Submitted:
04 August 2025
Posted:
05 August 2025
You are already at the latest version
Abstract
Keywords:
Significance
1. Introduction
1.1.
1.2.
1.3.
1.4.
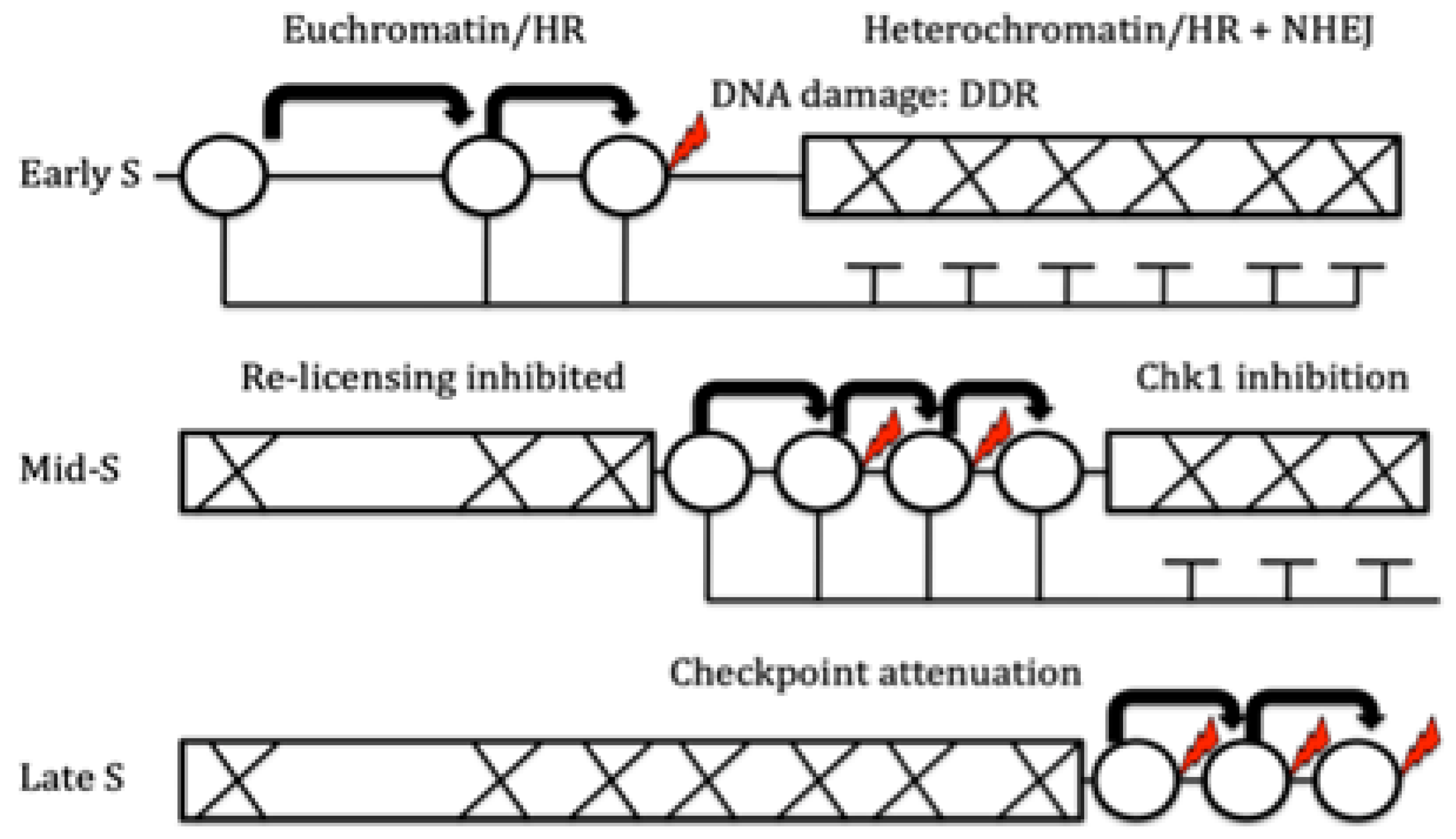
2.1. RT and Replication Origins
3.2. RT and the Regulation of Gene Transcription
3.3. RT and Protein Folding Versus Protein Function
4.1. RT and Genome Stability
4.2. RT and Introns as Adaptations to DNA Damage
4.3. RT and DNA Repair
5.1. RT and Genome Evolution
5.2. RT and the Correlation between dN and dS
6.1. RT and Transposable Elements
6.3. Genome Stability and Life History Traits
7. Conclusions
- 1)
- genome (in)stability drives genome evolution by either increasing or decreasing rates of karyotype evolution and rates of change in genome size and chromatin organization.
- 2)
- genome evolution significantly influences rates of speciation and therefore species richness; for example, by serving as a source of the standing genomic and allelic diversity on which ecological speciation can act.
References
- Bush, G. L. , Case, S. M., Wilson, A. C., & Patton, J. L. Rapid speciation and chromosomal evolution in mammals. Proceedings of the National Academy of Sciences 1977, 74, 3942–3946. [Google Scholar]
- Maxson, L. E. , & Wilson, A. C. Rates of molecular and chromosomal evolution in salamanders. C. Rates of molecular and chromosomal evolution in salamanders. Evolution 1979, 734–740. [Google Scholar]
- Levin, D. A. , & Wilson, A. C. Rates of evolution in seed plants: net increase in diversity of chromosome numbers and species numbers through time. Proceedings of the National Academy of sciences 1976, 73, 2086–2090. [Google Scholar] [CrossRef]
- Prachumwat, A. , & Li, W. H. Gene number expansion and contraction in vertebrate genomes with respect to invertebrate genomes. Genome research 2008, 18, 221–232. [Google Scholar] [CrossRef]
- Martinez, P. A. , Jacobina, U. P., Fernandes, R. V., Brito, C., Penone, C., Amado, T. F.,... & Bidau, C. J. A comparative study on karyotypic diversification rate in mammals. Heredity 2017, 118, 366–373. [Google Scholar]
- Griffin, D. K. , Kretschmer, R., Srikulnath, K., Singchat, W., O’Connor, R. E., & Romanov, M. N. Insights into avian molecular cytogenetics—with reptilian comparisons. Molecular cytogenetics 2024, 17, 24. [Google Scholar]
- Simakov, O. , Marlétaz, F., Yue, J. X., O’Connell, B., Jenkins, J., Brandt, A.,... & Rokhsar, D. S. Deeply conserved synteny resolves early events in vertebrate evolution. Nature ecology & evolution 2020, 4, 820–830. [Google Scholar]
- Martin, A. P. , & Palumbi, S. R. Body size, metabolic rate, generation time, and the molecular clock. Proceedings of the National Academy of Sciences 1993, 90, 4087–4091. [Google Scholar] [CrossRef]
- Sarich, V. M. , & Wilson, A. C. Immunological time scale for hominid evolution. Science 1967, 158, 1200–1203. [Google Scholar] [CrossRef]
- Bininda-Emonds, O. R. Fast genes and slow clades: comparative rates of molecular evolution in mammals. Evolutionary Bioinformatics 2007, 3, 117693430700300008. [Google Scholar] [CrossRef]
- Hedges, S. B. , Marin, J., Suleski, M., Paymer, M., & Kumar, S. Tree of life reveals clock-like speciation and diversification. Molecular biology and evolution 2015, 32, 835–845. [Google Scholar]
- Lancaster, L. T. Molecular evolutionary rates predict both extinction and speciation in temperate angiosperm lineages. BMC evolutionary biology 2010, 10, 1. [Google Scholar] [CrossRef]
- Goldie, X. , Lanfear, R., & Bromham, L. Diversification and the rate of molecular evolution: no evidence of a link in mammals. BMC evolutionary biology 2011, 11, 1–12. [Google Scholar]
- McPeek, M. A. , & Brown, J. M. Clade age and not diversification rate explains species richness among animal taxa. The American Naturalist 2007, 169, E97–E106. [Google Scholar] [CrossRef] [PubMed]
- Rundle, H. D. , & Nosil, P. Ecological speciation. Ecology letters 2005, 8, 336–352. [Google Scholar]
- Futuyma, D. The origin of species by means of ecological selection. Current Biology 2013, 23, R217–R219. [Google Scholar] [CrossRef]
- Jablonski, D. , & Edie, S. M. Mass extinctions and their rebounds: a macroevolutionary framework. Paleobiology 2025, 1–14. [Google Scholar]
- Scholl, J. P. , & Wiens, J. J. Diversification rates and species richness across the Tree of Life. Proceedings of the Royal Society B: Biological Sciences 2016, 283, 20161334. [Google Scholar] [CrossRef]
- DeMalach, N. , Ke, P. J., & Fukami, T. The effects of ecological selection on species diversity and trait distribution: predictions and an empirical test. Ecology 2022, 103, e03567. [Google Scholar] [CrossRef]
- Wellborn, G. A. , & Langerhans, R. B. Ecological opportunity and the adaptive diversification of lineages. Ecology and evolution 2015, 5, 176–195. [Google Scholar] [CrossRef]
- Hillebrand, H. On the generality of the latitudinal diversity gradient. The American Naturalist 2004, 163, 192–211. [Google Scholar] [CrossRef]
- Willig, M. R. , Kaufman, D. M., & Stevens, R. D. Latitudinal gradients of biodiversity: pattern, process, scale, and synthesis. Annual review of ecology, evolution, and systematics 2003, 34, 273–309. [Google Scholar]
- Schemske, D. W. , & Mittelbach, G. G. “Latitudinal gradients in species diversity”: reflections on Pianka’s 1966 article and a look forward. The American Naturalist 2017, 189, 599–603. [Google Scholar] [CrossRef]
- Bureš, P. , Elliott, T. L., Veselý, P., Šmarda, P., Forest, F., Leitch, I. J.,... & Zedek, F. The global distribution of angiosperm genome size is shaped by climate. New Phytologist 2024, 242, 744–759. [Google Scholar] [CrossRef] [PubMed]
- Pianka, E. R. Latitudinal gradients in species diversity: a review of concepts. The American Naturalist 1966, 100, 33–46. [Google Scholar] [CrossRef]
- Yu, D. , & Wiens, J. J. The causes of species richness patterns among clades. Proceedings of the Royal Society B 2024, 291, 20232436. [Google Scholar] [CrossRef]
- Wiens, J. J. Trait-based species richness: Ecology and macroevolution. Biological Reviews 2023, 98, 1365–1387. [Google Scholar] [CrossRef] [PubMed]
- Eiserhardt, W. L. , Hansen, L. E. S., Couvreur, T. L., Dransfield, J., Ferreira, P. D. L., Rakotoarinivo, M.,... & Baker, W. J. Explaining extreme differences in species richness among co-occurring palm clades in Madagascar. Evolutionary Journal of the Linnean Society 2024, 3, kzae026. [Google Scholar]
- Tietje, M. , Antonelli, A., Baker, W. J., Govaerts, R., Smith, S. A., & Eiserhardt, W. L. Global variation in diversification rate and species richness are unlinked in plants. Proceedings of the National Academy of Sciences 2022, 119, e2120662119. [Google Scholar]
- Castro-Insua, Adrián, Carola Gómez-Rodríguez, John J. Wiens, and Andrés Baselga. “Climatic niche divergence drives patterns of diversification and richness among mammal families.” Scientific Reports 8, no. 1 (2018): 8781.
- Title, P. O. , & Burns, K. J. Rates of climatic niche evolution are correlated with species richness in a large and ecologically diverse radiation of songbirds. Ecology Letters 2015, 18, 433–440. [Google Scholar]
- Smyčka, J. , Toszogyova, A., & Storch, D. The relationship between geographic range size and rates of species diversification. Nature Communications 2023, 14, 5559. [Google Scholar]
- Kozak, K. H. , & Wiens, J. J. Accelerated rates of climatic-niche evolution underlie rapid species diversification. Ecology letters 2010, 13, 1378–1389. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H. , Peterson, A. T., Myers, C. E., Yang, Q., & Saupe, E. E. Ecological niche conservatism spurs diversification in response to climate change. Nature Ecology & Evolution 2024, 8, 729–738. [Google Scholar]
- Kozak, K. H. , & Wiens, J. J. Climatic zonation drives latitudinal variation in speciation mechanisms. Proceedings of the Royal Society B: Biological Sciences 2007, 274, 2995–3003. [Google Scholar] [CrossRef]
- Kapusta, A. , Suh, A., & Feschotte, C. Dynamics of genome size evolution in birds and mammals. Proceedings of the National Academy of Sciences 2017, 114, E1460–E1469. [Google Scholar] [CrossRef]
- Elliott, T. A. , & Gregory, T. R. What’s in a genome? The C-value enigma and the evolution of eukaryotic genome content. Philosophical Transactions of the Royal Society B: Biological Sciences 2015, 370, 20140331. [Google Scholar] [CrossRef]
- Decena-Segarra, L. P. , Bizjak-Mali, L., Kladnik, A., Sessions, S. K., & Rovito, S. M. Miniaturization, genome size, and biological size in a diverse clade of salamanders. The American Naturalist 2020, 196, 634–648. [Google Scholar] [CrossRef]
- Martin, C. C. , & Gordon, R. Differentiation trees, a junk DNA molecular clock, and the evolution of neoteny in salamanders. Journal of Evolutionary Biology 1995, 8, 339–354. [Google Scholar]
- Nuzhdin, S. V. , & Mackay, T. The genomic rate of transposable element movement in Drosophila melanogaster. Molecular Biology and Evolution 1995, 12, 180–181. [Google Scholar] [CrossRef]
- Oliveira, T. D. D. , & Freitas, T. R. D. Investigating the evolutionary dynamics of diploid number variation in Ctenomys (Ctenomyidae, Rodentia). Genetics and Molecular Biology 2023, 46 Suppl 1, e20230180. [Google Scholar] [CrossRef]
- Ferguson-Smith, M. A. , & Trifonov, V. Mammalian karyotype evolution. Nature Reviews Genetics 2007, 8, 950–962. [Google Scholar] [CrossRef] [PubMed]
- Escudero, M. , Arroyo, J. M., Sánchez-Ramírez, S., & Jordano, P. Founder events and subsequent genetic bottlenecks underlie karyotype evolution in the Ibero-North African endemic Carex helodes. Annals of Botany 2024, 133, 871–882. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. The neutral theory of molecular evolution: a review of recent evidence. The Japanese Journal of Genetics 1991, 66, 367–386. [Google Scholar] [CrossRef]
- Saini, A. , Usmanova, D. R., & Vitkup, D. Integrated model of the protein molecular clock across mammalian species. bioRxiv 2025. [Google Scholar] [CrossRef]
- Wu, C. S. , & Chaw, S. M. Large-scale comparative analysis reveals the mechanisms driving plastomic compaction, reduction, and inversions in conifers II (Cupressophytes). Genome biology and evolution 2016, 8, 3740–3750. [Google Scholar] [CrossRef]
- Marais, G. , Domazet-Lošo, T., Tautz, D., & Charlesworth, B. Correlated evolution of synonymous and nonsynonymous sites in Drosophila. Journal of molecular evolution 2004, 59, 771–779. [Google Scholar] [CrossRef]
- Wyckoff, G. J. , Malcom, C. M., Vallender, E. J., & Lahn, B. T. A highly unexpected strong correlation between fixation probability of nonsynonymous mutations and mutation rate. Trends in Genetics 2005, 21, 381–385. [Google Scholar] [CrossRef]
- Stoletzki, N. , & Eyre-Walker, A. The positive correlation between d N/d S and d S in mammals is due to runs of adjacent substitutions. Molecular biology and evolution 2011, 28, 1371–1380. [Google Scholar] [CrossRef]
- Guisinger, M. M. , Kuehl, J. V., Boore, J. L., & Jansen, R. K. Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions. Proceedings of the National Academy of Sciences 2008, 105, 18424–18429. [Google Scholar] [CrossRef]
- Garamszegi, L. Z. , de Groot, N. G., & Bontrop, R. E. Correlated evolution of nucleotide substitution rates and allelic variation in Mhc-DRB lineages of primates. BMC Evolutionary Biology 2009, 9, 1–18. [Google Scholar] [CrossRef]
- Comeron, J. M. , & Kreitman, M. The correlation between synonymous and nonsynonymous substitutions in Drosophila: mutation, selection or relaxed constraints? Genetics 1998, 150, 767–775. [Google Scholar] [CrossRef] [PubMed]
- Chuang, J. H. , & Li, H. Functional bias and spatial organization of genes in mutational hot and cold regions in the human genome. PLoS biology 2004, 2, e29. [Google Scholar] [CrossRef]
- Cummins, M. , Watson, C., Edwards, R. J., & Mattick, J. S. The evolution of ultraconserved elements in vertebrates. Molecular biology and evolution 2024, 41, msae146. [Google Scholar]
- Makałowski, W. , & Boguski, M. S. Evolutionary parameters of the transcribed mammalian genome: an analysis of 2,820 orthologous rodent and human sequences. Proceedings of the National Academy of Sciences 1998, 95, 9407–9412. [Google Scholar] [CrossRef]
- Hodgkinson, A. , & Eyre-Walker, A. Variation in the mutation rate across mammalian genomes. Nature reviews genetics 2011, 12, 756–766. [Google Scholar]
- Agier, N. , & Fischer, G. The mutational profile of the yeast genome is shaped by replication. Molecular biology and evolution 2012, 29, 905–913. [Google Scholar] [CrossRef]
- Chen, C. L. , Rappailles, A., Duquenne, L., Huvet, M., Guilbaud, G., Farinelli, L.,... & Thermes, C. Impact of replication timing on non-CpG and CpG substitution rates in mammalian genomes. Genome research 2010, 20, 447–457. [Google Scholar] [CrossRef]
- Weber, C. C. , Pink, C. J., & Hurst, L. D. Late-replicating domains have higher divergence and diversity in Drosophila melanogaster. Molecular biology and evolution 2012, 29, 873–882. [Google Scholar] [CrossRef]
- Staunton, P. M. , Peters, A. J., & Seoighe, C. Somatic mutations inferred from RNA-seq data highlight the contribution of replication timing to mutation rate variation in a model plant. Genetics 2023, 225, iyad128. [Google Scholar] [CrossRef]
- Stamatoyannopoulos, J. A. , Adzhubei, I., Thurman, R. E., Kryukov, G. V., Mirkin, S. M., & Sunyaev, S. R. Human mutation rate associated with DNA replication timing. Nature genetics 2009, 41, 393–395. [Google Scholar]
- Gaboriaud, J. , & Wu, P. Y. J. Insights into the link between the organization of DNA replication and the mutational landscape. Genes 2019, 10, 252. [Google Scholar]
- Rhind, N. DNA replication timing: Biochemical mechanisms and biological significance. BioEssays 2022, 44, 2200097. [Google Scholar] [CrossRef] [PubMed]
- Boos, D. , & Ferreira, P. Origin firing regulations to control genome replication timing. Genes 2019, 10, 199. [Google Scholar] [PubMed]
- Bechhoefer, J. , & Rhind, N. Replication timing and its emergence from stochastic processes. Trends in Genetics 2012, 28, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Ge, X. Q. , & Blow, J. J. Chk1 inhibits replication factory activation but allows dormant origin firing in existing factories. Journal of Cell biology 2010, 191, 1285–1297. [Google Scholar]
- Goldar, A. , Marsolier-Kergoat, M. C., & Hyrien, O. Universal temporal profile of replication origin activation in eukaryotes. PLoS One 2009, 4, e5899. [Google Scholar]
- Yang, S. C. H. , Rhind, N. , & Bechhoefer, J. Modeling genome-wide replication kinetics reveals a mechanism for regulation of replication timing. Molecular systems biology 2010, 6, 404. [Google Scholar] [CrossRef]
- Yoshida, K. , Bacal, J., Desmarais, D., Padioleau, I., Tsaponina, O., Chabes, A.,... & Pasero, P. The histone deacetylases sir2 and rpd3 act on ribosomal DNA to control the replication program in budding yeast. Molecular cell 2014, 54, 691–697. [Google Scholar] [CrossRef]
- Richards, L. , Das, S., & Nordman, J. T. Rif1-dependent control of replication timing. Genes 2022, 13, 550. [Google Scholar] [CrossRef]
- Murat, P. , Perez, C., Crisp, A., van Eijk, P., Reed, S. H., Guilbaud, G., & Sale, J. E. DNA replication initiation shapes the mutational landscape and expression of the human genome. Science Advances 2022, 8, eadd3686. [Google Scholar] [CrossRef]
- Drummond, D. A. , Bloom, J. D., Adami, C., Wilke, C. O., & Arnold, F. H. Why highly expressed proteins evolve slowly. Proceedings of the National Academy of Sciences 2005, 102, 14338–14343. [Google Scholar] [CrossRef]
- Rhind, N. DNA replication timing: random thoughts about origin firing. Nature cell biology 2006, 8, 1313–1316. [Google Scholar] [CrossRef] [PubMed]
- Dimitrova, D. S. , & Gilbert, D. M. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Molecular cell 1999, 4, 983–993. [Google Scholar] [CrossRef] [PubMed]
- Sima, J. , & Gilbert, D. M. Complex correlations: replication timing and mutational landscapes during cancer and genome evolution. Current opinion in genetics & development 2014, 25, 93–100. [Google Scholar] [CrossRef]
- Rhind, N. , & Gilbert, D. M. DNA replication timing. Cold Spring Harbor perspectives in biology 2013, 5, a010132. [Google Scholar] [CrossRef]
- Tian, M. , Wang, Z., Su, Z., Shibata, E., Shibata, Y., Dutta, A., & Zang, C. Integrative analysis of DNA replication origins and ORC-/MCM-binding sites in human cells reveals a lack of overlap. Elife 2024, 12, RP89548. [Google Scholar] [CrossRef]
- Gilbert, D. M. Replication timing and transcriptional control: beyond cause and effect. Current opinion in cell biology 2002, 14, 377–383. [Google Scholar] [CrossRef]
- Sequeira-Mendes, J. , Díaz-Uriarte, R., Apedaile, A., Huntley, D., Brockdorff, N., & Gómez, M. Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS genetics 2009, 5, e1000446. [Google Scholar] [CrossRef]
- Müller, C. A. , & Nieduszynski, C. A. DNA replication timing influences gene expression level. Journal of Cell Biology 2017, 216, 1907–1914. [Google Scholar] [CrossRef]
- Das, S. P. , Borrman, T., Liu, V. W., Yang, S. C. H., Bechhoefer, J., & Rhind, N. Replication timing is regulated by the number of MCMs loaded at origins. Genome Research 2015, 25, 1886–1892. [Google Scholar] [CrossRef]
- Liu, Y. , Ai, C., Gan, T., Wu, J., Jiang, Y., Liu, X.,... & Hu, J. Transcription shapes DNA replication initiation to preserve genome integrity. Genome biology 2021, 22, 176. [Google Scholar] [CrossRef]
- Santos, M. M. , Johnson, M. C., Fiedler, L., & Zegerman, P. Global early replication disrupts gene expression and chromatin conformation in a single cell cycle. Genome Biology 2022, 23, 217. [Google Scholar]
- Smith, J. J. , Putta, S., Zhu, W., Pao, G. M., Verma, I. M., Hunter, T.,... & Voss, S. R. Genic regions of a large salamander genome contain long introns and novel genes. BMC genomics 2009, 10, 1–11. [Google Scholar]
- Farlow, A. , Meduri, E., & Schlötterer, C. DNA double-strand break repair and the evolution of intron density. Trends in Genetics 2011, 27, 1–6. [Google Scholar] [CrossRef]
- Swinburne, I. A. , & Silver, P. A. Intron delays and transcriptional timing during development. Developmental cell 2008, 14, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Heyn, P. , Kalinka, A. T., Tomancak, P., & Neugebauer, K. M. Introns and gene expression: cellular constraints, transcriptional regulation, and evolutionary consequences. Bioessays 2015, 37, 148–154. [Google Scholar] [CrossRef]
- Buffenstein R, Lewis KN, Gibney PA, Narayan V, Grimes KM, Smith M, Lin TD, Brown-Borg HM. Probing pedomorphy and prolonged lifespan in naked mole-rats and dwarf mice. Physiology 2020, 35:96-111. [CrossRef]
- De Groef, B. , Grommen, S. V., & Darras, V. M. Forever young: endocrinology of paedomorphosis in the Mexican axolotl (Ambystoma mexicanum). General and comparative endocrinology 2018, 266, 194–201. [Google Scholar] [CrossRef]
- Versteeg, R. , van Schaik, B. D., van Batenburg, M. F., Roos, M., Monajemi, R., Caron, H.,... & van Kampen, A. H. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome research 2003, 13, 1998–2004. [Google Scholar] [CrossRef]
- Castillo-Davis, C. I. , Mekhedov, S. L., Hartl, D. L., Koonin, E. V., & Kondrashov, F. A. Selection for short introns in highly expressed genes. Nature genetics 2002, 31, 415–418. [Google Scholar] [CrossRef]
- Francis, W. R. , & Wörheide, G. Similar ratios of introns to intergenic sequence across animal genomes. Genome biology and evolution 2017, 9, 1582–1598. [Google Scholar] [CrossRef]
- Haddrill, P. R. , Charlesworth, B., Halligan, D. L., & Andolfatto, P. Patterns of intron sequence evolution in Drosophila are dependent upon length and GC content. Genome biology 2005, 6, 1–8. [Google Scholar] [CrossRef]
- Agier, N. , Delmas, S., Zhang, Q., Fleiss, A., Jaszczyszyn, Y., Van Dijk, E.,... & Fischer, G. The evolution of the temporal program of genome replication. Nature communications 2018, 9, 2199. [Google Scholar] [PubMed]
- de Moura, A. , & Karschau, J. Mathematical model for the distribution of DNA replication origins. Physical Review E 2024, 110, 034408. [Google Scholar] [CrossRef]
- Bracci, Alexa N., Anissa Dallmann, Qiliang Ding, Melissa J. Hubisz, Madison Caballero, and Amnon Koren. “The evolution of the human DNA replication timing program.” Proceedings of the National Academy of Sciences 120, no. 10 (2023): e2213896120. [CrossRef]
- Yang, J. R. , Liao, B. Y., Zhuang, S. M., & Zhang, J. Protein misinteraction avoidance causes highly expressed proteins to evolve slowly. Proceedings of the National Academy of Sciences 2012, 109, E831–E840. [Google Scholar] [CrossRef]
- Drummond, D. A. , & Wilke, C. O. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 2008, 134, 341–352. [Google Scholar] [CrossRef]
- Syljuåsen, R. G. , Sørensen, C. S., Hansen, L. T., Fugger, K., Lundin, C., Johansson, F.,... & Bartek, J. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Molecular and cellular biology 2005. [Google Scholar] [CrossRef]
- Petermann, E. , Woodcock, M., & Helleday, T. Chk1 promotes replication fork progression by controlling replication initiation. Proceedings of the National Academy of Sciences 2010, 107, 16090–16095. [Google Scholar] [CrossRef]
- Maya-Mendoza, A. , Petermann, E., Gillespie, D. A., Caldecott, K. W., & Jackson, D. A. Chk1 regulates the density of active replication origins during the vertebrate S phase. The EMBO journal 2007, 26, 2719–2731. [Google Scholar] [CrossRef]
- Ge, X. Q. , Jackson, D. A., & Blow, J. J. Dormant origins licensed by excess Mcm2–7 are required for human cells to survive replicative stress. Genes & development 2007, 21, 3331–3341. [Google Scholar] [CrossRef]
- Alver, R. C. , Chadha, G. S., & Blow, J. J. The contribution of dormant origins to genome stability: from cell biology to human genetics. DNA repair 2014, 19, 182–189. [Google Scholar] [CrossRef]
- Poli, J. , Tsaponina, O., Crabbé, L., Keszthelyi, A., Pantesco, V., Chabes, A.,... & Pasero, P. dNTP pools determine fork progression and origin usage under replication stress. The EMBO journal 2012, 31, 883–894. [Google Scholar] [CrossRef] [PubMed]
- Hand, R. Regulation of DNA replication on subchromosomal units of mammalian cells. The Journal of cell biology 1975, 64, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Conti, C. , Sacca, B., Herrick, J., Lalou, C., Pommier, Y., & Bensimon, A. Replication fork velocities at adjacent replication origins are coordinately modified during DNA replication in human cells. Molecular biology of the cell 2007, 18, 3059–3067. [Google Scholar] [CrossRef]
- Ma, E. , Hyrien, O., & Goldar, A. Do replication forks control late origin firing in Saccharomyces cerevisiae? Nucleic acids research 2012, 40, 2010–2019. [Google Scholar] [CrossRef]
- Carmel, L. , Rogozin, I. B., Wolf, Y. I., & Koonin, E. V. Evolutionarily conserved genes preferentially accumulate introns. Genome Research 2007, 17, 1045–1050. [Google Scholar] [CrossRef]
- Keane, P. A. , & Seoighe, C. Intron length coevolution across mammalian genomes. Molecular Biology and Evolution 2016, 33, 2682–2691. [Google Scholar] [CrossRef]
- Blommaert, J. Genome size evolution: towards new model systems for old questions. Proceedings of the Royal Society B 2020, 287, 20201441. [Google Scholar] [CrossRef]
- Sun, C. , López Arriaza, J. R., & Mueller, R. L. Slow DNA loss in the gigantic genomes of salamanders. Genome biology and evolution 2012, 4, 1340–1348. [Google Scholar] [CrossRef]
- Macheret, M. , & Halazonetis, T. D. Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress. Nature 2018, 555, 112–116. [Google Scholar]
- Bonnet, A. , Grosso, A. R., Elkaoutari, A., Coleno, E., Presle, A., Sridhara, S. C.,... & Palancade, B. Introns protect eukaryotic genomes from transcription-associated genetic instability. Molecular cell 2017, 67, 608–621. [Google Scholar] [CrossRef]
- Ge, X. Q. , Han, J., Cheng, E. C., Yamaguchi, S., Shima, N., Thomas, J. L., & Lin, H. Embryonic stem cells license a high level of dormant origins to protect the genome against replication stress. Stem Cell Reports 2015, 5, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Woodward, A. M. , Göhler, T., Luciani, M. G., Oehlmann, M., Ge, X., Gartner, A.,... & Blow, J. J. Excess Mcm2–7 license dormant origins of replication that can be used under conditions of replicative stress. The Journal of cell biology 2006, 173, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Sun, C. , Shepard, D. B., Chong, R. A., López Arriaza, J., Hall, K., Castoe, T. A.,... & Mueller, R. L. LTR retrotransposons contribute to genomic gigantism in plethodontid salamanders. Genome biology and evolution 2012, 4, 168–183. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z. , Bozzella, M., Seluanov, A., & Gorbunova, V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA repair 2008, 7, 1765–1771. [Google Scholar] [CrossRef] [PubMed]
- Shrivastav, M. , De Haro, L. P., & Nickoloff, J. A. Regulation of DNA double-strand break repair pathway choice. Cell research 2008, 18, 134–147. [Google Scholar]
- Brandsma, I. , & van Gent, D. C. Pathway choice in DNA double strand break repair: observations of a balancing act. Genome integrity 2012, 3, 1–10. [Google Scholar] [CrossRef]
- Ben Yamin, B. , Ahmed-Seghir, S., Tomida, J., Despras, E., Pouvelle, C., Yurchenko, A.,... & Kannouche, P. L. DNA polymerase zeta contributes to heterochromatin replication to prevent genome instability. The EMBO journal 2021, 40, e104543. [Google Scholar] [CrossRef]
- Guirouilh-Barbat, J. , Huck, S., Bertrand, P., Pirzio, L., Desmaze, C., Sabatier, L., & Lopez, B. S. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Molecular cell 2004, 14, 611–623. [Google Scholar] [CrossRef]
- Scully, R. , Panday, A., Elango, R., & Willis, N. A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nature reviews Molecular cell biology 2019, 20, 698–714. [Google Scholar] [CrossRef]
- Takahashi, S. , Miura, H., Shibata, T., Nagao, K., Okumura, K., Ogata, M.,... & Hiratani, I. Genome-wide stability of the DNA replication program in single mammalian cells. Nature genetics 2019, 51, 529–540. [Google Scholar]
- Kelly, T. , & Callegari, A. J. Dynamics of DNA replication in a eukaryotic cell. Proceedings of the National Academy of Sciences 2019, 116, 4973–4982. [Google Scholar] [CrossRef]
- Zhang, Q. , Bassetti, F., Gherardi, M., & Lagomarsino, M. C. Cell-to-cell variability and robustness in S-phase duration from genome replication kinetics. Nucleic acids research 2017, 45, 8190–8198. [Google Scholar] [CrossRef] [PubMed]
- Nakatani, T. , Schauer, T., Altamirano-Pacheco, L., Klein, K. N., Ettinger, A., Pal, M.,... & Torres-Padilla, M. E. Emergence of replication timing during early mammalian development. Nature 2024, 625, 401–409. [Google Scholar] [PubMed]
- Massip, F. , Laurent, M., Brossas, C., Fernández-Justel, J. M., Gómez, M., Prioleau, M. N.,... & Picard, F. Evolution of replication origins in vertebrate genomes: rapid turnover despite selective constraints. Nucleic acids research 2019, 47, 5114–5125.doi. [Google Scholar] [PubMed]
- Ryba, T. , Hiratani, I., Lu, J., Itoh, M., Kulik, M., Zhang, J.,... & Gilbert, D. M. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome research 2010, 20, 761–770. [Google Scholar] [CrossRef]
- Yaffe, E. , Farkash-Amar, S., Polten, A., Yakhini, Z., Tanay, A., & Simon, I. Comparative analysis of DNA replication timing reveals conserved large-scale chromosomal architecture. PLoS genetics 2010, 6, e1001011. [Google Scholar] [CrossRef]
- Goldberg, M. E. , & Harris, K. Mutational signatures of replication timing and epigenetic modification persist through the global divergence of mutation spectra across the great ape phylogeny. Genome Biology and Evolution 2022, 14, evab104. [Google Scholar] [CrossRef]
- Lu, B. Evolutionary insights into the relationship of frogs, salamanders, and caecilians ad their adaptive traits, with an emphasis on salamander regeneration and longevity. Animals 2023, 13, 3449. [Google Scholar] [CrossRef]
- Rios-Carlos, H. , Segovia-Ramírez, M. G., Fujita, M. K., & Rovito, S. M. Genomic gigantism is not associated with reduced selection efficiency in neotropical salamanders. Journal of Molecular Evolution 2024, 92, 371–380. [Google Scholar] [CrossRef]
- Roddy, A. B. , Alvarez-Ponce, D., & Roy, S. W. Mammals with small populations do not exhibit larger genomes. Molecular Biology and Evolution 2021, 38, 3737–3741. [Google Scholar] [CrossRef]
- Mohlhenrich, E. R. , & Mueller, R. L. Genetic drift and mutational hazard in the evolution of salamander genomic gigantism. Evolution 2016, 70, 2865–2878. [Google Scholar]
- Sessions, S. K. Evolutionary cytogenetics in salamanders. Chromosome Research 2008, 16, 183–201. [Google Scholar] [CrossRef] [PubMed]
- Liedtke, H. C. , Gower, D. J., Wilkinson, M., & Gomez-Mestre, I. Macroevolutionary shift in the size of amphibian genomes and the role of life history and climate. Nature Ecology & Evolution 2018, 2, 1792–1799. [Google Scholar]
- Venditti, C. , & Pagel, M. Speciation as an active force in promoting genetic evolution. Trends in Ecology & Evolution 2010, 25, 14–20. [Google Scholar] [CrossRef]
- Lefébure, Tristan, Claire Morvan, Florian Malard, Clémentine François, Lara Konecny-Dupré, Laurent Guéguen, Michèle Weiss-Gayet et al. “Less effective selection leads to larger genomes.” Genome Research 27, no. 6 (2017): 1016-1028. [CrossRef]
- Fuselli, S. , Greco, S., Biello, R., Palmitessa, S., Lago, M., Meneghetti, C.,... & Bertorelle, G. Relaxation of natural selection in the evolution of the giant lungfish genomes. Molecular biology and evolution 2023, 40, msad193. [Google Scholar]
- Lynch, M. Evolution of the mutation rate. TRENDS in Genetics 2010, 26, 345–352. [Google Scholar] [CrossRef]
- Lam, D. K. , Frantz, A. C., Burke, T., Geffen, E., & Sin, S. Y. W. Both selection and drift drive the spatial pattern of adaptive genetic variation in a wild mammal. Evolution 2023, 77, 221–238. [Google Scholar] [CrossRef]
- Kabi, M. , & Filion, G. J. Heterochromatin: did H3K9 methylation evolve to tame transposons? Genome Biology 2021, 22, 1–3. [Google Scholar] [CrossRef]
- Janssen, A. , Colmenares, S. U., & Karpen, G. H. Heterochromatin: guardian of the genome. Annual review of cell and developmental biology 2018, 34, 265–288. [Google Scholar] [CrossRef]
- Wintersberger, E. Why is there late replication? Chromosoma 2000, 109, 300–307. [Google Scholar] [CrossRef]
- Herrick, J. Genetic variation and DNA replication timing, or why is there late replicating DNA? Evolution 2011, 65, 3031–3047. [Google Scholar] [CrossRef]
- Al Mamun, M. , Albergante, L., Moreno, A., Carrington, J. T., Blow, J. J., & Newman, T. J. Inevitability and containment of replication errors for eukaryotic genome lengths spanning megabase to gigabase. Proceedings of the National Academy of Sciences 2016, 113, E5765–E5774. [Google Scholar] [CrossRef]
- Fortuny, A. , & Polo, S. E. The response to DNA damage in heterochromatin domains. Chromosoma 2018, 127, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Al Mamun, M. , Albergante, L., Blow, J. J., & Newman, T. J. 3 tera-basepairs as a fundamental limit for robust DNA replication. Physical Biology 2020, 17, 046002. [Google Scholar] [CrossRef]
- Moreno, A. , Carrington, J. T., Albergante, L., Al Mamun, M., Haagensen, E. J., Komseli, E. S.,... & Blow, J. J. Unreplicated DNA remaining from unperturbed S phases passes through mitosis for resolution in daughter cells. Proceedings of the National Academy of Sciences 2016, 113, E5757–E5764. [Google Scholar] [CrossRef]
- Angileri, K. M. , Bagia, N. A., & Feschotte, C. Transposon control as a checkpoint for tissue regeneration. Development 2022, 149, dev191957. [Google Scholar] [CrossRef]
- Zuntini, A. R. , Carruthers, T., Maurin, O., Bailey, P. C., Leempoel, K., Brewer, G. E.,... & Knapp, S. Phylogenomics and the rise of the angiosperms. Nature 2024, 629, 843–850. [Google Scholar]
- Borowska-Zuchowska, N. , Senderowicz, M., Trunova, D., & Kolano, B. Tracing the evolution of the angiosperm genome from the cytogenetic point of view. Plants 2022, 11, 784. [Google Scholar]
- Schubert, I. , & Vu, G. T. Genome stability and evolution: attempting a holistic view. Trends in plant science 2016, 21, 749–757. [Google Scholar] [CrossRef]
- Giannattasio, M. , & Branzei, D. S-phase checkpoint regulations that preserve replication and chromosome integrity upon dNTP depletion. Cellular and Molecular Life Sciences 2017, 74, 2361–2380. [Google Scholar] [CrossRef]
- Voskarides, K. , & Giannopoulou, N. The role of TP53 in adaptation and evolution. Cells 2023, 12, 512. [Google Scholar] [CrossRef]
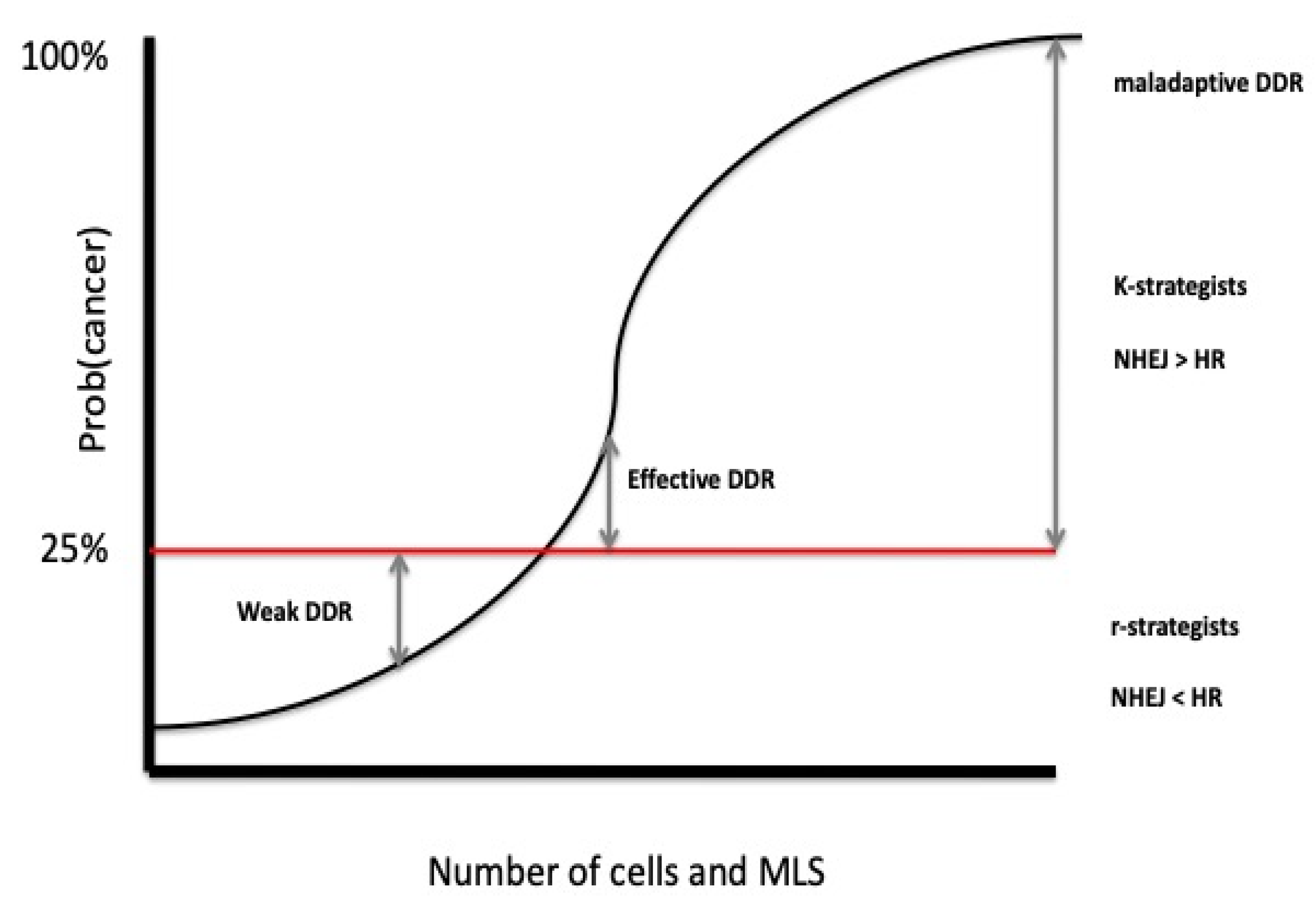
- Roche, B. , Hochberg, M. E., Caulin, A. F., Maley, C. C., Gatenby, R. A., Misse, D., & Thomas, F. Natural resistance to cancers: a Darwinian hypothesis to explain Peto’s paradox. BMC cancer 2012, 12, 1–4. [Google Scholar] [CrossRef]
- Vincze, O. , Colchero, F., Lemaître, J. F., Conde, D. A., Pavard, S., Bieuville, M.,... & Giraudeau, M. Cancer risk across mammals. Nature 2022, 601, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Dart, A. Peto’s paradox put to the test. Nature Reviews Cancer 2022, 22, 129–129. [Google Scholar] [CrossRef]
- Bowman, J. , & Lynch, V. J. Rapid evolution of genes with anti-cancer functions during the origins of large bodies and cancer resistance in elephants. bioRxiv 2024. [Google Scholar] [CrossRef]
- Maciak, S. Cell size, body size and Peto’s paradox. BMC ecology and evolution 2022, 22, 142. [Google Scholar] [CrossRef]
- Compton, Z. T. , Mellon, W., Harris, V. K., Rupp, S., Mallo, D., Kapsetaki, S. E.,... & Boddy, A. M. Cancer prevalence across vertebrates. Cancer discovery 2025, 15, 227–244. [Google Scholar] [CrossRef]
- Sulak, M. , Fong, L., Mika, K., Chigurupati, S., Yon, L., Mongan, N. P.,... & Lynch, V. J. TP53 copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. elife 2016, 5, e11994. [Google Scholar] [CrossRef]
- Butler, G. , Baker, J., Amend, S. R., Pienta, K. J., & Venditti, C. No evidence for Peto’s paradox in terrestrial vertebrates. Proceedings of the National Academy of Sciences 2025, 122, e2422861122. [Google Scholar] [CrossRef]
- Tian, X. , Firsanov, D., Zhang, Z., Cheng, Y., Luo, L., Tombline, G.,... & Gorbunova, V. SIRT6 is responsible for more efficient DNA double-strand break repair in long-lived species. Cell 2019, 177, 622–638. [Google Scholar] [CrossRef]
- Popov, A. A. , Petruseva, I. O., & Lavrik, O. I. Activity of DNA Repair Systems in the Cells of Long-Lived Rodents and Bats. Biochemistry (Moscow) 2024, 89, 1014–1023. [Google Scholar] [CrossRef]
- Cagan, A. , Baez-Ortega, A., Brzozowska, N., Abascal, F., Coorens, T. H., Sanders, M. A.,... & Martincorena, I. Somatic mutation rates scale with lifespan across mammals. Nature 2022, 604, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Johannes, F. Allometric scaling of somatic mutation and epimutation rates in trees. Evolution 2025, 79, 1–5. [Google Scholar] [CrossRef] [PubMed]
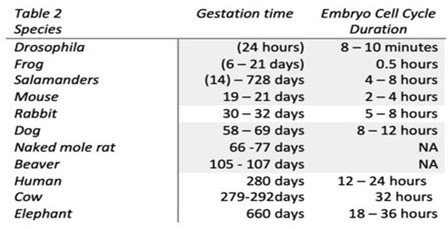
- Singh, V. P. , & Singh, P. Linking DNA damage and senescence to gestation period and lifespan in placental mammals. Frontiers in Cell and Developmental Biology 2024, 12, 1480695. [Google Scholar] [CrossRef]
- Damas, J. , Corbo, M., Kim, J., Turner-Maier, J., Farré, M., Larkin, D. M.,... & Lewin, H. A. Evolution of the ancestral mammalian karyotype and syntenic regions. Proceedings of the National Academy of Sciences 2022, 119, e2209139119. [Google Scholar] [CrossRef]
- Carta, A. , & Escudero, M. Karyotypic diversity: a neglected trait to explain angiosperm diversification? Evolution 2023, 77, 1158–1164. [Google Scholar] [CrossRef]
- Zhan, S. H. , Otto, S. P., & Barker, M. S. Broad variation in rates of polyploidy and dysploidy across flowering plants is correlated with lineage diversification. S. Broad variation in rates of polyploidy and dysploidy across flowering plants is correlated with lineage diversification. bioRxiv 2021, 2021–03. [Google Scholar] [CrossRef]
- Zhang, R. G. , Liu, H., Shang, H. Y., Shu, H., Liu, D. T., Yang, H.,... & Ma, Y. Convergent patterns of karyotype evolution underlying karyotype uniformity in conifers. Advanced Science 2025, 12, 2411098. [Google Scholar] [CrossRef]
- Afonso Silva, A. C. , Maliet, O., Aristide, L., Nogués-Bravo, D., Upham, N., Jetz, W., & Morlon, H. Negative global-scale association between genetic diversity and speciation rates in mammals. Nature communications 2025, 16, 1796. [Google Scholar] [CrossRef]
- Bergeron, L. A. , Besenbacher, S., Zheng, J., Li, P., Bertelsen, M. F., Quintard, B.,... & Zhang, G. Evolution of the germline mutation rate across vertebrates. Nature 2023, 615, 285–291. [Google Scholar] [CrossRef]
- Futuyma, D. J. Evolutionary constraint and ecological consequences. Evolution 2010, 64, 1865–1884. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, B. , Pothof, J., Vijg, J., & Hoeijmakers, J. H. The central role of DNA damage in the ageing process. Nature 2021, 592, 695–703. [Google Scholar] [CrossRef]
- Sahm, A. , Cherkasov, A., Liu, H., Voronov, D., Siniuk, K., Schwarz, R.,... & Hoffmann, S. The Greenland shark (Somniosus microcephalus) genome provides insights into extreme longevity. bioRxiv 2024. [Google Scholar] [CrossRef]
- Yang, K. , Nishiwaki, K., Mizobata, H., Asakawa, S., Yoshitake, K., Watanabe, Y. Y.,... & Kinoshita, S. The Greenland shark genome: insights into deep-sea ecology and lifespan extremes. bioRxiv 2025. [Google Scholar] [CrossRef]
- Kilili, H. , Padilla-Morales, B., Castillo-Morales, A., Monzón-Sandoval, J., Díaz-Barba, K., Cornejo-Paramo, P.,... & Urrutia, A. O. Maximum lifespan and brain size in mammals are associated with gene family size expansion related to immune system functions. Scientific Reports 2025, 15, 15087. [Google Scholar] [CrossRef]
- Rolland, J. , Henao-Diaz, L. F., Doebeli, M., Germain, R., Harmon, L. J., Knowles, L. L.,... & Schluter, D. Conceptual and empirical bridges between micro-and macroevolution. Nature Ecology & Evolution 2023, 7, 1181–1193. [Google Scholar]
- Bertucci-Richter, E. M. , & Parrott, B. B. The rate of epigenetic drift scales with maximum lifespan across mammals. Nature Communications 2023, 14, 7731. [Google Scholar]
- Zhou, W. , Liang, G., Molloy, P. L., & Jones, P. A. DNA methylation enables transposable element-driven genome expansion. Proceedings of the National Academy of Sciences 2020, 117, 19359–19366. [Google Scholar]
- Lynch, M. , & Conery, J. S. The origins of genome complexity. science 2003, 302, 1401–1404. [Google Scholar]
- Marino Alba, Debaecker Gautier, Fiston-Lavier Anna-Sophie, Haudry Annabelle, Nabholz Benoit (2024) Effective population size does not explain long-term variation in genome size and transposable element content in animals eLife 13:RP100574. [CrossRef]
- Lynch, M. , Ackerman, M. S., Gout, J. F., Long, H., Sung, W., Thomas, W. K., & Foster, P. L. Genetic drift, selection and the evolution of the mutation rate. Nature Reviews Genetics 2016, 17, 704–714. [Google Scholar]
- Bengtsson, B. O. Rates of karyotype evolution in placental mammals. Hereditas 1980, 92, 37–47. [Google Scholar] [CrossRef]
| Adj R2 | P | |
| MLS vs Body Mass (+) | 0.73 | 2 x 10-16 |
| MLS vs C-value (+) | 0,007 | 0,5 |
| SR vs Body Mass (-) | 0.56 | 0.01 |
| MLS vs Synteny (+) | 0.48 | 0.03 |
| Synteny vs SR (-) | 0.18 | 0.1 |
| MLS vs SR (+) | 0.59 | 0.016 |
| rKD Macro vs SR (+) | 0.42 | 3 x 10-10 |
| rKD Micro vs SR (+) | 0.07 | 0.06 |
 0,5 0,5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





