Submitted:
30 June 2025
Posted:
01 July 2025
You are already at the latest version
Abstract
Keywords:
1. Fusarium: A Genus of Global Agricultural and Phytopathological Significance
2. Taxonomy and Phylogenetics of Fusarium Species
2.1. From Morphology to Genomics: Evolution of Fusarium Classification
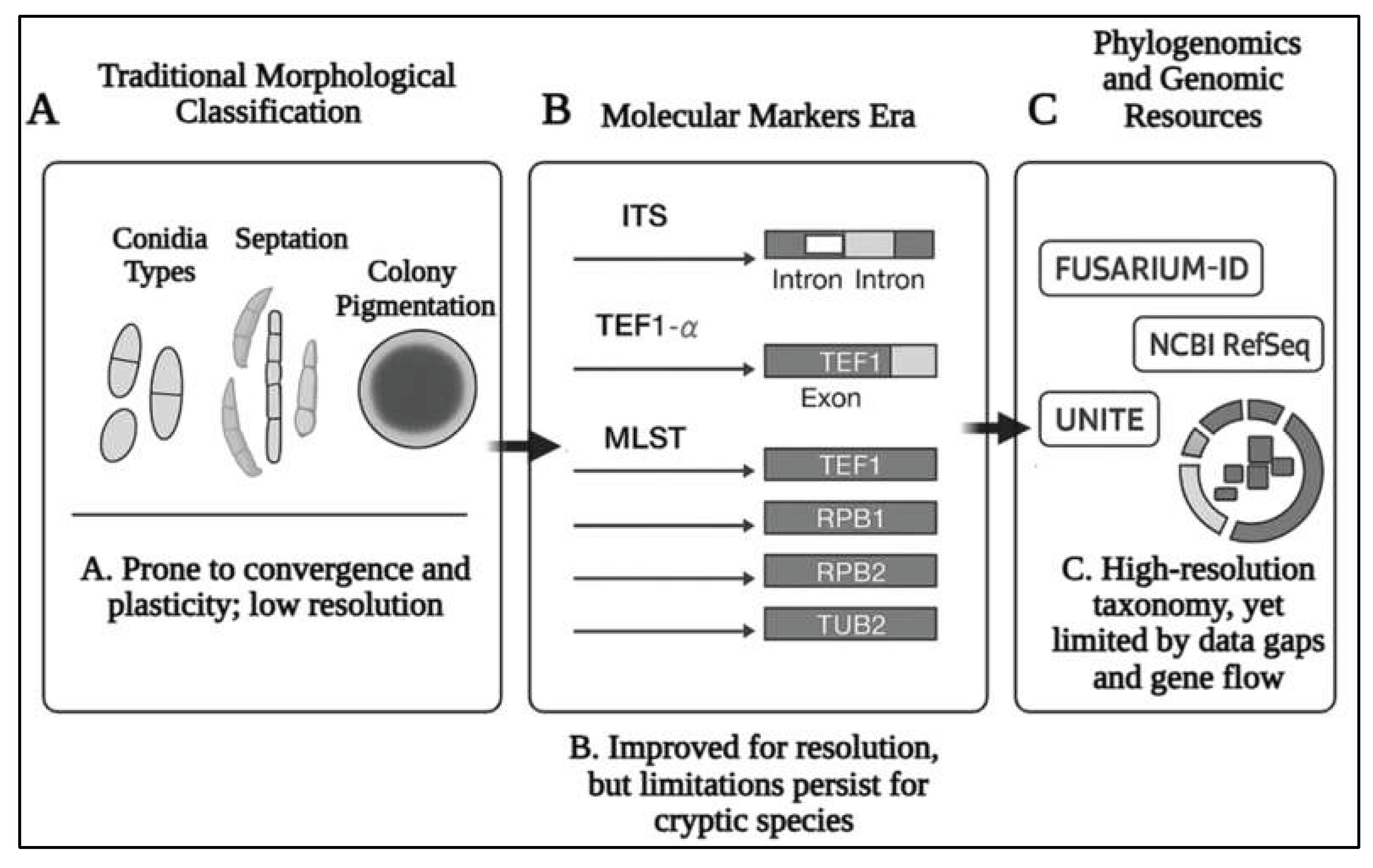
2.2. Rise and Relevance of Fusarium Species Complexes: Evolutionary Divergence, Taxonomic Challenges, Intraspecific Variability and Pathogenic Implications
2.3. The Neocosmospora Debate: Taxonomy Reshuffled, or Genus Redefined?
2.4. Emerging Fusarium Pathogens in Niche Crops and Geographies
2.5. Phylogenomics of Fusarium: Insights from Whole-Genome Data
3. Genomic Insights and Functional Genomics: Exploring Recent Genomic Studies and Their Implications for Understanding Pathogenicity and Resistance Mechanisms
3.1. Genomic Architecture of Fusarium spp.
3.2. Secondary Metabolites, Genome Plasticity, and Host Specialization
3.2.1. Functional Genomics and Pathogenicity Mechanisms
3.2.2 Proteomics and Secreted Virulence Factors
3.3. Resistance Mechanisms in Host Plants
3.4. Phylogenomics and Comparative Genome Analysis Across Species Complexes
4. Pathogenicity and Host Interactions
4.1. Fusarium spp. Pathogenicity
4.2. Host Specificity and Formae speciales in Fusarium Species
4.3. Plant Immune Responses and Fusarium Counter-Defenses
5. Toxins Produced by Fusarium: Global Burden in Major Staple Crops
5.1. Overview of Major Fusarium-derived Toxins in Staple Crops
5.2. Regional Analysis of Mycotoxin Burden and Exposure Risk
5.3. Mycotoxin Detection
6. Management and Diagnostic Strategies of Fusarium-Induced Plant Diseases
- a)
- Crop Rotation: Prior to the widespread use of synthetic fertilizers and pesticides in the 1950s, crop rotation played a particularly important role in the management of diseases and pests [216]. Because Fusarium inoculum is widely distributed in soil, on plant parts, and on debris, crop rotation has been the foundational basis for limiting its accumulation [217,218]. Crop rotation lowers the risk of disease development and pressure by using a nonhost crop to disrupt a Fusarium species’ life cycle [219,220,221]
- b)
- Chemical Control: The use of fungicides remains a common component of integrated management strategies aimed at mitigating diseases caused by Fusarium species. In large-scale cropping systems, this approach is often favored for its efficiency, ease of application, and relatively rapid suppression of disease symptoms [234]. The choice of fungicide depends on the species and the disease location - aboveground (foliar fungicide application) or soil/stubble borne (seed treatment) [235,236,214,237,155].
- c)
- Biocontrol: This offers a sustainable alternative to chemical fungicides for the management of plant diseases caused by Fusarium species, reducing the impact of associated environmental and human health risks [275, 276]. Several biological control agents (BCAs), including strains of Trichoderma species and Bacillus velezensis have demonstrated efficacy in suppressing Fusarium-induced plant diseases across both controlled and field environments [277,275,278,279].
- d)
- Use of Resistant Cultivars: Host resistance remains one of the most effective and sustainable Fusarium management strategies. Conventional breeding has identified several quantitative trait loci (QTLs) conferring resistance to Fusarium diseases, like FHB in wheat and barley, and Fusarium wilt in grain legumes [299,300,301,302]. However, conventional breeding methods are labor-intensive, can take a long time, and require enough genetic variation in the breeding material [303,304].
6.1. Molecular diagnostics
7. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aoki, T. , O'Donnell, K., & Geiser, D. M. (2014). Systematics of key phytopathogenic Fusarium species: Current status and future challenges. Journal of General Plant Pathology, 80, 189–201. [CrossRef]
- O'Donnell, K. , Whitaker, B. K., Laraba, I., Proctor, R. H., Brown, D. W., Broders, K., Kim, H.-S., McCormick, S. P., Busman, M., Aoki, T. (2022). DNA sequence-based identification of Fusarium: A work in progress. Plant Disease, 106, 1597–1609. [CrossRef]
- Kamil, D. , Mishra, A. K., Das, A., & Nishmitha, K. (2025). Genus Fusarium and Fusarium species complexes. In Biodiversity, bioengineering, and biotechnology of fungi (pp. 209-225). Academic Press. [CrossRef]
- Torbati, M. , Arzanlou, M., & da Silva Santos, A. C. (2021). Fungicolous Fusarium species: Ecology, diversity, isolation, and identification. Current Microbiology, 78, 2850–2859. [CrossRef]
- Armer, V. J. , Kroll, E., Darino, M., Smith, D. P., Urban, M., & Hammond-Kosack, K. E. (2024). Navigating the Fusarium species complex: Host-range plasticity and genome variations. Fungal Biology, 128, 2439–2459. [CrossRef]
- Khuna, S., Kumla, J., Thitla, T., Nuangmek, W., Lumyong, S., & Suwannarach, N. (2022). Morphology, molecular identification, and pathogenicity of two novel Fusarium species associated with postharvest fruit rot of cucurbits in Northern Thailand. Journal of Fungi, 8(11), 1135. [CrossRef]
- Moparthi, S. , Burrows, M., Mgbechi-Ezeri, J., & Agindotan, B. (2021). Fusarium spp. associated with root rot of pulse crops and their cross-pathogenicity to cereal crops in Montana. Plant Disease, 105 548–557. [CrossRef]
- Moparthi, S. , Perez-Hernandez, O., Burrows, M. E., Bradshaw, M. J., Bugingo, C., Brelsford, M., & McPhee, K. (2024). Identification of Fusarium spp. associated with chickpea root rot in Montana. Agriculture, 14, 974. [CrossRef]
- Dean, R. , van Kan, J. A. L., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., Rudd, J. J., Dickman, M., Kahmann, R., & Ellis, J. (2012). The top 10 fungal pathogens in molecular plant pathology. Molecular Plant Pathology, 13 414–430. [CrossRef]
- Ekwomadu, T. I., Akinola, S. A., & Mwanza, M. (2021). Fusarium mycotoxins, their metabolites (free, emerging, and masked), food safety concerns, and health impacts. International Journal of Environmental Research and Public Health, 18(22), 11741. [CrossRef]
- Okungbowa, F. I. , & Shittu, H. O. (2012). Fusarium wilts: An overview. Environmental Research Journal, 6, 83–102.
- Arie, T. (2019). Fusarium diseases of cultivated plants, control, diagnosis, and molecular and genetic studies. Journal of Pesticide Sciences, 44(4), 275–281. [CrossRef]
- Ma, L.-J., Geiser, D. M., Proctor, R. H., Rooney, A. P., O'Donnell, K., Trail, F., Gardiner, D. M., Manners, J. M., & Kazan, K. (2013). Fusarium pathogenomics. Annual Review of Microbiology, 67, 399–416. [CrossRef]
- Coleman, J. J. (2016). The Fusarium solani species complex: Ubiquitous pathogens of agricultural importance. Molecular Plant Pathology, 17(2), 146–158. [CrossRef]
- Chandra, N. S. , Wulff, E. G., Udayashankar, A. C., Nandini, B. P., Niranjana, S. R., Mortensen, C. N., & Prakash, H. S. (2011). Prospects of molecular markers in Fusarium species diversity. Applied Microbiology and Biotechnology, 90(5), 1625–1639. [CrossRef]
- Infantino, A. , Grottoli, A., Bergamaschi, V., Oufensou, S., Burgess, L. W., & Balmas, V. (2023). FusaHelp: A web site program for the morphological identification of Fusarium species. Journal of Plant Pathology, 105(2), 429–436. [CrossRef]
- O'Donnell, K. , Ward, T. J., Robert, V. A., Crous, P. W., Geiser, D. M., & Kang, S. (2015). DNA sequence-based identification of Fusarium: Current status and future directions. Phytoparasitica, 43(5), 583–595. [CrossRef]
- Desai, S. , Dubey, S. C., & Prasad, R. D. (2020). Impacts of climate change on Fusarium species vis-à-vis adaptation strategies. Indian Phytopathology, 73(4), 593–603. [CrossRef]
- Buxton, E. W. (1955). The taxonomy and variation in culture of Fusarium oxysporum from gladiolus. Transactions of the British Mycological Society, 38(3), 202–208. [CrossRef]
- Snyder, W. C., & Hansen, H. N. (1940). The species concept in Fusarium. American Journal of Botany, 27(1), 64–67. [CrossRef]
- Booth, C. (1971). The genus Fusarium. Commonwealth Mycological Institute.
- O'Donnell, K. , Sutton, D. A., Rinaldi, M. G., Sarver, B. A., Balajee, S. A., Schroers, H.-J., Summerbell, R. C., Robert, V. A., Crous, P. W., & Zhang, N. (2010). Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. Journal of Clinical Microbiology, 48(10), 3708–3718. [CrossRef]
- Watanabe, M. , Yonezawa, T., Lee, K.-i., Kumagai, S., Sugita-Konishi, Y., Goto, K., & Hara-Kudo, Y. (2011). Molecular phylogeny of the higher and lower taxonomy of the Fusarium genus and differences in the evolutionary histories of multiple genes. BMC Evolutionary Biology, 11(1), 322. [CrossRef]
- Lizcano Salas, A. F. , Duitama, J., Restrepo, S., & Celis Ramírez, A. M. (2024). Phylogenomic approaches reveal a robust time-scale phylogeny of the Terminal Fusarium Clade. IMA Fungus, 15(1), 13. [CrossRef]
- Schoch, C. L., Seifert, K. A., Huhndorf, S., Robert, V., Spouge, J. L., Levesque, C. A., Chen, W., & Fungal Barcoding Consortium. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences, 109(16), 6241–6246. [CrossRef]
- Singha, I. M., Kakoty, Y., Unni, B. G., Das, J., & Kalita, M. C. (2016). Identification and characterization of Fusarium spp. using ITS and RAPD causing fusarium wilt of tomato isolated from Assam, Northeast India. Journal of Genetic Engineering and Biotechnology, 14(1), 99–105. [CrossRef]
- Zarrin, M. , Ganj, F., & Faramarzi, S. (2016). Analysis of the rDNA internal transcribed spacer region of the Fusarium species by polymerase chain reaction-restriction fragment length polymorphism. Biomedical Reports, 4(4), 471–474. [CrossRef]
- O’Donnell, K. , Ward, T. J., Robert, V. A., Crous, P. W., Geiser, D. M., & Kang, S. (2015). DNA sequence-based identification of Fusarium: Current status and future directions. Phytoparasitica, 43(5), 583–595. [CrossRef]
- Crous, P. W. , Lombard, L., Sandoval-Denis, M., Seifert, K. A., Schroers, H.-J., Chaverri, P., Gené, J., Guarro, J., Hirooka, Y., & Bensch, K. (2021). Fusarium: More than a node or a foot-shaped basal cell. Studies in Mycology, 98, 100116. [CrossRef]
- van Diepeningen, A. D. , Feng, P., Ahmed, S., Sudhadham, M., Bunyaratavej, S., & de Hoog, G. S. (2015). Spectrum of Fusarium infections in tropical dermatology evidenced by multilocus sequencing typing diagnostics. Mycoses, 58(1), 48–57. [CrossRef]
- Oliveira, L. J. M. G. , Rodrigues, A. A. C., Silva, E. K. C., Oliveira, A. C. S., Barros, M. C., Fraga, E. C., Nascimento, I. O., & Silva, M. R. M. (2021). Morphological and phylogenetic characterization of Fusarium Link. Australian Journal of Crop Science, 15(12), 1406–1415. [CrossRef]
- Yörük, E. , & Yli-Mattila, T. (2024). Translation elongation factor 1-alpha sequencing provides reliable tool for identification of Fusarium graminearum species complex members. Diversity, 16(8), 481. [CrossRef]
- Geiser, D. M. , Al-Hatmi, A. M., Aoki, T., Arie, T., Balmas, V., Barnes, I., Bergstrom, G. C., Bhattacharyya, M. K., Blomquist, C. L., & Bowden, R. L. (2021). Phylogenomic analysis of a 55.1-kb 19-gene dataset resolves a monophyletic Fusarium that includes the Fusarium solani species complex. Phytopathology, 111(7), 1064–1079. [CrossRef]
- Sandoval-Denis, M. , Lombard, L., & Crous, P. W. (2019). Back to the roots: A reappraisal of Neocosmospora. Persoonia, 43(1), 90–185. [CrossRef]
- Maryani, N. , Sandoval-Denis, M., Lombard, L., Crous, P. W., & Kema, G. (2019). New endemic Fusarium species hitch-hiking with pathogenic Fusarium strains causing Panama disease in small-holder banana plots in Indonesia. Persoonia, 43(1), 48–69. [CrossRef]
- O’Donnell, K., Rooney, A. P., Proctor, R. H., Brown, D. W., McCormick, S. P., Ward, T. J., Frandsen, R. J., Lysøe, E., Rehner, S. A., & Aoki, T. (2013). Phylogenetic analyses of RPB1 and RPB2 support a middle Cretaceous origin for a clade comprising all agriculturally and medically important fusaria. Fungal Genetics and Biology, 52, 20–31. [CrossRef]
- Manukyan, I., Khatsaeva, R., Kosyrev, S., & Nagam, M. A. A. (2024). Species diversity of Fusarium fungi on grain crops in the conditions of the foothill zone of the Central Caucasus. BIO Web of Conferences, 118, 01018. [CrossRef]
- Wang, M. , Crous, P. W., Sandoval-Denis, M., Han, S., Liu, F., Liang, J., Duan, W., & Cai, L. (2022). Fusarium and allied genera from China: Species diversity and distribution. Persoonia, 48(1), 1–53. [CrossRef]
- Laurence, M. H., Summerell, B. A., Burgess, L. W., & Liew, E. C. (2014). Genealogical concordance phylogenetic species recognition in the Fusarium oxysporum species complex. Fungal Biology, 118(4), 374–384. [CrossRef]
- Sandoval-Denis, M. , Lombard, L., & Crous, P. W. (2019). Back to the roots: A reappraisal of Neocosmospora. Persoonia, 43(1), 90–185. [CrossRef]
- Mi, Z. , Su, J., Yu, L., & Zhang, T. (2024). Comparative mitochondrial genomics of Thelebolaceae in Antarctica: Insights into their extremophilic adaptations and evolutionary dynamics. IMA Fungus, 15(1), 33. [CrossRef]
- Peck, L. D. , Llewellyn, T., Bennetot, B., O'Donnell, S., Nowell, R. W., Ryan, M. J., Flood, J., de la Vega, R. C. R., Ropars, J., & Giraud, T. (2024). Horizontal transfers between fungal Fusarium species contributed to successive outbreaks of coffee wilt disease. PLoS Biology, 22(12), e3002480. [CrossRef]
- Lombard, L. , Sandoval-Denis, M., Cai, L., & Crous, P. W. (2019). Changing the game: Resolving systematic issues in key Fusarium species complexes. Persoonia, 43(1), i-ii. [CrossRef]
- O'Donnell, K. , Humber, R. A., Geiser, D. M., Kang, S., Park, B., Robert, V. A., Crous, P. W., Johnston, P. R., Aoki, T., & Rooney, A. P. (2012). Phylogenetic diversity of insecticolous fusaria inferred from multilocus DNA sequence data and their molecular identification via FUSARIUM-ID and Fusarium MLST. Mycologia, 104(2), 427-445. [CrossRef]
- Nilsson, R. H. , Larsson, K.-H., Taylor, A. F. S., Bengtsson-Palme, J., Jeppesen, T. S., Schigel, D., Kennedy, P., Picard, K., Glöckner, F. O., & Tedersoo, L. (2019). The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Research, 47(D1), D259-D264. [CrossRef]
- Villani, A. , Proctor, R. H., Kim, H.-S., Brown, D. W., Logrieco, A. F., Amatulli, M. T., Moretti, A., & Susca, A. (2019). Variation in secondary metabolite production potential in the Fusarium incarnatum-equiseti species complex revealed by comparative analysis of 13 genomes. BMC Genomics, 20(1), 314. [CrossRef]
- Ha, M. S., Ryu, H., Ju, H. J., & Choi, H.-W. (2023). Diversity and pathogenic characteristics of the Fusarium species isolated from minor legumes in Korea. Scientific Reports, 13(1), 22516. [CrossRef]
- Summerell, B. A. (2019). Resolving Fusarium: Current status of the genus. Annual Review of Phytopathology, 57(1), 323-339. [CrossRef]
- Kamil, D. , Mishra, A. K., Das, A., & Nishmitha, K. (2025). Genus Fusarium and Fusarium species complexes. In Biodiversity, bioengineering, and biotechnology of fungi (pp. 209-225). Academic Press. [CrossRef]
- Gao, Y., Zhang, Z., Ji, M., Ze, S., Wang, H., Yang, B., Hu, L., & Zhao, N. (2025). Identification and pathogenicity of Fusarium species from herbaceous plants on grassland in Qiaojia County, China. Microorganisms, 13(1), 113. [CrossRef]
- Harish, J. , Jambhulkar, P. P., Bajpai, R., Arya, M., Babele, P. K., Chaturvedi, S. K., Kumar, A., & Lakshman, D. K. (2023). Morphological characterization, pathogenicity screening, and molecular identification of Fusarium spp. isolates causing post-flowering stalk rot in maize. Frontiers in Microbiology, 14, 1121781. [CrossRef]
- Pothiraj, G. , Hussain, Z., Singh, A. K., Solanke, A. U., Aggarwal, R., Ramesh, R., & Shanmugam, V. (2021). Characterization of Fusarium spp. inciting vascular wilt of tomato and its management by a Chaetomium-based biocontrol consortium. Frontiers in Plant Science, 12, 748013. [CrossRef]
- Zhang, L., Hou, M., Zhang, X., Cao, Y., Sun, S., Zhu, Z., Han, S., Chen, Y., Ku, L., & Duan, C. (2023). Integrative transcriptome and proteome analysis reveals maize responses to Fusarium verticillioides infection inside the stalks. Molecular Plant Pathology, 24(7), 693-710. [CrossRef]
- Achari, S. R., Kaur, J., Dinh, Q., Mann, R., Sawbridge, T., Summerell, B. A., & Edwards, J. (2020). Phylogenetic relationship between Australian Fusarium oxysporum isolates and resolving the species complex using the multispecies coalescent model. BMC Genomics, 21(1), 1-20. [CrossRef]
- Brankovics, B. , van Dam, P., Rep, M., de Hoog, G. S., van der Lee, T. A. J., Waalwijk, C., & van Diepeningen, A. D. (2017). Mitochondrial genomes reveal recombination in the presumed asexual Fusarium oxysporum species complex. BMC Genomics, 18(1), 1-14. [CrossRef]
- Coleman, J. J. , Rounsley, S. D., Rodriguez-Carres, M., Kuo, A., Wasmann, C. C., Grimwood, J., Schmutz, J., Taga, M., White, G. J., & Zhou, S. (2009). The genome of Nectria haematococca: Contribution of supernumerary chromosomes to gene expansion. PLoS Genetics, 5(8), e1000618. [CrossRef]
- Ma, L. J., Geiser, D. M., Proctor, R. H., Rooney, A. P., O'Donnell, K., Trail, F., Gardiner, D. M., Manners, J. M., & Kazan, K. (2013). Fusarium pathogenomics. Annual Review of Microbiology, 67, 399-416. [CrossRef]
- Homa, M. , Galgóczy, L., Manikandan, P., Narendran, V., Sinka, R., Csernetics, Á., Vágvölgyi, C., Kredics, L., & Papp, T. (2018). South Indian isolates of the Fusarium solani species complex from clinical and environmental samples: Identification, antifungal susceptibilities, and virulence. Frontiers in Microbiology, 9, 1052. [CrossRef]
- Šišić, A. , Baćanović-Šišić, J., Al-Hatmi, A. M., Karlovsky, P., Ahmed, S. A., Maier, W., de Hoog, G. S., & Finckh, M. R. (2018). The 'forma specialis' issue in Fusarium: A case study in Fusarium solani f. sp. pisi. Scientific Reports, 8(1), 1252. [CrossRef]
- Moussa, T. A. A., Al-Zahrani, H. S., Kadasa, N. M. S., Ahmed, S. A., de Hoog, G. S., & Al-Hatmi, A. M. S. (2017). Two new species of the Fusarium fujikuroi species complex isolated from the natural environment. Antonie van Leeuwenhoek, 110(6), 819-832. [CrossRef]
- Geiser, D. M. , Aoki, T., Bacon, C. W., Baker, S. E., Bhattacharyya, M. K., Brandt, M. E., Brown, D. W., Burgess, L. W., Chulze, S., & Coleman, J. J. (2013). One fungus, one name: defining the genus Fusarium in a scientifically robust way that preserves longstanding use. Phytopathology, 103(5), 400-408. [CrossRef]
- Inami, K. , Yoshioka-Akiyama, C., Morita, Y., Yamasaki, M., Teraoka, T., & Arie, T. (2012). A genetic mechanism for emergence of races in Fusarium oxysporum f. sp. lycopersici: Inactivation of avirulence gene AVR1 by transposon insertion. PLOS ONE, 7 e44101. [CrossRef]
- Armenta-López, S. E. , Valenzuela-Solano, C., & Hernández-Martínez, R. (2021). Identification and molecular analysis of races of Fusarium oxysporum f. sp. lycopersici isolated from tomato in Baja California, Mexico. Mexican Journal of Phytopathology, 39 266–288. [CrossRef]
- Haegi, A. , De Felice, S., Scotton, M., Luongo, L., & Belisario, A. (2017). Fusarium oxysporum f. sp. melonis-melon interaction: Effect of grafting combination on pathogen gene expression. European Journal of Plant Pathology, 149 1–17. [CrossRef]
- Armer, V.J.; Kroll, E.; Darino, M.; Smith, D.P.; Urban, M.; Hammond-Kosack, K.E. Navigating the Fusarium Species Complex: Host-Range Plasticity and Genome Variations. Fungal Biology 2024, 128, 2439–2459. [Google Scholar] [CrossRef]
- O’Donnell, K. , Al-Hatmi, A. M., Aoki, T., Brankovics, B., Cano-Lira, J. F., Coleman, J. J., de Hoog, G. S., Di Pietro, A., Frandsen, R. J., & Geiser, D. M. (2020). No to Neocosmospora: phylogenomic and practical reasons for continued inclusion of the Fusarium solani species complex in the genus Fusarium. mSphere, 5(5), 10.1128/msphere. 00810-00820. [CrossRef]
- Ulaszewski, B. , Sandoval-Denis, M., Groenewald, J. Z., Costa, M. M., Mishra, B., Ploch, S., Crous, P. W., & Thines, M. (2025). Genomic features and evolution of lifestyles support the recognition of distinct genera among fusarioid fungi. Mycological Progress, 24(1), 20. [CrossRef]
- van Dam, P., Fokkens, L., Schmidt, S. M., Linmans, J. H., Kistler, H. C., Ma, L. J., & Rep, M. (2016a). Effector profiles distinguish formae speciales of Fusarium oxysporum. Environmental Microbiology, 18(11), 4087-4102. [CrossRef]
- Li, Y. , Steenwyk, J. L., Chang, Y., Wang, Y., James, T. Y., Stajich, J. E., Spatafora, J. W., Groenewald, M., Dunn, C. W., & Hittinger, C. T. (2021). A genome-scale phylogeny of the kingdom Fungi. Current Biology, 31(8), 1653-1665. e1655. [CrossRef]
- Stajich, J. E. (2017). Fungal genomes and insights into the evolution of the kingdom. Microbiology spectrum, 5(4), 10.1128/microbiolspec. funk-0055-2016. [CrossRef]
- O’Donnell, K., Ward, T. J., Aberra, D., Kistler, H. C., Aoki, T., Orwig, N., Kimura, M., Bjørnstad, Å., & Klemsdal, S. S. (2008). Multilocus genotyping and molecular phylogenetics resolve a novel head blight pathogen within the Fusarium graminearum species complex from Ethiopia. Fungal Genetics and Biology, 45(11), 1514-1522. [CrossRef]
- Lizcano Salas, A.F. , Duitama, J., Restrepo, S. and Celis Ramírez, A.M., (2024). Phylogenomic approaches reveal a robust time-scale phylogeny of the Terminal Fusarium Clade. IMA fungus, 15(1), p.13. [CrossRef]
- Jiao, J. , Zhong, S., Zhao, L., Yang, X., Tang, G., & Li, P. (2025). Genome-wide characterization of effector proteins in Fusarium zanthoxyli and their effects on plant’s innate immunity responses. BMC Plant Biology, 25(1), 298. [CrossRef]
- Zhou, X., O’Donnell, K., Aoki, T., Smith, J. A., Kasson, M. T., & Cao, Z.-M. (2016). Two novel Fusarium species that cause canker disease of prickly ash (Zanthoxylum bungeanum) in northern China form a novel clade with Fusarium torreyae. Mycologia, 108(4), 668-681. [CrossRef]
- Zhou, X. , Kerry, O. D., Hye-Seon, K., H., P. R., Gail, D., & and Cao, Z.-M. (2018). Heterothallic sexual reproduction in three canker-inducing tree pathogens within the Fusarium torreyae species complex. Mycologia, 110(4), 710-725. [CrossRef]
- Ruan, Z., Jiao, J., Zhao, J., Liu, J., Liang, C., Yang, X., Sun, Y., Tang, G., & Li, P. (2024). Genome sequencing and comparative genomics reveal insights into pathogenicity and evolution of Fusarium zanthoxyli, the causal agent of stem canker in prickly ash. BMC genomics, 25(1), 502. [CrossRef]
- Dita, M. , Barquero, M., Heck, D., Mizubuti, E.S., & Staver, C.P. (2018). Fusarium wilt of banana: current knowledge on epidemiology and research needs toward sustainable disease management. Frontiers in Plant Science 9, 1468. [CrossRef]
- Ordóñez, N. , Seidl, M.F., Waalwijk, C., Drenth, A., Kilian, A., Thomma, B.P.H.J., Ploetz, R.C., & Kema, G.H.J. (2015). Worse comes to worst: bananas and Panama disease—when plant and pathogen clones meet. PLoS Pathogens, 11, e1005197. [CrossRef]
- Liu, S., Li, J., Zhang, Y., Liu, N., Viljoen, A., Mostert, D., Zuo, C., Hu, C., Bi, F., Gao, H., & Sheng, O. (2020). Fusaric acid instigates the invasion of banana by Fusarium oxysporum f. sp. cubense TR4. New Phytologist, 225(2), 913–929. [CrossRef]
- Afzalinia, S. , Mehrabi-Koushki, M., & Farokhinejad, R. (2024). Two new species of Fusarium in the F. incarnatum-equiseti species complex from Oryza sativa in Iran. Antonie van Leeuwenhoek, 118(1), 5. [CrossRef]
- Matić, S. , Tabone, G., Guarnaccia, V., Gullino, M. L., & Garibaldi, A. (2020). Emerging leafy vegetable crop diseases caused by the Fusarium incarnatum-equiseti species complex. Phytopathol. Mediterr, 59(2), 2.
- Wang, M., Chen, Q., Diao, Y., Duan, W., & Cai, L. (2019). Fusarium incarnatum-equiseti complex from China. Persoonia-Molecular Phylogeny and Evolution of Fungi, 43(1), 70-89. [CrossRef]
- Avila, C. F., Moreira, G. M., Nicolli, C. P., Gomes, L. B., Abreu, L. M., Pfenning, L. H., Haidukowski, M., Moretti, A., Logrieco, A., & Del Ponte, E. M. (2019). Fusarium incarnatum-equiseti species complex associated with Brazilian rice: Phylogeny, morphology and toxigenic potential. International Journal of Food Microbiology, 306, 108267. [CrossRef]
- Moreira, G. M. , Nicolli, C. P., Gomes, L. B., Ogoshi, C., Scheuermann, K. K., Silva-Lobo, V. L., Schurt, D. A., Ritieni, A., Moretti, A., Pfenning, L. H., & Del Ponte, E. M. (2020). Nationwide survey reveals high diversity of Fusarium species and related mycotoxins in Brazilian rice: 2014 and 2015 harvests. Food Control, 113, 107171. [CrossRef]
- Lu, Y. , Qiu, J., Wang, S., Xu, J., Ma, G., Shi, J., & Bao, Z. (2021). Species diversity and toxigenic potential of Fusarium incarnatum-equiseti species complex isolates from rice and soybean in China. Plant Disease, 105(9), 2628-2636. [CrossRef]
- Petrović, K., Orzali, L., Krsmanović, S., Valente, M. T., Tolimir, M., Pavlov, J., & Riccioni, L. (2024). Genetic diversity and pathogenicity of the Fusarium species complex on soybean in Serbia. Plant Disease, 108(6), 1851-1860. [CrossRef]
- Liu, Y., Tian, Y., Zhao, X., Yue, L., Uwaremwe, C., Zhou, Q., Wang, Y., Zhang, Y., Dun, Z., Cui, Z., & Wang, R. (2022). Identification of Pathogenic Fusarium spp. Responsible for Root Rot of Angelica sinensis and Characterization of Their Biological Enemies in Dingxi, China. Plant Disease, 106(7), 1898-1910. [CrossRef]
- Niyongabo Turatsinze, A. , Xie, X., Ye, A., Chen, G., Wang, Y., Yue, L., Zhou, Q., Wu, L., Zhang, M., & Zhang, Z. (2025). Fusarium cross-infection in medicinal herbs alters rhizosphere microbiomes and disrupts mycorrhizal functions under soil physicochemical imbalances. Plant and soil, 1-31. [CrossRef]
- Wang, Y., Wang, R., & Sha, Y. (2022). Distribution, pathogenicity and disease control of Fusarium tricinctum. Front Microbiol, 13, 939927. [CrossRef]
- Senatore, M., Ward, T., Cappelletti, E., Beccari, G., McCormick, S., Busman, M., Laraba, I., O'Donnell, K., & Prodi, A. (2021). Species diversity and mycotoxin production by members of the Fusarium tricinctum species complex associated with Fusarium head blight of wheat and barley in Italy. International Journal of Food Microbiology, 358, 109298. [CrossRef]
- Moparthi, S.; Burrows, M.; Mgbechi-Ezeri, J.; Agindotan, B. (2021). Fusarium spp. Associated with Root Rot of Pulse Crops and Their Cross-Pathogenicity to Cereal Crops in Montana. Plant Dis. 105, 548–557. [CrossRef]
- Niyongabo Turatsinze, A., Xie, X., Chen, G., Ye, A., Yue, L., Wang, Y., Zhou, Q., Zhang, M., Zhang, Z., Zhao, J., Zhang, Y., & Wang, R. (2024). First Report of Fusarium avenaceum Causing Root Rot of Raspberry (Rubus corchorifolius) in China. Plant Disease, 108(10), 3194. [CrossRef]
- Okello, P. N., & Mathew, F. M. (2019). Cross-pathogenicity studies show South Dakota isolates of Fusarium acuminatum, F. equiseti, F. graminearum, F. oxysporum, F. proliferatum, F. solani, and F. subglutinans from either soybean or corn are pathogenic to both crops. Plant Health Progress, *20*(1), 44-49. [CrossRef]
- Bugingo, C. , Brelsford, M., Fonseka, D., Pasche, J., & Burrows, M. (2025). Unveiling the diversity and virulence of seedborne Fusarium species in lentil production: Insights from a two-year study in the Northern Great Plains. Plant Health Progress. [CrossRef]
- Jambhulkar, P. P., Bajpai, R., Reddy, H. J., Tripathy, P. S., Varun, P., Rout, A. K., Behera, B. K., Lakshman, D. K., & Nanjundappa, M. (2024). Assessment of Genetic Diversity and the Population Structure of Species from the Fusarium fujikuroi Species Complex Causing Fusarium Stalk Rot of Maize. Journal of Fungi, 10(8), 574. [CrossRef]
- Dewing, C. (2020). Comparative Genomics Reveal Processes Implicated in Host-Specificity in Species Within the American Clade of the Fusarium Fujikuroi Species Complex University of Pretoria (South Africa)]. https://www.proquest.com/dissertations-theses/comparative-genomics-reveal-processes-implicated/docview/3122660831/se-2.
- Witte, T. E. , Villeneuve, N., Boddy, C. N., & Overy, D. P. (2021). Accessory Chromosome-Acquired Secondary Metabolism in Plant Pathogenic Fungi: The Evolution of Biotrophs Into Host-Specific Pathogens [Review]. Frontiers in Microbiology, Volume 12 - 2021. [CrossRef]
- Li, X. , Xu, S., Zhang, J., & Li, M. (2021). Assembly and annotation of whole-genome sequence of Fusarium equiseti. Genomics, 113(4), 2870-2876. [CrossRef]
- Xie, S.-Y. , Ma, T., Zhao, N., Zhang, X., Fang, B., & Huang, L. (2022). Whole-genome sequencing and comparative genome analysis of Fusarium solani-melongenae causing Fusarium root and stem rot in sweetpotatoes. Microbiology spectrum, 10(4), e00683-00622. [CrossRef]
- Hoogendoorn, K., Barra, L., Waalwijk, C., Dickschat, J. S., Van der Lee, T. A., & Medema, M. H. (2018). Evolution and diversity of biosynthetic gene clusters in Fusarium. Frontiers in Microbiology, 9, 1158. [CrossRef]
- Fokkens, L. , Shahi, S., Connolly, L. R., Stam, R., Schmidt, S. M., Smith, K. M., Freitag, M., & Rep, M. (2018). The multi-speed genome of Fusarium oxysporum reveals association of histone modifications with sequence divergence and footprints of past horizontal chromosome transfer events. BioRxiv, 465070. [CrossRef]
- Bates, H. J., Pike, J., Price, R. J., Jenkins, S., Connell, J., Legg, A., Armitage, A., Harrison, R. J., & Clarkson, J. P. (2024). Comparative genomics and transcriptomics reveal differences in effector complement and expression between races of Fusarium oxysporum f.sp. lactucae [Original Research]. Frontiers in Plant Science, Volume 15 - 2024. [CrossRef]
- van Dam, P., Fokkens, L., Schmidt, S. M., Linmans, J. H. J., Kistler, H. C., Ma, L.-J., & Rep, M. (2016b). Effector profiles distinguish formae specialesof Fusarium oxysporum. Environmental Microbiology, 18(11), 4087-4102. [CrossRef]
- Crous, P. W. , Wingfield, M. J., Burgess, T. I., Carnegie, A. J., Hardy, G., Smith, D., Summerell, B. A., Cano-Lira, J. F., Guarro, J., & Houbraken, J. (2017). Fungal Planet description sheets: 625–715. Persoonia: Molecular Phylogeny and Evolution of Fungi, 39, 270. [CrossRef]
- Navale, V. D., Sawant, A. M., Gowda, V. U., & Vamkudoth, K. R. (2022). Assembly, annotation, and comparative whole genome sequence of Fusarium verticillioides isolated from stored maize grains. Pathogens, 11(7), 810. [CrossRef]
- Urbaniak, M. , Waśkiewicz, A., Trzebny, A., Koczyk, G., & Stępień, Ł. (2020). Cyclodepsipeptide biosynthesis in hypocreales fungi and sequence divergence of the non-ribosomal peptide synthase genes. Pathogens, 9(7), 552. [CrossRef]
- Navasca, A., Singh, J., Rivera-Varas, V., Gill, U., Secor, G., & Baldwin, T. (2025). Dispensable genome and segmental duplications drive the genome plasticity in Fusarium solani. Frontiers in Fungal Biology, 6, 1432339. [CrossRef]
- van Westerhoven, A. C. , Aguilera-Galvez, C., Nakasato-Tagami, G., Shi-Kunne, X., Martinez de la Parte, E., Chavarro-Carrero, E., Meijer, H. J., Feurtey, A., Maryani, N., & Ordóñez, N. (2024). Segmental duplications drive the evolution of accessory regions in a major crop pathogen. New Phytologist, 242(2), 610-625. [CrossRef]
- Ma, L. J. (2014). Horizontal chromosome transfer and rational strategies to manage Fusarium vascular wilt diseases. Molecular Plant Pathology, 15(8), 763. [CrossRef]
- Yang, H., Yu, H., & Ma, L.-J. (2020). Accessory chromosomes in Fusarium oxysporum. Phytopathology, 110(9), 1488-1496. [CrossRef]
- Wiemann, P. , Sieber, C.M., Von Bargen, K.W., Studt, L., Niehaus, E.M., Espino, J.J., Huss, K., Michielse, C.B., Albermann, S., Wagner, D. and Bergner, S.V. (2013). Deciphering the cryptic genome: genome-wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS pathogens, 9(6), p.e1003475. [CrossRef]
- Rampersad, S. N. (2020). Pathogenomics and Management of Fusarium Diseases in Plants. Pathogens (Basel, Switzerland), 9(5), 340. [CrossRef]
- Thatcher, L. F., Gardiner, D. M., Kazan, K., & Manners, J. M. (2012). A highly conserved effector in Fusarium oxysporum is required for full virulence on Arabidopsis. Molecular plant-microbe interactions: MPMI, 25(2), 180–190. [CrossRef]
- Henry, P. M., Pincot, D. D. A., Jenner, B. N., Borrero, C., Aviles, M., Nam, M.-H., Epstein, L., Knapp, S. J., & Gordon, T. R. (2021). Horizontal chromosome transfer and independent evolution drive diversification in Fusarium oxysporum f. sp. fragariae. New Phytologist, 230(1), 327-340. [CrossRef]
- Epstein, L., Kaur, S., & Henry, P. M. (2022). The emergence of Fusarium oxysporum f. sp. apii race 4 and Fusarium oxysporum f. sp. coriandrii highlights major obstacles facing agricultural production in coastal California in a warming climate: A case study. Frontiers in Plant Science, 13, 921516. [CrossRef]
- Ma, H., Liu, Y., Zhao, X., Zhang, S., & Ma, H. (2022). Exploring and applying genes to enhance the resistance to Fusarium head blight in wheat. Frontiers in Plant Science, 13, 1026611. [CrossRef]
- Niehaus, E.-M., Münsterkötter, M., Proctor, R. H., Brown, D. W., Sharon, A., Idan, Y., Oren-Young, L., Sieber, C. M., Novák, O., Pěnčík, A., Tarkowská, D., Hromadová, K., Freeman, S., Maymon, M., Elazar, M., Youssef, S. A., El-Shabrawy, E. S. M., Shalaby, A. B. A., Houterman, P., Brock, N. L., Burkhardt, I., Tsavkelova, E. A., Dickschat, J. S., Galuszka, P., Güldener, U., & Tudzynski, B. (2016). Comparative "omics" of the Fusarium fujikuroi species complex highlights differences in genetic potential and metabolite synthesis. Genome Biology and Evolution, 8(11), 3574–3599. [CrossRef]
- Armitage, A. D., Taylor, A., Sobczyk, M. K., Baxter, L., Greenfield, B. P. J., Bates, H. J., Wilson, F., Jackson, A. C., Ott, S., Harrison, R. J., & Clarkson, J. P. (2018). Characterisation of pathogen-specific regions and novel effector candidates in Fusarium oxysporum f. sp. cepae. Scientific Reports, 8, 13530. [CrossRef]
- Lanubile, A., Ferrarini, A., Maschietto, V., Delledonne, M., Marocco, A., & Bellin, D. (2014). Functional genomic analysis of constitutive and inducible defense responses to Fusarium verticillioides infection in maize genotypes with contrasting ear rot resistance. BMC Genomics, 15, 710. [CrossRef]
- Lightfoot, J. D., & Fuller, K. K. (2019). CRISPR/Cas9-Mediated Gene Replacement in the Fungal Keratitis Pathogen Fusarium solani var. petroliphilum. Microorganisms, 7(10), 457. [CrossRef]
- Pokhrel, A. , Seo, S., Wang, Q., Coleman, J. J. (2022). Targeted Gene Disruption Via CRISPR/Cas9 Ribonucleoprotein Complexes in Fusarium oxysporum. In: Coleman, J. (eds) Fusarium wilt. Methods in Molecular Biology, vol 2391. Humana, New York, NY. [CrossRef]
- van Dijk, A. , Wilson, A. M., Marx, B., Hough, B., Swalarsk-Parry, B., De Vos, L., Wingfield, M. J., Wingfield, B. D., & Steenkamp, E. T. (2025). CRISPR-Cas9 genome editing reveals that the Pgs gene of Fusarium circinatum is involved in pathogenicity, growth and sporulation. Fungal Genetics and Biology, 177, 103970. [CrossRef]
- de Vega-Bartol, J. J. , Martín-Dominguez, R., Ramos, B., García-Sánchez, M. A., & Díaz-Mínguez, J. M. (2011). New virulence groups in Fusarium oxysporum f. sp. phaseoli: the expression of the gene coding for the transcription factor ftf1 correlates with virulence. Phytopathology, 101, 470–479. [CrossRef]
- Niño-Sánchez, J. , Casado-Del Castillo, V., Tello, V., De Vega-Bartol, J. J., Ramos, B., Sukno, S. A., & Díaz Mínguez, J. M. (2016). The FTF gene family regulates virulence and expression of SIX effectors in Fusarium oxysporum. Molecular plant pathology, 17; 1124–1139. [Google Scholar] [CrossRef]
- Ma, L.J.; Geiser, D.M.; Proctor, R.H.; Rooney, A.P.; O'Donnell, K.; Trail, F.; Gardiner, D.M.; Manners, J.M.; Kazan, K. Fusarium Pathogenomics. Annu. Rev. Microbiol. 2013, 67, 399–416. [Google Scholar] [CrossRef]
- Hao, G., Proctor, R. H., Brown, D. W., Rhoades, N. A., Naumann, T. A., Kim, H., Gutiėrrez, S., & McCormick, S. P. (2024). TRI14 Is Critical for Fusarium graminearum Infection and Spread in Wheat. Applied Microbiology, 4(2), 839-855. [CrossRef]
- Rep, M., van der Does, H. C., Meijer, M., van Wijk, R., Houterman, P. M., Dekker, H. L., de Koster, C. G., & Cornelissen, B. J. (2004). A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Molecular microbiology, 53(5), 1373–1383. [CrossRef]
- Jenner, B. N., & Henry, P. M. (2022). Pathotypes of Fusarium oxysporum f. sp. fragariae express discrete repertoires of accessory genes and induce distinct host transcriptional responses during root infection. Environmental microbiology, 24(10), 4570–4586. [CrossRef]
- Brown, N. A., Evans, J., Mead, A., & Hammond-Kosack, K. E. (2017). A spatial temporal analysis of the Fusarium graminearum transcriptome during symptomless and symptomatic wheat infection. Molecular plant pathology, 18(9), 1295–1312. [CrossRef]
- Pimentel, M. F., Rocha, L. F., Subedi, A., Bond, J. P., & Fakhoury, A. M. (2025). Dual RNA-seq reveals transcriptome changes during Fusarium virguliforme-Trichoderma afroharzianum interactions. PloS one, 20(1), e0310850. [CrossRef]
- Seong, K.-Y., Pasquali, M., Zhou, X., Song, J., Hilburn, K., McCormick, S., Dong, Y., Xu, J.-R., & Kistler, H. C. (2009). Global gene regulation by Fusarium transcription factors Tri6 and Tri10 reveals adaptations for toxin biosynthesis. Molecular Microbiology, 72(2), 354-367. [CrossRef]
- Nasmith, C. G., Walkowiak, S., Wang, L., Leung, W. W. Y., Gong, Y., Johnston, A., Harris, L. J., Guttman, D. S., & Subramaniam, R. (2011). Tri6 is a global transcription regulator in the phytopathogen Fusarium graminearum. PLoS Pathogens, 7(9), e1002266. [CrossRef]
- Caracuel, Z., Roncero, M. I., Espeso, E. A., González-Verdejo, C. I., García-Maceira, F. I., & Di Pietro, A. (2003). The pH signaling transcription factor PacC controls virulence in the plant pathogen Fusarium oxysporum. Molecular microbiology, 48(3), 765–779. [CrossRef]
- van der Does, H. C., Fokkens, L., Yang, A., Schmidt, S. M., Langereis, L., Lukasiewicz, J. M., Hughes, T. R., & Rep, M. (2016). Transcription factors encoded on core and accessory chromosomes of Fusarium oxysporum induce expression of effector genes. PLoS Genetics, 12(11), e1006401. [CrossRef]
- Michielse, C. B., van Wijk, R., Reijnen, L., Manders, E. M. M., Boas, S., Olivain, C., Alabouvette, C., & Rep, M. (2009). The nuclear protein Sge1 of Fusarium oxysporum is required for parasitic growth. PLoS Pathogens, 5(10), e1000637. [CrossRef]
- Kim, H.-K., Lee, S., Jo, S.-M., McCormick, S. P., Butchko, R. A. E., Proctor, R. H., & Yun, S.-H. (2013). Functional roles of FgLaeA in controlling secondary metabolism, sexual development, and virulence in Fusarium graminearum. PLoS ONE, 8(7), e68441. [CrossRef]
- Studt, L. , Rösler, S. M., Burkhardt, I., Arndt, B., Freitag, M., Humpf, H.-U., Dickschat, J. S., & Tudzynski, B. (2016). Knock-down of the methyltransferase Kmt6 relieves H3K27me3 and results in induction of cryptic and otherwise silent secondary metabolite gene clusters in Fusarium fujikuroi. Environmental Microbiology, 18, 4037–4054. [CrossRef]
- Mihlan, M., Homann, V., Liu, T.-W. D., & Tudzynski, B. (2003). AREA directly mediates nitrogen regulation of gibberellin biosynthesis in Gibberella fujikuroi, but its activity is not affected by NMR. Molecular Microbiology, 47(4), 975-991. [CrossRef]
- Bae, H. , Kim, M. S., Sicher, R. C., Bae, H.-J., & Bailey, B. A. (2006). Necrosis- and ethylene-inducing peptide from Fusarium oxysporum induces a complex cascade of transcripts associated with signal transduction and cell death in Arabidopsis. Plant Physiology, 141, 1056–1067. [CrossRef]
- Ravalason, H., Grisel, S., Chevret, D., Favel, A., Berrin, J.-G., Sigoillot, J.-C., & Herpoël-Gimbert, I. (2012). Fusarium verticillioides secretome as a source of auxiliary enzymes to enhance saccharification of wheat straw. Bioresource Technology, 114, 589-596. [CrossRef]
- Gawehns, F. , Houterman, P. M., Ait Ichou, F., Michielse, C. B., Hijdra, M., Cornelissen, B. J. C., Rep, M., & Takken, F. L. W. (2014). The Fusarium oxysporum effector Six6 contributes to virulence and suppresses I-2-mediated cell death. *Molecular Plant-Microbe Interactions, 27*(4), 336-348. [CrossRef]
- Su, Z. , Bernardo, A., Tian, B., Chen, H., Wang, S., Ma, H., Cai, S., Liu, D., Zhang, D., Li, T., Trick, H., St. Amand, P., Yu, J., Zhang, Z., & Bai, G. A deletion mutation in TaHRC confers Fhb1 resistance to Fusarium head blight in wheat. >Nature Genetics 2019, 51, 1099–1105. [Google Scholar] [CrossRef]
- Catanzariti, A. M., Do, H. T. T., Bru, P., de Sain, M., Thatcher, L. F., Rep, M., & Jones, D. A. (2017). The tomato I gene for Fusarium wilt resistance encodes an atypical leucine-rich repeat receptor-like protein whose function is nevertheless dependent on SOBIR1 and SERK3/BAK1. Plant Journal, 89(6), 1195-1209. [CrossRef]
- Kumar, J. M., Dikshit, H. K., Aski, M., Gupta, S., Singh, A., Tripathi, A., Thribhuvan, R., & Chourasia, N. K. (2023). Molecular mapping of a gene conferring Fusarium wilt resistance in lentil (Lens culinaris Medikus subsp. culinaris) using bulked-segregant analysis. Legume Research, 46(6), 801-805. [CrossRef]
- Bugingo, C., Brelsford, M., McPhee, K., & Burrows, M. (2024). Pathotype characterization of Fusarium oxysporum f. sp. lentis (Fol) isolates in North America. Plant Health Progress. Advance online publication. [CrossRef]
- Srour, A., Afzal, A. J., Blahut-Beatty, L., Hemmati, N., Simmonds, D. H., Li, W., Liu, M., Town, C. D., Sharma, H., & Lightfoot, D. A. (2012). The receptor-like kinase at *Rhg1-a/Rfs2* caused pleiotropic resistance to sudden death syndrome and soybean cyst nematode as a transgene by altering signaling responses. BMC Genomics, 13, 368. [CrossRef]
- Ma, P., Liu, E., Zhang, Z., Li, T., Zhou, Z., Yao, W., Chen, J., Wu, J., Xu, Y., & Zhang, H. (2023). Genetic variation in ZmWAX2 confers maize resistance to Fusarium verticillioides. Plant biotechnology journal, 21(9), 1812–1826. [CrossRef]
- Xu, Y., Fan, Y., Liu, L., Cao, J., Zhou, J., Liu, E., Li, R., Ma, P., Yao, W., Wu, J., Li, T., & Zhang, H. (2025a). Enhancing maize resistance to Fusarium verticillioides through modulation of cell wall structure and components by ZmXYXT2. Journal of Advanced Research. Advance online publication. [CrossRef]
- Chen, A., Sun, J., Martin, G., Gray, L.-A., Hřibová, E., Christelová, P., Yahiaoui, N., Rounsley, S., Lyons, R., Batley, J., Chen, N., Hamill, S., Rai, S. K., Coin, L., Uwimana, B., D’Hont, A., Doležel, J., Edwards, D., Swennen, R., & Aitken, E. A. B. (2023). Identification of a Major QTL-Controlling Resistance to the Subtropical Race 4 of Fusarium oxysporum f. sp. cubense in Musa acuminata ssp. malaccensis. Pathogens, 12(2), 289. [CrossRef]
- Wang, P., Zhou, L., Jamieson, P., Zhang, L., Zhao, Z., Babilonia, K., Shao, W., Wu, L., Mustafa, R., Amin, I., Diomaiuti, A., Pontiggia, D., Ferrari, S., Hou, Y., He, P., & Shan, L. (2020). The cotton wall-associated kinase GhWAK7A mediates responses to fungal wilt pathogens by complexing with the chitin sensory receptors. The Plant Cell, 32(12), 3978-4001. [CrossRef]
- Dong, J., Xu, J., Xu, X., Xu, Q., & Chen, X. (2019). Inheritance and quantitative trait locus mapping of Fusarium wilt resistance in cucumber. Frontiers in Plant Science, 10, 1425. [CrossRef]
- Bartholomew, E. S., Xu, S., Zhang, Y., Yin, S., Feng, Z., Chen, S., Sun, L., Yang, S., Wang, Y., Liu, P., Ren, H., & Liu, X. (2022). A chitinase CsChi23 promoter polymorphism underlies cucumber resistance against Fusarium oxysporum f. sp. cucumerinum. New Phytologist, 236(4), 1471–1486. [CrossRef]
- Dean, R. , Van Kan, J. A. L., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., Rudd, J. J., Dickman, M.,... Foster, G. D. (2012). The Top 10 fungal pathogens in molecular plant pathology. Molecular Plant Pathology, 13(4), 414–430. [CrossRef]
- Yang, D. , Zhang, X., Ming, Y., Liu, C., Zhang, X., Liu, S., & Zhu, L. (2024). Characterization of the High-Quality Genome Sequence and Virulence Factors of Fusarium oxysporum f. sp. vasinfectum Race 7. Journal of Fungi, 10(4), 242. [CrossRef]
- Zhang, Q. , Shi, X., Gao, T., Xing, Y., Jin, H., Hao, J., Liu, X., Liu, X., & Liu, P. (2024). Precision management of Fusarium fujikuroi in rice through seed coating with an enhanced nanopesticide using a tannic acid-ZnII formulation. Journal of Nanobiotechnology, 22, 717. [CrossRef]
- Beccari, G. , Hao, G., & Liu, H. (2022). Editorial: Fusarium pathogenesis: Infection mechanisms and disease progression in host plants. Frontiers in Plant Science, 13, 1020404. [CrossRef]
- Sperschneider, J. , Gardiner, D. M., Thatcher, L. F., Lyons, R., Singh, K. B., Manners, J. M., & Taylor, J. M. (2015). Genome-Wide Analysis in Three Fusarium Pathogens Identifies Rapidly Evolving Chromosomes and Genes Associated with Pathogenicity. Genome biology and evolution, 7(6), 1613–1627. [CrossRef]
- Wang, H. , Yao, G., Chen, W. et al. A gap-free genome assembly of Fusarium oxysporum f. sp. conglutinans, 2024; 11, 925. [Google Scholar] [CrossRef]
- Huang, X.-Q. , Lu, X.-H., Sun, M.-H., Guo, R.-J., van Diepeningen, A. D., & Li, S.-D. (2019). Transcriptome analysis of virulence-differentiated Fusarium oxysporum f. sp. cucumerinum isolates during cucumber colonisation reveals pathogenicity profiles. BMC Genomics, 20, 570. [CrossRef]
- Armstrong, G. M., & Armstrong, J. K. (1981). Formae specialesand races of Fusarium oxysporum causing wilt disease. In P. E. Nelson, T. A. Toussoun, & R. J. Cook (Eds.), Fusarium: Disease, biology, and taxonomy (pp. 391–399). Pennsylvania State University Press. https://www.scirp.org/reference/referencespapers?referenceid=399958.
- Gordon, T. R., & Martyn, R. D. (1997). The evolutionary biology of Fusarium oxysporum. Annual review of phytopathology, 35, 111–128. [CrossRef]
- Lievens, B., Rep, M., & Thomma, B. P. (2008). Recent developments in the molecular discrimination of formae specialesof Fusarium oxysporum. Pest management science, 64(8), 781–788. [CrossRef]
- O'Donnell, K. , Sutton, D. A., Rinaldi, M. G., Magnon, K. C., Cox, P. A., Revankar, S. G., Sanche, S., Geiser, D. M., Juba, J. H., Van Burik, J. A. H., Padhye, A., Anaissie, E. J., Francesconi, A., Walsh, T. J., & Robinson, J. S. (2004). Genetic diversity of human pathogenic members of the Fusarium oxysporum complex inferred from multilocus DNA sequence data and amplified fragment length polymorphism analyses: Evidence for the recent dispersion of a geographically widespread clonal lineage and nosocomial origin. Journal of clinical microbiology, 42(11), 5109-5120. [CrossRef]
- Summerell, B. A., Laurence, M. H., Liew, E. C. Y., & Leslie, J. F. (2010). Biogeography and phylogeography of Fusarium: A review. Fungal Diversity, 44, 3–13. [CrossRef]
- Babilonia, K., Wang, P., Liu, Z., Jamieson, P., Mormile, B., Rodrigues, O., Zhang, L., Lin, W., Clement, C. D., Menezes de Moura, S., Alves-Ferreira, M., Finlayson, S. A., Nichols, R. L., Wheeler, T. A., Dever, J. K., Shan, L., & He, P. (2021). A nonproteinaceous Fusarium cell wall extract triggers receptor-like protein-dependent immune responses in Arabidopsis and cotton. New Phytologist, 230(1), 275–289. [CrossRef]
- Dong, J., Wang, Y., Xian, Q., Chen, X., & Xu, J. (2020). Transcriptome analysis reveals ethylene-mediated defense responses to Fusarium oxysporum f. sp. cucumerinum infection in Cucumis sativus L. BMC Plant Biology, 20(1), 334. [CrossRef]
- Mierziak, J., & Wojtasik, W. (2024). Epigenetic weapons of plants against fungal pathogens. BMC Plant Biology, 24(1), 175. [CrossRef]
- Liu, S., Wu, J., Sun, Y., Xu, Y., Zhou, S., Luo, P., Wang, Z., Chen, D., Liang, X., Kang, Z., & Zheng, L. (2025). A novel key virulence factor, FoSSP71, inhibits plant immunity and promotes pathogenesis in Fusarium oxysporum f. sp. cubense. Microbiology Spectrum, 13(2), e02940-24. [CrossRef]
- Wang, C., Huang, Z., Duan, Z., Zhu, L., Di, R., Bao, Y., Powell, C. A., Hu, Q., Chen, B., Zhang, M., & Yao, W. (2023). Pectate lyase from Fusarium sacchari induces plant immune responses and contributes to virulence. Microbiology Spectrum, 11(3), e00165-23. [CrossRef]
- Munkvold, G. P. (2021). Fusarium species and their associated mycotoxins. In A. Moretti & A. Susca (Eds.), Mycotoxigenic fungi: Methods and protocols (pp. 123–139). Springer. [CrossRef]
- Bräse, S., Encinas, A., Keck, J., & Nising, C. F. (2009). Chemistry and biology of mycotoxins and related fungal metabolites. Chemical Reviews, 109(9), 3903–3990. [CrossRef]
- Steyn, P. S. (1995). Mycotoxins, general view, chemistry and structure. Toxicology Letters, 82–83, 843–851. [CrossRef]
- International Agency for Research on Cancer. (1993). Some naturally occurring substances: Food items and constituents, heterocyclic aromatic amines and mycotoxins (IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, Vol. 56). World Health Organization.
- Pietragallo, Z., Kovalsky, P., Nagl, V., Schwartz-Zimmermann, H. E., Moll, W.-D., Schatzmayr, G., & Krska, R. (2021). Mycotoxin exposure and human health risk in rural sub-Saharan Africa. Toxins, 13(9), 618. [CrossRef]
- Alexander, N. J. , Hohn, T. M., & McCormick, S. P. (1999). The TRI11 gene of Fusarium sporotrichioides encodes a cytochrome P450 monooxygenase required for C-15 hydroxylation in trichothecene biosynthesis. Applied and Environmental Microbiology, 65(12), 5621-5625. [CrossRef]
- Brown, D. W., McCormick, S. P., Alexander, N. J., Proctor, R. H., & Desjardins, A. E. (2005). Inactivation of a Fusarium verticillioides FUM gene cluster prevents fumonisin production. Journal of Agricultural and Food Chemistry, 53(14), 5666-5673. [CrossRef]
- Chen, Y.; Kistler, H.C.; Ma, Z. (2019).Fusarium graminearum trichothecene mycotoxins: Biosynthesis, regulation, and management. Annu. Rev. Phytopathol. 57, 15–39. [CrossRef]
- Wang, J.-H., Ward, T. J., Lu, Y., Luo, C. X., Li, X., & Jin, J. (2022). Genetic basis of variation in trichothecene biosynthesis and pathogenicity in Fusarium graminearum. Frontiers in Microbiology, 13, 814270. [CrossRef]
- Schaarschmidt, S., and Fauhl-Hassek, C. 2019. Mycotoxins during the processes of nixtamalization and tortilla production. Toxins, 11, 227. [CrossRef]
- Food and Agriculture Organization of the United Nations. (2023). FAOSTAT statistical database.
- Food and Agriculture Organization of the United Nations. (2022). World food and agriculture—Statistical yearbook 2022. https://www.fao.org/statistics/yearbook.
- Logrieco, A., Battilani, P., Leggieri, M. C., Jiang, D., Haque, M. M., & Mahmud, N. U. (2021). Mycotoxins in maize: A global concern. Toxins, 13(7), 494. [CrossRef]
- Bottalico, A., & Perrone, G. (2002). Toxigenic Fusarium species and mycotoxins associated with head blight in small-grain cereals in Europe. European Journal of Plant Pathology, 108(7), 611–624. [CrossRef]
- Lee, H. J., & Ryu, D. (2017). Worldwide occurrence of mycotoxins in cereals and cereal-derived food products: Public health perspectives of their co-occurrence. Journal of Agricultural and Food Chemistry, 65(36), 7034–7051. [CrossRef]
- Yoshizawa, T. (1999). Mycotoxins in rice. International Journal of Food Microbiology, 59(2), 139–146. [CrossRef]
- Zhou, H., Fink-Gremmels, J., & van Egmond, H. P. (2008). Occurrence of Fusarium mycotoxins in rice. Food Additives and Contaminants, 25(10), 1124–1130. [CrossRef]
- Eskola, M., Kos, G., Elliott, C. T., Hajšlová, J., Mayar, S., & Krska, R. (2020). Worldwide contamination of food-crops with mycotoxins: Validity of the widely cited 'FAO estimate' of 25%. Critical Reviews in Food Science and Nutrition, 60(16), 2773–2789. [CrossRef]
- Magan, N., Medina, A., & Aldred, D. (2011). Possible climate-change effects on mycotoxin contamination of food crops pre- and postharvest. Plant Pathology, 60(1), 150–163. [CrossRef]
- Battilani, P., Toscano, P., Van der Fels-Klerx, H. J., Moretti, A., Camardo Leggieri, M., Brera, C., Rortais, A., Goumperis, T., & Robinson, T. (2016). Aflatoxin B1 contamination in maize in Europe increases due to climate change. Scientific Reports, 6, 24328. [CrossRef]
- Wagacha, J. M., & Muthomi, J. W. (2008). Mycotoxin problem in Africa: Current status, implications to food safety and health and possible management strategies. International Journal of Food Microbiology, 124(1), 1–12. [CrossRef]
- Torres, O., Matute, J., Gelineau-van Waes, J., Maddox, J. R., Gregory, S. G., Ashley-Koch, A. E., Showker, J. L., Voss, K. A., & Riley, R. T. (2007). Human dietary exposure to fumonisin B1 in a high-risk maize area of Guatemala. Food Additives & Contaminants: Part A, 24(4), 429–437. [CrossRef]
- Liu, Y., & Wu, F. (2010). Global burden of aflatoxin-induced hepatocellular carcinoma: A risk assessment. Environmental Health Perspectives, 118(6), 818–824. [CrossRef]
- Yoshizawa, T., Yamamoto, S., Sakamoto, S., & Watanabe, T. (2004). Natural occurrence of Fusarium mycotoxins (trichothecenes, zearalenone and fumonisins) in cereals collected in Nepal. Mycopathologia, 157(2), 233–241. [CrossRef]
- Medina, Á., Rodríguez, A., & Magan, N. (2014). Effect of climate change on Aspergillus flavus and aflatoxin B1 production. Frontiers in Microbiology, 5, 348. [CrossRef]
- Bhat, R. V., Rai, R. N., Karimullakhan, M., & Rao, V. S. (2003). Food safety in food security and food trade: Mycotoxin food safety risk in developing countries. International Food Policy Research Institute. https://www.ifpri.org/publication/mycotoxin-food-safety-risk-developing-countries.
- Udomkun, P., Wiredu, A. N., Nagle, M., Müller, J., Vanlauwe, B., & Bandyopadhyay, R. (2017). Mycotoxins in sub-Saharan Africa: Present situation, socio-economic impact, awareness, and outlook. Food Control, 72, 110–122. [CrossRef]
- Gómez-Osorio, L.-M. (2023). Addressing the gaps in mycotoxin management: Challenges and strategies for South America's poultry industry. MycotoxinSite. https://mycotoxinsite.com/addressing-gaps-mycotoxin-management-challenges-strategies-south-americas-poultry-industry/?lang=en.
- Juan-García, A. , & Jakšić, D. (2025). Novel Approaches in Mycotoxins Research: Detection, Prevention and Mode of Action. Toxins, 17(4), 161. [CrossRef]
- Li, R. , Wen, Y., Wang, F., & He, P. (2021). Recent advances in immunoassays and biosensors for mycotoxins detection in feedstuffs and foods. Journal of Animal Science and Biotechnology, 12(108). [CrossRef]
- Kamle, M., Mahato, D. K., Devi, S., Kumar, P., Patil, R. K., Kang, S. G., & Kumar, A. (2019). Fumonisins: Impact on agriculture, food, and human health and their management strategies. Toxins, 11(6), 328. [CrossRef]
- International Agency for Research on Cancer (IARC). (2002). Fumonisin B₁. In IARC monographs on the evaluation of carcinogenic risks to humans (Vol. 82). IARC Press.
- Mafe, A. N. , & Büsselberg, D. (2024). Mycotoxins in food: Cancer risks and strategies for control. Foods, 13, 3502. [CrossRef]
- Pestka, J. J. , & Smolinski, A. T. (2005). Deoxynivalenol: Toxicology and potential effects on humans. Journal of Toxicology and Environmental Health, Part B, 8, 39–69. [CrossRef]
- Kamle, M., Devi, S., Mahato, D. K., Mishra, S., & Kumar, A. (2022). Occurrence, toxicity, and risk assessment of deoxynivalenol and its modified forms in food. Toxins, 14(3), 187. [CrossRef]
- European Food Safety Authority (EFSA). (2017). Scientific opinion on the risks for animal and public health related to the presence of T-2 and HT-2 toxin in food and feed. EFSA Journal, 15(8), e04655. [CrossRef]
- European Food Safety Authority (EFSA). Scientific opinion on the risks for public health related to the presence of zearalenone in food. EFSA Journal 2011, 9(6), 2197. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA). Risks to human and animal health related to the presence of deoxynivalenol (DON) in food and feed. EFSA Journal 2017, 15(9), e04718. [Google Scholar] [CrossRef]
- Streit, E. , Schatzmayr, G., Tassis, P., Tzika, E., Marin, D., Taranu, I., Puel, O., & Oswald, I. P. (2013). Current situation of mycotoxin contamination and co-occurrence in animal feed—Focus on Europe. Toxins, 5, 895–921. [CrossRef]
- Pestka, J. J. (2010). Deoxynivalenol: Mechanisms of action, human exposure, and toxicological relevance. Archives of Toxicology, 84, 663–679. [CrossRef]
- Qie, Y. , Liu, C., Yang, C., Zhang, H., Wang, J., & Shen, C. (2024). Occurrence and toxicity of beauvericin: A review. Toxins, 16, 45. [CrossRef]
- Jestoi, M. (2008). Emerging Fusarium mycotoxins fusaproliferin, beauvericin, enniatins, and moniliformin—A review. Critical Reviews in Food Science and Nutrition, 48, 21–49. [CrossRef]
- Zinedine, A. , Soriano, J. M., Moltó, J. C., & Mañes, J. (2007). Review on the toxicity, occurrence, metabolism, detoxification, regulations and intake of zearalenone: An oestrogenic mycotoxin. Food and Chemical Toxicology, 45, 1–18. [CrossRef]
- Beres, B. L. , Brûlé-Babel, A. L., Ye, Z., Graf, R. J., Turkington, T. K., Harding, M. W., Kutcher, H. R., & Hooker, D. C. (2018). Exploring genotype × environment × management synergies to manage Fusarium head blight in wheat. Canadian Journal of Plant Pathology, 40, 179–188. [CrossRef]
- Marburger, D. A. , Venkateshwaran, M., Conley, S. P., Esker, P. D., Lauer, J. G., & Ané, J. M. (2015). Crop rotation and management effect on Fusarium spp. populations. Crop Science, 55, 365–376. [CrossRef]
- Rojas, E. C. , Jørgensen, H. J., Jensen, B., & Collinge, D. B. (2018). Fusarium diseases: Biology and management perspectives. In Integrated disease management of wheat and barley (pp. 43-66). Burleigh Dodds Science Publishing.
- University of Wisconsin-Madison. (n.d.). What do we know about crop rotations? Wisconsin Corn Agronomy. Retrieved May 6, 2025, from https://corn.agronomy.wisc.edu/WCM/W110.aspx.
- Leslie, J. F., Pearson, C. A., Nelson, P. E., & Toussoun, T. (1990). Fusarium spp. from corn, sorghum, and soybean fields in the central and eastern United States. Ecological Studies, 44, 66.
- Nelson, P. E., Toussoun, T. A., & Marasas, W. F. O. (1983). Fusarium species: An illustrated manual for identification. Pennsylvania State University Press.
- Islam, M. N., Banik, M., Sura, S., Tucker, J. R., & Wang, X. (2022). Implications of crop rotation and fungicide on Fusarium and mycotoxin spectra in Manitoba barley, 2017–2019. Toxins, 14(7), 463. [CrossRef]
- Islam, T. (2022). An integrated pest management program for managing Fusarium head blight disease in cereals. Journal of Integrative Agriculture, 21(12), 3434–3444. [CrossRef]
- Krupinsky, J. M., Bailey, K. L., McMullen, M. P., Gossen, B. D., & Turkington, T. K. (2002). Managing plant disease risk in diversified cropping systems. Agronomy Journal, 94(2), 198–209. [CrossRef]
- Arias, M. D., Munkvold, G. P., Ellis, M. L., & Leandro, L. F. (2013). Distribution and frequency of Fusarium species associated with soybean roots in Iowa. Plant Disease, 97(12), 1557–1562. [CrossRef]
- Broders, K. D. , Lipps, P. E., Paul, P. A., & Dorrance, A. E. Evaluation of Fusarium graminearum associated with corn and soybean seed and seedling disease in Ohio. Plant Disease 2007, 91(9), 1155–1160. [Google Scholar] [CrossRef]
- Okello, P. N. , Petrović, K., Kontz, B., Ali, S., Marek, L. F., & Mathew, F. M. Root rot caused by species of Fusarium on Brassica carinata in South Dakota. Plant Health Progress 2018, 19(3), 188–192. [Google Scholar] [CrossRef]
- Okello, P. N. , & Mathew, F. M. (2019). Cross-pathogenicity studies show South Dakota isolates of Fusarium acuminatum, F. equiseti, F. graminearum, F. oxysporum, F. proliferatum, F. solani, and F. subglutinans from either soybean or corn are pathogenic to both crops. Plant Health Progress, 20, 44–49. [CrossRef]
- Okello, P. N. , Petrovic, K., Singh, A. K., Kontz, B., & Mathew, F. M. Characterization of species of Fusarium causing root rot of soybean (Glycine max L.) in South Dakota, USA. Canadian Journal of Plant Pathology 2020, 42(4), 560–571. [Google Scholar] [CrossRef]
- Pioli, R. N., Mozzoni, L., & Morandi, E. N. (2004). First report of pathogenic association between Fusarium graminearum and soybean. Plant Disease, 88(2), 220. [CrossRef]
- Xue, A. G., Cober, E., Voldeng, H. D., Babcock, C., & Clear, R. M. (2007). Evaluation of the pathogenicity of Fusarium graminearum and Fusarium pseudograminearum on soybean seedlings under controlled conditions. Canadian Journal of Plant Pathology, 29(1), 35–40. [CrossRef]
- Xu, F., Liu, W., Song, Y., Zhou, Y., Xu, X., Yang, G., Wang, J., Zhang, J., & Liu, L. (2021). The distribution of Fusarium graminearum and Fusarium asiaticum causing Fusarium head blight of wheat in relation to climate and cropping system. Plant Disease, 105(10), 2830–2835. [CrossRef]
- Cotten, T. K., & Munkvold, G. P. (1998). Survival of Fusarium moniliforme, F. proliferatum, and F. subglutinans in maize stalk residue. Phytopathology, 88(6), 550–555. [CrossRef]
- Inch, S. A., & Gilbert, J. (2003). Survival of Gibberella zeae in Fusarium-damaged wheat kernels. Plant Disease, 87(3), 282–287. [CrossRef]
- Paulitz, T. C., Smiley, R. W., & Cook, R. J. (2002). Insights into the prevalence and management of soilborne cereal pathogens under direct seeding in the Pacific Northwest, USA. Canadian Journal of Plant Pathology, 24(4), 416–428. [CrossRef]
- Bernhoft, A., Torp, M., Clasen, P. E., Løes, A. K., & Kristoffersen, A. B. (2012). Influence of agronomic and climatic factors on Fusarium infestation and mycotoxin contamination of cereals in Norway. Food Additives & Contaminants: Part A, 29(7), 1129–1140. [CrossRef]
- Panth, M., Hassler, S. C., & Baysal-Gurel, F. (2020). Methods for management of soilborne diseases in crop production. Agriculture, 10(1), 16. [CrossRef]
- Bugingo, C., Ali, S., Yabwalo, D., & Byamukama, E. (2025). Optimizing fungicide seed treatments for early foliar disease management in wheat under Northern Great Plains conditions. Agronomy, 15(2), 291. [CrossRef]
- Lamichhane, J. R. , You, M. P., Laudinot, V., Barbetti, M. J., & Aubertot, J. N. Revisiting sustainability of fungicide seed treatments for field crops. Plant Disease 2020, 104(3), 610–623. [Google Scholar] [CrossRef]
- Munkvold, G. P. , & O'Mara, J. K. Laboratory and growth chamber evaluation of fungicidal seed treatments for maize seedling blight caused by Fusarium species. Plant Disease 2002, 86(2), 143–150. [Google Scholar] [CrossRef]
- Cromey, M. G. , Lauren, D. R., Parkes, R. A., Sinclair, K. I., Shorter, S. C., & Wallace, A. R. Control of Fusarium head blight of wheat with fungicides. Australasian Plant Pathology 2001, 30(4), 301–308. [Google Scholar] [CrossRef]
- Hollingsworth, C. R. , Motteberg, C. D., Assistant, R., & Thompson, W. G. Assessing fungicide efficacies for the management of Fusarium head blight on spring wheat and barley. Plant Health Progress 2006, 7(1), 14. [Google Scholar] [CrossRef]
- Ioos, R., Belhadj, A., Menez, M., & Faure, A. (2005). The effects of fungicides on Fusarium spp. and Microdochium nivale and their associated trichothecene mycotoxins in French naturally-infected cereal grains. Crop Protection, 24(10), 894–902. [CrossRef]
- Jones, R. K. (2000). Assessments of Fusarium head blight of wheat and barley in response to fungicide treatment. Plant Disease, 84, 1030. [CrossRef]
- Mesterhazy, A. , Bartok, T., & Lamper, C. Influence of wheat cultivar, species of Fusarium, and isolate aggressiveness on the efficacy of fungicides for control of Fusarium head blight. Fusarium 2003, 87(9), 1107–1115. [Google Scholar] [CrossRef]
- Wegulo, S. N. , Bockus, W. W., Nopsa, J. H., De Wolf, E. D., Eskridge, K. M., Peiris, K. H., & Dowell, F. E. Effects of integrating cultivar resistance and fungicide application on Fusarium head blight and deoxynivalenol in winter wheat. Fusarium 2011, 95(5), 554–560. [Google Scholar] [CrossRef]
- Willyerd, K. T. , Li, C., Madden, L. V., Bradley, C. A., Bergstrom, G. C., Sweets, L. E., McMullen, M., Ransom, J. K., Grybauskas, A., Osborne, L., & Wegulo, S. N. Efficacy and stability of integrating fungicide and cultivar resistance to manage Fusarium head blight and deoxynivalenol in wheat. Plant Disease 2012, 96(7), 957–967. [Google Scholar] [CrossRef] [PubMed]
- Kandel, Y. R. , Bradley, C. A., Chilvers, M. I., Mathew, F. M., Tenuta, A. U., Smith, D. L., Wise, K. A., & Mueller, D. S. Effect of seed treatment and foliar crop protection products on sudden death syndrome and yield of soybean. Plant Disease 2019, 103(7), 1712–1720. [Google Scholar] [CrossRef] [PubMed]
- Bugingo, C. , Brelsford, M., & Burrows, M. Fungicide sensitivity of Fusarium oxysporum f. sp. lentis and Fusarium acuminatum affecting lentil in the Northern Great Plains. Plant Disease 2024, 108(2), 286–290. [Google Scholar] [PubMed]
- Ellis, M. L. , Broders, K. D., Paul, P. A., & Dorrance, A. E. Infection of soybean seed by Fusarium graminearum and effect of seed treatments on disease under controlled conditions. Plant Disease 2011, 95(4), 401–407. [Google Scholar] [CrossRef]
- Capo, L. , Zappino, A., Reyneri, A., & Blandino, M. (2020). Role of the fungicide seed dressing in controlling seed-borne Fusarium spp. infection and in enhancing the early development and grain yield of maize. Agronomy, 10, 784. [CrossRef]
- Edwards, S. G. (2022). Pydiflumetofen co-formulated with prothioconazole: A novel fungicide for Fusarium head blight and deoxynivalenol control. Toxins, 14, 34. [CrossRef]
- Hou, Y. , Guo, Y., Wang, L., He, S., Zheng, W., Liu, S., & Xu, J. (2023). Impact of phenamacril on the growth and development of Fusarium pseudograminearum and control of crown rot of wheat. Plant Disease 107(12), 3843–3850. [CrossRef]
- Jayawardana, M. A. , & Fernando, W. G. (2024). The mechanisms of developing fungicide resistance in Fusarium graminearum causing Fusarium head blight and fungicide resistance management. Pathogens 13(11), 1012. [CrossRef]
- Ghosal, D., Tarafder, M., & Datta, B. (2020). Fusarium oxysporum isolates showed varied benzimidazole fungicides sensitivity under low temperature and heavy metal stress. Rhizosphere, 15, 100227. [CrossRef]
- Miao, J. , Li, Y., Hu, S., Li, G., Gao, X., Dai, T., & Liu, X. Resistance risk, resistance mechanism and the effect on DON production of a new SDHI fungicide cyclobutrifluram in Fusarium graminearum. Pesticide Biochemistry and Physiology 2024, 199, 105795. [Google Scholar] [CrossRef]
- Ceresini, P. C. , Silva, T. C., Vicentini, S. N., Leite Júnior, R. P., Moreira, S. I., Castro-Ríos, K., Garcés-Fiallos, F. R., Krug, L. D., Moura, S. S., Silva, A. G., & Custódio, A. A. (2024). Strategies for managing fungicide resistance in the Brazilian tropical agroecosystem: Safeguarding food safety, health, and the environmental quality. Tropical Plant Pathology, 49, 36–70. [CrossRef]
- Lucas, J. A. , Hawkins, N. J., & Fraaije, B. A. The evolution of fungicide resistance. Advances in Applied Microbiology 2015, 90, 29–92. [Google Scholar] [CrossRef]
- Naqvi, S. A., Farhan, M., Ahmad, M., Kiran, R., Shahbaz, M., Abbas, A., Hakim, F., Shabbir, M., Tan, Y. S., & Sathiya Seelan, J. S. (2025). Fungicide resistance in Fusarium species: Exploring environmental impacts and sustainable management strategies. Archives of Microbiology, 207(2), 1-24. [CrossRef]
- Shang, N., Yang, Y., Xiao, Y., Wu, Y., Li, K., Jiang, X., Sanganyado, E., Zhang, Q., & Xia, X. (2024). Exposure levels and health implications of fungicides, neonicotinoid insecticides, triazine herbicides and their associated metabolites in pregnant women and men. Environmental Pollution, 342, 123069. [CrossRef]
- Bartlett, D. W. , Clough, J. M., Godwin, J. R., Hall, A. A., Hamer, M., & Parr-Dobrzanski, B.. The strobilurin fungicides. Pest Management Science 2002, 58(7), 649–662. [Google Scholar] [CrossRef]
- Degani, O. , & Kalman, B. Assessment of commercial fungicides against onion (Allium cepa) basal rot disease caused by Fusarium oxysporum f. sp. cepae and Fusarium acutatum. Journal of Fungi 2021, 7(3), 235. [Google Scholar] [CrossRef]
- Ramirez, M. L. , Chulze, S., & Magan, N. Impact of environmental factors and fungicides on growth and deoxynivalenol production by Fusarium graminearum isolates from Argentinian wheat. Crop Protection 2004, 23(2), 117–125. [Google Scholar] [CrossRef]
- Dahal, N. , & Shrestha, R. K. Evaluation of efficacy of fungicides against Fusarium oxysporum f. sp. lentis in vitro at Lamjung, Nepal. Journal of the Institute of Agriculture and Animal Science 2018, 35(1), 105–112. [Google Scholar] [CrossRef]
- Lal, D. K. , Wagan, K. H., Mushatque, S., Ehsan, S., & Chaudhary, N. Investigations on seed-borne fungi associated with maize varieties and their control. Journal of Microbiological Sciences 2025, 4, 1–2. [Google Scholar] [CrossRef]
- Rajput, A. Q. , Arain, M. H., Pathan, M. A., Jiskani, M. M., & Lodhi, A. M. (2006). Efficacy of different fungicides against Fusarium wilt of cotton caused by Fusarium oxysporum f. sp. vasinfectum. Pakistan Journal of Botany 2006, 38(3), 875. [Google Scholar]
- Xu, C., Guo, M., Han, X., Ren, C., Liu, C., Fu, W., Qi, J., Ge, Z., Ma, Z., & Chen, Y. (2025). Fungal pathogen diversity and fungicide resistance assessment in Fusarium crown rot of wheat in the Huanghuai region of China. Journal of Agricultural and Food Chemistry. [CrossRef]
- Allen, T. W., Enebak, S. A., & Carey, W. A. (2004). Evaluation of fungicides for control of species of Fusarium on longleaf pine seed. Crop Protection, 23(10), 979-982. 10. [CrossRef]
- Hudec, K. (2006). Influence of seed treatment, temperature and origin of inocula on pathogenicity of Fusarium species to wheat and barley seedlings. Cereal Research Communications, 34, 1059–1066. [CrossRef]
- Müllenborn, C. Effect of fungicides on the complex of Fusarium species and saprophytic fungi colonizing wheat kernels. European Journal of Plant Pathology 2008, 120, 157–166. [Google Scholar] [CrossRef]
- Shin, J. H., Han, J. H., Lee, J. K., & Kim, K. S. (2014). Characterization of the maize stalk rot pathogens Fusarium subglutinans and F. temperatum and the effect of fungicides on their mycelial growth and colony formation. The Plant Pathology Journal, 30(4), 397-406. [CrossRef]
- Giedrojć, W. , Pluskota, W. E., & Wachowska, U. (2025). Fusarium graminearum in wheat—Management strategies in Central Europe. Pathogens, 14, 265. [CrossRef]
- Paul, P. A. , Lipps, P. E., Hershman, D. E., McMullen, M. P., Draper, M. A., & Madden, L. V. (2008). Efficacy of triazole-based fungicides for Fusarium head blight and deoxynivalenol control in wheat: A multivariate meta-analysis. Phytopathology, 98, 999–1011. [CrossRef]
- Buttar, H. S. , Singh, A., Sirari, A., Anupam, Kaur, K., Kumar, A., Lal, M. K., Tiwari, R. K., & Kumar, R. Investigating the impact of fungicides and mungbean genotypes on the management of pod rot disease caused by Fusarium equiseti and Fusarium chlamydosporum. Frontiers in Plant Science 2023, 14, 1164245. [Google Scholar] [CrossRef]
- Vatankhah, M., Saberi-Riseh, R., Moradzadeh Eskandari, M., & Afzali, H. (2019). Evaluation of some fungicides for the control of Fusarium dry rot of potato. Journal of Crop Protection, *8*(3), 275-285.
- Chala, A., Weinert, J., & Wolf, G. A. (2003). An integrated approach to the evaluation of the efficacy of fungicides against Fusarium culmorum, the cause of head blight of wheat. Journal of Phytopathology, 151(11-12), 673-678. [CrossRef]
- Weems, J. D. , Haudenshield, J. S., Bond, J. P., Hartman, G. L., Ames, K. A., & Bradley, C. A. (2015). Effect of fungicide seed treatments on Fusarium virguliforme infection of soybean and development of sudden death syndrome. Canadian Journal of Plant Pathology, 37, 435–447. [CrossRef]
- Boulahouat, S. , Cherif-Silini, H., Silini, A., Bouket, A. C., Luptakova, L., Alenezi, F. N., & Belbahri, L. (2023). Biocontrol efficiency of rhizospheric Bacillus against the plant pathogen Fusarium oxysporum: A promising approach for sustainable agriculture. Microbiology Research, 14, 892–908. [CrossRef]
- Mon, Y. Y., Bidabadi, S. S., Oo, K. S., & Zheng, S. J. (2021). The antagonistic mechanism of rhizosphere microbes and endophytes on the interaction between banana and Fusarium oxysporum f. sp. cubense. Physiological and Molecular Plant Pathology, 116, 101733. [CrossRef]
- Alukumbura, A. S., Bigi, A., Sarrocco, S., Fernando, W. D., Vannacci, G., Mazzoncini, M., & Bakker, M. G. (2022). Minimal impacts on the wheat microbiome when Trichoderma gamsii T6085 is applied as a biocontrol agent to manage Fusarium head blight disease. Frontiers in Microbiology, 13, 972016. [CrossRef]
- Liang, N., Charron, J. B., & Jabaji, S. (2023). Comparative transcriptome analysis reveals the biocontrol mechanism of Bacillus velezensis E68 against Fusarium graminearum DAOMC 180378, the causal agent of Fusarium head blight. PLOS ONE, 18(1), e0277983. [CrossRef]
- Yeo, Y. J., Park, A. R., Vuong, B. S., & Kim, J. C. (2024). Biocontrol of Fusarium head blight in rice using Bacillus velezensis JCK-7158. Frontiers in Microbiology, 15, 1358689. [CrossRef]
- Raaijmakers, J. M., Leeman, M., Van Oorschot, M. M., Van der Sluis, I., Schippers, B., & Bakker, P. A. (1995). Dose-response relationships in biological control of Fusarium wilt of radish by Pseudomonas spp. Phytopathology, 85(10), 1075–1080.
- Kiesewalter, H. T., Lozano-Andrade, C. N., Wibowo, M., Strube, M. L., Maróti, G., Snyder, D., Jørgensen, T. S., Larsen, T. O., Cooper, V. S., Weber, T., & Kovács, Á. T. (2021). Genomic and chemical diversity of Bacillus subtilis secondary metabolites against plant pathogenic fungi. mSystems, 6(1), e00770-20. [CrossRef]
- Sivasithamparam, K., & Ghisalberti, E. (2002). Secondary metabolism in Trichoderma. In Trichoderma and Gliocladium (Vol. 1, pp. 139–191). Taylor & Francis.
- Compant, S., Duffy, B., Nowak, J., Clément, C., & Barka, E. A. (2005). Use of plant growth-promoting bacteria for biocontrol of plant diseases: Principles, mechanisms of action, and future prospects. Applied and Environmental Microbiology, 71(9), 4951–4959. [CrossRef]
- Petrucci, A., Khairullina, A., Sarrocco, S., Jensen, D. F., Jensen, B., Jørgensen, H. J., & Collinge, D. B. (2023). Understanding the mechanisms underlying biological control of Fusarium diseases in cereals. European Journal of Plant Pathology, 167(4), 453–476. [CrossRef]
- Gimeno, A. , Kägi, A., Drakopoulos, D., Bänziger, I., Lehmann, E., Forrer, H. R., Keller, B., & Vogelgsang, S. (2020). From laboratory to the field: Biological control of Fusarium graminearum on infected maize crop residues. Journal of Applied Microbiology, 129, 680–694. [CrossRef]
- Xue, A. G. , Chen, Y., Voldeng, H. D., Fedak, G., Savard, M. E., Längle, T., Zhang, J., & Harman, G. E. (2014). Concentration and cultivar effects on efficacy of CLO-1 biofungicide in controlling Fusarium head blight of wheat. Biological Control, 73, 2–7. [CrossRef]
- Abaya, A., Serajazari, M., & Hsiang, T. (2021). Control of Fusarium head blight using the endophytic fungus, Simplicillium lamellicola, and its effect on the growth of Triticum aestivum. Biological Control, 160, 104684. [CrossRef]
- Bhardwaj, M., Kailoo, S., Khan, R. T., Khan, S. S., & Rasool, S. (2023). Harnessing fungal endophytes for natural management: A biocontrol perspective. Frontiers in Microbiology, 14, 1280258. [CrossRef]
- Noel, Z. A., Roze, L. V., Breunig, M., & Trail, F. (2022). Endophytic fungi as a promising biocontrol agent to protect wheat from Fusarium graminearum head blight. Plant Disease, 106(2), 595–602. [CrossRef]
- Thangavelu, R., & Gopi, M. (2015). Field suppression of Fusarium wilt disease in banana by the combined application of native endophytic and rhizospheric bacterial isolates possessing multiple functions. Phytopathologia Mediterranea, 54(2), 241–252. [CrossRef]
- De Lamo, F. J., & Takken, F. L. (2020). Biocontrol by Fusarium oxysporum using endophyte-mediated resistance. Frontiers in Plant Science, 11, 37. [CrossRef]
- Rojas, E. C., Jensen, B., Jørgensen, H. J., Latz, M. A., Esteban, P., Ding, Y., & Collinge, D. B. (2020). Selection of fungal endophytes with biocontrol potential against Fusarium head blight in wheat. Biological Control, 144, 104222. [CrossRef]
- Bosco, S., Prencipe, S., Mezzalama, M., & Spadaro, D. (2024). Screening and characterization of bacterial and fungal endophytes as potential biocontrol agents for rice seed dressing against Fusarium fujikuroi. Biological Control, 196, 105580. [CrossRef]
- Nawaz, M. E., Malik, K., & Hassan, M. N. (2022). Rice-associated antagonistic bacteria suppress the Fusarium fujikuroi causing rice bakanae disease. BioControl, 67(1), 101–109. [CrossRef]
- O’Sullivan, C. A., Roper, M. M., Myers, C. A., & Thatcher, L. F. (2021). Developing actinobacterial endophytes as biocontrol products for Fusarium pseudograminearum in wheat. Frontiers in Bioengineering and Biotechnology, 9, 691770. [CrossRef]
- Abd-Elgawad, M. M., & Askary, T. H. (2020). Factors affecting success of biological agents used in controlling the plant-parasitic nematodes. Egyptian Journal of Biological Pest Control, 30(1), 17. [CrossRef]
- He, D. C., He, M. H., Amalin, D. M., Liu, W., Alvindia, D. G., & Zhan, J. (2021). Biological control of plant diseases: An evolutionary and eco-economic consideration. Pathogens, 10(10), 1311. [CrossRef]
- Teixidó, N., Usall, J., & Torres, R. (2022). Insight into a successful development of biocontrol agents: Production, formulation, packaging, and shelf life as key aspects. Horticulturae, 8(4), 305. [CrossRef]
- Alisaac, E., & Mahlein, A. K. (2023). Fusarium head blight on wheat: Biology, modern detection and diagnosis and integrated disease management. Toxins, 15(3), 192. [CrossRef]
- Figlan, S., & Mwadzingeni, L. (2022). Breeding tools for assessing and improving resistance and limiting mycotoxin production by Fusarium graminearum in wheat. Plants, 11(15), 1933. [CrossRef]
- Poudel, B. (2024). *CRISPR/Cas9 based gene editing in wheat and potato to enhance multiple disease resistance* [PhD thesis, McGill University].
- Hameed, A. , Poznanski, P., Noman, M., Ahmed, T., Iqbal, A., Nadolska-Orczyk, A., & Orczyk, W. (2022). Barley resistance to Fusarium graminearum infections: From transcriptomics to field with food safety concerns. Journal of Agricultural and Food Chemistry, 70, 14571–14587. [CrossRef]
- Jha, U. C., Bohra, A., Pandey, S., & Parida, S. K. (2020). Breeding, genetics, and genomics approaches for improving Fusarium wilt resistance in major grain legumes. Frontiers in Genetics, 11, 1001. [CrossRef]
- Yin, K., & Qiu, J. L. (2019). Genome editing for plant disease resistance: Applications and perspectives. Philosophical Transactions of the Royal Society B, 374(1767), 20180322. [CrossRef]
- Ollier, M., Talle, V., Brisset, A. L., Le Bihan, Z., Duerr, S., Lemmens, M., Goudemand, E., Robert, O., Hilbert, J. L., & Buerstmayr, H. (2020). QTL mapping and successful introgression of the spring wheat-derived QTL Fhb1 for Fusarium head blight resistance in three European triticale populations. Theoretical and Applied Genetics, 133, 457–477. [CrossRef]
- Waites, J., Achary, V. M., Syombua, E. D., Hearne, S. J., & Bandyopadhyay, A. (2025). CRISPR-mediated genome editing of wheat for enhancing disease resistance. Frontiers in Genome Editing, 7, 1542487. [CrossRef]
- Zhang, F., Shi, Y., Ali, J., Xu, J., & Li, Z. (2021). Breeding by selective introgression: Theory, practices, and lessons learned from rice. The Crop Journal, 9(3), 646–657. [CrossRef]
- Ma, H., Zhang, X., Yao, J., & Cheng, S. (2019). Breeding for the resistance to Fusarium head blight of wheat in China. Frontiers of Agricultural Science and Engineering, 6(3), 251–264. [CrossRef]
- Mapari, A. R., & Mehandi, S. (2024). Enhancing crop resilience: Advances and challenges in marker-assisted selection for disease resistance. Journal of Advances in Biology & Biotechnology, 27(7), 569–580. [CrossRef]
- Gaire, R., de Arruda, M. P., Mohammadi, M., Brown-Guedira, G., Kolb, F. L., & Rutkoski, J. (2022). Multi-trait genomic selection can increase selection accuracy for deoxynivalenol accumulation resulting from Fusarium head blight in wheat. The Plant Genome, 15(1), e20188. [CrossRef]
- Steiner, B., Buerstmayr, M., Michel, S., Schweiger, W., Lemmens, M., & Buerstmayr, H. (2017). Breeding strategies and advances in line selection for Fusarium head blight resistance in wheat. Tropical Plant Pathology, 42, 165–174. [CrossRef]
- Guo-Liang, J. (2013). Molecular markers and marker-assisted breeding in plants. In A. Sven Bode (Ed.), Plant Breeding from Laboratories to Fields (pp. Ch.3). IntechOpen.
- Luo, K., Guo, J., He, D., Li, G., & Ouellet, T. (2023). Deoxynivalenol accumulation and detoxification in cereals and its potential role in wheat–Fusarium graminearum interactions. Abiotech, 4(2), 155–171. [CrossRef]
- Jiang, F., & Doudna, J. A. (2017). CRISPR–Cas9 structures and mechanisms. Annual Review of Biophysics, 46(1), 505–529. [CrossRef]
- Langner, T., Kamoun, S., & Belhaj, K. (2018). CRISPR crops: Plant genome editing toward disease resistance. Annual Review of Phytopathology, 56(1), 479–512. [CrossRef]
- Wolter, F. , Schindele, P., & Puchta, H. (2019). Plant breeding at the speed of light: The power of CRISPR/Cas to generate directed genetic diversity at multiple sites. BMC Plant Biology, 19, 176. [CrossRef]
- Pixley, K. V. , Falck-Zepeda, J. B., Paarlberg, R. L., Phillips, P. W., Slamet-Loedin, I. H., Dhugga, K. S., Campos, H., & Gutterson, N. (2022). Genome-edited crops for improved food security of smallholder farmers. Nature Genetics, 54, 364–367. [CrossRef]
- Borrelli, V. M. , Brambilla, V., Rogowsky, P., Marocco, A., & Lanubile, A. (2018). The enhancement of plant disease resistance using CRISPR/Cas9 technology. Frontiers in Plant Science, 9, 1245. [CrossRef]
- Nekrasov, V. , Wang, C., Win, J., Lanz, C., Weigel, D., & Kamoun, S. (2017). Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Scientific Reports, 7, 482. [CrossRef]
- Paul, N. C. , Park, S. W., Liu, H., Choi, S., Ma, J., MacCready, J. S., Chilvers, M. I., & Sang, H. (2021). Plant and fungal genome editing to enhance plant disease resistance using the CRISPR/Cas9 system. Frontiers in Plant Science, 12, 700925. [CrossRef]
- Tyagi, S. , Kumar, R., Kumar, V., Won, S. Y., & Shukla, P. (2021). Engineering disease resistant plants through CRISPR-Cas9 technology. GM Crops & Food, 12, 125–144. [CrossRef]
- Zhang, M. , Liu, Q., Yang, X., Xu, J., Liu, G., Yao, X., Ren, R., Xu, J., & Lou, L. (2020). CRISPR/Cas9-mediated mutagenesis of Clpsk1 in watermelon to confer resistance to Fusarium oxysporum f. sp. niveum. Plant Cell Reports, 39, 589–595. [CrossRef]
- Brauer, E. K. , Balcerzak, M., Rocheleau, H., Leung, W., Schernthaner, J., Subramaniam, R., & Ouellet, T. (2020). Genome editing of a deoxynivalenol-induced transcription factor confers resistance to Fusarium graminearum in wheat. *Molecular Plant-Microbe Interactions, 33*(3), 553–560. [CrossRef]
- Zaidi, S. S. , Mahas, A., Vanderschuren, H., & Mahfouz, M. M. (2020). Engineering crops of the future: CRISPR approaches to develop climate-resilient and disease-resistant plants. Genome Biology, 21, 289. [CrossRef]
- Ahmad, I. , Mazhar, K., Atiq, M., Khalaf, A. K., Rashid, M. H., Asif, M., Ahmed, S., Adil, Z., Fayyaz, A., Al-Sadoon, M. K., & Al-Otaibi, H. S. Epidemiology and management of Fusarium wilt of Eucalyptus camaldulensis through systemic acquired resistance. PeerJ 2024, 12, e17022. [Google Scholar] [CrossRef]
- Ejaz, M. R. , Jaoua, S., Ahmadi, M., & Shabani, F. (2023). An examination of how climate change could affect the future spread of Fusarium spp. around the world, using correlative models to model the changes. Environmental Technology & Innovation, 31, 103177. [CrossRef]
- Liu, C. , Kogel, K. H., & Ladera-Carmona, M. (2024). Harnessing RNA interference for the control of Fusarium species: A critical review. Molecular Plant Pathology, 25, e70011. [CrossRef]
- Araújo, M. B. , & New, M. (2007). Ensemble forecasting of species distributions. Trends in Ecology & Evolution, 22, 42–47. [CrossRef]
- Alagendran, S., & Prasanna, G. S. (2024). Climate-adapted pathogen control through biological and chemical agents. In Plant pathology in the age of climate change (1st ed., pp. 244–263). Golden Leaf Publishers. ISBN: 978-93-48240-31-6.
- De Wolf, E. D. , Madden, L. V., & Lipps, P. E. (2003). Risk assessment models for wheat Fusarium head blight epidemics based on within-season weather data. Phytopathology, 93, 428–435. [CrossRef]
- Beernink, B. M. , Amanat, N., Li, V. H., Manchur, C. L., Whyard, S., & Belmonte, M. F. (2024). SIGS vs. HIGS: Opportunities and challenges of RNAi pest and pathogen control strategies. Canadian Journal of Plant Pathology, 46, 675–689. [CrossRef]
- Koch, A. , Biedenkopf, D., Furch, A., Weber, L., Rossbach, O., Abdellatef, E., Linicus, L., Johannsmeier, J., Jelonek, L., Goesmann, A., & Cardoza, V. (2016). An RNAi-based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathogens, 12, e1005901. [CrossRef]
- Padilla-Roji, I. , Ruiz-Jiménez, L., Bakhat, N., Vielba-Fernández, A., Pérez-García, A., & Fernández-Ortuño, D. (2023). RNAi technology: A new path for the research and management of obligate biotrophic phytopathogenic fungi. International Journal of Molecular Sciences, 24, 9082. [CrossRef]
- Machado, A. K. , Brown, N. A., Urban, M., Kanyuka, K., & Hammond-Kosack, K. E. RNAi as an emerging approach to control Fusarium head blight disease and mycotoxin contamination in cereals. Pest Management Science 2018, 74(4), 790–799. [Google Scholar] [CrossRef]
- Huang, X. N. , Wang, Y., Li, Y. T., et al. (2025). Trans-kingdom sRNA silencing in the prevention and control of crop Fusarium wilt disease. Phytopathol Res, 7, 18. [CrossRef]
- Ouyang, S. Q. , Ji, H. M., Feng, T., Luo, S. J., Cheng, L., & Wang, N. (2023). Artificial trans-kingdom RNAi of FolRDR1 is a potential strategy to control tomato wilt disease. PLoS Pathogens, 19, e1011463. [CrossRef]
- Qiao, L. , Lan, C., Capriotti, L., Ah-Fong, A., Nino Sanchez, J., Hamby, R., Heller, J., Zhao, H., Glass, N. L., Judelson, H. S., & Mezzetti, B. (2021). Spray-induced gene silencing for disease control is dependent on the efficiency of pathogen RNA uptake. Plant Biotechnology Journal, 19, 1756–1768. [CrossRef]
- Werner, B. T. , Gaffar, F. Y., Schuemann, J., Biedenkopf, D., & Koch, A. M. (2020). RNA-spray-mediated silencing of Fusarium graminearum AGO and DCL genes improve barley disease resistance. Frontiers in Plant Science, 11, 476. [CrossRef]
- Yang, W. , Wang, B., Lei, G., Chen, G., & Liu, D. (2022). Advances in nanocarriers to improve the stability of dsRNA in the environment. Frontiers in Bioengineering and Biotechnology 10, 974646. [CrossRef]
- Hanamasagar, Y. , Ramesha, N. M., Mahapatra, S., Panigrahi, C. K., Vidhya, C. S., Agnihotri, N., & Satapathy, S. N. (2024). Advancing RNAi for sustainable insect management: Targeted solutions for eco-friendly pest control. Journal of Experimental Agriculture International, 46, 740–745. [CrossRef]
- Lamichhane, S. , Neupane, S., Timsina, S., Chapagain, B., Paudel, P. P., & Rimal, A. (2024). Potato late blight caused by Phytophthora infestans: An overview on pathology, integrated disease management approaches, and forecasting models. Social Science Research Network. [CrossRef]
- Gerlach, W., & Nirenberg, H. (1982). The genus Fusarium: A pictorial atlas. Berlin-Dahlem, Germany.
- Burgess, L. W., Liddell, C. M., & Summerell, B. A. (1988). Laboratory manual for Fusarium research (2nd ed.). Fusarium Research Laboratory.
- Leslie, J. F., & Summerell, B. A. (Eds.). (2006). The Fusarium laboratory manual. Blackwell Publishing. [CrossRef]
- Infantino, A. , Grottoli, A., Bergamaschi, V., Oufensou, S., Burgess, L. W., & Balmas, V. (2023). FusaHelp: A web site program for the morphological identification of Fusarium species. Journal of Plant Pathology, 105, 429–436. [CrossRef]
- Mulè, G. , González-Jaén, M. T., Hornok, L., Nicholson, P., & Waalwijk, C. (2005). Advances in molecular diagnosis of toxigenic Fusarium species: A review. Food Additives & Contaminants, 22, 316–323. [CrossRef]
- Jurado, M. , Vázquez, C., Marín, S., Sanchis, V., & González-Jaén, M. T. (2006). PCR-based strategy to detect contamination with mycotoxigenic Fusarium species in maize. Systematic and Applied Microbiology, 29, 681–689. [CrossRef]
- Gutiérrez-Aguirre, I. , Rački, N., Dreo, T., & Ravnikar, M. (2015). Droplet digital PCR for absolute quantification of pathogens. In C. Lacomme (Ed.), Plant pathology: Methods in molecular biology (Vol. 1302, pp. 431–442). Humana Press. [CrossRef]
- Morcia, C., Tumino, G., Gasparo, G., Ceresoli, C., Fattorini, C., Ghizzoni, R., Carnevali, P., & Terzi, V. (2020). Moving from qPCR to Chip Digital PCR Assays for tracking of some Fusarium species causing Fusarium head blight in cereals. Microorganisms, 8(9), 1307. [CrossRef]
- Natarajan, S., Bucur, D., Kildea, S., & Doohan, F. (2025). Digital PCR assays for quantifying trichothecene-producing Fusarium species, including Fusarium langsethiae, F. poae, and F. sporotrichioides, in oats. Analytical and Bioanalytical Chemistry. [CrossRef]
- Li, B. , Du, J., Lan, C., Liu, P., Weng, Q., & Chen, Q. (2013). Development of a loop-mediated isothermal amplification assay for rapid and sensitive detection of Fusarium oxysporum f. sp. cubense race 4. European Journal of Plant Pathology, 135, 903–911. [CrossRef]
- Lovera, A. , Rodríguez, E., Simbaqueba, J., Burbano-David, D., Carmona, S. L., Izquierdo-García, L. F., Gómez-Correa, J. C., Leon-Pacheco, R. I., Parra-Fuentes, M., Rodríguez-Yzquierdo, G., Betancourt, M., Zuluaga, P., & Soto-Suárez, M. (2023). Development and application of a droplet digital PCR assay for the detection of Fusarium oxysporum f. sp. cubense tropical race 4 from different biological samples. Plant Pathology. [CrossRef]
- Notomi, T. , Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., & Hase, T. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Research, 28, e63. [CrossRef]
- Li, Y. , Fan, P., Zhou, S., & Zhang, L. (2017). Loop-mediated isothermal amplification (LAMP): A novel rapid detection platform for pathogens. Microbial Pathogenesis, 107, 54–61. [CrossRef]
- Ghosh, R. , Nagavardhini, A., Sengupta, A., & Sharma, M. (2015). Development of loop-mediated isothermal amplification (LAMP) assay for rapid detection of Fusarium oxysporum f. sp. ciceris - wilt pathogen of chickpea. BMC Research Notes, 8, Article 40. [CrossRef]
- Jiang, H. , Wu, N., Jin, S., Ahmed, T., Wang, H., Li, B., Wu, X., Bao, Y., Liu, F., & Zhang, J.-Z. (2021). Identification of rice seed-derived Fusarium spp. and development of LAMP assay against Fusarium fujikuroi. Pathogens, 10, 1. [CrossRef]
- Ghosh, D.K. , Kokane, S.B., Kokane, A.D., Warghane, A.J., Motghare, M.R., Bhose, S., et al. (2018) Development of a recombinase polymerase based isothermal amplification combined with lateral flow assay (HLB-RPA-LFA) for rapid detection of "Candidatus Liberibacter asiaticus". PLoS ONE 13(12): e0208530. [CrossRef]
- Hu, S. , Yan, C., Yu, H., Zhang, Y., & Zhang, C. Q. (2023). Establishment of the Recombinase Polymerase Amplification-Lateral Flow Dipstick Detection Technique for Fusarium oxysporum. Plant disease, 107, 2665–2672. [CrossRef]
- Liang, X., Zhang, X., Haseeb, H. A., Tang, T., Shan, J., Yin, B., & Guo, W. (2022). Development and evaluation of a novel visual and rapid detection assay for toxigenic Fusarium graminearum in maize based on recombinase polymerase amplification and lateral flow analysis. International Journal of Food Microbiology, 372, 109682. [CrossRef]
- Cobo-Díaz, J. F., Baroncelli, R., Le Floch, G., & Picot, A. (2019). A novel metabarcoding approach to investigate Fusarium species composition in soil and plant samples. FEMS Microbiology Ecology, 95(6), fiz084. [CrossRef]
- Walder, F., Schlaeppi, K., Wittwer, R., van der Heijden, M. G. A., & Vogelgsang, S. (2017). Community profiling of Fusarium and other plant-associated fungi in crops using SMRT sequencing. Frontiers in Plant Science, 8, 2019. [CrossRef]
- Karlsson, I., Persson, P., & Friberg, H. (2021). Fusarium head blight from a microbiome perspective. Frontiers in Microbiology, 12, 628373. [CrossRef]
- Pieczul, K., Świerczyńska, I., & Wójtowicz, A. (2025). Advanced rDNA-based detection of wheat pathogens in grain samples using NGS. Pathogens, 14(2), 164. [CrossRef]
- Martinez, A. , & Bennett, J. W. (2021). Fungal volatile organic compounds. In O. Zaragoza (Ed.), Encyclopedia of mycology (Vol. 1, pp. 239-245). Elsevier.
- Cui, S. , Ling, P., Zhu, H., & Keener, H. M. (2018). Plant pest detection using an artificial nose system: A review. Sensors, 18, 378. [CrossRef]
- Cellini, A., Blasioli, S., Biondi, E., Bertaccini, A., Braschi, I., & Spinelli, F. (2017). Potential applications and limitations of electronic nose devices for plant disease diagnosis. Sensors, 17(11), 2596. [CrossRef]
- Infantino, A., Costa, C., Aragona, M., Reverberi, M., Taiti, C., & Mancuso, S. (2017). Identification of different Fusarium spp. through mVOCs profiling by means of proton-transfer-reaction time-of-flight (PTR-ToF-MS) analysis. Journal of Plant Pathology, 99(4), 663-669. [CrossRef]
- Camardo Leggieri, M., Mazzoni, M., Bertuzzi, T., Moschini, M., Prandini, A., & Battilani, P. (2022). Electronic nose for the rapid detection of deoxynivalenol in wheat using classification and regression trees. Toxins, 14(9), 617. [CrossRef]
- Bu´sko, M., Gracka, A., Jele ´ n, H., Szablewska, K.S., Przybylska-Balcerek, A.;,Szwajkowska-Michałek, L., Góral, T., (2023) The effect of organic and conventional cultivation systems on the profile of Volatile Organic Compounds in winter wheat grain, including susceptibility to Fusarium Head Blight. Metabolites, 13, 1045. [CrossRef]
- Zhang, J., Zhang, B., Dong, J., Tian, Y., Lin, Y., Fang, G., & Wang, S. (2022). Identification of mouldy rice using an electronic nose combined with SPME-GC/MS. Journal of Stored Products Research, 95, 101921. [CrossRef]
- Wesoly, M., Daulton, E., Jenkins, S., van Amsterdam, S., Clarkson, J., & Covington, J. A. (2024). Early detection of Fusarium basal rot infection in onions and shallots based on VOC profiles analysis. Journal of Agricultural and Food Chemistry, 72(7), 3664–3672. [CrossRef]
- Rizzi, L., Galuppi, R., Simioli, M., Canestrari, G., Bonoli, C., & Tampieri, M. P. (2009). Grains colonised by moulds: fungal identification and headspace analysis of produced volatile metabolites. Italian Journal of Animal Science, 8(sup2), 334–336. [CrossRef]
- Infantino, A., Taiti, C., Grottoli, A., Mancuso, S., Costa, C., & Garzoli, S. (2023). Examination of volatile signatures of Fusarium bulb rot in garlic using PTR-ToF-MS and SPME-GC/MS. Separations, 10(11), 556. [CrossRef]
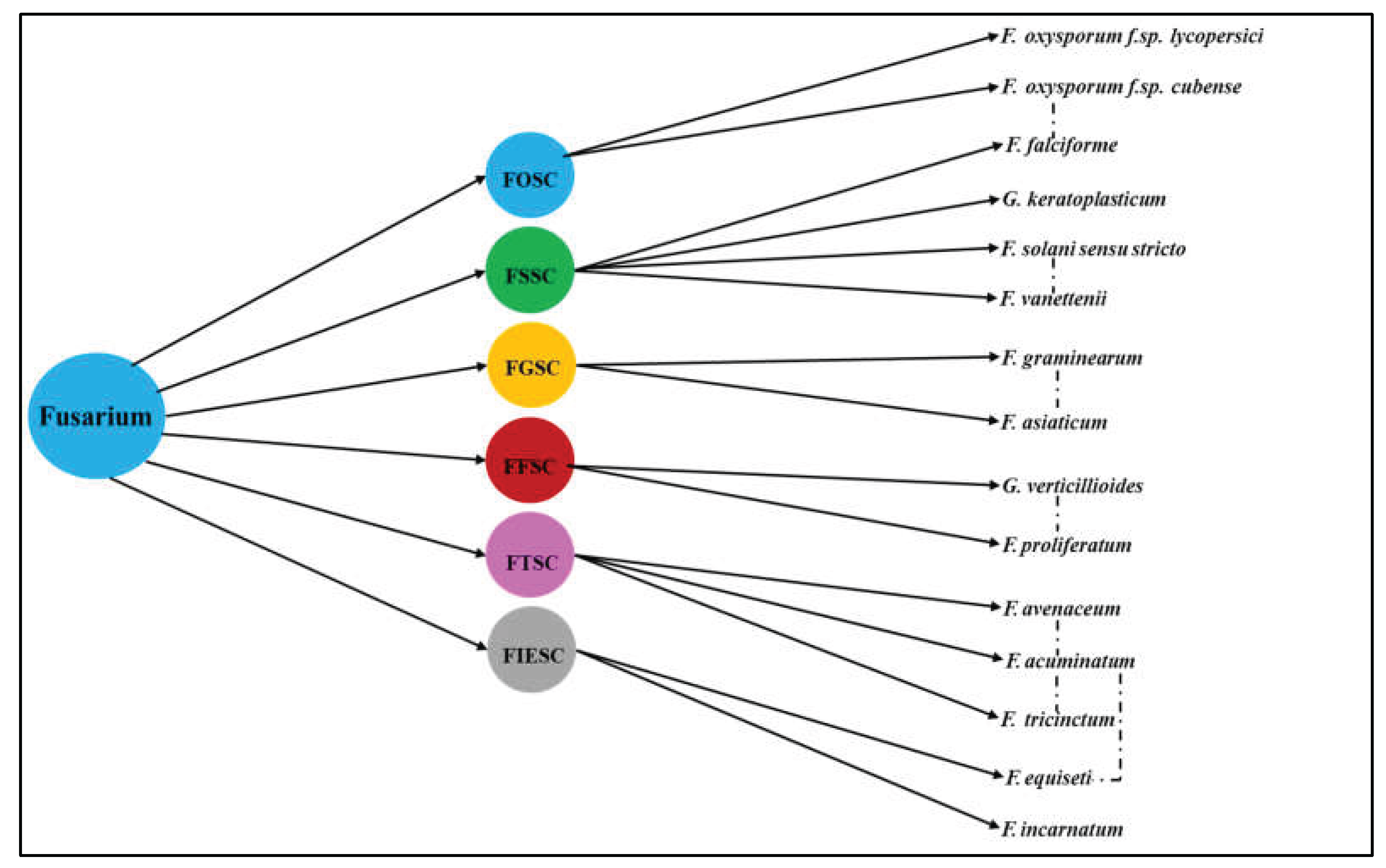
| Fusarium Species | Species Complex | Primary Host(s) | Crop Type | Region | References |
|---|---|---|---|---|---|
| F. zanthoxyli | FTOSC | Zanthoxylum bungeanum | Spice crop (woody) | Northern China | [73,74] |
| F. torreyae | FTOSC | Torreya taxifolia | Tree (conifer) | North America | [75] |
| F. oxysporum f. sp. cubense TR4 | FOSC | Banana (Musa spp.) | Fruit crop (perennial herb) | Southeast Asia, Middle East, Africa, Latin America | [77,78,79] |
| Novel FIESC spp. | FIESC | Various herbs & crops | Medicinal, legumes | China | [82] |
| FIESC spp. | FIESC | Rice, soybean, legumes | Cereals, legumes | South America, Africa, Europe | [86,83,85] |
| F. acuminatum, F. avenaceum, F. tricinctum | FTSC | Soybean, medicinal herbs, fruits, cereals | Multiple | Asia, Europe | [86,88,93] |
| F. avenaceum | FTSC | Raspberry, pulses, cereals, soybean | Berries, legumes | Europe, North-Estern China | [86,91,94,92] |
| Transcription Factor | Fusarium species | Function | Regulates | Reference |
|---|---|---|---|---|
| Tri6 | F. graminearum | Zn(II)2Cys6 TF; pathway-specific regulator | Trichothecene biosynthesis gene cluster (TRI genes); also acts as global regulator | [131,132] |
| Tri10 | F. graminearum | Transcriptional activator | Activates TRI gene expression in coordination with Tri6 | [131] |
| PacC | F. oxysporum | pH-responsive TF | Virulence gene expression under neutral-alkaline pH; adaptation to host environment | [133] |
| Ftf1 | F. oxysporum | Zn(II)2Cys6 TF encoded on accessory chromosome | Effector gene expression and host-specific pathogenicity | [134] |
| Sge1 | F. oxysporum | Global TF involved in effector regulation | SIX gene expression (secreted in xylem proteins) | [135] |
| FgLaeA (part of VeA complex) | F. graminearum | Global regulator of secondary metabolism, sexual development, and virulence | Regulates the expression of trichothecene biosynthetic genes (TRI6, ZEB2), sexual development genes, and virulence factors via interaction with FgVeA | [136] |
| Table 2 continued. | ||||
| Transcription Factor | Fusarium species | Function | Regulates | Reference |
| Kmt6 (H3K27 methyltransferase) | F. fujikuroi | Epigenetic regulator of secondary metabolism via histone modification | Controls the activation of silent secondary metabolite gene clusters by modifying chromatin accessibility through H3K27me3 marks | [137] |
| AreA | F. fujikuroi | Nitrogen metabolism TF | Gibberellin biosynthesis genes (in response to nitrogen availability) | [138] |
| Fusarium species | Host crop | Disease | Resistance gene/locus | Reference |
|---|---|---|---|---|
| F. graminearum | Wheat | Fusarium head blight | Fhb1 | [142] |
| F. oxysporum f. sp. lycopersici | Tomato | Fusarium wilt | I, I-2, I-3 | [143] |
| F. oxysporum f. sp. lentis | Lentil | Fusarium wilt/root rot | Fw Gene | [144,145] |
| F. virguliforme (syn. F. solani f. sp. Glycines) | Soybean | Sudden death syndrome | Rhg1, Rhg4 | [146] |
| F. verticillioides | Maize | Ear rot, seedling blight | ZmWAX2, ZmXYXT2, | [147,148] |
| F. oxysporum f. sp. cubense tropical race 4 (Tr4) | Banana | Fusarium wilt (Panama disease) | QTLs (Ma848, Ma851) | [149] |
| F. oxysporum f. sp. vasinfectum | Cotton | Fusarium wilt | GhWAK7A | [150] |
| F. oxysporum f. sp. cucumerinum | Cucumber | Fusarium wilt | QTLs CsChi23 |
[151,151] |
| Fusarium Species | Known for Formae Speciales? | Notable Formae speciales (f. sp.) |
|---|---|---|
| F. oxysporum | Yes – over 100 | f. sp. lycopersici (tomato), cubense (banana), vasinfectum (cotton), cepae (onion), melonis (melon), pisi (pea) |
| F. solani (FSSC) | Yes, but being revised | f. sp. pisi (pea), phaseoli (bean), cucurbitae (cucurbits), mori (mulberry) |
| F. graminearum | No | Not applicable – broad host range |
| F. verticillioides | No | Not applicable – infects maize, sorghum, and others broadly |
| F. proliferatum | No | Not applicable – opportunistic across many hosts |
| F. avenaceum | No | Not applicable – generalist necrotrophy |
| Fungicide (Common Name) | Commercial Name (if specificied) | FRAC Category / Chemical Group | Mode of Application | Target Fusarium Species / Disease a | Reference |
|---|---|---|---|---|---|
| Azoxystrobin | Azimut, Amistar, Dynasty, Ortiva | Strobilurin (QoI, FRAC group 11) | Seed coating, Culture plates, Seed treatment, Foliar spray, in vitro | F. acutatum, F. oxysporum f. sp. cepae, Fusarium spp., F. virguliforme, F. subglutinans, F. temperatum, F. graminearum, F. oxysporum, F. pseudograminearum | [258,223,259,260] |
| Carbendazim | Antracol | Benzimidazole (Systemic, Broad-spectrum) | Seed treatment, Soil mixture, Culture media | Fusarium spp., F. oxysporum f. sp. vasinfectum, F. oxysporum f. sp. lentis, F. oxysporum (maize), F. equiseti, F. chlamydosporum, F. pseudograminearum | [261,262,263] |
| Fungicide (Common Name) | Commercial Name (if specificied) | FRAC Category / Chemical Group | Mode of Application | Target Fusarium Species / Disease a | Reference |
| Cyclobutrifluram | - | SDHI (FRAC group 7) | In vitro | F. pseudograminearum | [264] |
| Difenoconazole | Dividend XL RTA | Azole (DMI) | Culture plates, Seed treatment, in vitro | F. solani, F. proliferatum, F. oxysporum, F. circinatum, F. avenaceum, F. culmorum, F. poae, F. sporotrichioides, F. subglutinans, F. temperatum, F. pseudograminearum | [265,266,267,268] |
| Fludioxonil | Vibrance, Maxim 4 FS, MaximQuattro, Celest XL, Celest Quattro | Phenylpyrrole | Seed coating, Culture plates, Seed treatment | F. acutatum, F. oxysporum f. sp. cepae, Rhizoctonia, Fusarium spp., F. graminearum, F. virguliforme, F. solani, F. oxysporum (dry rot), Sclerotinia sclerotiorum, F. verticillioides, F. pseudograminearum | [223,247,237] |
| Fluopyram | ILeVO | SDHI (FRAC group 7) | Seed treatment | F. virguliforme | [245] |
| Phenamacril | - | Myosin inhibitor | In vitro | F. pseudograminearum | [250,264] |
| Prochloraz | Sportak | Azole (DMI) | Seed coating, Potted sprout irrigation, Culture plates | F. oxysporum f. sp. cepae, F. acutatum, F. subglutinans, F. temperatum, F. oxysporum (banana wilt), F. graminearum (FHB), F. culmorum (FHB), F. oxysporum f. sp. lycopersici, F. pseudograminearum | [259,268] |
| Prothioconazole | Redigo, Proline, Prosaro | Azole (DMI, FRAC group 3) | Seed treatment, Foliar fungicide, in vitro | Fusarium spp., F. virguliforme, F. graminearum (FHB), F. poae, F. pseudograminearum | [269,246,270] |
| Pyraclostrobin | Stamina, BAS 580 | Strobilurin (QoI, FRAC group 11) | Seed treatment, Foliar fungicide, in vitro | Fusarium spp., F. virguliforme, F. graminearum, F. pseudograminearum | [247,269,264] |
| Tebuconazole | Orius 25, Azimut, Raxil 250 FL, Raxil MD, Raxil T, Folicur, Nativo SC300, Twinstar 75 WG, Prosaro | Azole (DMI, FRAC group 3) | Culture plates, Seed coating, In vitro, Foliar spray | F. acutatum, F. oxysporum f. sp. cepae, F. subglutinans, F. temperatum, F. graminearum (FHB), F. culmorum (FHB), F. poae, Fusarium equiseti, F. chlamydosporum, Fusarium spp., F. pseudograminearum | [271,267,268] |
| Thiabendazole | Mertect 340F, Rival, Tecto, MaximQuattro, Trilex AL (part of) | Benzimidazole (FRAC group 1) | Culture plates, Seed treatment, In vitro, In vivo, In situ | F. solani, Fusarium spp., F. oxysporum, F. graminearum (part of combination), F. verticillioides (part of combination) | [265,272] |
| Trifloxystrobin | Trilex, Fortix | Strobilurin (QoI, FRAC group 11) | Seed treatment, Foliar fungicide, in vitro | Fusarium spp., F. graminearum, F. virguliforme, F. chlamydosporum, F. asiaticum | [271,273,267] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





