Submitted:
27 June 2025
Posted:
01 July 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Preparation of Faecal Slurry
2.2. MiGut Model Setup
2.3. Choline-d9 Fermentations
2.3.1. Experiment 1 Timeline
2.3.1. Experiment 2 Timeline
2.4. Measurement of Choline-d9 and TMA-d9
2.5. Microbiome Analysis
2.5.1. DNA Extraction and Sequencing
2.5.2. Metagenomic Sequence Analysis
2.7. Data Analysis and Statistics
3. Results
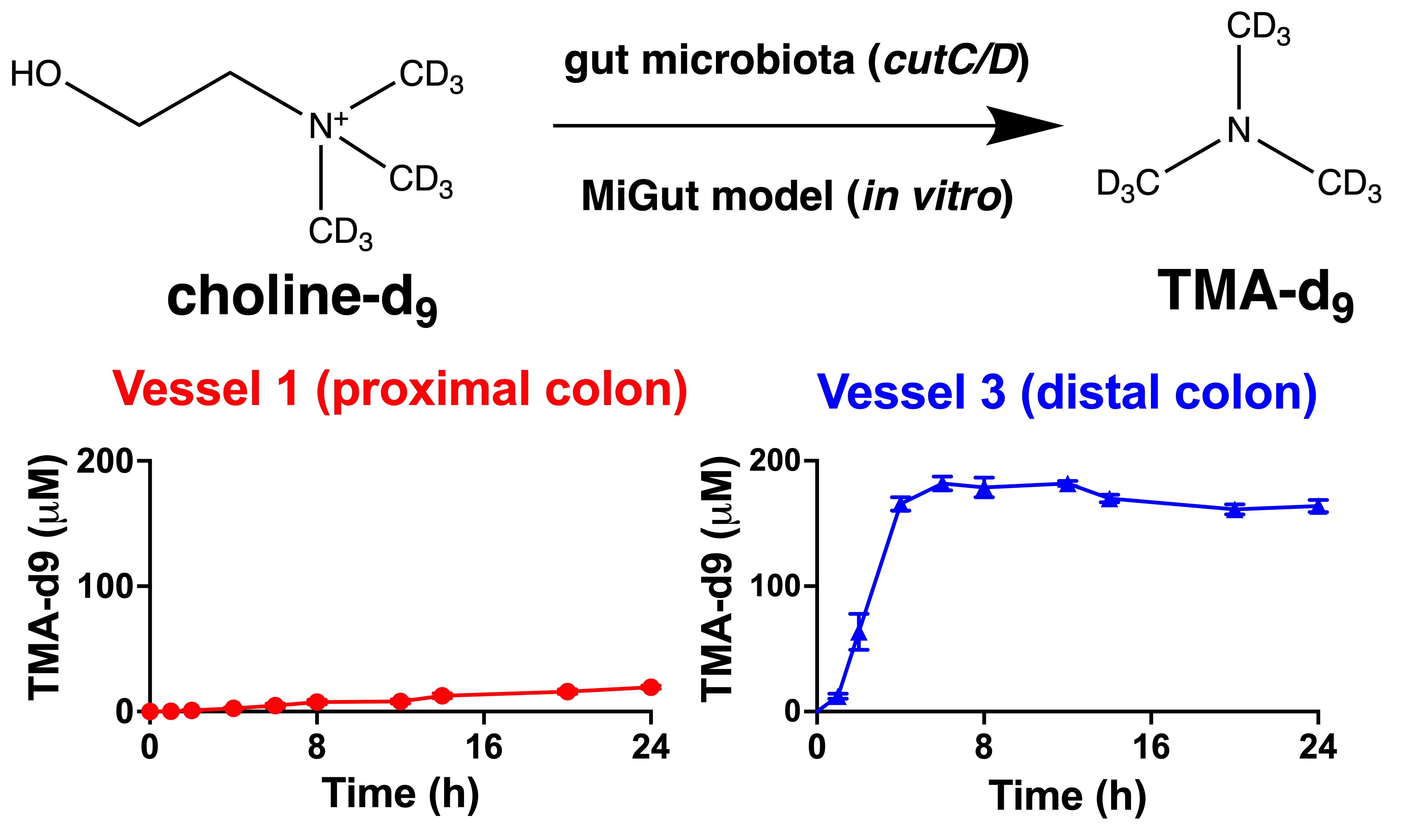
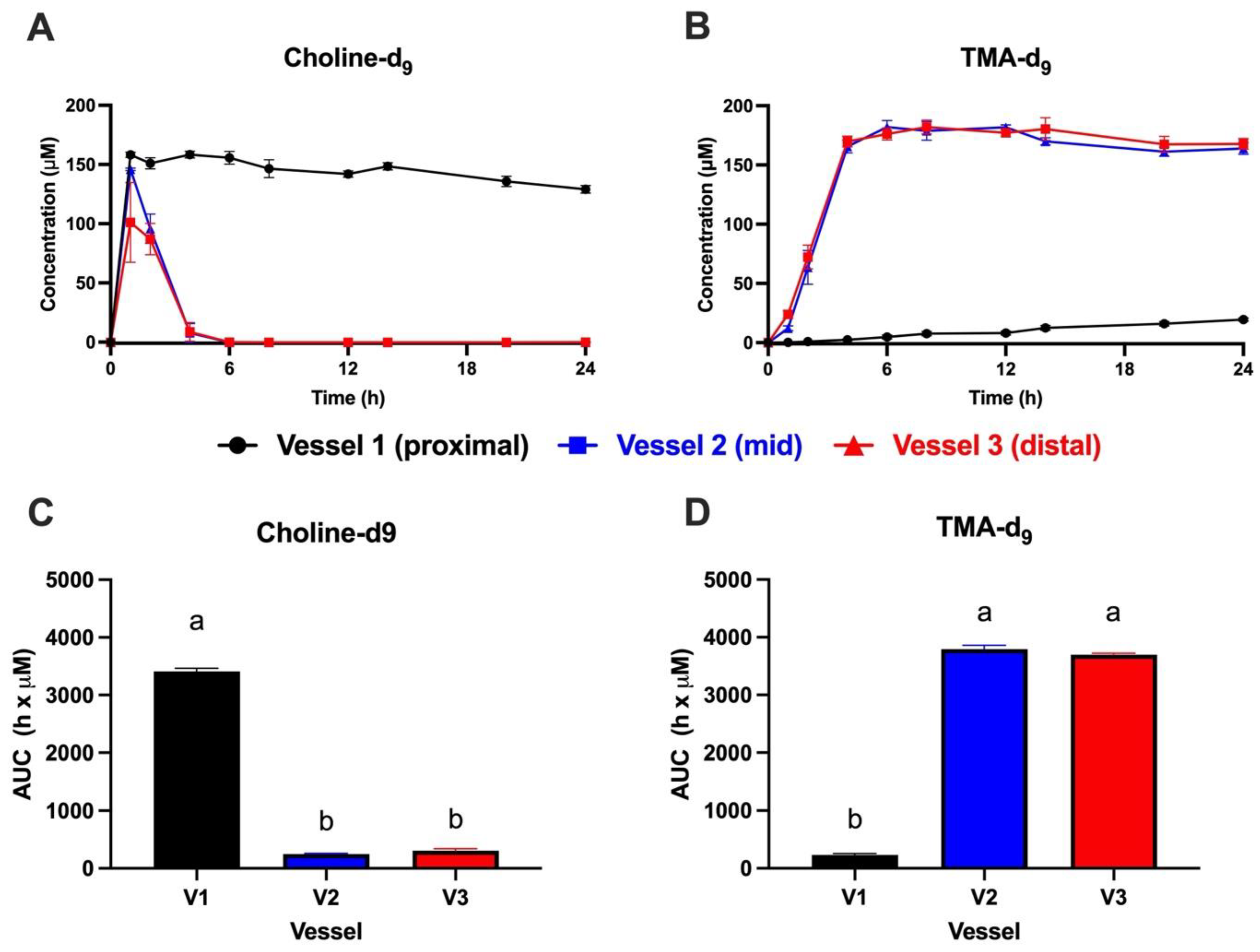
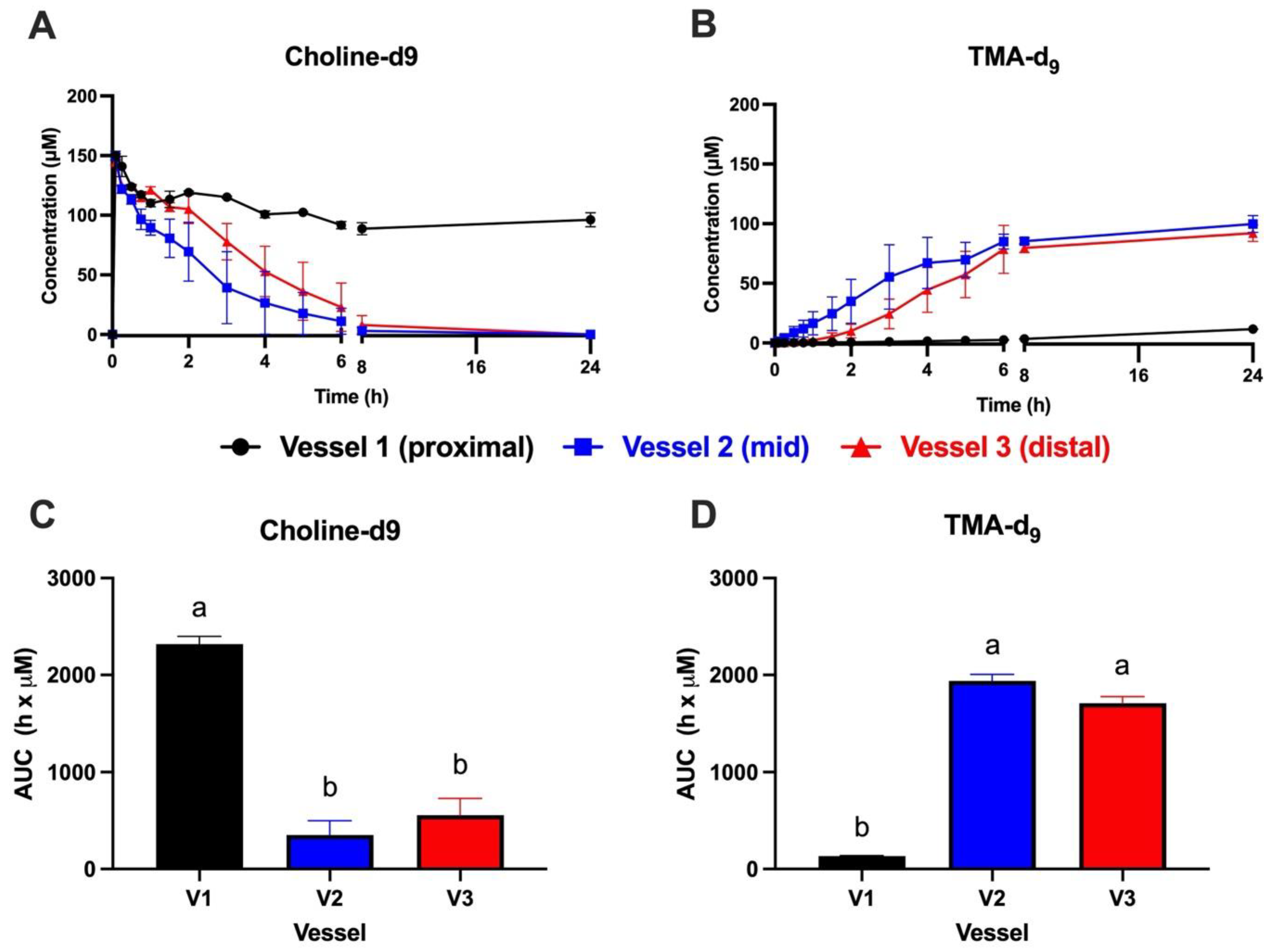
3.1. Choline-d9 Conversion to TMA-d9 Differs by Simulated Colon Region
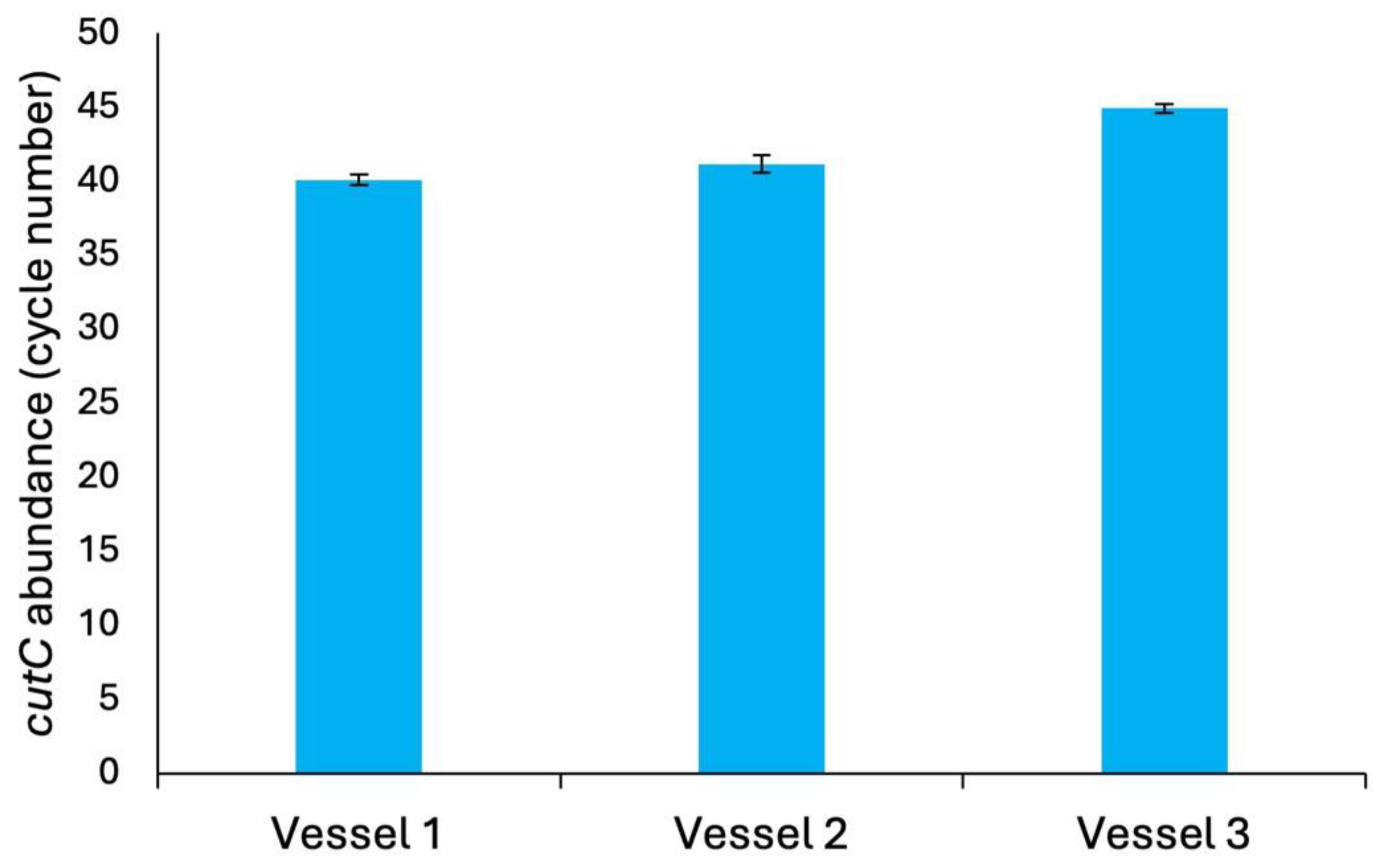
3.2. Microbiome Characterization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding Sources
Acknowledgements
Conflicts of Interest
References
- Amini, M.; Zayeri, F.; Salehi, M. Trend Analysis of Cardiovascular Disease Mortality, Incidence, and Mortality-to-Incidence Ratio: Results from Global Burden of Disease Study 2017. BMC Public Health 2021, 21, 401. [Google Scholar] [CrossRef] [PubMed]
- Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Ammirati, E.; Baddour, L.M.; Barengo, N.C.; Beaton, A.Z.; Benjamin, E.J.; Benziger, C.P.; et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990–2019: Update From the GBD 2019 Study. J. Am. Coll. Cardiol. 2020, 76, 2982–3021. [Google Scholar] [CrossRef] [PubMed]
- Calazans, J.A.; Permanyer, I. Levels, Trends, and Determinants of Cause-of-Death Diversity in a Global Perspective: 1990–2019. BMC Public Health 2023, 23, 650. [Google Scholar] [CrossRef] [PubMed]
- Witkowski, M.; Weeks, T.L.; Hazen, S.L. Gut Microbiota and Cardiovascular Disease. Circ. Res. 2020, 127, 553–570. [Google Scholar] [CrossRef]
- Talmor-Barkan, Y.; Bar, N.; Shaul, A.A.; Shahaf, N.; Godneva, A.; Bussi, Y.; Lotan-Pompan, M.; Weinberger, A.; Shechter, A.; Chezar-Azerrad, C.; et al. Metabolomic and Microbiome Profiling Reveals Personalized Risk Factors for Coronary Artery Disease. Nat. Med. 2022, 28, 295–302. [Google Scholar] [CrossRef]
- Kazemian, N.; Mahmoudi, M.; Halperin, F.; Wu, J.C.; Pakpour, S. Gut Microbiota and Cardiovascular Disease: Opportunities and Challenges. Microbiome 2020, 8, 36. [Google Scholar] [CrossRef]
- Wang, Z.; Klipfell, E.; Bennett, B.J.; Koeth, R.; Levison, B.S.; DuGar, B.; Feldstein, A.E.; Britt, E.B.; Fu, X.; Chung, Y.-M. Gut Flora Metabolism of Phosphatidylcholine Promotes Cardiovascular Disease. Nature 2011, 472, 57–63. [Google Scholar] [CrossRef]
- Tang, W.W.; Wang, Z.; Levison, B.S.; Koeth, R.A.; Britt, E.B.; Fu, X.; Wu, Y.; Hazen, S.L. Intestinal Microbial Metabolism of Phosphatidylcholine and Cardiovascular Risk. N. Engl. J. Med. 2013, 368, 1575–1584. [Google Scholar] [CrossRef]
- Jameson, E.; Quareshy, M.; Chen, Y. Methodological Considerations for the Identification of Choline and Carnitine-Degrading Bacteria in the Gut. Methods 2018, 149, 42–48. [Google Scholar] [CrossRef]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L. Intestinal Microbiota Metabolism of L-Carnitine, a Nutrient in Red Meat, Promotes Atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef]
- Koeth, R.A.; Levison, B.S.; Culley, M.K.; Buffa, J.A.; Wang, Z.; Gregory, J.C.; Org, E.; Wu, Y.; Li, L.; Smith, J.D.; et al. γ-Butyrobetaine Is a Proatherogenic Intermediate in Gut Microbial Metabolism of L-Carnitine to TMAO. Cell Metab. 2014, 20, 799–812. [Google Scholar] [CrossRef] [PubMed]
- Seim, H.; Löster, H.; Claus, R.; Kleber, H.-P.; Strack, E. Formation of γ-Butyrobetaine and Trimethylamine from Quaternary Ammonium Compounds Structure-Related to l-Carnitine and Choline by Proteus Vulgaris. FEMS Microbiol. Lett. 1982, 13, 201–205. [Google Scholar] [CrossRef]
- Meyer, K.A.; Shea, J.W. Dietary Choline and Betaine and Risk of CVD: A Systematic Review and Meta-Analysis of Prospective Studies. Nutrients 2017, 9, 711. [Google Scholar] [CrossRef] [PubMed]
- Cashman, J.R.; Camp, K.; Fakharzadeh, S.S.; Fennessey, P.V.; Hines, R.N.; Mamer, O.A.; Mitchell, S.C.; Preti, G.; Schlenk, D.; Smith, R.L. Biochemical and Clinical Aspects of the Human Flavin-Containing Monooxygenase Form 3 (FMO3) Related to Trimethylaminuria. Curr. Drug Metab. 2003, 4, 151–170. [Google Scholar] [CrossRef]
- Shimizu, M.; Koibuchi, N.; Mizugaki, A.; Hishinuma, E.; Saito, S.; Hiratsuka, M.; Yamazaki, H. Genetic Variants of Flavin-Containing Monooxygenase 3 (FMO3) in Japanese Subjects Identified by Phenotyping for Trimethylaminuria and Found in a Database of Genome Resources. Drug Metab. Pharmacokinet. 2021, 38, 100387. [Google Scholar] [CrossRef]
- Roberts, A.B.; Gu, X.; Buffa, J.A.; Hurd, A.G.; Wang, Z.; Zhu, W.; Gupta, N.; Skye, S.M.; Cody, D.B.; Levison, B.S.; et al. Development of a Gut Microbe–Targeted Nonlethal Therapeutic to Inhibit Thrombosis Potential. Nat. Med. 2018, 24, 1407–1417. [Google Scholar] [CrossRef]
- Wang, Z.; Roberts, A.B.; Buffa, J.A.; Levison, B.S.; Zhu, W.; Org, E.; Gu, X.; Huang, Y.; Zamanian-Daryoush, M.; Culley, M.K. Non-Lethal Inhibition of Gut Microbial Trimethylamine Production for the Treatment of Atherosclerosis. Cell 2015, 163, 1585–1595. [Google Scholar] [CrossRef]
- Iglesias-Carres, L.; Krueger, E.S.; Herring, J.A.; Tessem, J.S.; Neilson, A.P. Potential of Phenolic Compounds and Their Gut Microbiota-Derived Metabolites to Reduce TMA Formation: Application of an In Vitro Fermentation High-Throughput Screening Model. J. Agric. Food Chem. 2022, 70, 3207–3218. [Google Scholar] [CrossRef]
- Iglesias-Carres, L.; Essenmacher, L.A.; Racine, K.C.; Neilson, A.P. Development of a High-Throughput Method to Study the Inhibitory Effect of Phytochemicals on Trimethylamine Formation. Nutrients 2021, 13, 1466. [Google Scholar] [CrossRef]
- Iglesias-Carres, L.; Bruno, A.; D’Antuono, I.; Linsalata, V.; Cardinali, A.; Neilson, A.P. In Vitro Evidences of the Globe Artichoke Antioxidant, Cardioprotective and Neuroprotective Effects. J. Funct. Foods 2023, 107, 105674. [Google Scholar] [CrossRef]
- Iglesias-Carres, L.; Racine, K.C.; Neilson, A.P. Phenolic-Rich Beverages Reduce Bacterial TMA Formation in an Ex Vivo–in Vitro Colonic Fermentation Model. Food Funct. 2022, 13, 8022–8037. [Google Scholar] [CrossRef] [PubMed]
- Bresciani, L.; Dall’Asta, M.; Favari, C.; Calani, L.; Rio, D.D.; Brighenti, F. An in Vitro Exploratory Study of Dietary Strategies Based on Polyphenol-Rich Beverages, Fruit Juices and Oils to Control Trimethylamine Production in the Colon. Food Funct. 2018, 9, 6470–6483. [Google Scholar] [CrossRef] [PubMed]
- Day-Walsh, P.; Shehata, E.; Saha, S.; Savva, G.M.; Nemeckova, B.; Speranza, J.; Kellingray, L.; Narbad, A.; Kroon, P.A. The Use of an In-Vitro Batch Fermentation (Human Colon) Model for Investigating Mechanisms of TMA Production from Choline, l-Carnitine and Related Precursors by the Human Gut Microbiota. Eur. J. Nutr. 2021, 60, 3987–3999. [Google Scholar] [CrossRef]
- Orman, M.; Bodea, S.; Funk, M.A.; Campo, A.M.-D.; Bollenbach, M.; Drennan, C.L.; Balskus, E.P. Structure-Guided Identification of a Small Molecule That Inhibits Anaerobic Choline Metabolism by Human Gut Bacteria. J. Am. Chem. Soc. 2019, 141, 33–37. [Google Scholar] [CrossRef]
- Venema, K. The TNO In Vitro Model of the Colon (TIM-2). In The Impact of Food Bioactives on Health: in vitro and ex vivo models; Verhoeckx, K., Cotter, P., López-Expósito, I., Kleiveland, C., Lea, T., Mackie, A., Requena, T., Swiatecka, D., Wichers, H., Eds.; Springer: Cham (CH), 2015 ISBN 978-3-319-15791-7.
- Van de Wiele, T.; Van den Abbeele, P.; Ossieur, W.; Possemiers, S.; Marzorati, M. The Simulator of the Human Intestinal Microbial Ecosystem (SHIME®). In The Impact of Food Bioactives on Health: in vitro and ex vivo models; Verhoeckx, K., Cotter, P., López-Expósito, I., Kleiveland, C., Lea, T., Mackie, A., Requena, T., Swiatecka, D., Wichers, H., Eds.; Springer: Cham (CH), 2015 ISBN 978-3-319-15791-7.
- Davis Birch, W.A.; Moura, I.B.; Ewin, D.J.; Wilcox, M.H.; Buckley, A.M.; Culmer, P.R.; Kapur, N. MiGut: A Scalable in Vitro Platform for Simulating the Human Gut Microbiome—Development, Validation and Simulation of Antibiotic-Induced Dysbiosis. Microb. Biotechnol. 2023, 16, 1312–1324. [Google Scholar] [CrossRef]
- Wang, Z.; Hazen, J.; Jia, X.; Org, E.; Zhao, Y.; Osborn, L.J.; Nimer, N.; Buffa, J.; Culley, M.K.; Krajcik, D.; et al. The Nutritional Supplement L-Alpha Glycerylphosphorylcholine Promotes Atherosclerosis. Int. J. Mol. Sci. 2021, 22, 13477. [Google Scholar] [CrossRef]
- Rath, S.; Heidrich, B.; Pieper, D.H.; Vital, M. Uncovering the Trimethylamine-Producing Bacteria of the Human Gut Microbiota. Microbiome 2017, 5, 1–14. [Google Scholar] [CrossRef]
- Jameson, E.; Doxey, A.C.; Airs, R.; Purdy, K.J.; Murrell, J.C.; Chen, Y. Metagenomic Data-Mining Reveals Contrasting Microbial Populations Responsible for Trimethylamine Formation in Human Gut and Marine Ecosystems. Microb. Genomics 2016, 2, e000080. [Google Scholar] [CrossRef]
- Ramireddy, L.; Tsen, H.-Y.; Chiang, Y.-C.; Hung, C.Y.; Chen, F.-C.; Yen, H.-T. The Gene Expression and Bioinformatic Analysis of Choline Trimethylamine-Lyase (CutC) and Its Activating Enzyme (CutD) for Gut Microbes and Comparison with Their TMA Production Levels. Curr. Res. Microb. Sci. 2021, 2, 100043. [Google Scholar] [CrossRef]
- Ferrell, M.; Bazeley, P.; Wang, Z.; Levison, B.S.; Li, X.S.; Jia, X.; Krauss, R.M.; Knight, R.; Lusis, A.J.; Garcia-Garcia, J.C. Fecal Microbiome Composition Does Not Predict Diet-Induced TMAO Production in Healthy Adults. J. Am. Heart Assoc. 2021, 10, e021934. [Google Scholar] [CrossRef]
- Kashyap, J.; Ringiesn, J.R.; Schwab, N.; Ferguson, D.J. Isolation and Characterization of a Novel Choline Degrading Citrobacter Amalonaticus Strain from the Human Gut. Curr. Res. Microb. Sci. 2022, 3, 100157. [Google Scholar] [CrossRef] [PubMed]
- Ramezani, A.; Nolin, T.D.; Barrows, I.R.; Serrano, M.G.; Buck, G.A.; Regunathan-Shenk, R.; West, R.E.; Latham, P.S.; Amdur, R.; Raj, D.S. Gut Colonization with Methanogenic Archaea Lowers Plasma Trimethylamine N-Oxide Concentrations in Apolipoprotein E−/− Mice. Sci. Rep. 2018, 8, 14752. [Google Scholar] [CrossRef] [PubMed]
- de la Cuesta-Zuluaga, J.; Spector, T.D.; Youngblut, N.D.; Ley, R.E. Genomic Insights into Adaptations of Trimethylamine-Utilizing Methanogens to Diverse Habitats, Including the Human Gut. mSystems 2021, 6, 10.1128–msystems.00939. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, K.M.; Smithson, A.T.; Ickes, A.K.; Neilson, A.P. Pan-Colonic Pharmacokinetics of Catechins and Procyanidins in Male Sprague–Dawley Rats. J. Nutr. Biochem. 2015, 26, 1007–1014. [Google Scholar] [CrossRef]
- Casso, A.G.; VanDongen, N.S.; Gioscia-Ryan, R.A.; Clayton, Z.S.; Greenberg, N.T.; Ziemba, B.P.; Hutton, D.A.; Neilson, A.P.; Davy, K.P.; Seals, D.R.; et al. Initiation of 3,3-Dimethyl-1-Butanol at Midlife Prevents Endothelial Dysfunction and Attenuates in Vivo Aortic Stiffening with Ageing in Mice. J. Physiol. 2022, 600, 4633–4651. [Google Scholar] [CrossRef]
- Winslow, C.J.; Nichols, B.L.B.; Novo, D.C.; Mosquera-Giraldo, L.I.; Taylor, L.S.; Edgar, K.J.; Neilson, A.P. Cellulose-Based Amorphous Solid Dispersions Enhance Rifapentine Delivery Characteristics in Vitro. Carbohydr. Polym. 2018, 182, 149–158. [Google Scholar] [CrossRef]
- Gilley, A.D.; Arca, H.C.; Nichols, B.L.B.; Mosquera-Giraldo, L.I.; Taylor, L.S.; Edgar, K.J.; Neilson, A.P. Novel Cellulose-Based Amorphous Solid Dispersions Enhance Quercetin Solution Concentrations in Vitro. Carbohydr. Polym. 2017, 157, 86–93. [Google Scholar] [CrossRef]
- Goodrich, K.M.; Neilson, A.P. Simultaneous UPLC-MS/MS Analysis of Native Catechins and Procyanidins and Their Microbial Metabolites in Intestinal Contents and Tissues. J. Chromatogr. B.
- Martínez-del Campo, A.; Bodea, S.; Hamer, H.A.; Marks, J.A.; Haiser, H.J.; Turnbaugh, P.J.; Balskus, E.P. Characterization and Detection of a Widely Distributed Gene Cluster That Predicts Anaerobic Choline Utilization by Human Gut Bacteria. MBio 2015, 6, e00042–15. [Google Scholar] [CrossRef]
| Compound | MW | MS/MS transition |
CV (V) |
CE (eV) |
| Choline-d9 | 113.2 | 113.3>69.1 | 40 | 16 |
| Choline-1-13C-1,1,2,2-d4 | 109.2 | 109.3>60.3 | 36 | 18 |
| Ethyl betaine-d9a | 155.2 | 155.3>127.2 | 34 | 20 |
| Ethyl betaine-13C3-15Na | 150.2 | 150.3>122.2 | 34 | 18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





