Submitted:
12 June 2025
Posted:
13 June 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
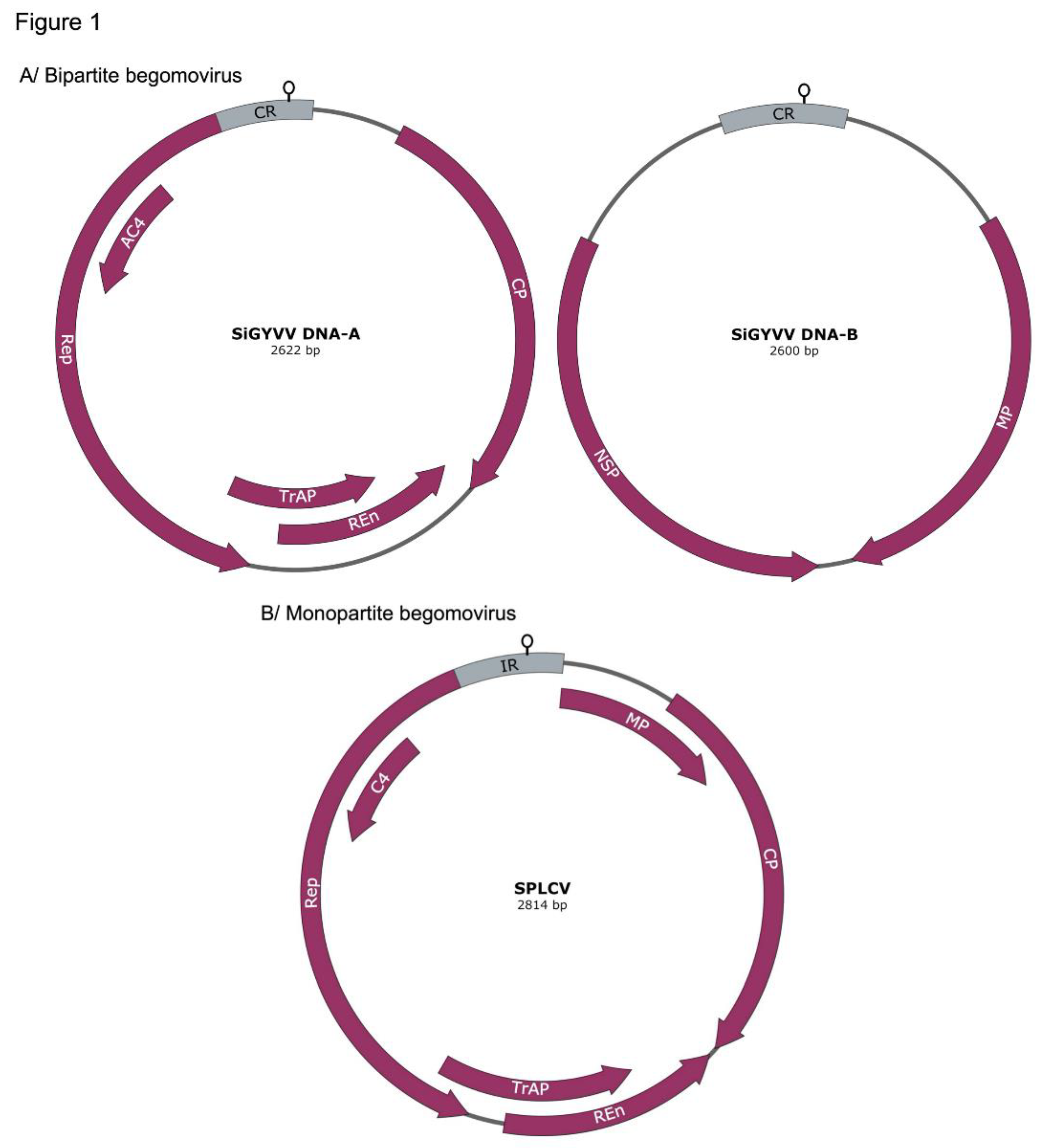
1.1. Structure and Function of Begomovirus Satellite Molecules
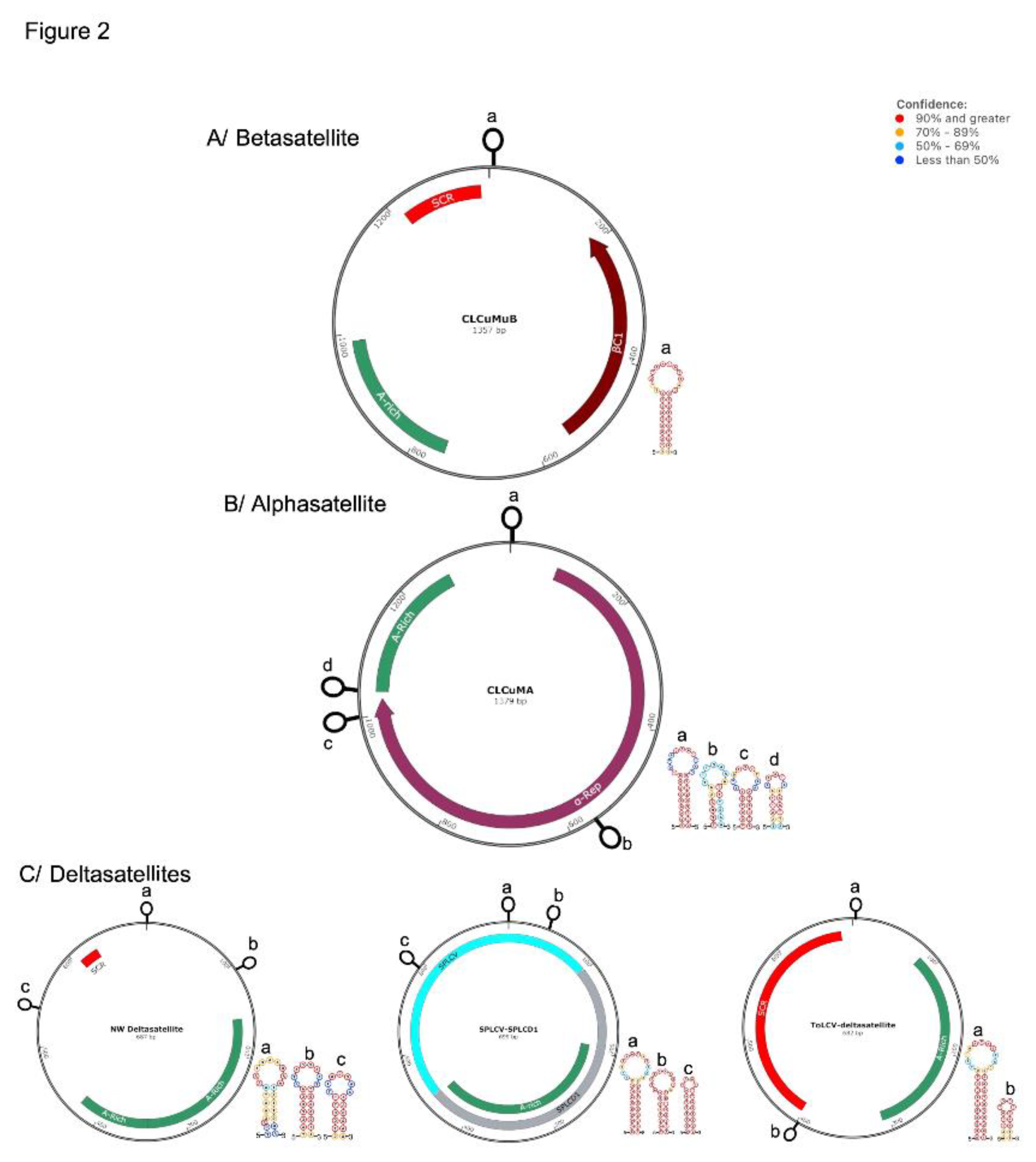
1.2. Betasatellites (family Tolecusatellitidae)
1.3. Alphasatellites (family Alphasatellitidae)
1.4. Deltasatellites (family Tolecusatellitidae)
2. Role of DNA Satellites on Virus Disease Development
2.1. Interaction between betasatellites and associated helper begomoviruses is synergistic
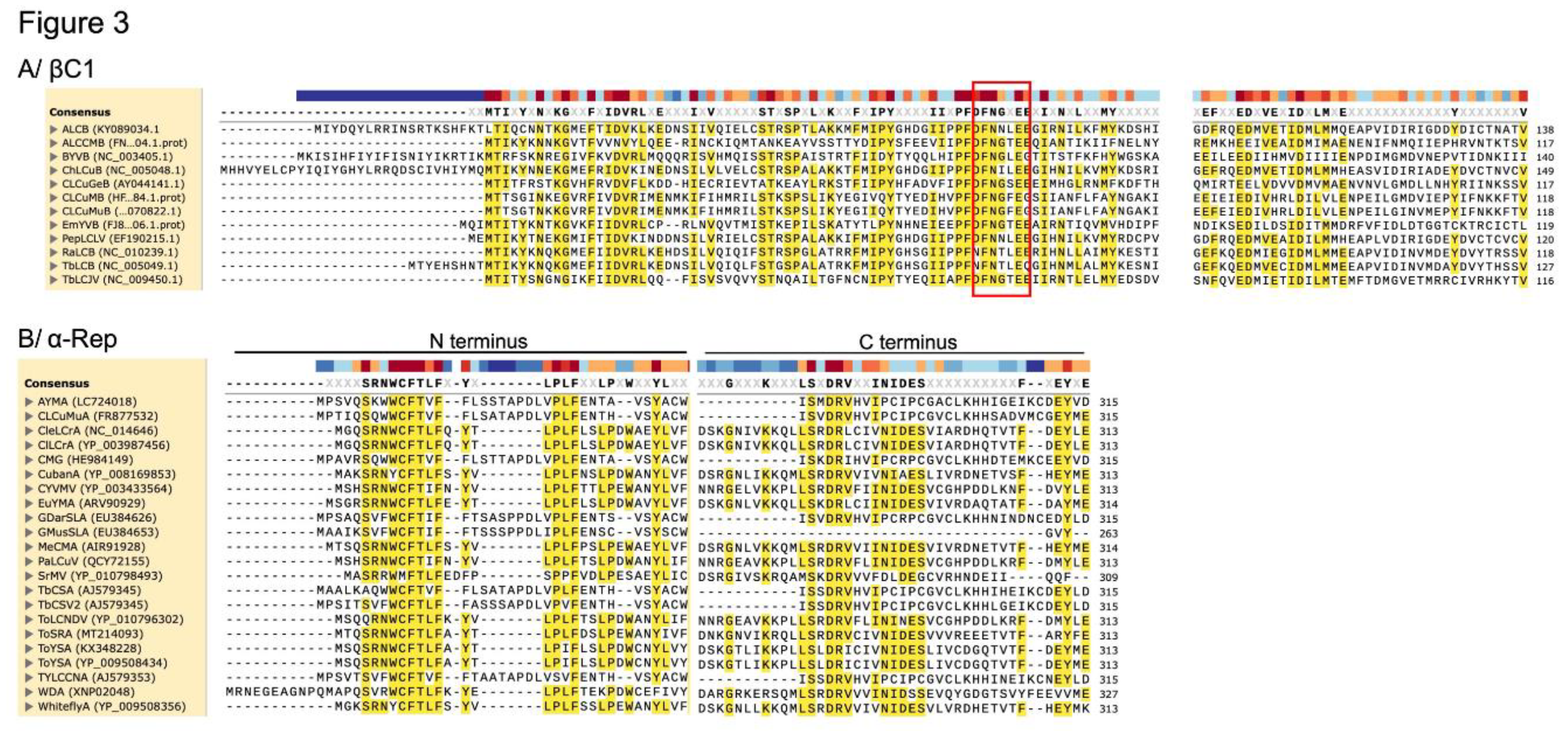
2.1.1. The βC1 RNA silencing suppression function contributes to betasatellite enhancement of helper virus symptoms
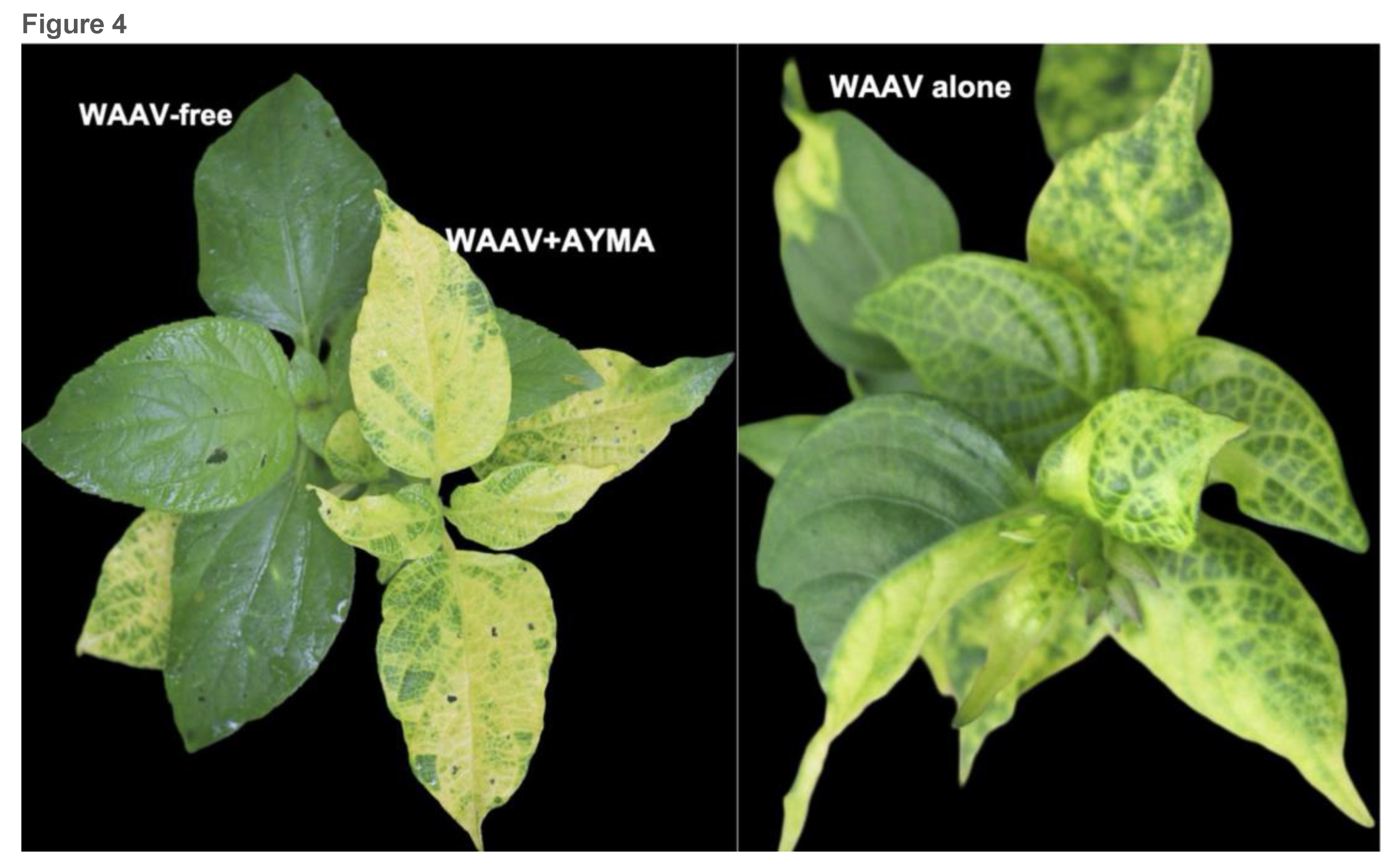
2.2. Alphasatellites tend to attenuate symptoms in coinfections with helper viruses.
2.2.1. The ability of alphasatellites to attenuate begomovirus disease symptoms may be due to α
2.3. Deltasatellites do not enhance helper virus DNA levels or symptom severity
2.4. DNA satellites complementation of begomoviruses from different regions may lead to new disease epidemics
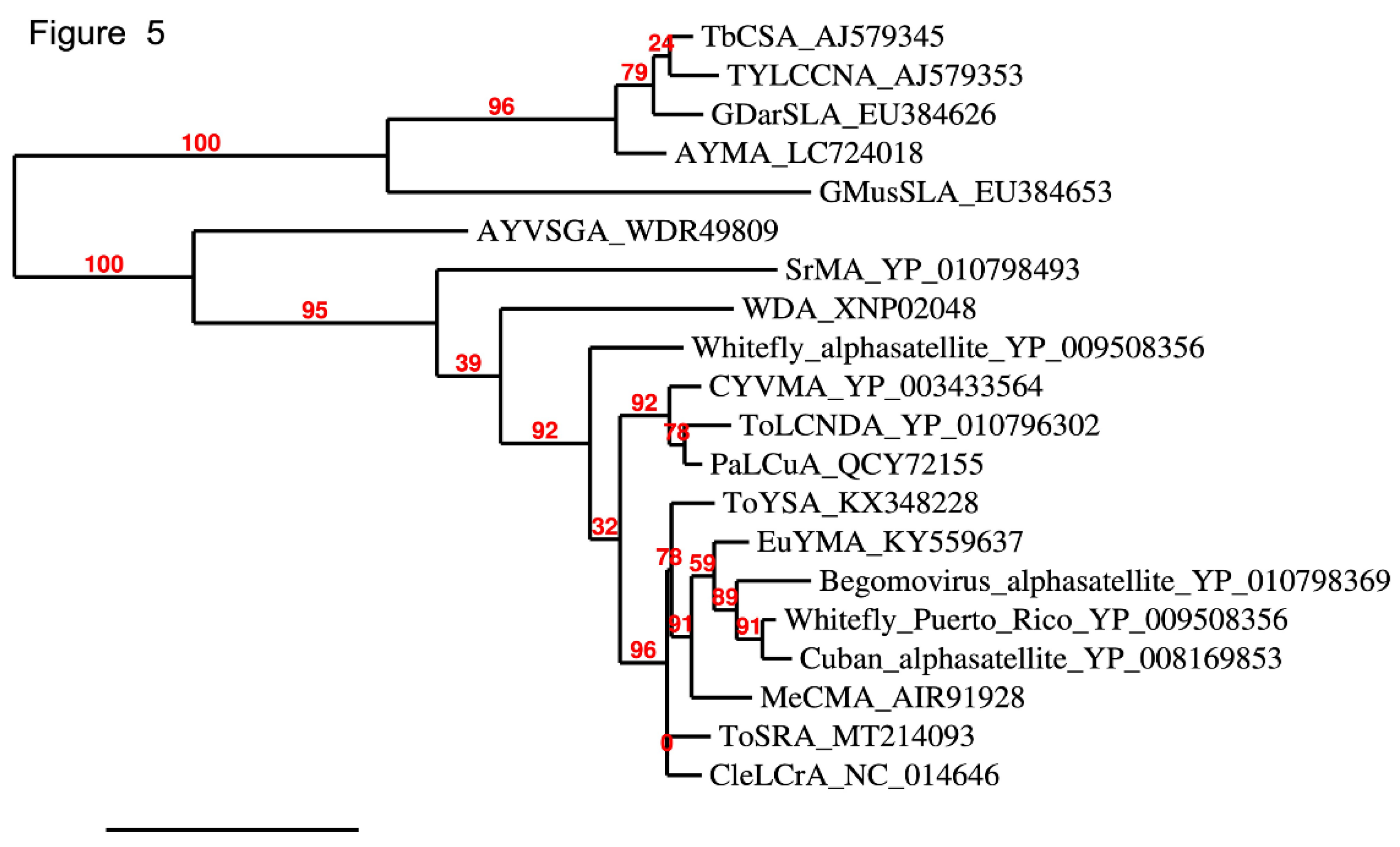
3. Recombination Between Helper Viruses and DNA Satellites Contributes to Virus and Satellite Evolution
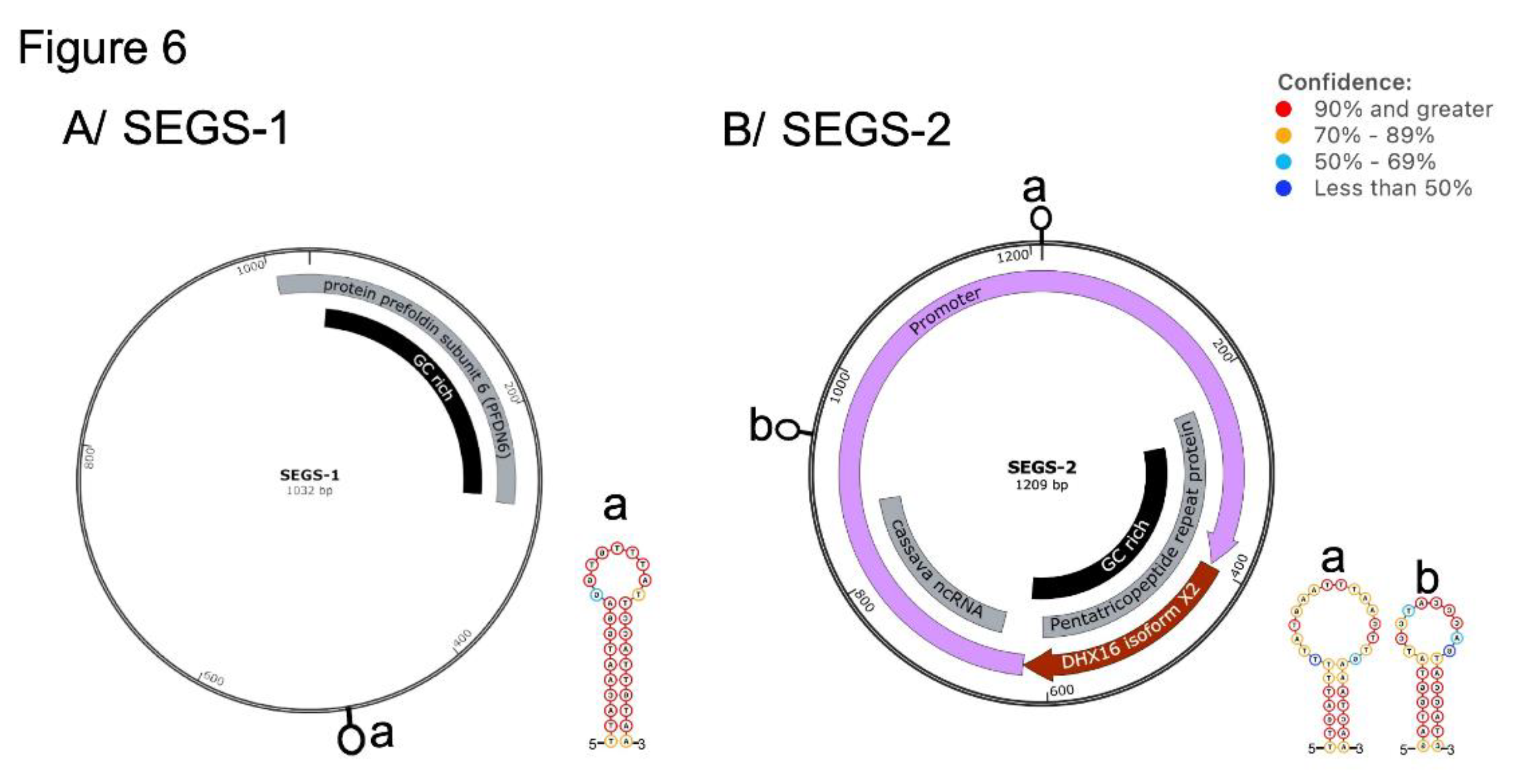
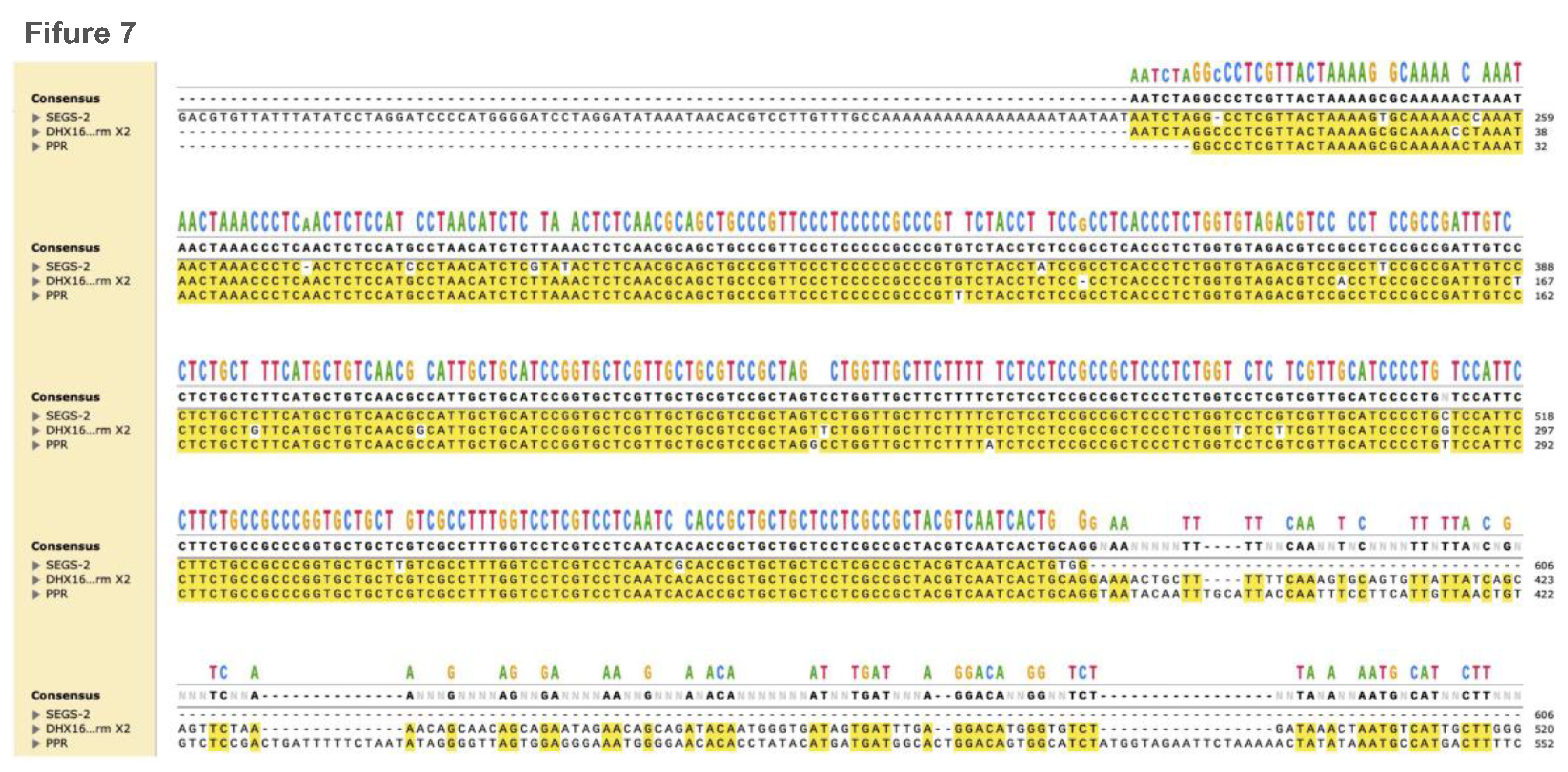
4. Cassava Geminivirus Associated SEGS-1 and SEGS-2 Episomes are Novel DNA Satellites
5. Outlook
5.1. Knowledge gaps that need addressing on the begomovirus diseases
5.2. DNA satellites and biotechnological management of begomovirus diseases
Acknowledgments
References
- Zerbini, F.M.; Briddon, R.W.; Idris, A.; Martin, D.P.; Moriones, E.; Navas-Castillo, J.; Rivera-Bustamante, R.; Roumagnac, P.; Varsani, A. ICTV Virus Taxonomy Profile: Geminiviridae. J. Gen. Virol. 2017, 98, 131–133. [Google Scholar] [CrossRef] [PubMed]
- Stenger, D.C.; Revington, G.N.; Stevenson, M.C.; Bisaro, D.M. Replicational Release of Geminivirus Genomes from Tandemly Repeated Copies: Evidence for Rolling-Circle Replication of a Plant Viral DNA. Proc. Natl. Acad. Sci. 1991, 88, 8029–8033. [Google Scholar] [CrossRef]
- Orozco, B.M.; Hanley-Bowdoin, L. A DNA Structure Is Required for Geminivirus Replication Origin Function. J. Virol. 1996, 70, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Fondong, V.N. Geminivirus Protein Structure and Function. Mol. Plant Pathol. 2013, 14, 635–649. [Google Scholar] [CrossRef]
- Fiallo-Olivé, E.; Lett, J.-M.; Martin, D.P.; Roumagnac, P.; Varsani, A.; Zerbini, F.M.; Navas-Castillo, J. ICTV Virus Taxonomy Profile: Geminiviridae 2021. J. Gen. Virol. 2021, 102, 001696. [Google Scholar] [CrossRef] [PubMed]
- Laufs, J.; Jupin, I.; David, C.; Schumacher, S.; Heyraud-Nitschke, F.; Gronenborn, B. Geminivirus Replication: Genetic and Biochemical Characterization of Rep Protein Function, a Review. Biochimie 1995, 77, 765–773. [Google Scholar] [CrossRef]
- Roossinck, M.J.; Sleat, D.; Palukaitis, P. Satellite RNAs of Plant Viruses: Structures and Biological Effects. Microbiol. Rev. 1992, 56, 265–279. [Google Scholar] [CrossRef]
- Simon, A.E. Satellite RNAs of Plant Viruses. Plant Mol. Biol. Report. 1988, 6, 240–252. [Google Scholar] [CrossRef]
- Dry, I.B.; Krake, L.R.; Rigden, J.E.; Rezaian, M.A. A Novel Subviral Agent Associated with a Geminivirus: The First Report of a DNA Satellite. Proc. Natl. Acad. Sci. 1997, 94, 7088–7093. [Google Scholar] [CrossRef]
- Haible, D.; Kober, S.; Jeske, H. Rolling Circle Amplification Revolutionizes Diagnosis and Genomics of Geminiviruses. J. Virol. Methods 2006, 135, 9–16. [Google Scholar] [CrossRef]
- Inoue-Nagata, A.K.; Albuquerque, L.C.; Rocha, W.B.; Nagata, T. A Simple Method for Cloning the Complete Begomovirus Genome Using the Bacteriophage Phi29 DNA Polymerase. J. Virol. Methods 2004, 116, 209–211. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Li, G.; Wang, D.; Hu, D.; Zhou, X. A Begomovirus DNAbeta-Encoded Protein Binds DNA, Functions as a Suppressor of RNA Silencing, and Targets the Cell Nucleus. J. Virol. 2005, 79, 10764–10775. [Google Scholar] [CrossRef]
- Gnanasekaran, P.; Ponnusamy, K.; Chakraborty, S. A Geminivirus Betasatellite Encoded βC1 Protein Interacts with PsbP and Subverts PsbP-mediated Antiviral Defence in Plants. Mol. Plant Pathol. 2019, 20, 943–960. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Tang, Y.; She, X.; Yu, L.; Lan, G.; Ding, S.; He, Z. Characterisation of a Betasatellite Associated With Tomato Yellow Leaf Curl Guangdong Virus and Discovery of an Unusual Modulation of Virus Infection Associated With C4 Protein. Mol. Plant Pathol. 2025, 26, e70051. [Google Scholar] [CrossRef]
- Mirzazadeh, M.; Heydarnejad, J.; Salari, A.; Massumi, H.; Esmaeili, M. Association of Tomato Leaf Curl Betasatellite (Betasatellite Solani) with the Turncurtovirus Sesame Curly Top Virus (Turncurtovirus Sesami) Increases the Severity of Symptoms in Watermelon Plants. Trop. Plant Pathol. 2025, 50, 5. [Google Scholar] [CrossRef]
- Saeed, M.; Zafar, Y.; Randles, J.W.; Rezaian, M.A. A Monopartite Begomovirus-Associated DNA Beta Satellite Substitutes for the DNA B of a Bipartite Begomovirus to Permit Systemic Infection. J. Gen. Virol. 2007, 88, 2881–2889. [Google Scholar] [CrossRef]
- Yang, J.-Y.; Iwasaki, M.; Machida, C.; Machida, Y.; Zhou, X.; Chua, N.-H. betaC1, the Pathogenicity Factor of TYLCCNV, Interacts with AS1 to Alter Leaf Development and Suppress Selective Jasmonic Acid Responses. Genes Dev. 2008, 22, 2564–2577. [Google Scholar] [CrossRef]
- Hu, T.; Song, Y.; Wang, Y.; Zhou, X. Functional Analysis of a Novel βV1 Gene Identified in a Geminivirus Betasatellite. Sci. China Life Sci. 2020, 63, 688–696. [Google Scholar] [CrossRef]
- Gupta, N.; Reddy, K.; Gnanasekaran, P.; Zhai, Y.; Chakraborty, S.; Pappu, H.R. Functional Characterization of a New ORF βV1 Encoded by Radish Leaf Curl Betasatellite. Front. Plant Sci. 2022, 13, 972386. [Google Scholar] [CrossRef]
- Maliano, M.R.; Melgarejo, T.A.; Rojas, M.R.; Barboza, N.; Gilbertson, R.L. The Begomovirus Species Melon Chlorotic Leaf Curl Virus Is Composed of Two Highly Divergent Strains That Differ in Their Genetic and Biological Properties. Plant Dis. 2021, 105, 3162–3170. [Google Scholar] [CrossRef]
- Briddon, R.W.; Bull, S.E.; Amin, I.; Idris, A.M.; Mansoor, S.; Bedford, I.D.; Dhawan, P.; Rishi, N.; Siwatch, S.S.; Abdel-Salam, A.M.; et al. Diversity of DNA Beta, a Satellite Molecule Associated with Some Monopartite Begomoviruses. Virology 2003, 312, 106–121. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Stanley, J. Subviral Agents Associated with Plant Single-Stranded DNA Viruses. Virology 2006, 344, 198–210. [Google Scholar] [CrossRef]
- Saunders, K.; Norman, A.; Gucciardo, S.; Stanley, J. The DNA Beta Satellite Component Associated with Ageratum Yellow Vein Disease Encodes an Essential Pathogenicity Protein (betaC1). Virology 2004, 324, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Saunders, K.; Briddon, R.W.; Stanley, J. Replication Promiscuity of DNA-Beta Satellites Associated with Monopartite Begomoviruses; Deletion Mutagenesis of the Ageratum Yellow Vein Virus DNA-Beta Satellite Localizes Sequences Involved in Replication. J. Gen. Virol. 2008, 89, 3165–3172. [Google Scholar] [CrossRef]
- Sattar, M.N.; Kvarnheden, A.; Saeed, M.; Briddon, R.W. Cotton Leaf Curl Disease - an Emerging Threat to Cotton Production Worldwide. J. Gen. Virol. 2013, 94, 695–710. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X. Advances in Understanding Begomovirus Satellites. Annu. Rev. Phytopathol. 2013, 51, 357–381. [Google Scholar] [CrossRef]
- Fiallo-Olivé, E.; Navas-Castillo, J. Begomoviruses: What Is the Secret(s) of Their Success? Trends Plant Sci. 2023, 28, 715–727. [Google Scholar] [CrossRef]
- Lozano, G.; Trenado, H.P.; Fiallo-Olivé, E.; Chirinos, D.; Geraud-Pouey, F.; Briddon, R.W.; Navas-Castillo, J. Characterization of Non-Coding DNA Satellites Associated with Sweepoviruses (Genus Begomovirus, Geminiviridae) – Definition of a Distinct Class of Begomovirus-Associated Satellites. Front. Microbiol. 2016, 7, 162. [Google Scholar] [CrossRef]
- Briddon, R.W.; Bull, S.E.; Amin, I.; Mansoor, S.; Bedford, I.D.; Rishi, N.; Siwatch, S.S.; Zafar, Y.; Abdel-Salam, A.M.; Markham, P.G. Diversity of DNA 1: A Satellite-like Molecule Associated with Monopartite Begomovirus-DNA Beta Complexes. Virology 2004, 324, 462–474. [Google Scholar] [CrossRef]
- Mansoor, S.; Khan, S.H.; Bashir, A.; Saeed, M.; Zafar, Y.; Malik, K.A.; Briddon, R.; Stanley, J.; Markham, P.G. Identification of a Novel Circular Single-Stranded DNA Associated with Cotton Leaf Curl Disease in Pakistan. Virology 1999, 259, 190–199. [Google Scholar] [CrossRef]
- Saunders, K.; Stanley, J. A Nanovirus-like DNA Component Associated with Yellow Vein Disease of Ageratum Conyzoides: Evidence for Interfamilial Recombination between Plant DNA Viruses. Virology 1999, 264, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Ferro, C.G.; Zerbini, F.M.; Navas-Castillo, J.; Fiallo-Olivé, E. Revealing the Complexity of Sweepovirus-Deltasatellite-Plant Host Interactions: Expanded Natural and Experimental Helper Virus Range and Effect Dependence on Virus-Host Combination. Microorganisms 2021, 9, 1018. [Google Scholar] [CrossRef] [PubMed]
- Fiallo-Olivé, E.; Martínez-Zubiaur, Y.; Moriones, E.; Navas-Castillo, J. A Novel Class of DNA Satellites Associated with New World Begomoviruses. Virology 2012, 426, 1–6. [Google Scholar] [CrossRef]
- Fiallo-Olivé, E.; Navas-Castillo, J. Molecular and Biological Characterization of a New World Mono-/Bipartite Begomovirus/Deltasatellite Complex Infecting Corchorus Siliquosus. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Bennett, A.; Mietzsch, M.; Agbandje-McKenna, M. Understanding Capsid Assembly and Genome Packaging for Adeno-Associated Viruses. Future Virol. 2017, 12, 283–297. [Google Scholar] [CrossRef]
- Bikard, D.; Loot, C.; Baharoglu, Z.; Mazel, D. Folded DNA in Action: Hairpin Formation and Biological Functions in Prokaryotes. Microbiol. Mol. Biol. Rev. MMBR 2010, 74, 570–588. [Google Scholar] [CrossRef] [PubMed]
- Musatov, S.; Roberts, J.; Pfaff, D.; Kaplitt, M. A Cis-Acting Element That Directs Circular Adeno-Associated Virus Replication and Packaging. J. Virol. 2002, 76, 12792–12802. [Google Scholar] [CrossRef]
- Russel, M.; Linderoth, N.A.; Sali, A. Filamentous Phage Assembly: Variation on a Protein Export Theme. Gene 1997, 192, 23–32. [Google Scholar] [CrossRef]
- Russel, M.; Model, P. Genetic Analysis of the Filamentous Bacteriophage Packaging Signal and of the Proteins That Interact with It. J. Virol. 1989, 63, 3284–3295. [Google Scholar] [CrossRef]
- Sun, S.; Rao, V.B.; Rossmann, M.G. Genome Packaging in Viruses. Curr. Opin. Struct. Biol. 2010, 20, 114–120. [Google Scholar] [CrossRef]
- Singh, A.; Chattopadhyay, B.; Chakraborty, S. Biology and Interactions of Two Distinct Monopartite Begomoviruses and Betasatellites Associated with Radish Leaf Curl Disease in India. Virol. J. 2012, 9, 43. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, D.; Gnanasekaran, P.; Kumar, R.K.; Kushwaha, N.K.; Sharma, V.K.; Yusuf, M.A.; Chakraborty, S. A Geminivirus Betasatellite Damages the Structural and Functional Integrity of Chloroplasts Leading to Symptom Formation and Inhibition of Photosynthesis. J. Exp. Bot. 2015, 66, 5881–5895. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Xie, Y.; Raja, P.; Li, S.; Wolf, J.N.; Shen, Q.; Bisaro, D.M.; Zhou, X. Suppression of Methylation-Mediated Transcriptional Gene Silencing by βC1-SAHH Protein Interaction during Geminivirus-Betasatellite Infection. PLOS Pathog. 2011, 7, e1002329. [Google Scholar] [CrossRef]
- Tahir, M.; Haider, M.S.; Briddon, R.W. Chili Leaf Curl Betasatellite Is Associated with a Distinct Recombinant Begomovirus, Pepper Leaf Curl Lahore Virus, in Capsicum in Pakistan. Virus Res. 2010, 149, 109–114. [Google Scholar] [CrossRef]
- Venkataravanappa, V.; Kodandaram, M.H.; Prasanna, H.C.; Reddy, M.K.; Reddy, C.N.L. Unraveling Different Begomoviruses, DNA Satellites and Cryptic Species of Bemisia Tabaci and Their Endosymbionts in Vegetable Ecosystem. Microb. Pathog. 2023, 174, 105892. [Google Scholar] [CrossRef]
- Leke, W.N.; Sattar, M.N.; Ngane, E.B.; Ngeve, J.M.; Kvarnheden, A.; Brown, J.K. Molecular Characterization of Begomoviruses and DNA Satellites Associated with Okra Leaf Curl Disease in Cameroon. Virus Res. 2013, 174, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Leke, W.N.; Brown, J.K.; Ligthart, M.E.; Sattar, N.; Njualem, D.K.; Kvarnheden, A. Ageratum Conyzoides: A Host to a Unique Begomovirus Disease Complex in Cameroon. Virus Res. 2012, 163, 229–237. [Google Scholar] [CrossRef]
- Kon, T.; Gilbertson, R.L. Two Genetically Related Begomoviruses Causing Tomato Leaf Curl Disease in Togo and Nigeria Differ in Virulence and Host Range but Do Not Require a Betasatellite for Induction of Disease Symptoms. Arch. Virol. 2012, 157, 107–120. [Google Scholar] [CrossRef]
- Li, Z.; Xie, Y.; Zhou, X. Tobacco Curly Shoot Virus DNAβ Is Not Necessary for Infection but Intensifies Symptoms in a Host-Dependent Manner. Phytopathology® 2005, 95, 902–908. [Google Scholar] [CrossRef]
- Chen, L.-F.; Rojas, M.; Kon, T.; Gamby, K.; Xoconostle-Cazares, B.; Gilbertson, R.L. A Severe Symptom Phenotype in Tomato in Mali Is Caused by a Reassortant between a Novel Recombinant Begomovirus (Tomato Yellow Leaf Curl Mali Virus) and a Betasatellite. Mol. Plant Pathol. 2009, 10, 415–430. [Google Scholar] [CrossRef]
- Tiendrébéogo, F.; Lefeuvre, P.; Hoareau, M.; Villemot, J.; Konaté, G.; Traoré, A.S.; Barro, N.; Traoré, V.S.; Reynaud, B.; Traoré, O.; et al. Molecular Diversity of Cotton Leaf Curl Gezira Virus Isolates and Their Satellite DNAs Associated with Okra Leaf Curl Disease in Burkina Faso. Virol. J. 2010, 7, 48. [Google Scholar] [CrossRef] [PubMed]
- Mollel, H.G.; Ndunguru, J.; Sseruwagi, P.; Alicai, T.; Colvin, J.; Navas-Castillo, J.; Fiallo-Olivé, E. A Novel East African Monopartite Begomovirus-Betasatellite Complex That Infects Vernonia Amygdalina. Arch. Virol. 2017, 162, 1079–1082. [Google Scholar] [CrossRef]
- Li, F.; Huang, C.; Li, Z.; Zhou, X. Suppression of RNA Silencing by a Plant DNA Virus Satellite Requires a Host Calmodulin-Like Protein to Repress RDR6 Expression. PLOS Pathog. 2014, 10, e1003921. [Google Scholar] [CrossRef] [PubMed]
- Eini, O. [A Betasatellite-Encoded Protein Regulates Key Components of Gene Silencing System in Plants]. Mol. Biol. (Mosk.) 2017, 51, 656–663. [Google Scholar] [CrossRef] [PubMed]
- Kumar P, P.; Usha, R.; Zrachya, A.; Levy, Y.; Spanov, H.; Gafni, Y. Protein-Protein Interactions and Nuclear Trafficking of Coat Protein and betaC1 Protein Associated with Bhendi Yellow Vein Mosaic Disease. Virus Res. 2006, 122, 127–136. [Google Scholar] [CrossRef]
- Fu, Y.; Xu, L.; Xu, B.; Yang, L.; Ling, Q.; Wang, H.; Huang, H. Genetic Interactions between Leaf Polarity-Controlling Genes and ASYMMETRIC LEAVES1 and 2 in Arabidopsis Leaf Patterning. Plant Cell Physiol. 2007, 48, 724–735. [Google Scholar] [CrossRef]
- Zubair, M.; Zaidi, S.S.-A.; Shakir, S.; Amin, I.; Mansoor, S. An Insight into Cotton Leaf Curl Multan Betasatellite, the Most Important Component of Cotton Leaf Curl Disease Complex. Viruses 2017, 9, 280. [Google Scholar] [CrossRef]
- Harimalala, M.; De Bruyn, A.; Hoareau, M.; Andrianjaka, A.; Ranomenjanahary, S.; Reynaud, B.; Lefeuvre, P.; Lett, J.-M. Molecular Characterization of a New Alphasatellite Associated with a Cassava Mosaic Geminivirus in Madagascar. Arch. Virol. 2013, 158, 1829–1832. [Google Scholar] [CrossRef]
- Kumar, M.; Zarreen, F.; Chakraborty, S. Roles of Two Distinct Alphasatellites Modulating Geminivirus Pathogenesis. Virol. J. 2021, 18, 249. [Google Scholar] [CrossRef]
- Romay, G.; Chirinos, D.; Geraud-Pouey, F.; Desbiez, C. Association of an Atypical Alphasatellite with a Bipartite New World Begomovirus. Arch. Virol. 2010, 155, 1843–1847. [Google Scholar] [CrossRef]
- Jeske, H.; Kober, S.; Schäfer, B.; Strohmeier, S. Circomics of Cuban Geminiviruses Reveals the First Alpha-Satellite DNA in the Caribbean. Virus Genes 2014, 49, 312–324. [Google Scholar] [CrossRef] [PubMed]
- Paprotka, T.; Metzler, V.; Jeske, H. The First DNA 1-like Alpha Satellites in Association with New World Begomoviruses in Natural Infections. Virology 2010, 404, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Rosario, K.; Marr, C.; Varsani, A.; Kraberger, S.; Stainton, D.; Moriones, E.; Polston, J.E.; Breitbart, M. Begomovirus-Associated Satellite DNA Diversity Captured Through Vector-Enabled Metagenomic (VEM) Surveys Using Whiteflies (Aleyrodidae). Viruses 2016, 8, 36. [Google Scholar] [CrossRef]
- Yoboué, A.A.N.; Kouakou, B.S.M.; Pita, J.S.; N’Zué, B.; Amoakon, W.J.-L.; Kouassi, K.M.; Vanié-Léabo, L.P.L.; Kouassi, N.K.; Sorho, F.; Zouzou, M. Emergence of Begomoviruses and DNA Satellites Associated with Weeds and Intercrops: A Potential Threat to Sustainable Production of Cassava in Côte d’Ivoire. Front. Plant Sci. 2025, 16. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wu, P.; Liu, P.; Gong, H.; Zhou, X. Characterization of Alphasatellites Associated with Monopartite Begomovirus/Betasatellite Complexes in Yunnan, China. Virol. J. 2010, 7, 178. [Google Scholar] [CrossRef]
- Zhao, L.; Che, X.; Wang, Z.; Zhou, X.; Xie, Y. Functional Characterization of Replication-Associated Proteins Encoded by Alphasatellites Identified in Yunnan Province, China. Viruses 2022, 14, 222. [Google Scholar] [CrossRef]
- Abbas, Q.; Amin, I.; Mansoor, S.; Shafiq, M.; Wassenegger, M.; Briddon, R.W. The Rep Proteins Encoded by Alphasatellites Restore Expression of a Transcriptionally Silenced Green Fluorescent Protein Transgene in Nicotiana Benthamiana. VirusDisease 2019, 30, 101–105. [Google Scholar] [CrossRef]
- Nawaz-Ul-Rehman, M.S.; Nahid, N.; Mansoor, S.; Briddon, R.W.; Fauquet, C.M. Post-Transcriptional Gene Silencing Suppressor Activity of Two Non-Pathogenic Alphasatellites Associated with a Begomovirus. Virology 2010, 405, 300–308. [Google Scholar] [CrossRef]
- Mar, T.B.; Mendes, I.R.; Lau, D.; Fiallo-Olivé, E.; Navas-Castillo, J.; Alves, M.S.; Murilo Zerbini, F. Interaction between the New World Begomovirus Euphorbia Yellow Mosaic Virus and Its Associated Alphasatellite: Effects on Infection and Transmission by the Whitefly Bemisia Tabaci. J. Gen. Virol. 2017, 98, 1552–1562. [Google Scholar] [CrossRef]
- Nogueira, A.M.; Nascimento, M.B.; Barbosa, T.M.C.; Quadros, A.F.F.; Gomes, J.P.A.; Orílio, A.F.; Barros, D.R.; Zerbini, F.M. The Association between New World Alphasatellites and Bipartite Begomoviruses: Effects on Infection and Vector Transmission. Pathogens 2021, 10, 1244. [Google Scholar] [CrossRef]
- Fiallo-Olivé, E.; Tovar, R.; Navas-Castillo, J. Deciphering the Biology of Deltasatellites from the New World: Maintenance by New World Begomoviruses and Whitefly Transmission. New Phytol. 2016, 212, 680–692. [Google Scholar] [CrossRef] [PubMed]
- Hassan, I.; Orílio, A.F.; Fiallo-Olivé, E.; Briddon, R.W.; Navas-Castillo, J. Infectivity, Effects on Helper Viruses and Whitefly Transmission of the Deltasatellites Associated with Sweepoviruses (Genus Begomovirus, Family Geminiviridae). Sci. Rep. 2016, 6, 30204. [Google Scholar] [CrossRef]
- Sattar, M.N.; Ligthart, M.; Kvarnheden, A. Compatibility and Interaction of Begomoviruses and DNA-Satellites Causing Leaf Curl Disease in Asia, Africa and Mediterranean Region. Eur. J. Plant Pathol. 2019, 155, 111–124. [Google Scholar] [CrossRef]
- Fouad, N.; Granier, M.; Blanc, S.; Thébaud, G.; Urbino, C. Demonstration of Insect Vector-Mediated Transfer of a Betasatellite between Two Helper Viruses. Viruses 2024, 16, 1420. [Google Scholar] [CrossRef]
- Varma, A.; Singh, M.K. The Role of Satellites in the Evolution of Begomoviruses. Viruses 2024, 16, 970. [Google Scholar] [CrossRef] [PubMed]
- Stanley, J.; Saunders, K.; Pinner, M.S.; Wong, S.M. Novel Defective Interfering DNAs Associated with Ageratum Yellow Vein Geminivirus Infection of Ageratum Conyzoides. Virology 1997, 239, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Fiallo-Olivé, E.; Navas-Castillo, J. The Role of Extensive Recombination in the Evolution of Geminiviruses. Curr. Top. Microbiol. Immunol. 2023, 439, 139–166. [Google Scholar] [CrossRef]
- Baig, M.S.; Akhtar, S.; Khan, J.A. Engineering Tolerance to CLCuD in Transgenic Gossypium Hirsutum Cv. HS6 Expressing Cotton Leaf Curl Multan Virus-C4 Intron Hairpin. Sci. Rep. 2021, 11, 14172. [Google Scholar] [CrossRef]
- Biswas, K.K.; Bhattacharyya, U.K.; Palchoudhury, S.; Balram, N.; Kumar, A.; Arora, R.; Sain, S.K.; Kumar, P.; Khetarpal, R.K.; Sanyal, A.; et al. Dominance of Recombinant Cotton Leaf Curl Multan-Rajasthan Virus Associated with Cotton Leaf Curl Disease Outbreak in Northwest India. PLoS ONE 2020, 15, e0231886. [Google Scholar] [CrossRef]
- Qadir, R.; Khan, Z.A.; Monga, D.; Khan, J.A. Diversity and Recombination Analysis of Cotton Leaf Curl Multan Virus: A Highly Emerging Begomovirus in Northern India. BMC Genomics 2019, 20, 274. [Google Scholar] [CrossRef]
- Amin, I.; Mansoor, S.; Amrao, L.; Hussain, M.; Irum, S.; Zafar, Y.; Bull, S.E.; Briddon, R.W. Mobilisation into Cotton and Spread of a Recombinant Cotton Leaf Curl Disease Satellite. Arch. Virol. 2006, 151, 2055–2065. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Amin, I.; Zaidi, S.S.-E.-A.; Rahman, S.U.; Farooq, M.; Fauquet, C.M.; Mansoor, S. Circular DNA Enrichment Sequencing Reveals the Viral/Satellites Genetic Diversity Associated with the Third Epidemic of Cotton Leaf Curl Disease. Biol. Methods Protoc. 2021, 6, bpab005. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Akbar, F.; Iqbal, Z.; Amrao, L.; Amin, I.; Saeed, M.; Mansoor, S. Effects of Genetic Changes to the Begomovirus/Betasatellite Complex Causing Cotton Leaf Curl Disease in South Asia Post-Resistance Breaking. Virus Res. 2014, 186, 114–119. [Google Scholar] [CrossRef]
- Mansoor, S.; Amin, I.; Iram, S.; Hussain, M.; Zafar, Y.; Malik, K.A.; Briddon, R.W. Breakdown of Resistance in Cotton to Cotton Leaf Curl Disease in Pakistan. Plant Pathol. 2003, 52, 784–784. [Google Scholar] [CrossRef]
- Aimone, C.D.; De León, L.; Dallas, M.M.; Ndunguru, J.; Ascencio-Ibáñez, J.T.; Hanley-Bowdoin, L. A New Type of Satellite Associated with Cassava Mosaic Begomoviruses. J. Virol. 2021, 95, e0043221. [Google Scholar] [CrossRef]
- Ndunguru, J.; De León, L.; Doyle, C.D.; Sseruwagi, P.; Plata, G.; Legg, J.P.; Thompson, G.; Tohme, J.; Aveling, T.; Ascencio-Ibáñez, J.T.; et al. Two Novel DNAs That Enhance Symptoms and Overcome CMD2 Resistance to Cassava Mosaic Disease. J. Virol. 2016, 90, 4160–4173. [Google Scholar] [CrossRef] [PubMed]
- Torkpo, S.K.; Amponsah, E.; Arhin, C.D.; Offei, S.K. Occurrence of Cassava Mosaic Begomovirus-Associated Satellites on Cassava in Ghana. Cogent Food Agric. 2021, 7, 1963929. [Google Scholar] [CrossRef]
- Borah, B.K.; Cheema, G.S.; Gill, C.K.; Dasgupta, I. A Geminivirus-Satellite Complex Is Associated with Leaf Deformity of Mentha (Mint) Plants in Punjab. Indian J. Virol. Off. Organ Indian Virol. Soc. 2010, 21, 103–109. [Google Scholar] [CrossRef]
- Geissler, S.; Siegers, K.; Schiebel, E. A Novel Protein Complex Promoting Formation of Functional Alpha- and Gamma-Tubulin. EMBO J. 1998, 17, 952–966. [Google Scholar] [CrossRef]
- Herranz-Montoya, I.; Park, S.; Djouder, N. A Comprehensive Analysis of Prefoldins and Their Implication in Cancer. iScience 2021, 24, 103273. [Google Scholar] [CrossRef]
- Li, X.; Sun, M.; Liu, S.; Teng, Q.; Li, S.; Jiang, Y. Functions of PPR Proteins in Plant Growth and Development. Int. J. Mol. Sci. 2021, 22, 11274. [Google Scholar] [CrossRef] [PubMed]
- Umate, P.; Tuteja, R.; Tuteja, N. Genome-Wide Analysis of Helicase Gene Family from Rice and Arabidopsis: A Comparison with Yeast and Human. Plant Mol. Biol. 2010, 73, 449–465. [Google Scholar] [CrossRef] [PubMed]
- Ashikawa, I. Gene-Associated CpG Islands in Plants as Revealed by Analyses of Genomic Sequences. Plant J. Cell Mol. Biol. 2001, 26, 617–625. [Google Scholar] [CrossRef]
- Deaton, A.M.; Bird, A. CpG Islands and the Regulation of Transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef] [PubMed]
- Stephens, K.E.; Miaskowski, C.A.; Levine, J.D.; Pullinger, C.R.; Aouizerat, B.E. Epigenetic Regulation and Measurement of Epigenetic Changes. Biol. Res. Nurs. 2013, 15, 373–381. [Google Scholar] [CrossRef]
- Zhang, X.; Yazaki, J.; Sundaresan, A.; Cokus, S.; Chan, S.W.-L.; Chen, H.; Henderson, I.R.; Shinn, P.; Pellegrini, M.; Jacobsen, S.E.; et al. Genome-Wide High-Resolution Mapping and Functional Analysis of DNA Methylation in Arabidopsis. Cell 2006, 126, 1189–1201. [Google Scholar] [CrossRef]
- Zilberman, D.; Gehring, M.; Tran, R.K.; Ballinger, T.; Henikoff, S. Genome-Wide Analysis of Arabidopsis Thaliana DNA Methylation Uncovers an Interdependence between Methylation and Transcription. Nat. Genet. 2007, 39, 61–69. [Google Scholar] [CrossRef]
- Chiunga, E.; Aimone, C.D.; Rajabu, C.; Dallas, M.M.; Ndunguru, J.; Ascencio-Ibáñez, J.T.; Ateka, E.M.; Hanley-Bowdoin, L. SEGS-1 Episomes Generated during Cassava Mosaic Disease Enhance Disease Severity. Front. Plant Sci. 2024, 15, 1469045. [Google Scholar] [CrossRef]
- Nagar, S.; Hanley-Bowdoin, L.; Robertson, D. Host DNA Replication Is Induced by Geminivirus Infection of Differentiated Plant Cells. Plant Cell 2002, 14, 2995–3007. [Google Scholar] [CrossRef]
- Ascencio-Ibáñez, J.T.; Sozzani, R.; Lee, T.-J.; Chu, T.-M.; Wolfinger, R.D.; Cella, R.; Hanley-Bowdoin, L. Global Analysis of Arabidopsis Gene Expression Uncovers a Complex Array of Changes Impacting Pathogen Response and Cell Cycle during Geminivirus Infection. Plant Physiol. 2008, 148, 436–454. [Google Scholar] [CrossRef]
- Gui, X.; Liu, C.; Qi, Y.; Zhou, X. Geminiviruses Employ Host DNA Glycosylases to Subvert DNA Methylation-Mediated Defense. Nat. Commun. 2022, 13, 575. [Google Scholar] [CrossRef] [PubMed]
- Rajabu, C.A.; Dallas, M.M.; Chiunga, E.; De León, L.; Ateka, E.M.; Tairo, F.; Ndunguru, J.; Ascencio-Ibanez, J.T.; Hanley-Bowdoin, L. SEGS-1 a Cassava Genomic Sequence Increases the Severity of African Cassava Mosaic Virus Infection in Arabidopsis Thaliana. Front. Plant Sci. 2023, 14, 1250105. [Google Scholar] [CrossRef]
- Fortes, I.M.; Pérez-Padilla, V.; Romero-Rodríguez, B.; Fernández-Muñoz, R.; Moyano, C.; Castillo, A.G.; De León, L.; Moriones, E. Begomovirus Tomato Leaf Curl New Delhi Virus Is Seedborne but Not Seed Transmitted in Melon. Plant Dis. 2023, 107, 473–479. [Google Scholar] [CrossRef]
- Moriones, E.; Praveen, S.; Chakraborty, S. Tomato Leaf Curl New Delhi Virus: An Emerging Virus Complex Threatening Vegetable and Fiber Crops. Viruses 2017, 9, 264. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Pendón, J.A.; Sánchez-Campos, S.; Fortes, I.M.; Moriones, E. Tomato Yellow Leaf Curl Sardinia Virus, a Begomovirus Species Evolving by Mutation and Recombination: A Challenge for Virus Control. Viruses 2019, 11, 45. [Google Scholar] [CrossRef]
- Leke, W.N.; Khatabi, B.; Mignouna, D.B.; Brown, J.K.; Fondong, V.N. Complete Genome Sequence of a New Bipartite Begomovirus Infecting Cotton in the Republic of Benin in West Africa. Arch. Virol. 2016, 161, 2329–2333. [Google Scholar] [CrossRef] [PubMed]
- Leke, W.N.; Mignouna, D.B.; Brown, J.K.; Fondong, V.N. First Report of Soybean Chlorotic Blotch Virus and West African Asystasia Virus 1 Infecting Cassava and a Wild Cassava Relative in Cameroon and Togo. New Dis. Rep. 2016, 33, 24–24. [Google Scholar] [CrossRef]
- Keita, G.K.; Kansaye, L.; Doumbia, L.; Keita, I.; Sangare, M.; Sinayoko, T.; Macalou, B.; Maiga, M.N.; Sanogo, N.P.; Koita, O. Molecular Identification and Characterization of a Begomovirus Associated with Okra Enation Leaf Curl Disease in Mali. Adv. Biosci. Biotechnol. 2023, 14, 429–438. [Google Scholar] [CrossRef]
- Baulcombe, D.C.; Saunders, G.R.; Bevan, M.W.; Mayo, M.A.; Harrison, B.D. Expression of Biologically Active Viral Satellite RNA from the Nuclear Genome of Transformed Plants. Nature 1986, 321, 446–449. [Google Scholar] [CrossRef]
- Cao, X.; Liu, S.; Yu, C.; Li, X.; Yuan, X. A New Strategy of Using Satellite RNA to Control Viral Plant Diseases: Post-inoculation with Satellite RNA Attenuates Symptoms Derived from Pre-infection with Its Helper Virus. Plant Biotechnol. J. 2019, 17, 1856–1858. [Google Scholar] [CrossRef]
- Cillo, F.; Finetti-Sialer, M.M.; Papanice, M.A.; Gallitelli, D. Analysis of Mechanisms Involved in the Cucumber Mosaic Virus Satellite RNA-Mediated Transgenic Resistance in Tomato Plants. Mol. Plant-Microbe Interactions® 2004, 17, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Gerlach, W.L.; Llewellyn, D.; Haseloff, J. Construction of a Plant Disease Resistance Gene from the Satellite RNA of Tobacco Ringspot Virus. Nature 1987, 328, 802–805. [Google Scholar] [CrossRef]
- Lin, K.-Y.; Lin, N.-S. Interfering Satellite RNAs of Bamboo Mosaic Virus. Front. Microbiol. 2017, 8, 787. [Google Scholar] [CrossRef]
- Simon, A.E.; Roossinck, M.J.; Havelda, Z. PLANT VIRUS SATELLITE AND DEFECTIVE INTERFERING RNAS: New Paradigms for a New Century. Annu. Rev. Phytopathol. 2004, 42, 415–437. [Google Scholar] [CrossRef] [PubMed]
- Tien, P.; Wu, G. Satellite RNA for the Biocontrol of Plant Disease. In Advances in Virus Research; Maramorosch, K., Murphy, F.A., Shatkin, A.J., Eds.; Academic Press, 1991; Vol. 39, pp. 321–339.
- Fondong, V.N. The Search for Resistance to Cassava Mosaic Geminiviruses: How Much We Have Accomplished, and What Lies Ahead. Front. Plant Sci. 2017, 8, 408. [Google Scholar] [CrossRef]
- Fondong, V.N.; Rey, C.; Fondong, V.N.; Rey, C. Recent Biotechnological Advances in the Improvement of Cassava; IntechOpen, 2018; ISBN 978-953-51-3741-2.
- Giudice, G.; Moffa, L.; Varotto, S.; Cardone, M.F.; Bergamini, C.; De Lorenzis, G.; Velasco, R.; Nerva, L.; Chitarra, W. Novel and Emerging Biotechnological Crop Protection Approaches. Plant Biotechnol. J. 2021, 19, 1495–1510. [Google Scholar] [CrossRef]
- Niraula, P.M.; Fondong, V.N. Development and Adoption of Genetically Engineered Plants for Virus Resistance: Advances, Opportunities and Challenges. Plants Basel Switz. 2021, 10, 2339. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-Replace Genome Editing without Double-Strand Breaks or Donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Baldrich, P.; Liu, A.; Meyers, B.C.; Fondong, V.N. An Atlas of Small RNAs from Potato. Plant Direct 2022, 6, e466. [Google Scholar] [CrossRef]
- Luan, W.; Dai, Y.; Li, X.-Y.; Wang, Y.; Tao, X.; Li, C.-X.; Mao, P.; Ma, X.-R. Identification of tRFs and phasiRNAs in Tomato (Solanum Lycopersicum) and Their Responses to Exogenous Abscisic Acid. BMC Plant Biol. 2020, 20, 320. [Google Scholar] [CrossRef]
- Khatabi, B.; Arikit, S.; Xia, R.; Winter, S.; Oumar, D.; Mongomake, K.; Meyers, B.C.; Fondong, V.N. High-Resolution Identification and Abundance Profiling of Cassava (Manihot Esculenta Crantz) microRNAs. BMC Genomics 2016, 17, 85. [Google Scholar] [CrossRef] [PubMed]
- Seo, E.; Kim, T.; Park, J.H.; Yeom, S.-I.; Kim, S.; Seo, M.-K.; Shin, C.; Choi, D. Genome-Wide Comparative Analysis in Solanaceous Species Reveals Evolution of microRNAs Targeting Defense Genes in Capsicum Spp. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2018, 25, 561–575. [Google Scholar] [CrossRef]
- Zhao, T.; Tao, X.; Li, M.; Gao, M.; Chen, J.; Zhou, N.; Mei, G.; Fang, L.; Ding, L.; Zhou, B.; et al. Role of phasiRNAs from Two Distinct Phasing Frames of GhMYB2 Loci in Cis- Gene Regulation in the Cotton Genome. BMC Plant Biol. 2020, 20, 219. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



