Submitted:
13 June 2025
Posted:
17 June 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
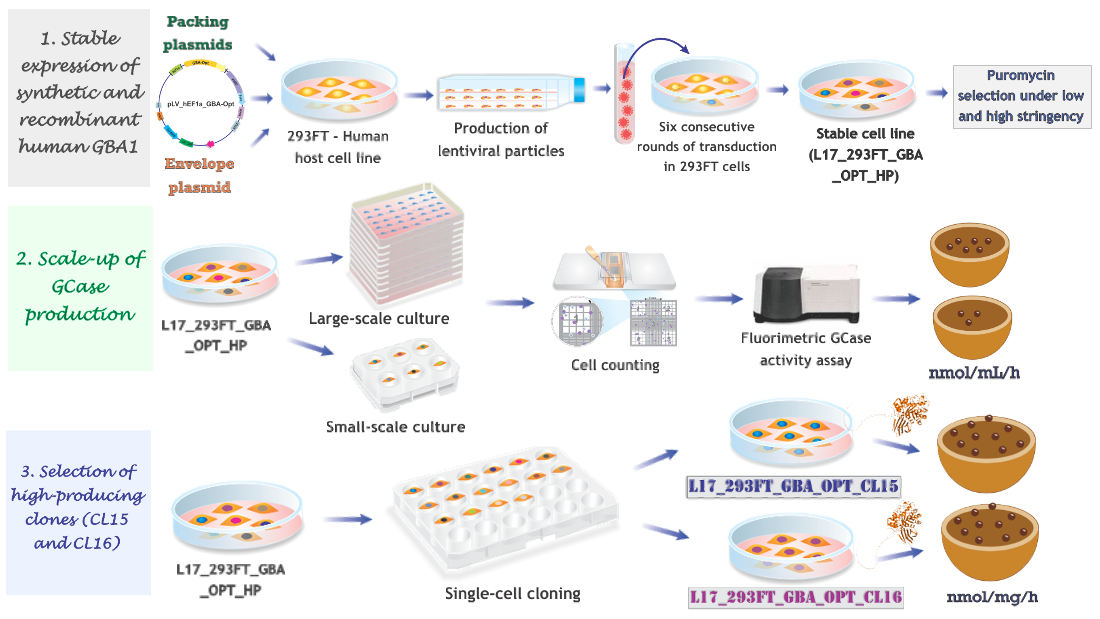
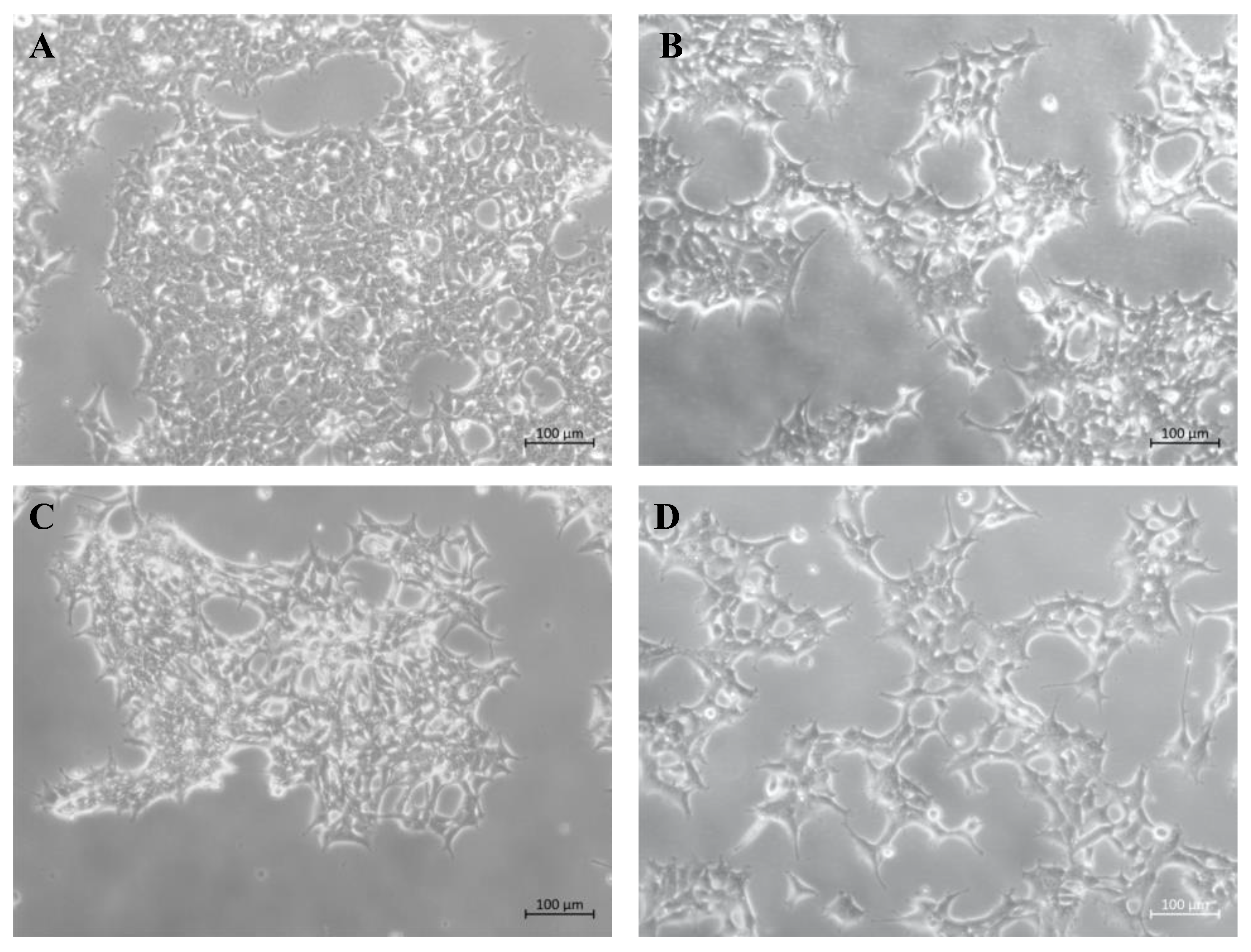
2.1. Production of Stable Lentiviral-Transduced Human Cell Lines
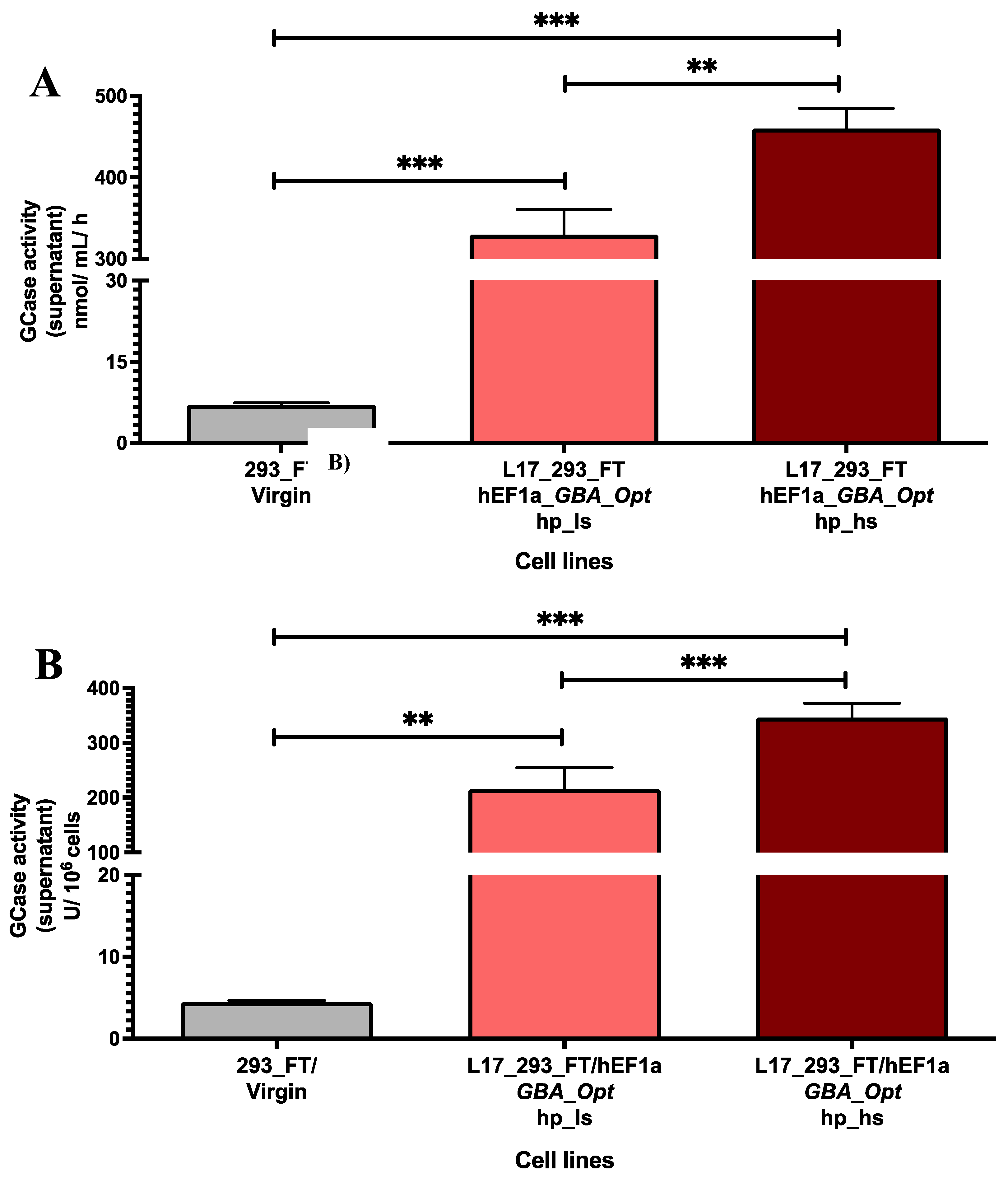
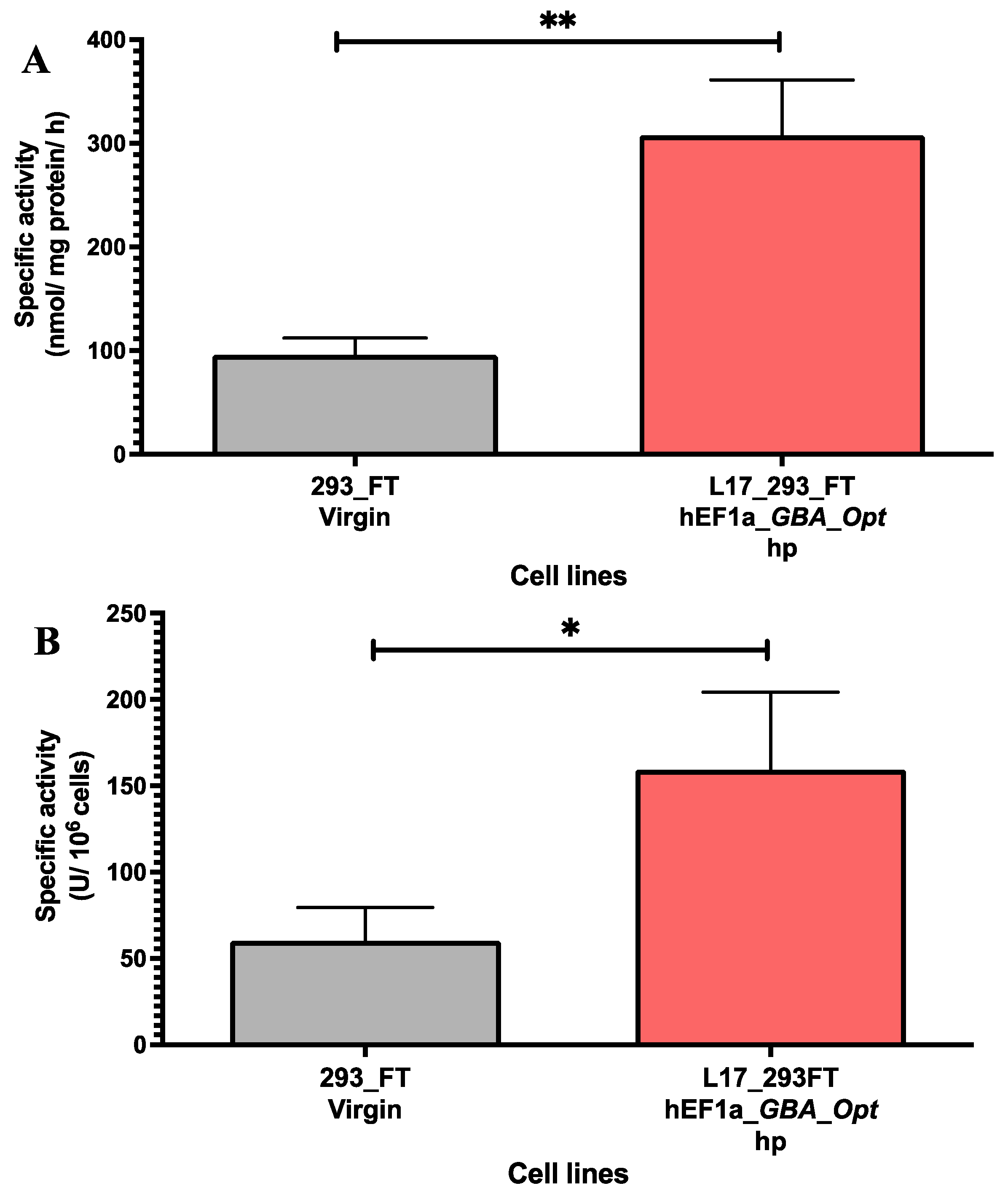
2.2. GCase Activity in Lentiviral Transduced and Puromycin-Selected L17_293FT_GBA_OPT_HP Cells
2.3. High-Producer Clone Selection from Puromycin-Selected Population
3. Discussion
4. Materials and Methods
4.1. Plasmid Constructs
4.2. Cell Culture
4.3. Production of Lentiviral Particles
4.4. Lentiviral Transduction and Establishment of Stable Transduced Cell Lines
4.5. Puromycin Treatment of L17_293FT_GBA_OPT_HP Heterogeneous Population
4.6. GCase Activity Analysis: Secreted and Intracellular (GCase-Specific Activity)
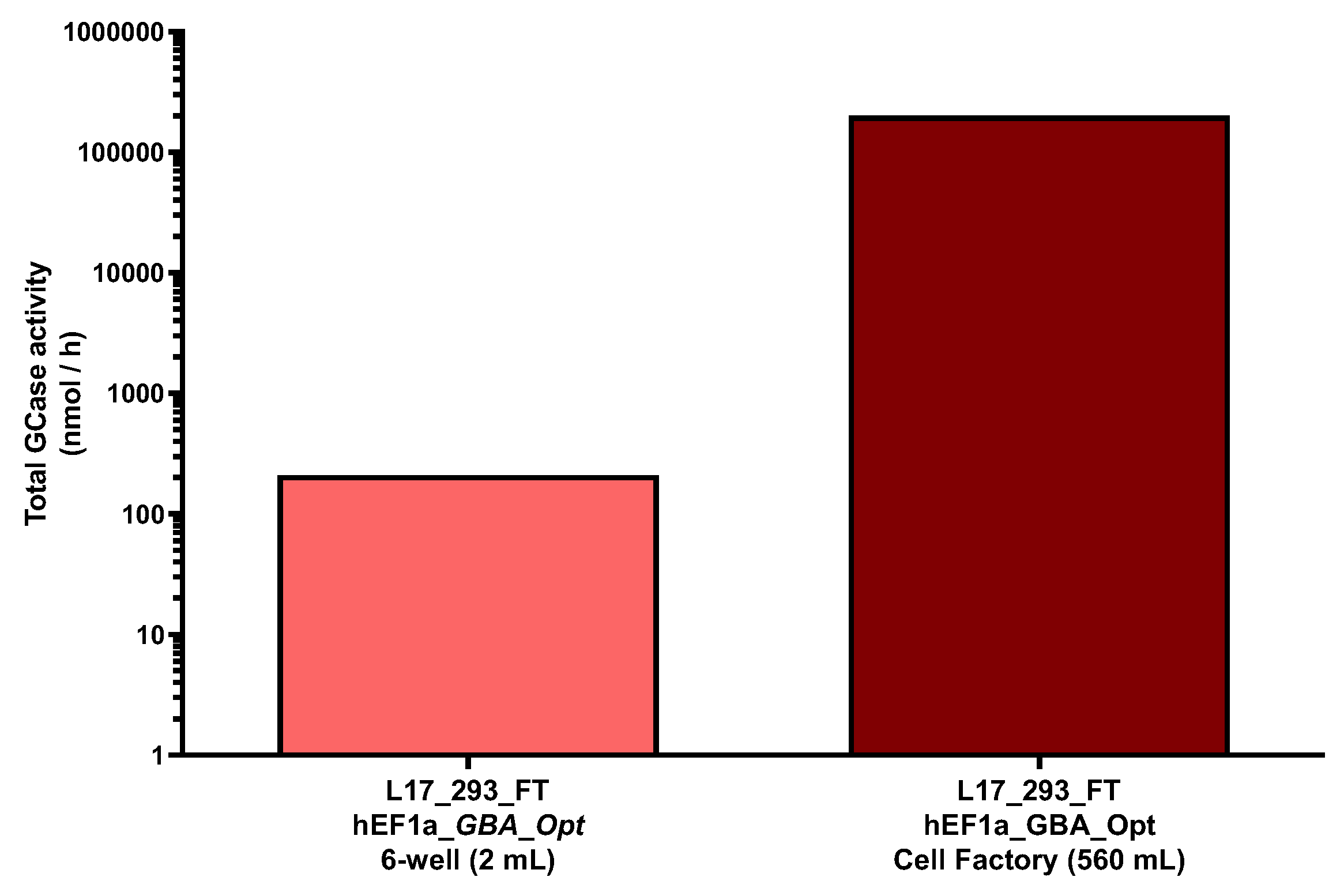
4.7. Scaling of GCase Production in L17_293FT_GBA_OPT_HP Cell Supernatants
4.8. Clone Cell Selection (Isolation)
4.9. Biological Activity by Fluorimetric Assay
4.10. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Abbreviation | Meaning |
| CHO | Chinese hamster ovary cell |
| DMEM | Dulbecco's Modified Eagle's Medium |
| ERT | Enzyme replacement therapy |
| ELISA | Enzyme-linked immunosorbent assay |
| FBS | Fetal bovine serum |
| FDA | Food and Drug Administration |
| GBA1 | Glucocerebrosidase, Glucosylceramidase beta 1 |
| GCase | Glucocerebrosidase, Glucosylceramidase beta 1 |
| GD | Gaucher disease |
| GlcCer | Glucosylceramide |
| 293_FT | Human embryonic kidney 293 cells |
| HP | Heterogeneous population |
| HS | High stringency |
| ICGG | International Collaborative Gaucher Group |
| L17 | Lineage 17 |
| LS | Low stringency |
| LV | Lentiviral vector |
| LTR | Long terminal repeat |
| MOI | Multiplicity of infection |
| MTX | Methotrexate |
| 4MU | 4-methylumbiliferone |
| 4MUG | 4-methylumbiliferon-β-D-glucopyranoside |
| qPCR | Real-time quantitative PCR |
| SUS | Unified Health System |
| TDC | Sodium taurodeoxycholate hydrate |
References
- Hughes DA, Pastores GM: Gaucher Disease. In GeneReviews((R)). Edited by Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Amemiya A. Seattle (WA); 1993.
- Brady RO, Kanfer JN, Shapiro D: Metabolism of Glucocerebrosides. Ii. Evidence of an Enzymatic Deficiency in Gaucher's Disease. Biochem Biophys Res Commun 1965, 18:221-225. [CrossRef]
- Elstein D, Belmatoug N, Bembi B, Deegan P, Fernandez-Sasso D, Giraldo P, Goker-Alpan O, Hughes D, Lau H, Lukina E, et al: Twelve Years of the Gaucher Outcomes Survey (GOS): Insights, Achievements, and Lessons Learned from a Global Patient Registry. J Clin Med 2024, 13. [CrossRef]
- Belinsky G, Ruan J, Fattahi N, Mehta S, Boddupalli CS, Mistry PK, Nair S: Modeling bone marrow microenvironment and hematopoietic dysregulation in Gaucher disease through VavCre mediated Gba deletion. Hum Mol Genet 2025. [CrossRef]
- Ducatez F, Berger MG, Pilon C, Plichet T, Lesueur C, Berger J, Belmatoug N, Marret S, Bekri S, Tebani A: Deciphering metabolic shifts in Gaucher disease type 1: a multi-omics study. J Mol Med (Berl) 2025, 103:187-203. [CrossRef]
- Grabowski GA: Phenotype, diagnosis, and treatment of Gaucher's disease. Lancet 2008, 372:1263-1271. [CrossRef]
- Cohen D, Levy Y, Bar-Ziv Y, Revel-Vilk S, Zimran A, Lebel E: Simultaneous Bilateral Femoral Osteonecrosis in Gaucher Disease. Life (Basel) 2023, 13. [CrossRef]
- Grabowski GA, Antommaria AHM, Kolodny EH, Mistry PK: Gaucher disease: Basic and translational science needs for more complete therapy and management. Mol Genet Metab 2021, 132:59-75. [CrossRef]
- Grabowski GA, Kishnani PS, Alcalay RN, Prakalapakorn SG, Rosenbloom BE, Tuason DA, Weinreb NJ: Challenges in Gaucher disease: Perspectives from an expert panel. Mol Genet Metab 2025, 145:109074. [CrossRef]
- Furderer ML, Hertz E, Lopez GJ, Sidransky E: Neuropathological Features of Gaucher Disease and Gaucher Disease with Parkinsonism. Int J Mol Sci 2022, 23. [CrossRef]
- Hertz E, Chen Y, Sidransky E: Gaucher disease provides a unique window into Parkinson disease pathogenesis. Nat Rev Neurol 2024, 20:526-540. [CrossRef]
- Imbalzano G, Ledda C, Romagnolo A, Covolo A, Lopiano L, Artusi CA: Neurological symptoms in adults with Gaucher disease: a systematic review. J Neurol 2024, 271:3897-3907. [CrossRef]
- Sidransky E, Nalls MA, Aasly JO, Aharon-Peretz J, Annesi G, Barbosa ER, Bar-Shira A, Berg D, Bras J, Brice A, et al: Multicenter analysis of glucocerebrosidase mutations in Parkinson's disease. N Engl J Med 2009, 361:1651-1661. [CrossRef]
- Motta I, Delbini P, Scaramellini N, Ghiandai V, Duca L, Nava I, Nascimbeni F, Lugari S, Consonni D, Trombetta E, et al: Enzyme replacement therapy improves erythropoiesis and iron dysregulation in Gaucher disease. Ann Hematol 2024, 103:5113-5121. [CrossRef]
- Deegan P, Lau H, Elstein D, Fernandez-Sasso D, Giraldo P, Hughes D, Zimran A, Istaiti M, Gadir N, Botha J, et al: Long-Term Treatment of Gaucher Disease with Velaglucerase Alfa in ERT-Naive Patients from the Gaucher Outcome Survey (GOS) Registry. J Clin Med 2024, 13. [CrossRef]
- Revel-Vilk S, Mansfield R, Feder-Krengel N, Machtiger-Azoulay N, Kuter D, Szer J, Rosenbaum H, Ferreira DC, Ruhrman-Shahar N, Wajnrajch M, Zimran A: Real-World Experiences with Taliglucerase Alfa Home Infusions for Patients with Gaucher Disease: A Global Cohort Study. J Clin Med 2023, 12. [CrossRef]
- Barton NW, Brady RO, Dambrosia JM, Di Bisceglie AM, Doppelt SH, Hill SC, Mankin HJ, Murray GJ, Parker RI, Argoff CE, et al.: Replacement therapy for inherited enzyme deficiency--macrophage-targeted glucocerebrosidase for Gaucher's disease. N Engl J Med 1991, 324:1464-1470. [CrossRef]
- Weinreb NJ, Camelo JS, Jr., Charrow J, McClain MR, Mistry P, Belmatoug N, International Collaborative Gaucher Group Gaucher Registry i: Gaucher disease type 1 patients from the ICGG Gaucher Registry sustain initial clinical improvements during twenty years of imiglucerase treatment. Mol Genet Metab 2021, 132:100-111. [CrossRef]
- Mistry PK, Belmatoug N, vom Dahl S, Giugliani R: Understanding the natural history of Gaucher disease. Am J Hematol 2015, 90 Suppl 1:S6-11. [CrossRef]
- Stepien KM, Kiec-Wilk B, Lampe C, Tangeraas T, Cefalo G, Belmatoug N, Francisco R, Del Toro M, Wagner L, Lauridsen AG, et al: Challenges in Transition From Childhood to Adulthood Care in Rare Metabolic Diseases: Results From the First Multi-Center European Survey. Front Med (Lausanne) 2021, 8:652358. [CrossRef]
- Borin MC, Alvares-Teodoro J, Acurcio FA, Guerra AA, Jr.: Gaucher disease in Brazil: a comprehensive 16 year retrospective study on survival, cost, and treatment insights. Front Pharmacol 2024, 15:1433970. [CrossRef]
- Grabowski GA, Barton NW, Pastores G, Dambrosia JM, Banerjee TK, McKee MA, Parker C, Schiffmann R, Hill SC, Brady RO: Enzyme therapy in type 1 Gaucher disease: comparative efficacy of mannose-terminated glucocerebrosidase from natural and recombinant sources. Ann Intern Med 1995, 122:33-39. [CrossRef]
- Zimran A, Elstein D, Levy-Lahad E, Zevin S, Hadas-Halpern I, Bar-Ziv Y, Foldes J, Schwartz AJ, Abrahamov A: Replacement therapy with imiglucerase for type 1 Gaucher's disease. Lancet 1995, 345:1479-1480. [CrossRef]
- Zimran A, Altarescu G, Philips M, Attias D, Jmoudiak M, Deeb M, Wang N, Bhirangi K, Cohn GM, Elstein D: Phase 1/2 and extension study of velaglucerase alfa replacement therapy in adults with type 1 Gaucher disease: 48-month experience. Blood 2010, 115:4651-4656. [CrossRef]
- Zimran A: Velaglucerase alfa: a new option for Gaucher disease treatment. Drugs Today (Barc) 2011, 47:515-529. [CrossRef]
- Zimran A, Brill-Almon E, Chertkoff R, Petakov M, Blanco-Favela F, Munoz ET, Solorio-Meza SE, Amato D, Duran G, Giona F, et al: Pivotal trial with plant cell-expressed recombinant glucocerebrosidase, taliglucerase alfa, a novel enzyme replacement therapy for Gaucher disease. Blood 2011, 118:5767-5773. [CrossRef]
- van Dussen L, Zimran A, Akkerman EM, Aerts JM, Petakov M, Elstein D, Rosenbaum H, Aviezer D, Brill-Almon E, Chertkoff R, et al: Taliglucerase alfa leads to favorable bone marrow responses in patients with type I Gaucher disease. Blood Cells Mol Dis 2013, 50:206-211. [CrossRef]
- BRASIL: Ministério da Saúde. Secretaria de Atenção à Saúde. Protocolo Clínico e Diretrizes Terapêuticas da Doença de Gaucher. Brasília, 2014 (atualizado em 2017). 2017.
- Liu C, Bahnson AB, Dunigan JT, Watkins SC, Barranger JA: Long-term expression and secretion of human glucocerebrosidase by primary murine and human myoblasts and differentiated myotubes. J Mol Med (Berl) 1998, 76:773-781. [CrossRef]
- Kim EY, Hong YB, Lai Z, Kim HJ, Cho YH, Brady RO, Jung SC: Expression and secretion of human glucocerebrosidase mediated by recombinant lentivirus vectors in vitro and in vivo: implications for gene therapy of Gaucher disease. Biochem Biophys Res Commun 2004, 318:381-390. [CrossRef]
- Scharenberg SG, Poletto E, Lucot KL, Colella P, Sheikali A, Montine TJ, Porteus MH, Gomez-Ospina N: Engineering monocyte/macrophage-specific glucocerebrosidase expression in human hematopoietic stem cells using genome editing. Nat Commun 2020, 11:3327. [CrossRef]
- Novo JB, Morganti L, Moro AM, Paes Leme AF, Serrano SM, Raw I, Ho PL: Generation of a Chinese hamster ovary cell line producing recombinant human glucocerebrosidase. J Biomed Biotechnol 2012, 2012:875383. [CrossRef]
- Naphatsamon U, Ohashi T, Misaki R, Fujiyama K: The Production of Human beta-Glucocerebrosidase in Nicotiana benthamiana Root Culture. Int J Mol Sci 2018, 19. [CrossRef]
- Uthailak N, Kajiura H, Misaki R, Fujiyama K: Production of recombinant beta-glucocerebrosidase in wild-type and glycoengineered transgenic Nicotiana benthamiana root cultures with different N-glycan profiles. J Biosci Bioeng 2022, 133:481-488. [CrossRef]
- Hia F, Yang SF, Shichino Y, Yoshinaga M, Murakawa Y, Vandenbon A, Fukao A, Fujiwara T, Landthaler M, Natsume T, et al: Codon bias confers stability to human mRNAs. EMBO Rep 2019, 20:e48220. [CrossRef]
- Wu G, Zheng Y, Qureshi I, Zin HT, Beck T, Bulka B, Freeland SJ: SGDB: a database of synthetic genes re-designed for optimizing protein over-expression. Nucleic Acids Res 2007, 35:D76-79. [CrossRef]
- Plotkin JB, Kudla G: Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet 2011, 12:32-42. [CrossRef]
- VandenDriessche T, Chuah MK: Hemophilia Gene Therapy: Ready for Prime Time? Hum Gene Ther 2017, 28:1013-1023. [CrossRef]
- Srivastava A, Abraham A, Aboobacker F, Singh G, Geevar T, Kulkarni U, Selvarajan S, Korula A, Dave RG, Shankar M, et al: Lentiviral Gene Therapy with CD34+ Hematopoietic Cells for Hemophilia A. N Engl J Med 2025, 392:450-457. [CrossRef]
- Poletti V, Charrier S, Corre G, Gjata B, Vignaud A, Zhang F, Rothe M, Schambach A, Gaspar HB, Thrasher AJ, Mavilio F: Preclinical Development of a Lentiviral Vector for Gene Therapy of X-Linked Severe Combined Immunodeficiency. Mol Ther Methods Clin Dev 2018, 9:257-269. [CrossRef]
- Banning A, Konig JF, Gray SJ, Tikkanen R: Functional Analysis of the Ser149/Thr149 Variants of Human Aspartylglucosaminidase and Optimization of the Coding Sequence for Protein Production. Int J Mol Sci 2017, 18. [CrossRef]
- Jeyakumar JM, Kia A, Tam LCS, McIntosh J, Spiewak J, Mills K, Heywood W, Chisari E, Castaldo N, Verhoef D, et al: Preclinical evaluation of FLT190, a liver-directed AAV gene therapy for Fabry disease. Gene Ther 2023, 30:487-502. [CrossRef]
- Sinclair G, Choy FY: Synonymous codon usage bias and the expression of human glucocerebrosidase in the methylotrophic yeast, Pichia pastoris. Protein Expr Purif 2002, 26:96-105. [CrossRef]
- Figueiredo LLS, Lau Jr W, Goncalves VWS, Ramos ES, D’Almeida V, Souza LEB, Orellana MD, Abraham KJ, Lichtenstein F, Bleicher L, et al: Engineering Synthetic and Recombinant Human Lysosomal β-Glucocerebrosidase for Enzyme Replacement Therapy for Gaucher Disease. Discover Applied Sciences 2024, 6. [CrossRef]
- Ungari S, Montepeloso A, Morena F, Cocchiarella F, Recchia A, Martino S, Gentner B, Naldini L, Biffi A: Design of a regulated lentiviral vector for hematopoietic stem cell gene therapy of globoid cell leukodystrophy. Mol Ther Methods Clin Dev 2015, 2:15038. [CrossRef]
- Pimentel N, Rodriguez-Lopez A, Diaz S, Losada JC, Diaz-Rincon DJ, Cardona C, Espejo-Mojica AJ, Ramirez AM, Ruiz F, Landazuri P, et al: Production and characterization of a human lysosomal recombinant iduronate-2-sulfatase produced in Pichia pastoris. Biotechnol Appl Biochem 2018, 65:655-664. [CrossRef]
- Smith MC, Belur LR, Karlen AD, Erlanson O, Podetz-Pedersen KM, McKenzie J, Detellis J, Gagnidze K, Parsons G, Robinson N, et al: Phenotypic Correction of Murine Mucopolysaccharidosis Type II by Engraftment of Ex Vivo Lentiviral Vector-Transduced Hematopoietic Stem and Progenitor Cells. Hum Gene Ther 2022, 33:1279-1292. [CrossRef]
- Pan X, Sands SA, Yue Y, Zhang K, LeVine SM, Duan D: An Engineered Galactosylceramidase Construct Improves AAV Gene Therapy for Krabbe Disease in Twitcher Mice. Hum Gene Ther 2019, 30:1039-1051. [CrossRef]
- Doerfler PA, Todd AG, Clement N, Falk DJ, Nayak S, Herzog RW, Byrne BJ: Copackaged AAV9 Vectors Promote Simultaneous Immune Tolerance and Phenotypic Correction of Pompe Disease. Hum Gene Ther 2016, 27:43-59. [CrossRef]
- Stok M, de Boer H, Huston MW, Jacobs EH, Roovers O, Visser TP, Jahr H, Duncker DJ, van Deel ED, Reuser AJJ, et al: Lentiviral Hematopoietic Stem Cell Gene Therapy Corrects Murine Pompe Disease. Mol Ther Methods Clin Dev 2020, 17:1014-1025. [CrossRef]
- Liang Q, Catalano F, Vlaar EC, Pijnenburg JM, Stok M, van Helsdingen Y, Vulto AG, van der Ploeg AT, van Til NP, Pijnappel W: IGF2-tagging of GAA promotes full correction of murine Pompe disease at a clinically relevant dosage of lentiviral gene therapy. Mol Ther Methods Clin Dev 2022, 27:109-130. [CrossRef]
- Ornaghi F, Sala D, Tedeschi F, Maffia MC, Bazzucchi M, Morena F, Valsecchi M, Aureli M, Martino S, Gritti A: Novel bicistronic lentiviral vectors correct beta-Hexosaminidase deficiency in neural and hematopoietic stem cells and progeny: implications for in vivo and ex vivo gene therapy of GM2 gangliosidosis. Neurobiol Dis 2020, 134:104667. [CrossRef]
- Hu M, Xu Q, Zhang F, Buckland KF, Gao Y, Du W, Ding Y, Zhou L, Sun X, Ma L, et al: Preclinical ex vivo IL2RG gene therapy using autologous hematopoietic stem cells as an effective and safe treatment for X-linked severe combined immunodeficiency disease. Genes Dis 2025, 12:101445. [CrossRef]
- Saleh AH, Rothe M, Barber DL, McKillop WM, Fraser G, Morel CF, Schambach A, Auray-Blais C, West ML, Khan A, et al: Persistent hematopoietic polyclonality after lentivirus-mediated gene therapy for Fabry disease. Mol Ther Methods Clin Dev 2023, 28:262-271. [CrossRef]
- Mangiameli E, Cecchele A, Morena F, Sanvito F, Matafora V, Cattaneo A, Della Volpe L, Gnani D, Paulis M, Susani L, et al: Human iPSC-based neurodevelopmental models of globoid cell leukodystrophy uncover patient- and cell type-specific disease phenotypes. Stem Cell Reports 2021, 16:1478-1495. [CrossRef]
- Ellison S, Buckland K, Learmonth Y, Day V, Kalra S, Howe L, Roman-Rodriguez FJ, Bonafont J, Booth L, Holley R, et al: Design and validation of a GMP stem cell manufacturing protocol for MPSII hematopoietic stem cell gene therapy. Mol Ther Methods Clin Dev 2024, 32:101271. [CrossRef]
- Dogan Y, Barese CN, Schindler JW, Yoon JK, Unnisa Z, Guda S, Jacobs ME, Oborski C, Maiwald T, Clarke DL, et al: Screening chimeric GAA variants in preclinical study results in hematopoietic stem cell gene therapy candidate vectors for Pompe disease. Mol Ther Methods Clin Dev 2022, 27:464-487. [CrossRef]
- Yilmaz BS, Gurung S, Perocheau D, Counsell J, Baruteau J: Gene therapy for inherited metabolic diseases. J Mother Child 2020, 24:53-64. [CrossRef]
- Zielske SP, Gerson SL: Cytokines, including stem cell factor alone, enhance lentiviral transduction in nondividing human LTCIC and NOD/SCID repopulating cells. Mol Ther 2003, 7:325-333. [CrossRef]
- Fontes AM, Davis BM, Encell LP, Lingas K, Covas DT, Zago MA, Loeb LA, Pegg AE, Gerson SL: Differential competitive resistance to methylating versus chloroethylating agents among five O6-alkylguanine DNA alkyltransferases in human hematopoietic cells. Mol Cancer Ther 2006, 5:121-128. [CrossRef]
- Fontes AM, Melo FU, Greene LJ, Faca VM, Lin Y, Gerson SL, Covas DT: Production of human factor VIII-FL in 293T cells using the bicistronic MGMT(P140K)-retroviral vector. Genet Mol Res 2012, 11:775-789. [CrossRef]
- Castilho-Fernandes A, Fontes AM, Abraham KJ, de Freitas MC, da Rosa NG, Picanco-Castro V, de Sousa Russo-Carbolante EM, Covas DT: Significant differences in integration sites of Moloney murine leukemia virus/Moloney murine sarcoma virus retroviral vector carrying recombinant coagulation factor IX in two human cell lines. Biotechnol Lett 2015, 37:991-1001. [CrossRef]
- Correa de Freitas MC, Fontes AM, de Castilho Fernandes A, Picanco-Castro V, de Sousa Russo EM, Covas DT: Murine leukemia virus-derived retroviral vector has differential integration patterns in human cell lines used to produce recombinant factor VIII. Rev Bras Hematol Hemoter 2014, 36:213-218. [CrossRef]
- Milone MC, O'Doherty U: Clinical use of lentiviral vectors. Leukemia 2018, 32:1529-1541. [CrossRef]
- Naldini L, Trono D, Verma IM: Lentiviral vectors, two decades later. Science 2016, 353:1101-1102. [CrossRef]
- Scotti C, Aiuti A, Naldini L: Challenges and solutions to the sustainability of gene and cell therapies. Nat Rev Genet 2025. [CrossRef]
- al Yacoub N, Romanowska M, Haritonova N, Foerster J: Optimized production and concentration of lentiviral vectors containing large inserts. J Gene Med 2007, 9:579-584. [CrossRef]
- Hirch T, Brander N, Schenk F, Pollmann SJ, Reichenbach J, Schubert R, Modlich U: Expression of a large coding sequence: Gene therapy vectors for Ataxia Telangiectasia. Sci Rep 2023, 13:19386. [CrossRef]
- Wang Z, Chen C, Ge X: Large T antigen mediated target gene replication improves site-specific recombination efficiency. Front Bioeng Biotechnol 2024, 12:1377167. [CrossRef]
- Abaandou L, Quan D, Shiloach J: Affecting HEK293 Cell Growth and Production Performance by Modifying the Expression of Specific Genes. Cells 2021, 10. [CrossRef]
- Thomas P, Smart TG: HEK293 cell line: a vehicle for the expression of recombinant proteins. J Pharmacol Toxicol Methods 2005, 51:187-200. [CrossRef]
- Perry C, Rayat A: Lentiviral Vector Bioprocessing. Viruses 2021, 13. [CrossRef]
- Jargalsaikhan BE, Muto M, Been Y, Matsumoto S, Okamura E, Takahashi T, Narimichi Y, Kurebayashi Y, Takeuchi H, Shinohara T, et al: The Dual-Pseudotyped Lentiviral Vector with VSV-G and Sendai Virus HN Enhances Infection Efficiency through the Synergistic Effect of the Envelope Proteins. Viruses 2024, 16. [CrossRef]
- Jia J, Hao Y, Zhang L, Cao X, An L, Wang H, Ma Q, Jin X, Ma X: Development and validation of optimized lentivirus-like particles for gene editing tool delivery with Gag-Only strategy. Eur J Med Res 2025, 30:242. [CrossRef]
- Cabrera A, Edelstein HI, Glykofrydis F, Love KS, Palacios S, Tycko J, Zhang M, Lensch S, Shields CE, Livingston M, et al: The sound of silence: Transgene silencing in mammalian cell engineering. Cell Syst 2022, 13:950-973. [CrossRef]
- Xu ZJ, Jia YL, Wang M, Yi DD, Zhang WL, Wang XY, Zhang JH: Effect of promoter, promoter mutation and enhancer on transgene expression mediated by episomal vectors in transfected HEK293, Chang liver and primary cells. Bioengineered 2019, 10:548-560. [CrossRef]
- Fumagalli F, Calbi V, Natali Sora MG, Sessa M, Baldoli C, Rancoita PMV, Ciotti F, Sarzana M, Fraschini M, Zambon AA, et al: Lentiviral haematopoietic stem-cell gene therapy for early-onset metachromatic leukodystrophy: long-term results from a non-randomised, open-label, phase 1/2 trial and expanded access. Lancet 2022, 399:372-383. [CrossRef]
- Liang Q, Vlaar EC, Pijnenburg JM, Rijkers E, Demmers JAA, Vulto AG, van der Ploeg AT, van Til NP, Pijnappel W: Lentiviral gene therapy with IGF2-tagged GAA normalizes the skeletal muscle proteome in murine Pompe disease. J Proteomics 2024, 291:105037. [CrossRef]
- da Rosa NG, Swiech K, Picanco-Castro V, Russo-Carbolante EM, Soares Neto MA, de Castilho-Fernandes A, Faca VM, Fontes AM, Covas DT: SK-HEP cells and lentiviral vector for production of human recombinant factor VIII. Biotechnol Lett 2012, 34:1435-1443. [CrossRef]
- Spencer HT, Denning G, Gautney RE, Dropulic B, Roy AJ, Baranyi L, Gangadharan B, Parker ET, Lollar P, Doering CB: Lentiviral vector platform for production of bioengineered recombinant coagulation factor VIII. Mol Ther 2011, 19:302-309. [CrossRef]
- Kutner RH, Zhang XY, Reiser J: Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat Protoc 2009, 4:495-505. [CrossRef]
- Fantacini DM, Fontes AM, de Abreu Neto MS, Covas DT, Picanco-Castro V: The F309S mutation increases factor VIII secretion in human cell line. Rev Bras Hematol Hemoter 2016, 38:135-140. [CrossRef]
- Matsui H, Shibata M, Brown B, Labelle A, Hegadorn C, Andrews C, Hebbel RP, Galipeau J, Hough C, Lillicrap D: Ex vivo gene therapy for hemophilia A that enhances safe delivery and sustained in vivo factor VIII expression from lentivirally engineered endothelial progenitors. Stem Cells 2007, 25:2660-2669. [CrossRef]
- Peters SP, Coyle P, Glew RH: Differentiation of beta-glucocerebrosidase from beta-glucosidase in human tissues using sodium taurocholate. Arch Biochem Biophys 1976, 175:569-582. [CrossRef]
- Muller KB, Rodrigues MD, Pereira VG, Martins AM, D'Almeida V: Reference values for lysosomal enzymes activities using dried blood spots samples - a Brazilian experience. Diagn Pathol 2010, 5:65. [CrossRef]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ: Protein measurement with the Folin phenol reagent. J Biol Chem 1951, 193:265-275.
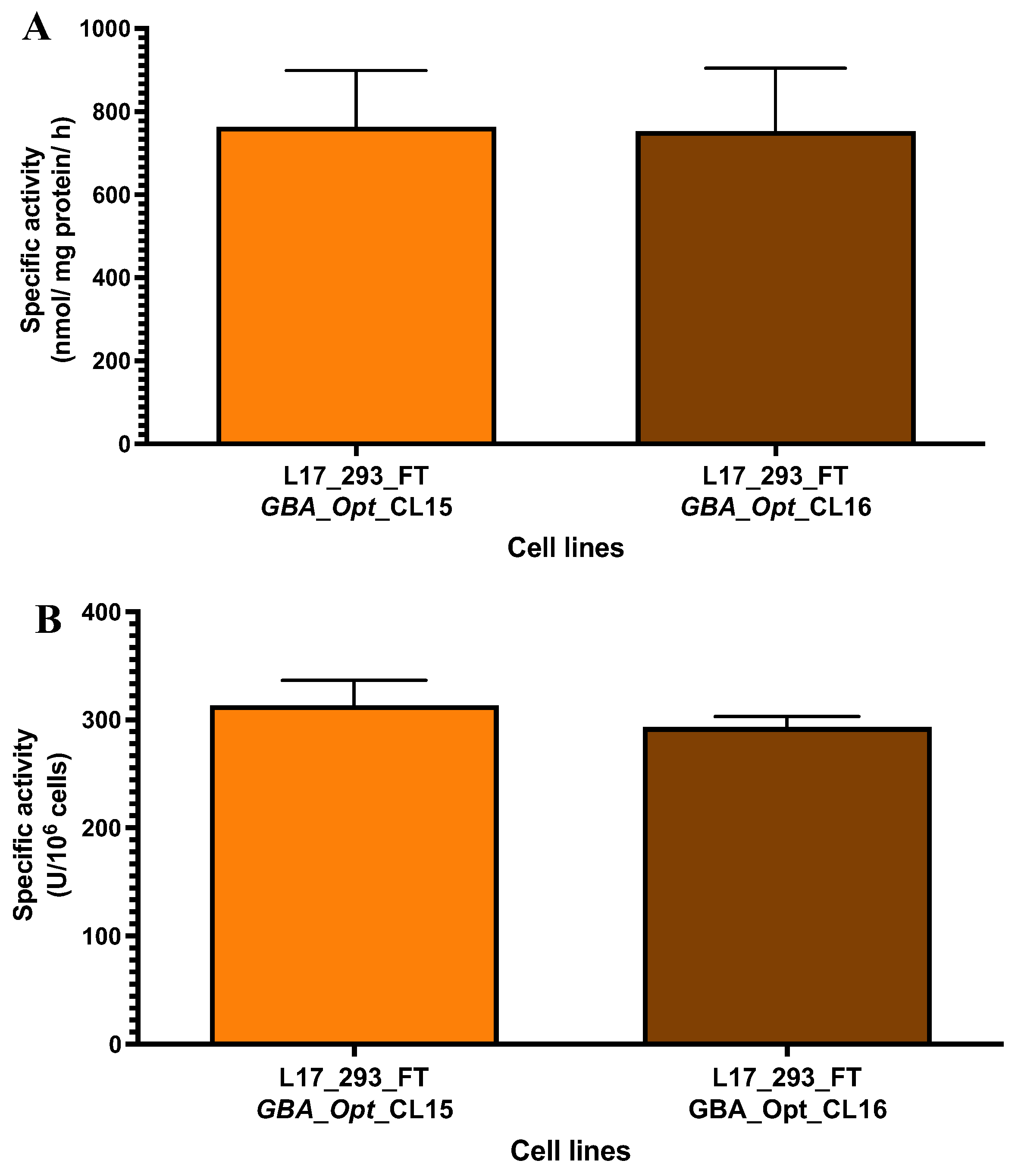
| Sample | GCase activity (nmol/mL/h) |
GCase activity (U/106cells) |
|---|---|---|
| L17_293FT_GBA_OPT_CL5 | 265.087 | 252.464 |
| L17_293FT_GBA_OPT_CL7 | 89.911 | 128.444 |
| L17_293FT_GBA_OPT_CL8 | 230.045 | 135.320 |
| L17_293FT_GBA_OPT_CL9 | 301.160 | 200.773 |
| L17_293FT_GBA_OPT_CL10 | 118.907 | 72.065 |
| L17_293FT_GBA_OPT_CL11 | 285.561 | 219.662 |
| L17_293FT_GBA_OPT_CL13 | 440.955 | 275.597 |
| L17_293FT_GBA_OPT_CL15 | 585.464 | 390.310 |
| L17_293FT_GBA_OPT_CL16 | 683.952 | 455.968 |
| L17_293FT_GBA_OPT_CL17 | 207.610 | 143.180 |
| L17_293FT_GBA_OPT_CL18 | 167.931 | 108.342 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





