I. Introduction
Skin diseases are among the most prevalent conditions worldwide, significantly affecting people's health and quality of life. With the continuous advancement of medical imaging technologies, a large number of skin disease images have been collected and stored[
1]. This provides a solid foundation for the development of computer-aided diagnosis systems. Among various auxiliary diagnostic techniques, image segmentation plays a crucial role as an essential pre-processing step in skin image analysis[
2]. It is vital for subsequent tasks such as feature extraction, lesion classification, and treatment planning. Accurately and efficiently separating skin lesions from the background can greatly improve the reliability and automation of diagnoses. Therefore, researching advanced semantic segmentation methods for skin disease images holds important theoretical and practical value.
Traditional image segmentation methods, such as thresholding, edge detection, and region growing, often show clear limitations when dealing with complex skin disease images. These images frequently feature blurry boundaries, complex textures, and significant color variations, making simple methods unable to produce satisfactory segmentation results[
3]. With the rise of deep learning, convolutional neural networks (CNNs) have demonstrated superior performance in semantic segmentation and feature extraction. Encoder-decoder network designs, in particular, can effectively capture both local and global information, improving segmentation accuracy. However, fully extracting and fusing multi-scale feature information remains a critical challenge when addressing multi-scale lesion areas.
Feature Pyramid Networks (FPN) provide an effective mechanism for multi-scale feature fusion. Through a top-down pathway and lateral connections, FPNs integrate feature maps across different levels, enhancing the model's ability to detect small objects and fine details. For skin disease images, where lesion areas vary greatly in scale and shape, using a feature pyramid structure can significantly improve the recognition of small lesions and complex backgrounds. However, traditional feature pyramid methods still suffer from information loss and semantic shifts during feature transmission and fusion. These issues limit their further development in high-precision segmentation tasks. Therefore, exploring an extended feature pyramid mechanism to better utilize multi-scale features and enhance semantic representation is of great significance for skin disease segmentation[
4,
5].
Against this background, research on skin disease segmentation algorithms based on extended feature pyramids has become a challenging yet promising direction. By introducing richer feature fusion strategies, such as dynamic convolution, deformable convolution, or attention mechanisms, it is possible to build more effective information transfer paths between different scales and semantic levels. This can improve the model's recognition accuracy for skin lesions. Furthermore, an extended feature pyramid structure can enhance the ability to capture fine-grained information, enabling more precise segmentation in images with blurry boundaries and complex lesion morphologies. This will not only drive technical progress in computer-aided skin disease diagnosis but also offer new ideas and methods for multi-scale feature modeling in medical image segmentation.
With the continuous advancement of intelligent healthcare and the rapid evolution of deep learning technologies, developing smarter, more robust, and more efficient skin disease image segmentation algorithms has become an important and urgent task. Segmentation methods based on extended feature pyramids offer superior performance in multi-scale perception, fine-grained feature expression, and adaptation to complex backgrounds. They have broad application potential and great value for wider adoption. Research in this area can improve diagnostic accuracy and efficiency, reduce errors caused by manual annotation and subjective judgment, and further promote the application of medical image analysis technologies in other disease detection and treatment assistance fields. Therefore, in-depth exploration of skin disease segmentation algorithms based on extended feature pyramids holds significant theoretical and practical value and deserves sustained research and innovation.
II. Background
In recent years, deep learning has significantly advanced the field of medical image segmentation, particularly for applications involving skin lesion analysis. UNet and its numerous variants have emerged as foundational architectures for semantic segmentation tasks due to their ability to capture both local details and global context through encoder-decoder structures. A comprehensive review of medical image segmentation techniques based on modified UNet architectures illustrates how enhancements in skip connections, attention modules, and multi-scale processing can lead to improved segmentation performance, especially in the domain of dermatological image analysis [
6].
Feature pyramid structures have gained prominence for their effectiveness in managing multi-scale information in medical images. These architectures allow the model to retain fine-grained details while integrating high-level semantic representations, making them well-suited for segmenting lesions of varying sizes and textures. Notably, recent studies have proposed lightweight yet effective multi-attention UNet variants that improve feature aggregation while maintaining computational efficiency, which is vital for real-world clinical applications [
7]. Similarly, collaborative learning frameworks have been introduced to jointly optimize segmentation and classification, leveraging shared features to boost overall performance [
8].
The exploration of attention mechanisms has also extended into other domains of deep learning, where spatial and channel-wise attention has been applied to tasks like gesture recognition [
9], few-shot text classification [
10], and recommendation systems [
11]. Although these studies are rooted in different fields, the underlying methodologies—such as transformer-based architectures, dual loss strategies, and semantic path modeling—provide valuable insights into efficient feature representation and contextual understanding, which are transferable to the medical imaging context.
Other areas of deep learning research have contributed techniques relevant to our segmentation framework. For instance, probabilistic modeling with mixture density networks has proven useful in capturing uncertain patterns, offering a probabilistic interpretation that could be beneficial in modeling lesion boundaries [
12]. Federated learning approaches propose secure and distributed training methods that could enhance privacy in collaborative medical datasets [
13]. Reinforcement learning has been leveraged for dynamic task scheduling and adaptive sampling in complex data environments, presenting a new frontier for learning-based optimization strategies in medical imaging [
14,
15].
Moreover, emerging models based on diffusion-transformer frameworks and capsule networks focus on adaptive and high-dimensional data representation, enabling richer modeling of lesion features and their spatial configurations [
16,
17]. Automated feature extraction combined with transformer-based modeling has also been effective for temporal and structural data, hinting at the potential integration of temporal dermoscopic image sequences in future segmentation studies [
18].
Together, these diverse studies underpin the methodological advancements that inform our proposed segmentation framework. By drawing on innovations from transformer models, attention mechanisms, probabilistic learning, and efficient network architectures, our enhanced feature pyramid approach seeks to push the boundaries of precision and robustness in skin lesion segmentation.
III. Method
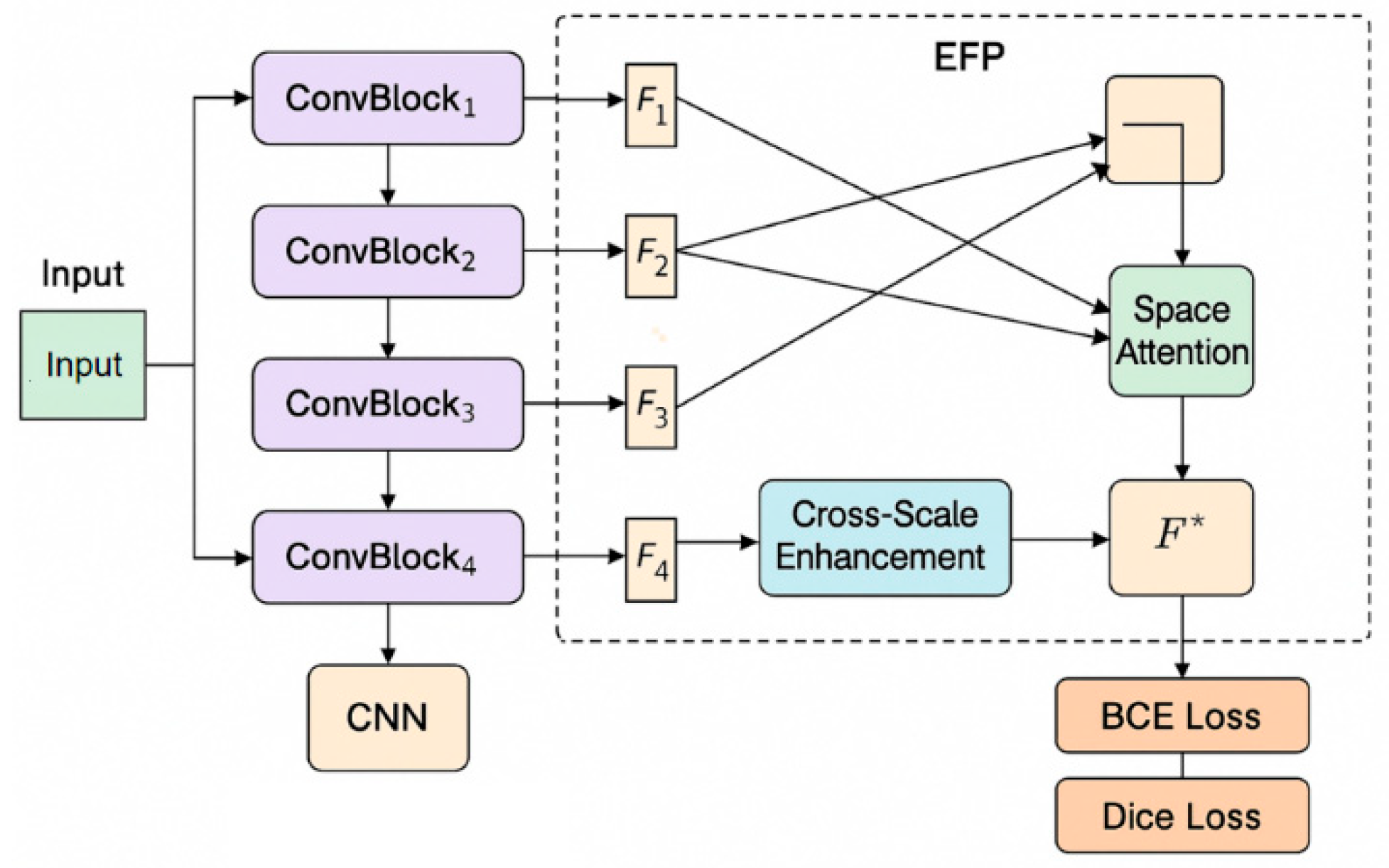
In this study, an enhanced feature pyramid (EFP) structure was first constructed for multi-scale feature extraction and fusion of skin disease images. The model architecture is shown in
Figure 1.
The proposed network begins by extracting hierarchical feature representations through a multi-layer convolutional encoder, designed to progressively capture both local textures and higher-level semantic information. This structure draws from the hierarchical fusion principles demonstrated by Yan et al., who emphasized the importance of multi-level feature integration for robust target representation in small object detection scenarios. Inspired by their work, our encoder is constructed to retain discriminative details across varying spatial resolutions. To further bridge the semantic gaps across feature layers, a cross-scale enhancement module is introduced, enabling efficient information exchange across different feature levels. This approach is conceptually influenced by the dynamic collaboration mechanism proposed by Yan et al. [
19], which supports the reinforcement of weak signals through enhanced inter-level feature interactions. Subsequently, a spatial attention mechanism is employed to refine the localization capability of the network. By referencing the multi-scale attention strategy outlined by Yang et al. [
20], we integrate spatial attention to amplify critical lesion regions while suppressing irrelevant background noise. This selective focus enhances the model’s discrimination between subtle lesion boundaries and surrounding tissue. Moreover, to ensure effective convergence and accurate segmentation, the final output feature maps are supervised using a composite loss function that combines Dice Loss and Binary Cross-Entropy (BCE) Loss. This formulation balances region-level similarity and pixel-level classification. The utility of such hybrid optimization strategies is also supported recent work on multimodal detection [
21], where attention-guided feature alignment was optimized through joint loss design to improve boundary precision and contextual consistency.
The basic feature extraction part uses a convolution encoder to perform multi-level feature mapping on the input image
to obtain feature sets
at different scales, where the resolution of each feature map is halved in turn. The basic operation of feature extraction can be expressed as:
represents a feature transformation module composed of convolution, batch normalization, and nonlinear activation. In order to enhance the expressiveness of pyramid features at different scales, a cross-scale enhancement module is introduced to establish associations between different scales through an adaptive weighting mechanism. Specifically, for any two features
of different scales, cross-scale fusion is expressed as:
where
is the learnable convolution weight and
is the nonlinear activation function. This mechanism not only captures local information, but also transmits cross-scale contextual information, thereby improving the model's perception of skin lesions of different sizes.
In order to further enhance the response of the key areas within the feature map, the spatial attention mechanism is applied to weighted optimize the fused features. For any fused feature map
, the calculation definition of its spatial attention weight
is as follows:
represents the feature concatenation operation,
and
represent the global average pooling and global maximum pooling respectively, and
represents the two-dimensional convolution operation. The final attention weighted feature
can be expressed as:
Where
represents the element-by-element multiplication operation. After processing by the extended feature pyramid and spatial attention module, the obtained feature map takes into account the information of different scales and key areas, which helps to improve the segmentation effect of skin lesion areas. In order to optimize the model training process, a weighted loss function is used to jointly supervise the boundary and regional information. The total loss function is defined as:
is the Dice coefficient loss, which emphasizes contour matching, is the binary cross entropy loss, which enhances pixel-level classification accuracy, and is the loss weight coefficient. Through the above method, the model can accurately segment skin lesions of different shapes and scales under complex backgrounds, improving the overall segmentation performance.
IV. Experimental Results
A. Dataset
In this study, the ISIC 2018 Skin Lesion Analysis dataset was selected as the source of experimental data. The dataset provides a large number of dermoscopic images labeled with precise lesion segmentation masks, covering a wide variety of skin disease types such as melanoma, nevus, and keratosis. All images are collected under standardized imaging protocols to ensure consistency in visual quality and facilitate model training and evaluation.
The dataset includes over 2,500 annotated skin lesion images with corresponding pixel-level segmentation ground truth. Each image varies in terms of lesion size, shape, color, and boundary definition, reflecting the complexity and diversity of real-world clinical cases. This variability makes the dataset particularly suitable for validating segmentation models that require robust multi-scale feature extraction and fine-grained localization abilities.
To prepare the data for training and evaluation, all images and corresponding masks were resized to a unified resolution. Standard preprocessing steps, such as intensity normalization and data augmentation techniques like rotation, flipping, and scaling, were applied to enhance model generalization. The dataset was split into training, validation, and testing subsets to ensure unbiased performance assessment across different stages of model development.
B. Experimental Results
In this section, this paper first gives the comparative experimental results of the proposed algorithm and other algorithms, as shown in
Table 1.
As shown in
Table 1, the baseline model UNet achieves moderate segmentation performance with an mIOU of 78.5%, an mDice of 84.2%, and an F1-Score of 83.8%. After incorporating attention mechanisms, UNet+Attention demonstrates noticeable improvements across all metrics, indicating that integrating attention can enhance the model's ability to focus on relevant lesion regions and capture more discriminative features.
Comparing transformer-based methods, SegFormer and MaskFormer achieve higher scores than convolutional network-based models. SegFormer attains an mIOU of 82.6% and MaskFormer slightly outperforms it with an mIOU of 83.2%, reflecting the effectiveness of transformer architectures in modeling global contextual information. Their corresponding Dice and F1-Score values also follow a similar trend, suggesting that advanced feature extraction and aggregation strategies contribute significantly to segmentation accuracy.
The proposed EFP model outperforms all compared methods, achieving the highest scores across all evaluation metrics, with an mIOU of 85.4%, an mDice of 89.6%, and an F1-Score of 89.2%. This improvement highlights the effectiveness of the enhanced feature pyramid structure and cross-scale feature fusion strategy, which enable the model to better capture multi-scale lesion information and refine spatial localization, ultimately leading to superior segmentation results.
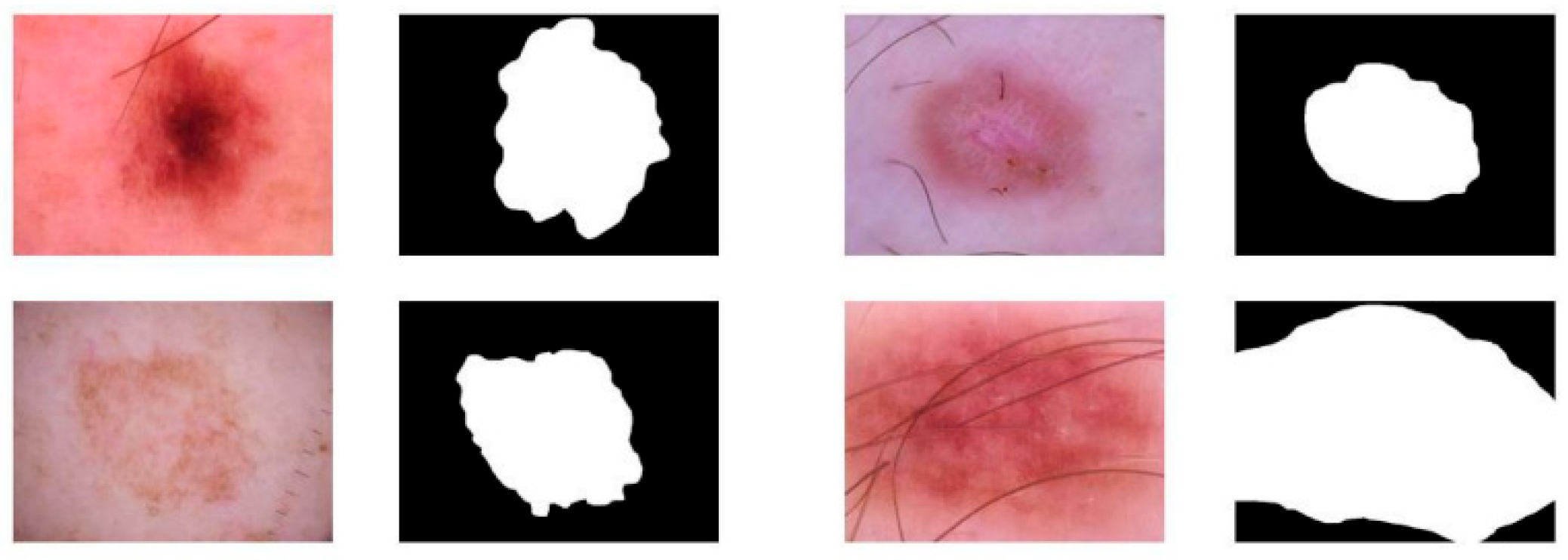
Furthermore, an intuitive segmentation result is given, as shown in
Figure 2.
As illustrated in
Figure 2, the intuitive segmentation results demonstrate that the proposed model is capable of accurately delineating lesion boundaries across a variety of skin disease images. Regardless of differences in lesion size, shape, or color distribution, the segmentation masks closely match the true lesion regions, effectively capturing both coarse and fine-grained details. This suggests that the model generalizes well to diverse appearances commonly seen in clinical practice.
The results further show that the model can effectively suppress irrelevant background noise while preserving the integrity of the lesion regions. Even in cases where the lesions have low contrast with the surrounding skin or are partially occluded by artifacts such as body hair, the model maintains consistent segmentation performance. This highlights the robustness of the enhanced feature extraction and fusion mechanisms in addressing complex real-world challenges.
Overall, the visualized examples validate the model's ability to generate precise and coherent segmentation outputs. The accurate delineation of lesion boundaries not only facilitates subsequent clinical analysis but also confirms that the architectural improvements, such as cross-scale enhancement and space attention, significantly contribute to enhancing spatial localization and boundary refinement in skin disease segmentation tasks.
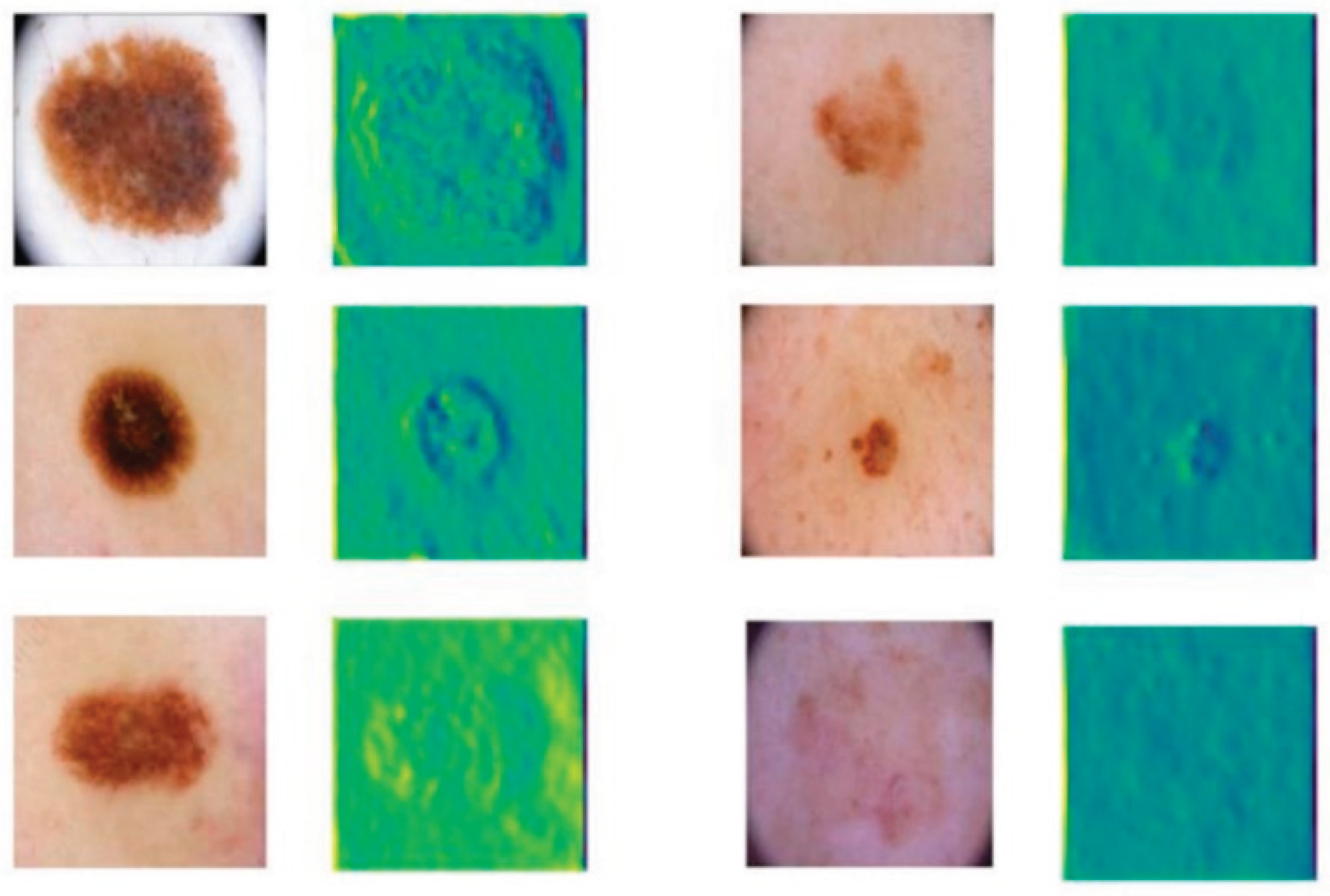
Finally, the feature map of the model is given, as shown in
Figure 3.
As shown in
Figure 3, the model output feature maps illustrate the effectiveness of the enhanced feature extraction process. The feature maps corresponding to different input images highlight lesion regions with strong activations while suppressing irrelevant background noise. This indicates that the model successfully captures the essential characteristics of skin lesions, enabling better discrimination between lesion and non-lesion areas.
The visualization results demonstrate that the feature representations maintain clear spatial localization, even when lesions vary significantly in size, shape, and texture. Fine-grained structures within the lesion areas are preserved in the feature maps, suggesting that the multi-scale feature fusion and space attention mechanisms effectively enhance the model's sensitivity to both coarse and subtle patterns present in the input images.
Overall, the output feature maps validate that the proposed architecture is capable of learning meaningful, task-specific representations crucial for accurate segmentation. By focusing on relevant lesion features while filtering out noise and redundant information, the model is better equipped to handle the variability and complexity inherent in clinical skin disease images, thereby contributing to improved overall performance.
V. Conclusion
In this study, an enhanced feature pyramid-based segmentation framework was proposed to address the challenges of accurate skin disease lesion segmentation. By incorporating cross-scale feature enhancement and spatial attention mechanisms, the model demonstrated superior performance in capturing multi-scale lesion information and refining spatial localization. Comparative experiments and intuitive visualizations confirmed that the proposed method achieves more precise and robust segmentation results compared to existing approaches.
The ability to effectively extract and integrate features across different scales proved critical for handling the variability of skin lesions in terms of size, shape, and appearance. Through the designed architecture, the model not only improves segmentation accuracy but also enhances its generalization capability across diverse clinical scenarios. The improved performance on a standard dermoscopic dataset indicates that the proposed method holds strong potential for integration into computer-aided diagnosis systems.
Looking toward future work, further exploration could involve incorporating transformer-based global context modeling into the feature pyramid structure to enhance long-range dependency learning. Additionally, extending the model to handle multi-modal inputs, such as combining dermoscopic and clinical photographs, may further improve segmentation performance. Research on lightweight model optimization could also facilitate the deployment of the proposed system in real-time clinical environments and mobile health applications.
The advances achieved in this study contribute to the broader field of medical image analysis by providing a more effective and scalable solution for lesion segmentation. Beyond skin disease diagnosis, the techniques developed here have the potential to be adapted to other medical imaging tasks such as tumor boundary delineation, wound analysis, and organ segmentation, promoting the development of intelligent healthcare systems and improving clinical decision-making processes.
References
- Z. Mirikharaji, et al., “A survey on deep learning for skin lesion segmentation,” Medical Image Analysis, vol. 88, pp. 102863, 2023.
- M. K. Hasan, et al., “A survey, review, and future trends of skin lesion segmentation and classification,” Computers in Biology and Medicine, vol. 155, pp. 106624, 2023.
- K. M. Hosny, et al., “Deep learning and optimization-based methods for skin lesions segmentation: a review,” IEEE Access, vol. 11, pp. 85467-85488, 2023.
- H. Wu, et al., “FAT-Net: Feature adaptive transformers for automated skin lesion segmentation,” Medical Image Analysis, vol. 76, pp. 102327, 2022.
- H. Basak, R. Kundu and R. Sarkar, “MFSNet: A multi focus segmentation network for skin lesion segmentation,” Pattern Recognition, vol. 128, pp. 108673, 2022.
- K. A. AnbuDevi and K. Suganthi, “Review of semantic segmentation of medical images using modified architectures of UNET,” Diagnostics, vol. 12, no. 12, pp. 3064, 2022.
- J. Ruan, et al., “Malunet: A multi-attention and light-weight unet for skin lesion segmentation,” Proceedings of the 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pp. [insert page numbers], 2022.
- Y. Wang, et al., “A collaborative learning model for skin lesion segmentation and classification,” Diagnostics, vol. 13, no. 5, pp. 912, 2023.
- T. Zhang, F. Shao, R. Zhang, Y. Zhuang and L. Yang, “DeepSORT-Driven Visual Tracking Approach for Gesture Recognition in Interactive Systems,” arXiv preprint. arXiv:2505.07110, 2025.
- X. Han, Y. Sun, W. Huang, H. Zheng and J. Du, “Towards Robust Few-Shot Text Classification Using Transformer Architectures and Dual Loss Strategies,” arXiv preprint. arXiv:2505.06145, 2025.
- H. Zheng, Y. Xing, L. Zhu, X. Han, J. Du and W. Cui, “Modeling Multi-Hop Semantic Paths for Recommendation in Heterogeneous Information Networks,” arXiv preprint. arXiv:2505.05989, 2025.
- L. Dai, W. Zhu, X. Quan, R. Meng, S. Cai and Y. Wang, “Deep Probabilistic Modeling of User Behavior for Anomaly Detection via Mixture Density Networks,” arXiv preprint. arXiv:2505.08220, 2025.
- Y. Zhang, J. Liu, J. Wang, L. Dai, F. Guo and G. Cai, “Federated Learning for Cross-Domain Data Privacy: A Distributed Approach to Secure Collaboration,” arXiv preprint. arXiv:2504.00282, 2025.
- Y. Wang, T. Tang, Z. Fang, Y. Deng and Y. Duan, “Intelligent Task Scheduling for Microservices via A3C-Based Reinforcement Learning,” arXiv preprint. arXiv:2505.00299, 2025.
- J. Liu, “Reinforcement Learning-Controlled Subspace Ensemble Sampling for Complex Data Structures,” Preprints, 2025.
- W. Cui and A. Liang, “Diffusion-Transformer Framework for Deep Mining of High-Dimensional Sparse Data,” Journal of Computer Technology and Software, vol. 4, no. 4, 2025.
- Y. Lou, “Capsule Network-Based AI Model for Structured Data Mining with Adaptive Feature Representation,” Transactions on Computational and Scientific Methods, vol. 4, no. 9, 2024.
- Y. Cheng, “Multivariate Time Series Forecasting through Automated Feature Extraction and Transformer-Based Modeling,” Journal of Computer Science and Software Applications, vol. 5, no. 5, 2025.
- X. Yan, J. Du, X. Li, X. Wang, X. Sun, P. Li and H. Zheng, “A Hierarchical Feature Fusion and Dynamic Collaboration Framework for Robust Small Target Detection,” IEEE Access, vol. 13, pp. 123456–123467, 2025.
- T. Yang, Y. Cheng, Y. Ren, Y. Lou, M. Wei and H. Xin, “A Deep Learning Framework for Sequence Mining with Bidirectional LSTM and Multi-Scale Attention,” arXiv preprint. arXiv:2504.15223, 2025.
- Y. Lou, “RT-DETR-Based Multimodal Detection with Modality Attention and Feature Alignment,” Journal of Computer Technology and Software, vol. 3, no. 5, 2024.
- M. Xiao, Y. Li, X. Yan, M. Gao, and W. Wang, "Convolutional neural network classification of cancer cytopathology images: taking breast cancer as an example," Proceedings of the 2024 7th International Conference on Machine Vision and Applications, pp. 145–149, Singapore, Singapore,2024.
- N. Das and S. Das, “Attention-UNet architectures with pretrained backbones for multi-class cardiac MR image segmentation,” Current Problems in Cardiology, vol. 49, no. 1, pp. 102129, 2024.
- E. Xie, et al., “SegFormer: Simple and efficient design for semantic segmentation with transformers,” Advances in Neural Information Processing Systems, vol. 34, pp. 12077-12090, 2021.
- X. Zhu, et al., “Refactored Maskformer: Refactor localization and classification for improved universal image segmentation,” Displays, pp. 102981, 2025.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).