Submitted:
26 June 2025
Posted:
27 June 2025
You are already at the latest version
Abstract
Keywords:
1. General Considerations on MIPs
2. MIP Fungal Diversity and Function
3. Case Study of Fungal XIPs – State of Art
3.1. State of Art
3.2. Diversity of the Fungal XIPs – Evolution and Topology
3.3. Fungal XIPs: A Relatively Recent Chapter in the Evolution of Fungal AQP
3.4. Fungal XIP – Outstanding Questions
4. Conclusion
Supplementary Materials
Author Contributions
Funding
Data availability
Acknowledgments
Conflicts of Interest
References
- Agre, P.; Sasaki, S.; Chrispeels, M.J. Aquaporins: a family of water channel proteins. American Journal of Physics 1993, 265, F461. [Google Scholar] [CrossRef] [PubMed]
- Zardoya, R. Phylogeny and evolution of the major intrinsic protein family. Biology of the Cell 2005, 97, 397–414. [Google Scholar] [CrossRef] [PubMed]
- Gazzarrini, S.; Kang, M.; Epimashko, S.; Van Etten, J.L.; Dainty, J.; Thiel, G.; et al. Chlorella virus MT325 encodes water and potassium channels that interact synergistically. Proceedings of the National Academy of Sciences of USA 2006, 103, 5355–5360. [Google Scholar] [CrossRef] [PubMed]
- Fujiyoshi, Y.; Mitsuoka, K.; de Groot, BL.; Philippsen, A.; Grubmüller, H.; Agre, P.; Engel, A. Structure and function of water channels. Current Opinion in Structural Biology 2002, 12, 509–515. [Google Scholar] [CrossRef]
- Ozu, M.; Galizia, L.; Acuña, C.; Amodeo, G. Aquaporins: More Than Functional Monomers in a Tetrameric Arrangement. Cells 2018, 7, 209. [Google Scholar] [CrossRef]
- Luu, D.T.; Maurel, C. Aquaporins in a challenging environment: molecular gears for adjusting plant water status. Plant, Cell & Environment 2005, 28, 85–96. [Google Scholar]
- Tajkhorshid, E.; Nollert, P.; Jensen, M.; Miercke, L.; O’Connell, J.; Stroud, R.M.; Schulten, K. Control of the selectivity of the aquaporin water channel family by global orientation tuning. Science 2002, 296, 525–530. [Google Scholar] [CrossRef]
- Sui, H.; Han, B.G.; Lee, J.K.; Walian, P.; Jap, B.K. Structural basis of water specific transport through the AQP1 water channel. Nature 2001, 414, 872–878. [Google Scholar] [CrossRef]
- Wallace, I.S.; Roberts, D.M. Homology modeling of representative subfamilies of Arabidopsis major intrinsic proteins. Classification based on the aromatic/arginine selectivity filter. Plant Physiology 2004, 135, 1059–1068. [Google Scholar] [CrossRef]
- Fu, D.; Libson, A.; Miercke, L.J. : Weitzman, C.; Nollert, P.; Krucinski, J.; Stroud, R.M. Structure of a glycerol-conducting channel and the basis for its selectivity. Science 2000, 290, 481–486. [Google Scholar] [CrossRef]
- Wang, Y.; Schulten, K.; Tajkhorshid, E. What makes an aquaporin a glycerol channel? A comparative study of AqpZ and GlpF. Structure 2005, 13, 1107–1118. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Beitz, E. Aquaporins with selectivity for unconventional permeants. Cellular and Molecular Life Sciences 2007, 64, 2413–2421. [Google Scholar] [CrossRef] [PubMed]
- Saboe, P. O,. Rapisarda, C,. Kaptan, S,. Hsiao, Y.S,. Summers, S.R,. De Zorzi, R,. et al. Role of Pore-Lining Residues in Defining the Rate of Water Conduction by Aquaporin-0. Biophysical Journal 2017, 112, 953–965. [Google Scholar] [CrossRef] [PubMed]
- Pluhackova, K.; Schittny, V.; Bürkner, P.C.; Siligan, C.; Horner, A. Multiple pore lining residues modulate water permeability of GlpF. Protein Science 2022, 31, e4431. [Google Scholar] [CrossRef]
- 15 Finn, R.N.; Cerdà, J. Evolution and functional diversity of aquaporins. Biol. Bull. 2015, 229, 6–23. [Google Scholar] [CrossRef]
- Johanson, U.; Karlsson, M.; Johansson, I.; Gustavsson, S.; Sjovall, S.; Fraysse, L.; Weig, A.R.; Kjellbom, P. The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiology 2001, 126, 358–1369. [Google Scholar] [CrossRef]
- Maurel, C. Plant aquaporins: Novel functions and regulation properties. FEBS Letters 2007, 581, 2227–2236. [Google Scholar] [CrossRef]
- Danielson, J.Å.H.; Johanson, U. Unexpected complexity of the aquaporin gene family in the moss Physcomitrella patens. BMC Plant Biology 2008, 8, 1–15. [Google Scholar] [CrossRef]
- Lopez, D.; Bronner, G.; Brunel, N.; Auguin, D.; Bourgerie, S.; Brignolas, F.; et al. Insights into Populus XIP aquaporins: evolutionary expansion, protein functionality, and environmental regulation. Journal of Experimental Botany 2012, 65, 2217–2230. [Google Scholar] [CrossRef]
- Anderberg, H.I.; Kjellbom, P.; Johanson, U. Annotation of Selaginella moellendorffii major intrinsic proteins and the evolution of the protein family in terrestrial plants. Front. Plant Sciences 2012, 3, 1–14. [Google Scholar] [CrossRef]
- Khabudaev, K.V.; Petrova, D.P.; Grachev, M.A.; Likhoshway, Y.V. A new subfamily LIP of the major intrinsic proteins. BMC Genomics 2014, 15, 173. [Google Scholar] [CrossRef] [PubMed]
- Abascal, F.; Irisarri, I.; Zardoya, R. Diversity and evolution of membrane intrinsic proteins. Biochimica et Biophysica Acta 2014, 1840, 1468–1481. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.K.; Prabh, N.D.; Sankararamakrishnan, R. New subfamilies of major intrinsic proteins in fungi suggest novel transport properties in fungal channels: Implications for the host-fungal interactions. BMC Ecology and Evolution 2014, 14, 173. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Cooke, J.E.K.; Zwiazek, J.J. Phylogenetic analysis of fungal aquaporins provide insight into their possible role in water transport of mycorrhizal associations. Botany 2013, 91, 495–504. [Google Scholar] [CrossRef]
- Ben Amira, M.; Mom, R.; Lopez, D.; Chaar, H.; Khouaja, A.; Pujade-Renaud, V.; Fumanal, B.; et al. MIP diversity from Trichoderma: Structural considerations and transcriptional modulation during mycoparasitic association with Fusarium solani olive trees. PLoS ONE 2018, 13, e0193760. [Google Scholar] [CrossRef]
- Purvis, A.; Hector, A. Getting the measure of biodiversity. Nature 2000, 405, 212–219. [Google Scholar] [CrossRef]
- Hawksworth, DL.; Luecking, R. Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol Spectrum 2017, 5, FUNK-0052-2016. [Google Scholar] [CrossRef]
- James, T.Y.; Stajich, J.E.; Hittinger, C.T.; Rokas, A. Toward a Fully Resolved Fungal Tree of Life. Annual Review of Microbiology 2020, 74, 291–313. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Al-Ani, L.; Tedersoo, L.; Haelewaters, D.; Rajeshkumar, K.C.; et al. Outline of Fungi and fungus-like taxa. Mycosphere 2020, 11, 1060–1456. [Google Scholar] [CrossRef]
- Baldrian, P.; Větrovský, T.; Lepinay, C.; Kohout, P. High-throughput sequencing view on the magnitude of global fungal diversity. Fungal Diversity 2021, 114, 539–547. [Google Scholar] [CrossRef]
- Li, Y.; Steenwyk, J.L.; Chang, Y.; Wang, Y.; James, T.Y.; Stajich, J.E.; et al. A genome-scale phylogeny of the kingdom Fungi. Curr Biol. 2021, 31, 1653–1665. [Google Scholar] [CrossRef] [PubMed]
- Grigoriev, I.V.; Nikitin, R.; Haridas, S.; Kuo, A.; Ohm, R.; Otillar, R.; et al. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Research 2014, 42, 699–704. [Google Scholar] [CrossRef] [PubMed]
- Pettersson, N.; Filipsson, C.; Becit, E.; Brive, L.; Hohmann, S. Aquaporins in yeasts and filamentous fungi. Biology of the Cell 2005, 97, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Dietz, S.; Bülow, J.V.; Beitz, E.; Nehls, U. The aquaporin gene family of the ectomycorrhizal fungus Laccaria bicolor: Lessons for symbiotic functions. New Phytologist 2011, 190, 927–940. [Google Scholar] [CrossRef]
- Nehls, U.; Dietz, S. Fungal aquaporins: Cellular functions and ecophysiological perspectives. Applied Microbiology and Biotechnology 2014, 98, 8835–8851. [Google Scholar] [CrossRef]
- Xu, H.; Zwiazek, JJ. Fungal Aquaporins in Ectomycorrhizal Root Water Transport. Frontiers in Plant Science 2020, 11, 302. [Google Scholar] [CrossRef]
- Hansen, M.; Kun, J.F.; Schultz, J.E.; Beitz, E. A single, bi-functional aquaglyceroporin in blood-stage Plasmodium falciparum malaria parasites. Journal of Biological Chemistry 2002, 277, 4874–4882. [Google Scholar] [CrossRef]
- Pavlovic-Djuranovic, S.; Schultz, J.E.; Beitz, E. A single aquaporin gene encodes a water/glycerol/urea facilitator in Toxoplasma gondii with similarity to plant tonoplast intrinsic proteins. FEBS Letters 2003, 555, 500–504. [Google Scholar] [CrossRef]
- Laize, V.; Tacnet, F.; Ripoche, P.; Hohmann, S. Polymorphism of Saccharomyces cerevisiae aquaporins. Yeast 2000, 16, 897–903. [Google Scholar] [CrossRef]
- Sidoux-Walter, F.; Pettersson, N.; Hohmann, S. The Saccharomyces cerevisiae aquaporin Aqy1 is involved in sporulation. Proceedings of the National Academy of Sciences, USA 2004, 101, 17422–17427. [Google Scholar] [CrossRef]
- Pettersson, N.; Hagstrom, J.; Bill, RM.; Hohmann, S. Expression of heterologous aquaporins for functional analysis in Saccharomyces cerevisiae. Current Genetics 2006, 50, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Xu, H. Major Intrinsic Proteins of Laccaria Bicolor: Characterization, Transcript Profiling and Functions in Ectomycorrhizal Associations with Picea glauca. Ph.D. thesis, University of Alberta, Edmonton. 2015, pp 99.
- Navarro-Ródenas, A.; Ruíz-Lozano, J.M.; Kaldenhoff, R.; Morte, A. The aquaporin TcAQP1 of the desert truffle Terfezia claveryi is a membrane pore for water and CO2 transport. Molecular Plant-Microbe Interactions 2012, 25, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Ródenas, A.; Bárzana, G.; Nicolás, E.; Carra, A.; Schubert, A.; Morte, A. Expression analysis of aquaporins from desert truffle mycorrhizal symbiosis reveals a fine-tuned regulation under drought. Molecular Plant-Microbe Interactions 2013, 26, 1068–1078. [Google Scholar] [CrossRef] [PubMed]
- Hacquard, S.; Tisserant, E.; Brun, A.; Lege, V.; Martin, F.; Kohler, A. Laser microdissection and microarray analysis of Tuber melanosporum ectomycorrhizas reveal functional heterogeneity between mantle and Hartig net compartments. Environmental Microbiology 2013, 15, 1853–1869. [Google Scholar] [CrossRef]
- Li, T.; Hu, Y.J.; Hao, Z.P.; Li, H.; Wang, Y.S.; Chen, B.D. First cloning and characterization of two functional aquaporin genes from an arbuscular mycorrhizal fungus Glomus intraradices. New Phytologist 2013, 197, 617–630. [Google Scholar] [CrossRef]
- An, B.; Li, B.; Li, H.; Zhang, Z.; Qin, G.; Tian, S. Aquaporin-8 regulates cellular development and reactive oxygen species production, a critical component of virulence in Botrytis cinerea. New Phytol. 2016, 209, 1668–1680. [Google Scholar] [CrossRef]
- Peter, M.; Kohler, A.; Ohm, R.A.; Kuo, A.; Krützmann, J.; Morin, E.; et al. Ectomycorrhizal ecology is imprinted in the genome of the dominant symbiotic fungus Cenococcum geophilum. Naure Communication 2016, 7, 1–15. [Google Scholar] [CrossRef]
- Ben Amira, M.; Faize, M.; Karlsson, M.; Dubey, M.; Frąc, M.; Panek, J.; et al. Fungal X-Intrinsic Protein aquaporin from Trichoderma atroviride: structural and functional considerations. Biomolecules 2021, 11, 338. [Google Scholar] [CrossRef]
- Laothanachareon, T.; Asin-Garcia, E.; Volkers, R.J.M.; Tamayo-Ramos, J.A.; Martins dos Santos, V.A.P.; Schaap, P.J. Identification of Aspergillus niger Aquaporins Involved in Hydrogen Peroxide Signaling. Journal of Fungi 2023, 9, 499. [Google Scholar] [CrossRef]
- Gupta, A.B.; Sankararamakrishnan, R. Genome-wide analysis of major intrinsic proteins in the tree plant Populus trichocarpa: characterization of XIP subfamily of aquaporins from evolutionary perspective. BMC Plant Biology 2009, 9, 134. [Google Scholar] [CrossRef]
- Laur, J.; Hacke, U.G. Exploring Picea glauca aquaporins in the context of needle water uptake and xylem refilling. New Phytologist 2014, 203, 388–400. [Google Scholar] [CrossRef] [PubMed]
- Diehn, T.A.; Pommerrenig, B.; Bernhardt, N.; Hartmann, A.; Bienert, G.P. Genome-wide identification of aquaporin encoding genes in Brassica oleracea and their phylogenetic sequence comparison to Brassica crops and Arabidopsis. Front. Plant Sciences 2015, 6, 166. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, J.; Yu, J.W.; Gaston, D.; et al. Molecular evolution and functional divergence of X-intrinsic protein genes in plants. Mol. Genet. Genomics 2015, 290, 443–460. [Google Scholar] [CrossRef] [PubMed]
- Maurel, C.; Boursiac, Y.; Luu, D.T.; Santoni, V.; Shahzad, Z.; Verdoucq, L. Aquaporins in Plants. Physiological Reviews 2015, 95, 1321–1358. [Google Scholar] [CrossRef]
- Bienert, G.P.; Bienert, M.D.; Jahn, T.P.; Boutry, M.; Chaumont, F. Solanaceae XIPs are plasma membrane aquaporins that facilitate the transport of many uncharged substrates. Plant J. 2011, 66, 306–317. [Google Scholar] [CrossRef]
- Zhang, DY.; Ali, Z.; Wang, CB.; Xu, L.; Yi, JX.; Xu, ZL.; et al. Genome-wide sequence characterization and expression analysis of major intrinsic proteins in Soybean (Glycine max L.). PLoS ONE 2013, 8, e56312. [Google Scholar] [CrossRef]
- Reuscher, S.; Akiyama, M.; Mori, C.; Aoki, K.; Shibata, D.; Shiratake, K. Genome-wide identification and expression analysis of aquaporins in tomato. PLoS ONE 2013, 8, e79052R. [Google Scholar] [CrossRef]
- Venkatesh, J.; Yu, J.W.; Park, S.W. Genome-wide analysis and expression profiling of the Solanum tuberosum aquaporins. Plant Physiology and Biochemistry 2013, 73, 392–404. [Google Scholar] [CrossRef]
- Yue, C.; Cao, H.; Wang, L.; Zhou, Y.; Hao, X.; Zeng, J.; Wang, X.; Yang, Y. Molecular cloning and expression analysis of tea plant aquaporin (AQP) gene family. Plant Physiology and Biochemistry 2014, 83, 65–76R. [Google Scholar] [CrossRef]
- de Paula Santos Martins, C.; Pedrosa, A.M.; Du, D.; Gonçalves, L.P.; Yu, Q.; Gmitter, F.G., Jr.; et al. Genome-Wide Characterization and Expression Analysis of Major Intrinsic Proteins during Abiotic and Biotic Stresses in Sweet Orange (Citrus sinensis L. Osb.). PLoS ONE 2015, 10, e0138786. [Google Scholar]
- Berbee, M.L.; James, T.Y.; Strullu-Derrien, C. Early diverging fungi: diversity and impact at the dawn of terrestrial life. Annual Review of Microbiology 2017, 71, 41–60. [Google Scholar] [CrossRef] [PubMed]
- Voigt, K.; James, T.Y.; Kirk, P.M.; et al. Early-diverging fungal phyla: taxonomy, species concept, ecology, distribution, anthropogenic impact, and novel phylogenetic proposals. Fungal Diversity 2021, 109, 59–98. [Google Scholar] [CrossRef] [PubMed]
- Jaramillo, V.D.A.; Sukno, S.A.; Thon, M.R. Identification of horizontally transferred genes in the genus Colletotrichum reveals a steady tempo of bacterial to fungal gene transfer. BMC Genomics 2015, 16, 2. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Sun, S.; Ge, W.; Zhao, L.; Hou, B.; Wang, K.; et al. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 2020, 368, eaba5435. [Google Scholar] [CrossRef]
- Tiwari, P.; Bae, H. Horizontal Gene Transfer and Endophytes: An Implication for the Acquisition of Novel Traits. Plants 2020, 9, 305. [Google Scholar] [CrossRef]
- Liu, F.; Wang, S.; Cheewangkoon, R.; Zhao, R. Uneven distribution of prokaryote-derived horizontal gene transfer in fungi: a lifestyle-dependent phenomenon. mBio 2025, 16, e02855-24. [Google Scholar] [CrossRef]
- Chaumont, F.; Moshelion, M.; Daniels, M.J. Regulation of plant aquaporin activity. Biology of the Cell 2005, 97, 749–764. [Google Scholar] [CrossRef]
- Day, R.E.; Kitchen, P.; Owen, D.S.; Bland, C.; Marshall, L.; Conner, A.C.; Bill, R.M.; Conner, M.T. Human aquaporins: regulators of transcellular water flow. Biochimica et Biophysica Acta 2014, 1840, 1492–1506. [Google Scholar] [CrossRef]
- De Groot, B.L.; Grubmüller, H. Water permeation across biological membranes: Mechanism and dynamics of aquaporin-1 and GlpF. Science 2001, 294, 2353–2357. [Google Scholar] [CrossRef]
- Hanson, K.; et al. Fungi use efficient algorithms for the exploration of microfluidic networks. Small 2006, 2, 1212–1220. [Google Scholar] [CrossRef]
- Nagy, L.G.; Kovács, G.M.; Krizsán, K. Complex multicellularity in fungi: evolutionary convergence, single origin, or both? Biological Reviews 2018, 93, 1778–1794. [Google Scholar] [CrossRef] [PubMed]
- László, G. Nagy, Torda Varga, Árpád Csernetics, Máté Virágh. Fungi took a unique evolutionary route to multicellularity: Seven key challenges for fungal multicellular life. Fungal Biology Reviews 2020, 34, 151–169. [Google Scholar]
- Silberfeld, T.; Leigh, J.W.; Verbruggen, H.; Cruaud, C.; de Reviers, B.; Rousseau, F. A multi-locus time-calibrated phylogeny of the brown algae (Heterokonta, Ochrophyta, Phaeophyceae): Investigating the evolutionary nature of the “brown algal crown radiation”. Molecular Phylogenetics and Evolution 2010, 56, 659–674. [Google Scholar] [CrossRef]
- Lutzoni, F.; Nowak, M.D.; Alfaro, M.E.; Reeb, V.; Miadlikowska, J.; Krug, M.; et al. Contemporaneous radiations of fungi and plants linked to symbiosis. Nature Communication 2018, 9, 5451. [Google Scholar] [CrossRef] [PubMed]
- Irisarri, I.; Lorente-Martínez, H.; Strassert, J.F.H.; Zardoya, A.A.R.; San Mauro, D.; de Vries, J. Early Diversification of Membrane Intrinsic Proteins (MIPs) in Eukaryotes. Genome Biology and Evolution 2024, 16, evae164. [Google Scholar] [CrossRef]
- Burki, F.; Roger, A.J.; Brown, M.W.; Simpson, A.G.B. The new tree of eukaryotes. Trends in Ecology & Evolution 2020, 35, 43–55. [Google Scholar] [CrossRef]
- Strassert, J.F.H.; Irisarri, I.; Williams, T.A.; Burki, F. A molecular timescale for eukaryote evolution with implications for the origin of red algal- derived plastids. Nature Communication 2021, 12, 1879. [Google Scholar] [CrossRef]
- Song, J.; Mak, E.; Wu, B.; Beitz, E. Parasite aquaporins: current developments in drug facilitation and resistance. Biochimica et Biophysica Acta 2014, 1840, 1566–1573. [Google Scholar] [CrossRef]
- Hongsanan, S.; Maharachchikumbura, S.S.N.; Hyde, K.D.; Samarakoon, M.C.; et al. An updated phylogeny of Sordariomycetes based on phylogenetic and molecular clock evidence. Fungal Diversity 2017, 84, 25–41. [Google Scholar] [CrossRef]
- Altschul, S.; Madden, T.; Schaffer, A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D. Gapped BLASt and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 1997, 25, 3389–3401. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Sonnhammer, E.L.L.; von Heijne, G.; Krogh, A. A hidden Markov model for predicting transmembrane helices in protein sequences. In Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology, Montreal, QC, Canada, 28 June–1 July 1998; AAAI Press: Menlo Park, CA, USA, 1998; pp. 175–182. [Google Scholar]
- Hirokawa, T.; Boon Chieng, S.; Mitaku, S. SOSUI: Classification and secondary structure prediction system for membrane proteins. Bioinformatics 1998, 14, 378–379. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Berka, K.; Hanák, O.; Sehnal, D.; Banáš, P.; Navratilova, V.; Jaiswal, D.; et al. MOLE online 2.0: Interactive web-based analysis of biomacromolecular channels. Nucleic Acids Research 2012, 40, W222–W227. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Molecular Biology and Evolution 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.; Wong, T.; et al. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Research 2019, 47, W256–CW259. [Google Scholar] [CrossRef]
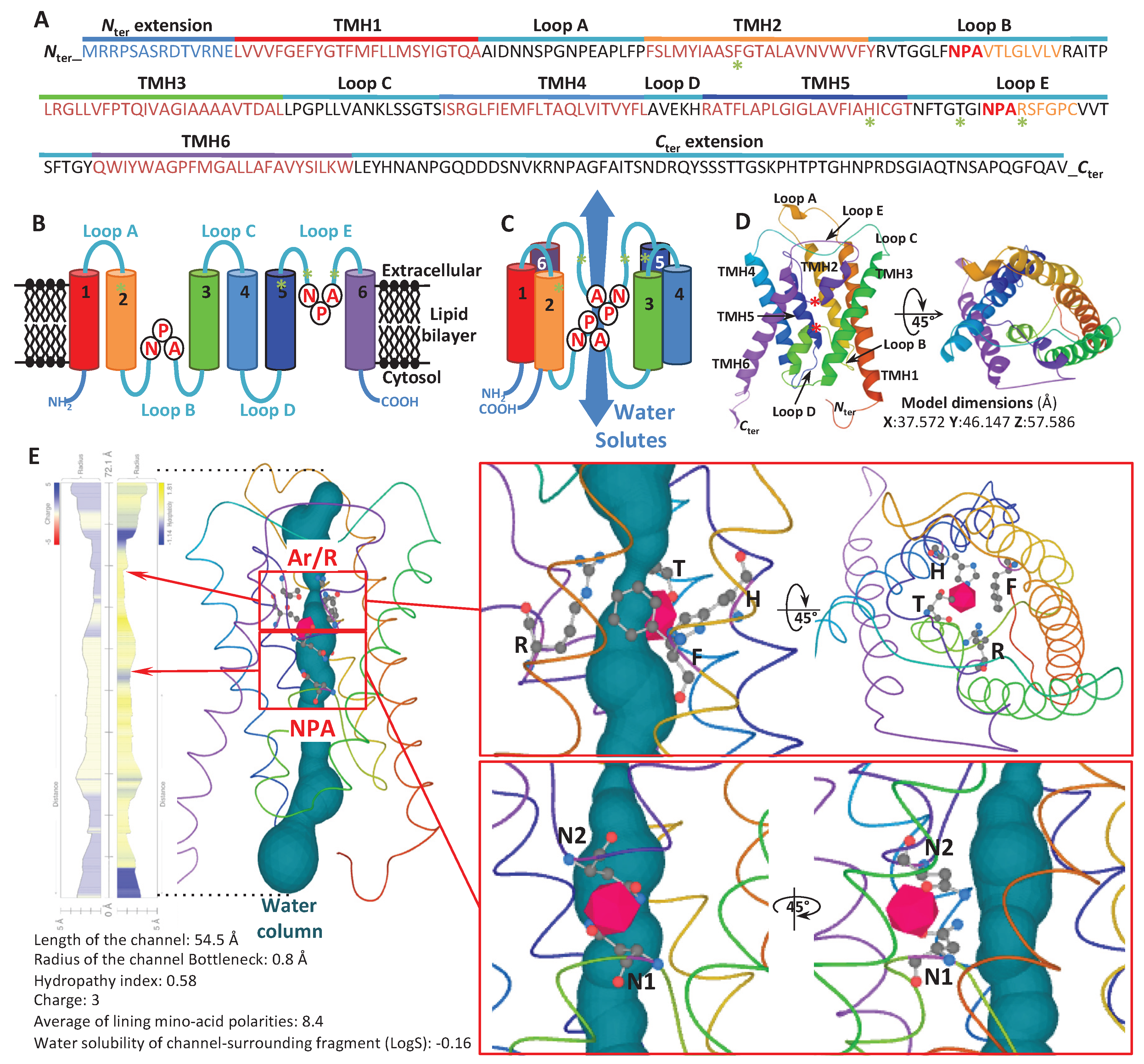
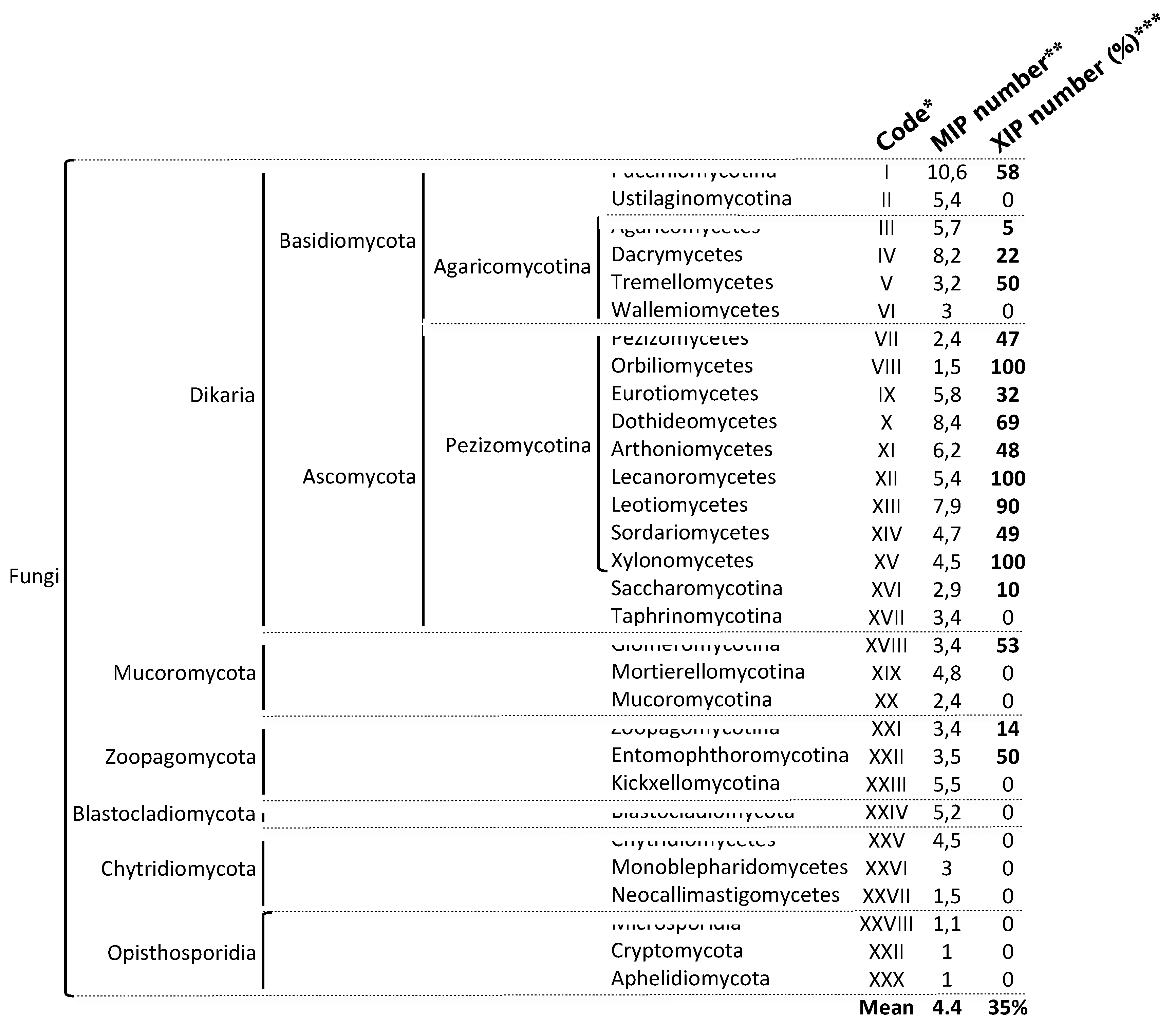
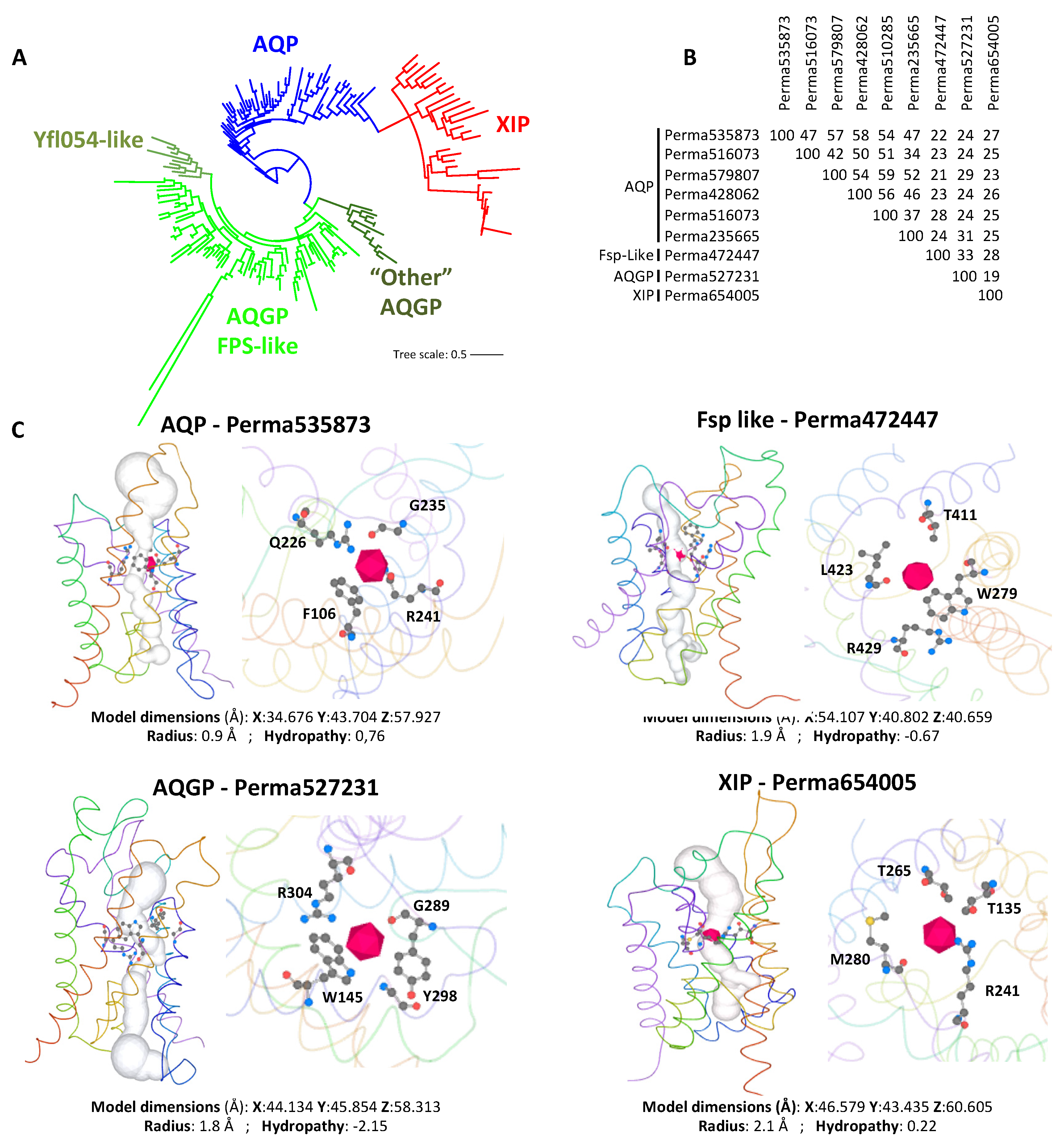
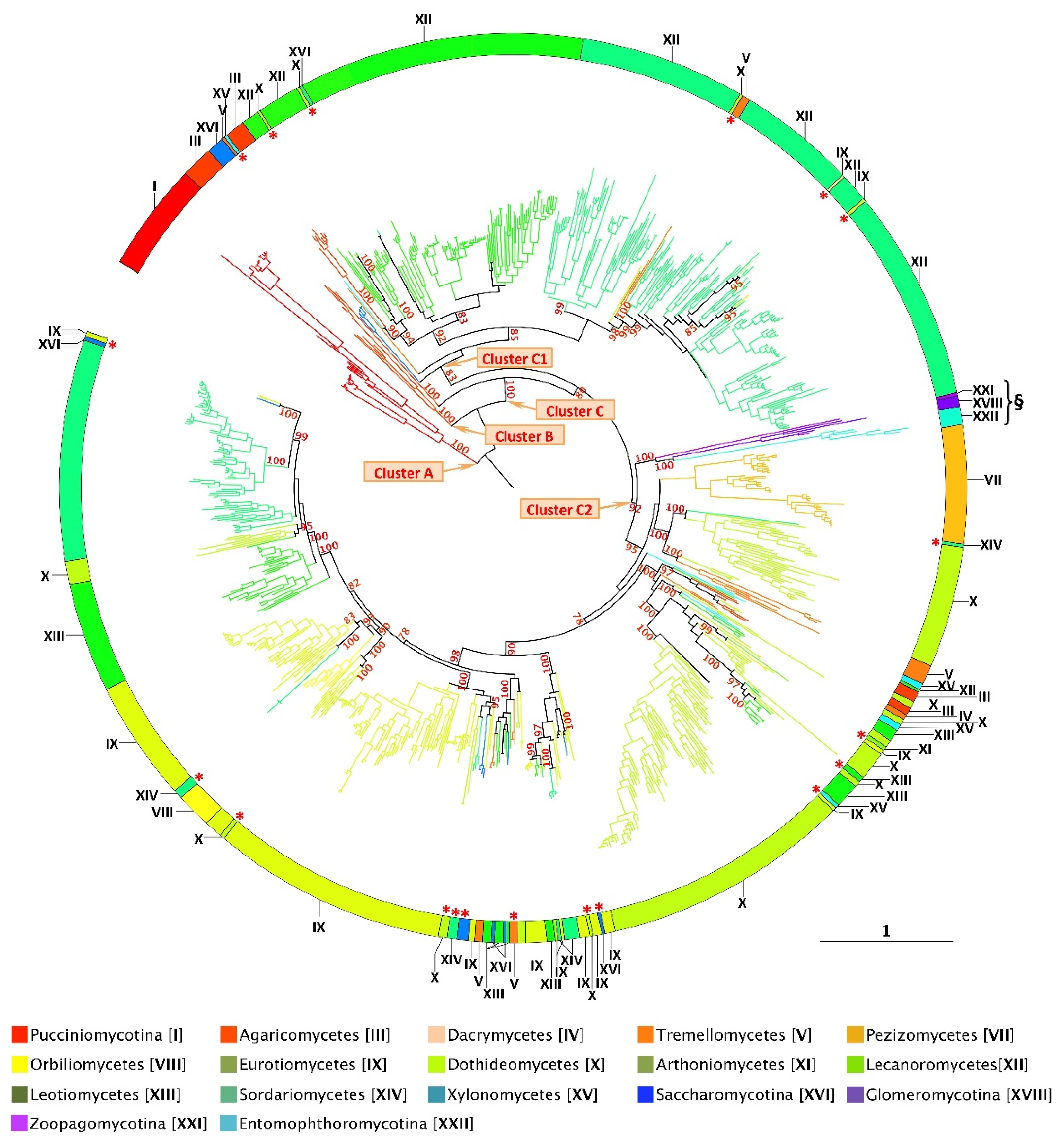
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





