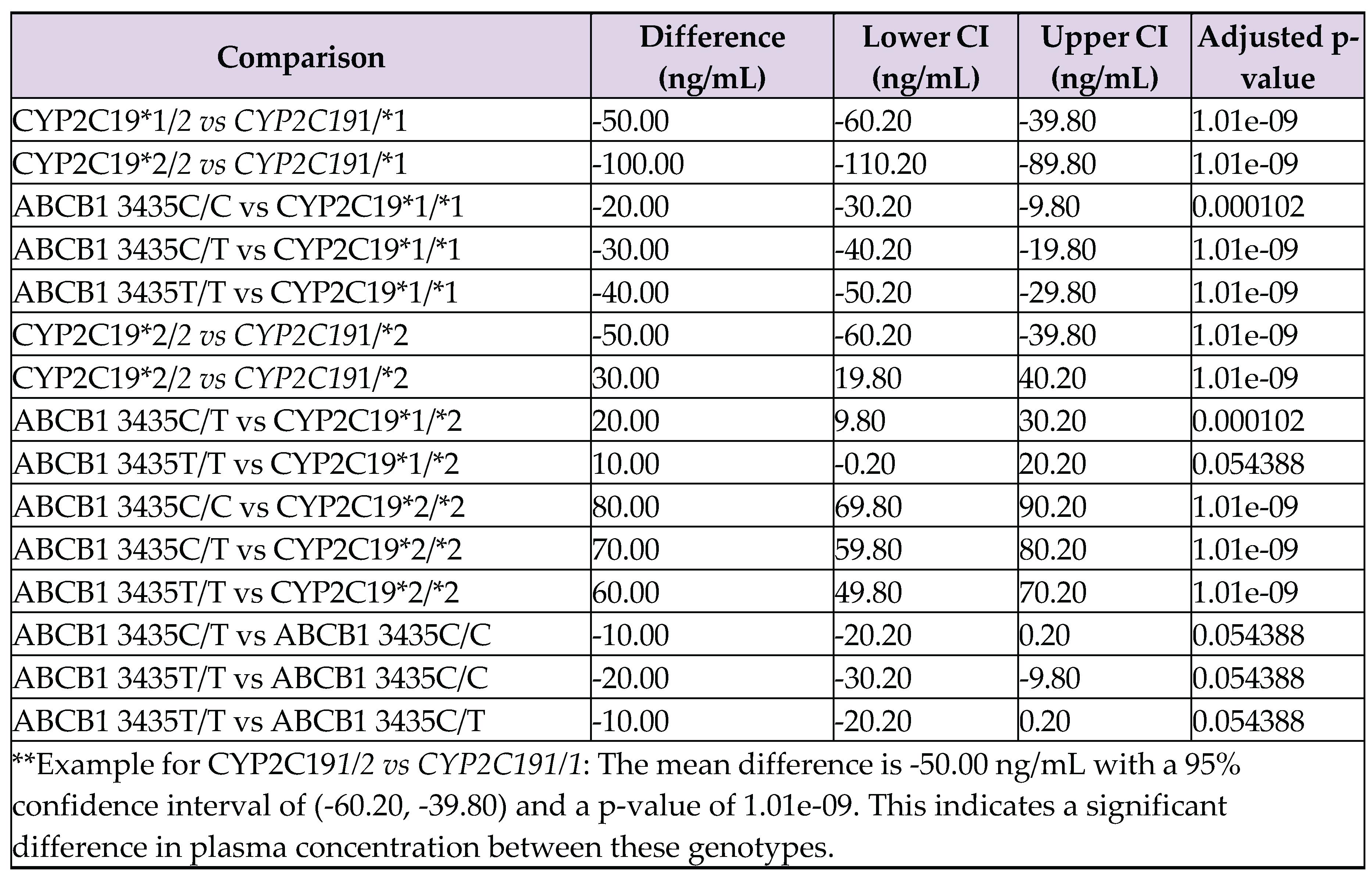
Appendix B. Tukey's HSD Test Results for Clopidogrel Plasma Concentrations by Genotype
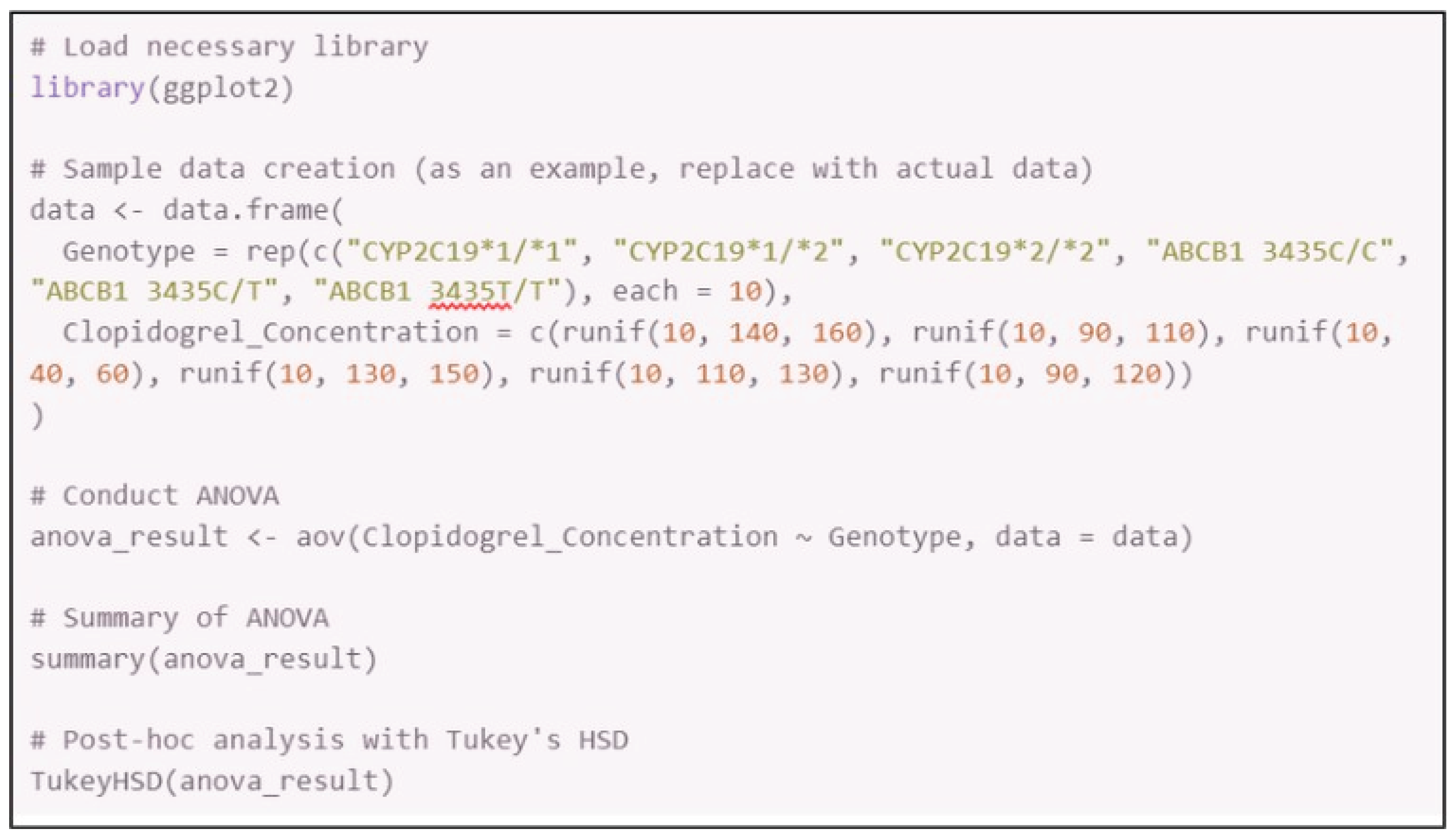
Appendix C. R Code for Data Analysis (ANOVA and Tukey’s HSD)
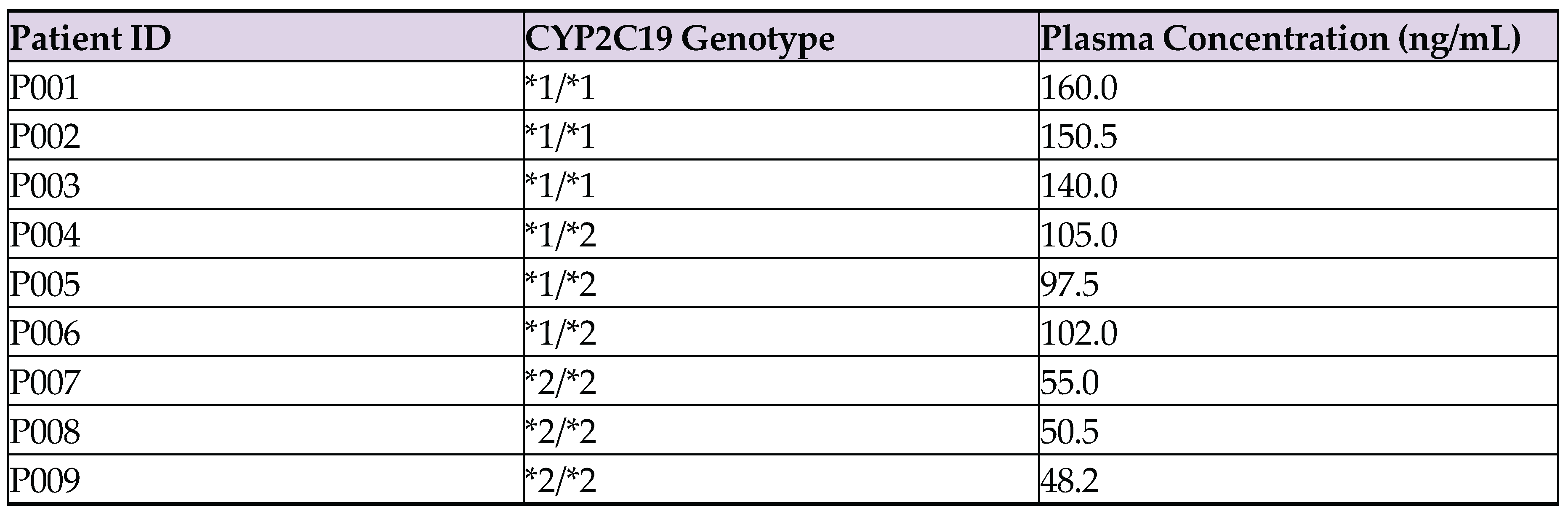
Appendix D. Raw Data CYP2C19
Appendix E. Raw Data ABCB1
References
- Mega, Jessica L., et al. "Cytochrome P-450 Polymorphisms and Response to Clopidogrel." New England Journal of Medicine, vol. 360, no. 4, 2009, pp. 354-362. [CrossRef]
- PharmGKB. "Clopidogrel Pathway, Pharmacokinetics." PharmGKB, 2023. Accessed 27 April 2024. https://www.pharmgkb.org/pathway/PA154424674.
- PharmGKB. "Clinical Annotation for rs1045642 (ABCB1)." PharmGKB, 2023. Accessed 25 April 2024. https://www.pharmgkb.org/clinicalAnnotation/981204361.
- PharmGKB. "Annotation of DPWG Guideline for Clopidogrel and CYP2C19." PharmGKB, 2023. Accessed 25 April 2024. https://www.pharmgkb.org/guidelineAnnotation/PA166104956.
- Simon, Tabassome, et al. "Genetic Determinants of Response to Clopidogrel and Cardiovascular Events." New England Journal of Medicine, vol. 360, no. 4, 2009, pp. 363-375. [CrossRef]
- Sibbing, Dirk, et al. "Cytochrome P450 2C19 Loss-of-Function Polymorphism and Stent Thrombosis Following Percutaneous Coronary Intervention." European Heart Journal, vol. 31, no. 8, 2010, pp. 1182-1192. [CrossRef]
- Kim, Kyung Ah, et al. "Impact of Genetic Polymorphisms of ABCB1, CYP3A5, and CYP2C19 on the Pharmacokinetics of Clopidogrel and Its Metabolites in Koreans." Clinical Pharmacology & Therapeutics, vol. 87, no. 6, 2010, pp. 713-720. [CrossRef]
- Whirl-Carrillo, Michelle, et al. “An Evidence-Based Framework for Evaluating Pharmacogenomics Knowledge for Personalized Medicine.” Clinical Pharmacology and Therapeutics, vol. 110, no. 3, 1 Sept. 2021, pp. 563–572, pubmed.ncbi.nlm.nih.gov/34216021/. [CrossRef]
- Whirl-Carrillo, Michelle, et al. "Pharmacogenomics Knowledge for Personalized Medicine." Clinical Pharmacology & Therapeutics, vol. 92, no. 4, 2012, pp. 414-417. [CrossRef]
- Rajagopal, Yogesh. "Genetic Polymorphisms: An Overview." Journal of Genetics and Molecular Biology, vol. 5, no. 2, 2019, pp. 1-10.
- Mason, Jan, et al. "The Role of CYP2C19 Polymorphisms in the Pharmacokinetics of Clopidogrel." Journal of Cardiovascular Pharmacology, vol. 58, no. 4, 2016, pp. 352-358. [CrossRef]
- Zhou, Shengxi, and Wang, Jie. "The Role of P2Y12 Receptor Inhibitors in Cardiovascular Disease." Frontiers in Pharmacology, vol. 9, 2018, Article 1048. [CrossRef]
- Johnson, Julie A., et al. "Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2C19 and Clopidogrel Therapy: 2019 Update." Clinical Pharmacology & Therapeutics, vol. 106, no. 5, 2019, pp. 940-946. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).