Submitted:
14 October 2025
Posted:
15 October 2025
You are already at the latest version
Abstract
Keywords:
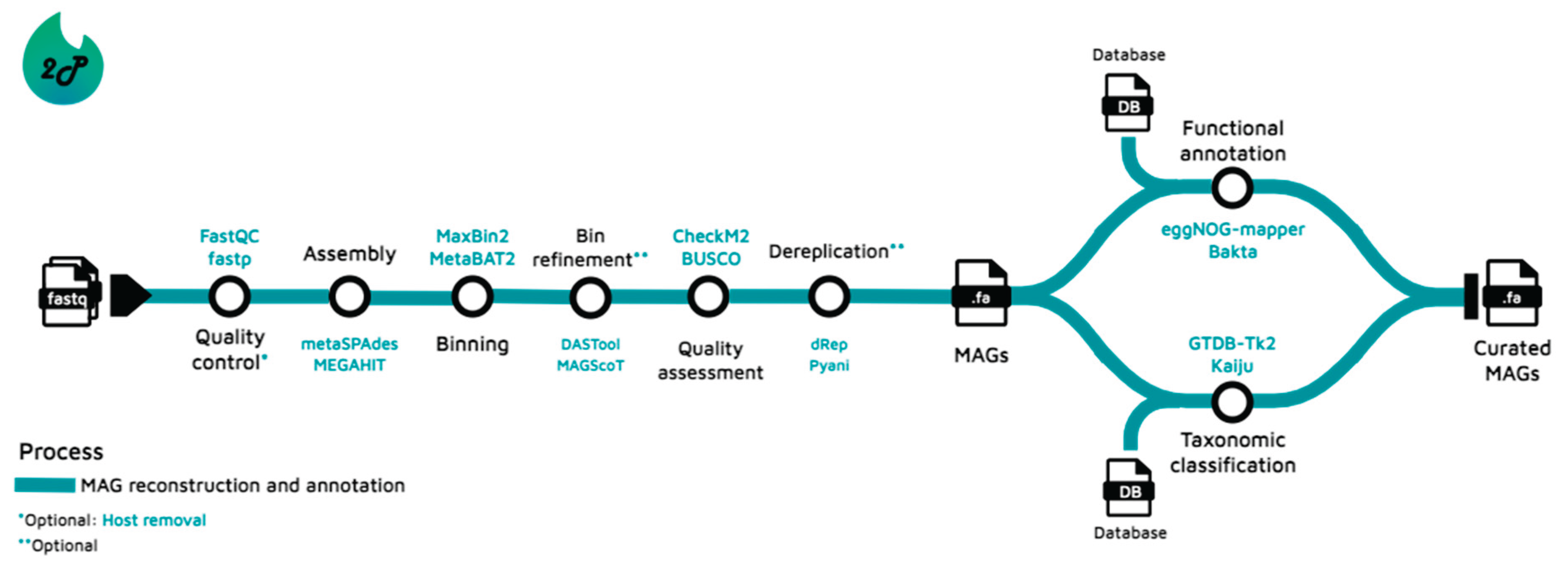
Introduction
- Pipeline workflow, tools and benchmarks
| Pipeline/Platform | Quality Control Preprocessing | Assembly* | Binning | Quality Assessment | Bin Refinement | Taxonomic Annotation** | Functional Annotation** | Other | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Ancient DNA [44] | FastQC [30], fastp [8], BBTools [20] | Bowtie2, MEGAHIT [36] | CONCOCT [45], MaxBin [46], MetaBAT [47] | CheckM [48] | DASTool [12] | GTDB-Tk [17] | mapDamage2 [49] | |
| 2 | Anvi'o△ [50] | Illumina-utils [51] | metaSPAdes [37], MEGAHIT, IBDA-UD [38] | MetaBAT2 [52], CONCOCT, MaxBin2 [53], BinSanity [54] | DASTool | KrakenUniq [34], Centrifuge [55] | DIAMOND [56] (NCBI COG [57]), Pyrodigal [58], HMMER [59] |
||
| 3 | Aviary [60] | FastQC, Filtlong [27], NanoPack2 [28], SingleM [61] | metaSPAdes, MEGAHIT, metaFlye [39], Unicycler [62] | MetaBAT2, MetaBAT, MaxBin2, VAMB [63], CONCOCT, Rosella [64] | CheckM, metaQUAST [65], CoverM [66] | DASTool | GTDB-Tk | Prodigal [67], DIAMOND(eggNOG [68]) | Lorikeet [69] |
| 4 | BugBuster [70] | fastp, Bowtie2 [22] | MEGAHIT | METABAT2, SemiBin2 [71], COMEBin [72] | CheckM2 [73] | MetaWRAP-native module [74] | GTDB-Tk2 [75] | Prodigal, MetaCerberus [76] | Kraken2 [26], Sourmash [77], deepARG [78] |
| 5 | BV-BRC△ [79] | TrimGalore [80], BBTools, BLAST [81] | metaSPAdes, MEGAHIT | PATRIC metagenome binning service [82] | EvalG and EvalCon [83] | RASTtk [84] | VIGOR4 [85], Mat_Peptide [86] | ||
| 6 | DATMA [87] | Trimmomatic [9], FastQC, FLASH2 [88], BWA [24] | metaSPAdes, Velvet [89], MEGAHIT | CLAME [90] | CheckM | BLAST, Kaiju [91] | Prodigal, GeneMark [92] | Krona [93] | |
| 7 | EasyMetagenome [94] | KneadData [21], HostPurge [94], FastQC | metaSPAdes, MEGAHIT | MetaWRAP-native [74] module | CoverM, CheckM2 | MetaWRAP-native module | GTDB-Tk2 | MetaProdigal [95], eggNOG-mapper [96] | dRep [97], Kraken2, Bracken [98], HUMAnN3 [21] |
| 8 | EasyNanoMeta [99] | fastp, Minimap2 [23], SAMtools [100], Porechop [29], BEDTools [101] | metaFlye, OPERA-MS [42], metaSPAdes, MetaPlatanus [102], NextPolish [103] | SemiBin2, MetaBAT2, MaxBin2, CONCOCT, VAMB | CheckM2 | GTDB-Tk2, PhyloPhlAn [104] | Prokka [105] | Kraken2, Centrifuge | |
| 9 | Eukfinder [106] | Bowtie2, Trimmomatic | metaSPAdes | MyCC [107], Metaxa2 [108] | Centrifuge, PLAST [109] | ||||
| 10 | EURYALE (MEDUSA) [110,111] | FastQC, fastp, Bowtie2, MultiQC [31] | MEGAHIT | Kaiju, Kraken2 | DIAMOND (NCBI nr [112]) | Krona | |||
| 11 | Galaxy△ [113] | FastQC, Seqtk [114], Trimmomatic | metaSPAdes | MaxBin2 | GTDB-Tk2, CAT [115] | Prokka | Kraken [25] | ||
| 12 | GEN-ERA [116] | fastp, FastQC | SPAdes [117], metaSPAdes, Canu [40], metaFlye, Pilon [118], RagTag [119] | MetaBAT2, CONCOCT | CheckM, GUNC [120], CheckM2, EukCC [121], BUSCO [122], Physeter [123], Kraken, QUAST [124] | AMAW [125], BRAKER2 [126], GTDB-Tk | Prodigal, Mantis [127], Anvi'o scripts (KEGG [128]) | OrthoFinder [129] | |
| 13 | HiFi-MAG [130] | MetaBAT2, SemiBin2 | CheckM2 | DASTool | GTDB-Tk2 | ||||
| 14 | IDseq△ [131] | Trimmomatic, STAR [132], Bowtie2, CD-HIT [133] | SPAdes, Bowtie2 | GSNAPL [134], RAPsearch2 [135] | |||||
| 15 | IMG/M△ [136] | SemiBin2 | CheckM | GTDB-Tk | Prodigal, GeneMarkS-2 [137], HMMER (NCBI COG, Pfam [138], TIGRFAMs [139]) | EukCC, SignalP [140], TMHMM [141] | |||
| 16 | JAMS [142] | Trimmomatic, Bowtie2 | MEGAHIT, SPAdes | Kraken2 | Prokka, InterProScan [143] |
Samtools, BEDTools |
|||
|
17 |
KBase△ [144] | FastQC, Trimmomatic, Cutadapt [19] |
metaSPAdes, MEGAHIT, IBDA-UD | MetaBAT2, CONCOCT, MaxBin2 | CheckM | DASTool | RASTtk, GTDB-Tk | Prokka, dbCAN3 [145], DRAM [146] | OMEGGA [147], ModelSEED2 [148], Kaiju, FastANI [149], dRep, FastTree2 [150], Muscle5 [151] |
| 18 | MAGNETO [152] | fastp, Bowtie2, FastQscreen [153] | MEGAHIT, Simka [154] | MetaBAT2 | CheckM | GTDB-Tk, | Prodigal, Linclust [155], CD-HIT, eggNOG-mapper | mOTUs [156], dRep | |
| 19 | MAGO [157] | FastQC, fastp | metaSPAdes, MEGAHIT, IBDA-UD | MaxBin2, MetaBAT, CONCOCT, BinSanity | CheckM | GTDB-Tk | Prokka | Roary [158], ezTree [159], FastANI | |
| 20 | Mapler [160] | FastQC | metaMDBG [161], hifiasm [41], metaFlye, OPERA-MS, Minimap2 | MetaBAT2 | CheckM2, metaQUAST | GTDB-Tk2, Kraken2 | KAT [162] | ||
| 21 | MetaGEM [163] | fastp | MEGAHIT, BWA | MetaBAT2, CONCOCT, MaxBin2 | MetaWRAP–native module | GTDB-Tk | Prokka | Roary, CarveMe [164], SMETANA [165], MEMOTE [166], GRiD [167] | |
| 22 | MetaGenePipe [168] | Trimmomatic, TrimGalore, FastQC | MEGAHIT | DIAMOND (SwissProt [169]) | Prodigal, HMMER [170] (KOfam [171]) | BLAST | |||
| 23 | Metagenome-Atlas [172] | BBTools | MEGAHIT, metaSPAdes | MetaBAT2, MaxBin2, VAMB | BUSCO, CheckM, CheckM2 | DASTool | GTDB-Tk | Prodigal, eggNOG, DRAM | dRep |
| 24 |
Metagenomics- Toolkit [173] |
fastp, Porechop, Filtlong, NanoPack2, KMC [174], Nonpareil [175] | metaFlye, metaSPAdes, MEGAHIT, Assembler Resource Estimator [173] | MetaBAT2, MetaCoAG [176], Metabinner [177] | CheckM | MAGScoT [13] | MMSeqs2 taxonomy [178], GTDB-Tk2 | Prodigal, Prokka, RGI [179] | CarveMe, SMETANA, MEMOTE, gapseq [180], Pyani [181], SANS [182] |
| 25 | Metaphor [183] | FastQC, fastp, MultiQC | MEGAHIT | VAMB, MetaBAT2, CONCOCT | metaQUAST | DASTool | DIAMOND (NCBI COG) | Prodigal, Prokka | |
| 26 | metagWGS [184] | FastQC, Cutadapt, Sickle [185], SAMtools, BWA | metaSPAdes, MEGAHIT, hifiasm, metaFlye | MetaBAT2, CONCOCT, MaxBin2 | metaQUAST | Binette [186] | GTDB-Tk2 | Prodigal, eggNOG-mapper | dRep, Kaiju |
| 27 | MetaWRAP [74] | FastQC, TrimGalore | metaSPAdes, MEGAHIT | MetaBAT2, CONCOCT, MaxBin2 | CheckM | MetaWRAP-native module | Kraken, BLAST | Prokka | Kraken, Blobology [187] |
| 28 | MG-TK [188] | Trimmomatic, Porechop, Kraken, Kraken2, SDM [189] | SPAdes, MEGAHIT, Flye [190], metaMDBG | MetaBAT2, SemiBin2, MetaDecoder [191] | CheckM, CheckM2 | GTDB-Tk | Prodigal, DIAMOND (KEGG CAZy [192], eggNOG) | mOTUs2 [193], MetaPhlAn [194], Freebayes [195], riboFinder [196], BCFtools [100] |
|
| 29 | MGnify△ [197] | Trimmomatic, Biopython [198] | metaSPAdes | DIAMOND (UniRef90 [199]) | Prodigal, FragGeneScan [200], InterProScan, eggNOG-mapper, HMMER [59] | mOTUs2, antiSMASH [201] | |||
| 30 | MOSHPIT△ [202] | Cutadapt, Bowtie2 | SPAdes, MEGAHIT | MetaBAT2 | QUAST, BUSCO | Sourmash | Kraken2, Kaiju | eggNOG-mapper, DIAMOND (eggNOG, CAZy) | |
| 31 | MUFFIN [203] | fastp, Filtlong | SPAdes, Flye, Unicycler | MetaBAT2, CONCOCT, MaxBin2 | CheckM | MetaWRAP-native module | Sourmash (GTDB [204]) | eggNOG-mapper | Salmon [205], Trinity [206] |
| 32 | NanoPhase [207] | Filtlong | metaFlye, Racon [208], medaka [209] | MetaBAT2, MaxBin2 | CheckM, QUAST | MetaWRAP-native module | GTDB-Tk | Prodigal, DIAMOND (UniProtKB [210]) | |
| 33 | nf-core/mag [211] | fastp, AdapterRemoval [212], Bowtie2, BBTools, Trimmomatic, FastQC, Porechop, Filtlong, NanoPack2 | MEGAHIT, metaSPAdes, Flye, metaMDBG, hybridSPAdes [43] | MetaBAT2, CONCOCT, MaxBin2 | BUSCO, CheckM, CheckM2, GUNC, QUAST | DASTool | GTDB-Tk2, CAT | Prodigal, Prokka, MetaEuk [213] | Kraken2, MultiQC, Centrifuge, PyDamage [214] geNomad [215], Tiara [216] |
| 34 |
ngs-preprocess MpGAp Bacannot [217] |
Porechop, Nanopack2, pycoQC [32], fastp | SPAdes, Flye, Canu, Unicycler, Shovill [218], HASLR [219], Raven [220], Shasta [221], wtdbg2 [222], Pilon | Prokka, antiSMASH, KOfamScan [171], KEGGDecoder [223], Bakta [16], Barrnap [224] | AMRFinderPlus [225], CARD-RGI, BEDTools, Phigaro [226], VFDB [227], PlasmidFinder [228], MLST [229], Platon [230], PHASTER [231], ARGminer [232], ResFinder [233] |
||||
| 35 | nIMP3 [234] | BWA, Samtools, BBTools, FastQC, Kraken2, SortMeRNA [235] |
MEGAHIT | mOTUs, MultiQC, MetaPhlAn4 [21], Salmon, gffquant [236], kallisto [237] | |||||
| 36 | SnakeMAGs [238] | Illumina-utils, Trimmomatic, Bowtie2 | MEGAHIT | MetaBAT2 | CheckM, GUNC, CoverM | GTDB-Tk2 | |||
| 37 | SPIRE [239] | NGLess [240] | MEGAHIT, BWA, Samtools | MetaBAT2 | CheckM2, GUNC | GTDB-Tk2 | Prodigal, eggNOG-mapper | Barrnap, RGI [179], ABRicate [241] (MEGARes [242], VFDB), Seqtk, Macrel [243], Mash [244] | |
| 38 | SqueezeMeta [245] | PRINSEQ [246], Trimmomatic, SAMtools |
MEGAHIT, SPAdes, Canu, Flye | MetaBAT2, CONCOCT, MaxBin2 | CheckM, CheckM2, CompareM [247] | DASTool | GTDB-Tk2 | Prodigal, MUMmer [248], HMMER, Barrnap | DIAMOND (NCBI COG, KEGG), SQMtools [249], POGENOM [250] |
| 39 | Sunbeam [251] | Trimmomatic, Cutadapt, Komplexity [251], BWA | MEGAHIT | Prodigal, BLAST, DIAMOND | Kraken | ||||
| 40 | VEBA [252] | KneadData, fastp, BBTools, Bowtie2, NanoPack2, Minimap2 | metaSPAdes, SPAdes, rnaSPAdes [253], MEGAHIT, Flye, metaFlye | MetaBAT2, CONCOCT, MaxBin2, SemiBin2 | CheckM, Tiara, CheckV [254], BUSCO, CoverM | Binette | GTDB-Tk2, MetaEuk, geNomad, VirFinder [255] | Prodigal, DIAMOND (UniRef50/90, MIBiG [256], VFDB, CAZy) HMMER (Pfam, NCBIfam-AMR [225], AntiFam [257], KOfam), MicrobeAnnotator [258] | antiSMASH, Muscle5, FastTree2, FastANI, sylph [259], HUMAnN3 |
| 41 | WGSA2+/LoRA△ [260] | KneadData, fastp, Kraken2 | metaSPAdes, metaFlye, MiniMap2, Samtools | MetaBAT2 | CheckM, CheckM2 | GTDB-Tk2 | Prodigal, eggNOG-mapper, MinPath [261] | SortMeRNA, Krona, Trinity, AMRFinderPlus |
- 2
- Practical and technical considerations for pipeline execution
- ONT: Oxford Nanopore Technology.
- PacBio: Pacific Biosciences.
- HPC: High Performance Cluster.
- CC: Cloud Computing.
- External: Pipelines controlled by the platform or suite and use external resources.
- CLI: Command Line Interface.
- GUI: Graphical User Interface.
- 3
- 2Pipe: It starts with a question
Conclusion
Consent for publication
Availability of data and materials
Competing interests
Contributions
Additional files
Acknowledgments
References
- Navgire, G.S.; et al. Analysis and Interpretation of metagenomics data: an approach. Biol. Proced. Online 2022, 24, 1–22. [Google Scholar] [CrossRef]
- Kim, N.; et al. Genome-resolved metagenomics: a game changer for microbiome medicine. Exp. Mol. Med. 2024, 56, 1501–1512. [Google Scholar] [CrossRef]
- Lemos, L.N.; Mendes, L.W.; Baldrian, P.; Pylro, V.S. Genome-Resolved Metagenomics Is Essential for Unlocking the Microbial Black Box of the Soil. Trends Microbiol. 2021, 29, 279–282. [Google Scholar] [CrossRef]
- Bowers, R.M.; et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 2017, 35, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Setubal, J.C. Metagenome-assembled genomes: concepts, analogies, and challenges. Biophys. Rev. 2021, 13, 905–909. [Google Scholar] [CrossRef]
- Yang, C.; et al. A review of computational tools for generating metagenome-assembled genomes from metagenomic sequencing data. Computational and Structural Biotechnology Journal 2021, 19, 6301–6314. [Google Scholar] [CrossRef]
- Ahmed, A.E.; et al. Design considerations for workflow management systems use in production genomics research and the clinic. Sci. Rep. 2021, 11, 1–18. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; et al. Benchmarking short-read metagenomics tools for removing host contamination. GigaScience 2025, 14, giaf004. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Wang, Z.; Zhu, S. Benchmarking metagenomic binning tools on real datasets across sequencing platforms and binning modes. Nat. Commun. 2025, 16, 2865. [Google Scholar] [CrossRef]
- Sieber, C.M.K.; et al. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 2018, 3, 836–843. [Google Scholar] [CrossRef]
- Christoph, M.; Rühlemann, R.; Wacker, E.M.; Ellinghaus, D.; Franke, A. MAGScoT: a fast, lightweight and accurate bin-refinement tool. Bioinformatics 2022, 38, 5430–5433. [Google Scholar]
- Cornet, L.; Baurain, D. Contamination detection in genomic data: more is not enough. Genome Biol. 2022, 23, 1–15. [Google Scholar] [CrossRef]
- Evans, J.T.; Denef, V.J. To Dereplicate or Not To Dereplicate? mSphere 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Schwengers, O.; et al. Bakta: Rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genomics 2021, 7, 000685. [Google Scholar] [CrossRef] [PubMed]
- Chaumeil, P.A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 2020, 36, 1925–1927. [Google Scholar] [CrossRef]
- Wajid, B.; et al. Music of metagenomics—a review of its applications, analysis pipeline, and associated tools. Funct. Integr. Genomics 2022, 22, 3–26. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bushnell, B. BBMap: A Fast, Accurate, Splice-Aware Aligner. LBL Publications, (2014).
- Beghini, F.; et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. eLife 2021, 10, e65088. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 2013. [Google Scholar] [CrossRef]
- Wood, D.E.; Salzberg, S.L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 1–13. [Google Scholar] [CrossRef]
- Haveman, N.J.; et al. Evaluating the lettuce metatranscriptome with MinION sequencing for future spaceflight food production applications. Npj Microgravity 2021, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- De Coster, W.; Rademakers, R. NanoPack2: population-scale evaluation of long-read sequencing data. Bioinformatics 2023, 39, btad311. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb. Genomics 2017, 3, e000132. [Google Scholar] [CrossRef]
- Simon, A. FastQC A Quality Control tool for High Throughput Sequence Data. FastQC [Online] 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Leger, A.; Leonardi, T. pycoQC, interactive quality control for Oxford Nanopore Sequencing. J. Open Source Softw. 2019, 4, 1236. [Google Scholar] [CrossRef]
- Shen, W.; et al. KMCP: accurate metagenomic profiling of both prokaryotic and viral populations by pseudo-mapping. Bioinformatics 2023, 39, btac845. [Google Scholar] [CrossRef]
- Breitwieser, F.P.; Baker, D.N.; Salzberg, S.L. KrakenUniq: confident and fast metagenomics classification using unique k-mer counts. Genome Biol. 2018, 19, 198. [Google Scholar] [CrossRef]
- Ayling, M.; Clark, M.D.; Leggett, R.M. New approaches for metagenome assembly with short reads. Brief. Bioinform. 2020, 21, 584–594. [Google Scholar] [CrossRef]
- Li, D.; et al. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 2016, 102, 3–11. [Google Scholar] [CrossRef]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. MetaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef]
- Peng, Y.; Leung, H.C.M.; Yiu, S.M.; Chin, F.Y.L. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef]
- Kolmogorov, M.; et al. metaFlye: scalable long-read metagenome assembly using repeat graphs. Nat. Methods 2020, 17, 1103–1110. [Google Scholar] [CrossRef]
- Koren, S.; et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Concepcion, G.T.; Feng, X.; Zhang, H.; Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 2021, 18, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, D.; et al. Hybrid metagenomic assembly enables high-resolution analysis of resistance determinants and mobile elements in human microbiomes. Nat. Biotechnol. 2019, 37, 937–944. [Google Scholar] [CrossRef]
- Antipov, D.; Korobeynikov, A.; McLean, J.S.; Pevzner, P.A. hybridSPAdes: an algorithm for hybrid assembly of short and long reads. Bioinformatics 2016, 32, 1009–1015. [Google Scholar] [CrossRef]
- Standeven, F.J.; Dahlquist-Axe, G.; Speller, C.F.; Meehan, C.J.; Tedder, A. An efficient pipeline for creating metagenomic-assembled genomes from ancient oral microbiomes. 2024. [Google Scholar] [CrossRef]
- Alneberg, J.; et al. Binning metagenomic contigs by coverage and composition. Nat. Methods 2014, 11, 1144–1146. [Google Scholar] [CrossRef]
- Wu, Y.-W.; Tang, Y.-H.; Tringe, S.G.; Simmons, B.A.; Singer, S.W. MaxBin: an automated binning method to recover individual genomes from metagenomes using an expectation-maximization algorithm. Microbiome 2014, 2, 26. [Google Scholar] [CrossRef]
- Kang, D.D.; Froula, J.; Egan, R.; Wang, Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 2015, 3, e1165. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Jónsson, H.; Ginolhac, A.; Schubert, M.; Johnson, P.L.F.; Orlando, L. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 2013, 29, 1682–1684. [Google Scholar] [CrossRef] [PubMed]
- Eren, A.M.; et al. Community-led, integrated, reproducible multi-omics with anvi’o. Nat. Microbiol. 2020, 6, 3–6. [Google Scholar] [CrossRef]
- Eren, A.M.; Vineis, J.H.; Morrison, H.G.; Sogin, M.L. A Filtering Method to Generate High Quality Short Reads Using Illumina Paired-End Technology. PLOS ONE 2013, 8, e66643. [Google Scholar] [CrossRef]
- Kang, D.D.; et al. MetaBAT 2: An adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 2019, 2019. [Google Scholar] [CrossRef]
- Wu, Y.W.; Simmons, B.A.; Singer, S.W. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 2016, 32, 605–607. [Google Scholar] [CrossRef]
- Graham, E.D.; Heidelberg, J.F.; Tully, B.J. BinSanity: unsupervised clustering of environmental microbial assemblies using coverage and affinity propagation. PeerJ 2017, 5, e3035. [Google Scholar] [CrossRef]
- Kim, D.; Song, L.; Breitwieser, F.P.; Salzberg, S.L. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res. 2016, 26, 1721–1729. [Google Scholar] [CrossRef]
- Buchfink, B.; Reuter, K.; Drost, H.G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef]
- Galperin, M.Y.; et al. COG database update 2024. Nucleic Acids Res. 2025, 53, D356–D363. [Google Scholar] [CrossRef]
- Larralde, M. Pyrodigal: Python bindings and interface to Prodigal, an efficient method for gene prediction in prokaryotes. J. Open Source Softw. 2022, 7, 4296. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Newell, R.J.P.; Aroney, S.T.N.; Zaugg, J.; Sternes, P.; Tyson, G.W.; & Woodcroft, B.J. Aviary: Hybrid assembly and genome recovery from metagenomes with Aviary (v0.12.0). Zenodo. 2025. [Google Scholar] [CrossRef]
- Woodcroft, B.J.; et al. Comprehensive taxonomic identification of microbial species in metagenomic data using SingleM and Sandpiper. Nat. Biotechnol. 2025; 1–6. [Google Scholar]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Nissen, J.N.; et al. Improved metagenome binning and assembly using deep variational autoencoders. Nat. Biotechnol. 2021, 39, 555–560. [Google Scholar] [CrossRef]
- Newell, R.J.P.; Tyson, G.W.; & Woodcroft, B.J. . Rosella: Metagenomic binning using UMAP and HDBSCAN (v0.5.3). Zenodo. 2024. [CrossRef]
- Mikheenko, A.; Saveliev, V.; Gurevich, A. MetaQUAST: evaluation of metagenome assemblies. Bioinformatics 2016, 32, 1088–1090. [Google Scholar] [CrossRef] [PubMed]
- Aroney, S.T.N.; et al. CoverM: read alignment statistics for metagenomics. Bioinformatics 2025, 41, btaf147. [Google Scholar] [CrossRef]
- Hyatt, D.; et al. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Newell, R.J.P.; McMaster, E.S.; Craig, P.; Boden, M.; Tyson, G.W.; & Woodcroft, B.J. Lorikeet: strain-resolved metagenome analysis using local reassembly (v0.8.2). Zenodo. 2023. [Google Scholar] [CrossRef]
- Fuentes-Santander, F.; Curiqueo, C.; Araos, R.; Ugalde, J.A. BugBuster: a novel automatic and reproducible workflow for metagenomic data analysis. Bioinforma. Adv. 2025, 5, vbaf152. [Google Scholar] [CrossRef]
- Pan, S.; Zhao, X.M.; Coelho, L.P. SemiBin2: self-supervised contrastive learning leads to better MAGs for short- and long-read sequencing. Bioinformatics 2023, 39, i21–i29. [Google Scholar] [CrossRef]
- Wang, Z.; et al. Effective binning of metagenomic contigs using contrastive multi-view representation learning. Nat. Commun. 2024, 15, 1–14. [Google Scholar] [CrossRef]
- Chklovski, A.; Parks, D.H.; Woodcroft, B.J.; Tyson, G.W. CheckM2: a rapid, scalable and accurate tool for assessing microbial genome quality using machine learning. Nat. Methods 2023, 20, 1203–1212. [Google Scholar] [CrossRef]
- Uritskiy, G.V.; Diruggiero, J.; Taylor, J. MetaWRAP - A flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 2018, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk v2: memory friendly classification with the genome taxonomy database. Bioinformatics 2022, 38, 5315–5316. [Google Scholar] [CrossRef]
- Figueroa III, J.L.; Dhungel, E.; Bellanger, M.; Brouwer, C.R.; White III, R.A. MetaCerberus: distributed highly parallelized HMM-based processing for robust functional annotation across the tree of life. Bioinformatics 2024, 40, btae119. [Google Scholar] [CrossRef]
- Irber, L.; et al. sourmash v4: A multitool to quickly search, compare, and analyze genomic and metagenomic data sets. J. Open Source Softw. 2024, 9, 6830. [Google Scholar] [CrossRef]
- Arango-Argoty, G.; et al. DeepARG: A deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome 2018, 6, 1–15. [Google Scholar] [CrossRef]
- Olson, R.D.; et al. Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): a resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023, 51, D678–D689. [Google Scholar] [CrossRef]
- Krueger, F. Source code for: A wrapper around Cutadapt and FastQC to consistently apply adapter and quality trimming to FastQ files, with extra functionality for RRBS data 2023. Available online: https://github.com/FelixKrueger/TrimGalore.
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Parrello, B.; Butler, R.; Chlenski, P.; Pusch, G.D.; Overbeek, R. Supervised extraction of near-complete genomes from metagenomic samples: A new service in PATRIC. PLOS ONE 2021, 16, e0250092. [Google Scholar] [CrossRef]
- Parrello, B.; et al. A machine learning-based service for estimating quality of genomes using PATRIC. BMC Bioinformatics 2019, 20, 486. [Google Scholar] [CrossRef]
- Brettin, T.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef]
- Wang, S.; Sundaram, J.P.; Spiro, D. VIGOR, an annotation program for small viral genomes. BMC Bioinformatics 2010, 11, 451. [Google Scholar] [CrossRef] [PubMed]
- Larsen, C.N.; et al. Mat_peptide: comprehensive annotation of mature peptides from polyproteins in five virus families. Bioinformatics 2020, 36, 1627–1628. [Google Scholar] [CrossRef]
- Benavides, A.; Sanchez, F.; Alzate, J.F.; Cabarcas, F. DATMA: Distributed Automatic Metagenomic Assembly and annotation framework. PeerJ, 2020; 8. [Google Scholar]
- Magoč, T.; Salzberg, S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Benavides, A.; Isaza, J.P.; Niño-García, J.P.; Alzate, J.F.; Cabarcas, F. CLAME: a new alignment-based binning algorithm allows the genomic description of a novel Xanthomonadaceae from the Colombian Andes. BMC Genomics, 2018; 19. [Google Scholar]
- Menzel, P.; Ng, K.L.; Krogh, A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat. Commun. 2016, 7, 11257. [Google Scholar] [CrossRef]
- Besemer, J.; Borodovsky, M. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005, 33, W451–W454. [Google Scholar] [CrossRef]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics 2011, 12, 1–10. [Google Scholar] [CrossRef]
- Bai, D.; et al. EasyMetagenome: A user-friendly and flexible pipeline for shotgun metagenomic analysis in microbiome research. iMeta 2025, 4, e70001. [Google Scholar] [CrossRef]
- Hyatt, D.; LoCascio, P.F.; Hauser, L.J.; Uberbacher, E.C. Gene and translation initiation site prediction in metagenomic sequences. Bioinformatics 2012, 28, 2223–2230. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Olm, M.R.; Brown, C.T.; Brooks, B.; Banfield, J.F. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 2017, 11, 2864–2868. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Breitwieser, F.P.; Thielen, P.; Salzberg, S.L. Bracken: Estimating species abundance in metagenomics data. PeerJ Comput. Sci. 2017, 2017, e104. [Google Scholar] [CrossRef]
- Peng, K.; et al. Benchmarking of analysis tools and pipeline development for nanopore long-read metagenomics. Sci. Bull. 2025, 70, 1591–1595. [Google Scholar] [CrossRef]
- Danecek, P.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Kajitani, R.; et al. MetaPlatanus: a metagenome assembler that combines long-range sequence links and species-specific features. Nucleic Acids Res. 2021, 49, e130. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Fan, J.; Sun, Z.; Liu, S. NextPolish: a fast and efficient genome polishing tool for long-read assembly. Bioinformatics 2020, 36, 2253–2255. [Google Scholar] [CrossRef] [PubMed]
- Asnicar, F.; et al. Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Zhao, D.; et al. Eukfinder: a pipeline to retrieve microbial eukaryote genome sequences from metagenomic data. mBio 2025, 16, e00699-25. [Google Scholar] [CrossRef]
- Lin, H.-H.; Liao, Y.-C. Accurate binning of metagenomic contigs via automated clustering sequences using information of genomic signatures and marker genes. Sci. Rep. 2016, 6, 24175. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; et al. metaxa2: improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Mol. Ecol. Resour. 2015, 15, 1403–1414. [Google Scholar] [CrossRef]
- Van Nguyen, H.; Lavenier, D. PLAST: parallel local alignment search tool for database comparison. BMC Bioinformatics 2009, 10, 329. [Google Scholar] [CrossRef]
- Cavalcante, J.V.F.; Dantas de Souza, I.; Morais, D.A.A.; Dalmolin, R.J.S. EURYALE: A versatile Nextflow pipeline for taxonomic classification and functional annotation of metagenomics data. in 2024 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB) 1–7 (2024).
- Morais, D.A.A.; Cavalcante, J.V.F.; Monteiro, S.S.; Pasquali, M.A.B.; Dalmolin, R.J.S. MEDUSA: A Pipeline for Sensitive Taxonomic Classification and Flexible Functional Annotation of Metagenomic Shotgun Sequences. Front. Genet. 2022; 13. [Google Scholar]
- NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2014, 42, D7–D17. [Google Scholar] [CrossRef] [PubMed]
- The Galaxy, Community; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 2022, 50, W345–W351. [Google Scholar] [CrossRef]
- Li, H. Source code for: Seqtk. 2025. Available online: https://github.com/lh3/seqtk.
- von Meijenfeldt, F.A.B.; Arkhipova, K.; Cambuy, D.D.; Coutinho, F.H.; Dutilh, B.E. Robust taxonomic classification of uncharted microbial sequences and bins with CAT and BAT. Genome Biol. 2019, 20, 217. [Google Scholar] [CrossRef]
- Cornet, L.; et al. The GEN-ERA toolbox: unified and reproducible workflows for research in microbial genomics. GigaScience 2022, 12, 1–10. [Google Scholar] [CrossRef]
- Bankevich, A.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Walker, B.J.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLOS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Alonge, M.; et al. Automated assembly scaffolding using RagTag elevates a new tomato system for high-throughput genome editing. Genome Biol. 2022, 23, 258. [Google Scholar] [CrossRef] [PubMed]
- Orakov, A.; et al. GUNC: detection of chimerism and contamination in prokaryotic genomes. Genome Biol. 2021, 22, 1–19. [Google Scholar] [CrossRef]
- Saary, P.; Mitchell, A.L.; Finn, R.D. Estimating the quality of eukaryotic genomes recovered from metagenomic analysis with EukCC. Genome Biol. 2020, 21, 244. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Zdobnov, E.M. BUSCO: Assessing Genomic Data Quality and Beyond. Curr. Protoc. 2021, 1, e323. [Google Scholar] [CrossRef] [PubMed]
- Cornet, L.; et al. Consensus assessment of the contamination level of publicly available cyanobacterial genomes. PLOS ONE 2018, 13, e0200323. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Meunier, L.; Baurain, D.; Cornet, L. AMAW: automated gene annotation for non-model eukaryotic genomes. F1000Research 2023, 12, 186. [Google Scholar] [CrossRef]
- Brůna, T.; Hoff, K.J.; Lomsadze, A.; Stanke, M.; Borodovsky, M. BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genomics Bioinforma. 2021, 3, lqaa108. [Google Scholar] [CrossRef] [PubMed]
- Queirós, P.; Delogu, F.; Hickl, O.; May, P.; Wilmes, P. Mantis: flexible and consensus-driven genome annotation. GigaScience 2021, 10, giab042. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Portik, D.M.; et al. Highly accurate metagenome-assembled genomes from human gut microbiota using long-read assembly, binning, and consolidation methods. 2024. [Google Scholar] [CrossRef]
- Kalantar, K.L.; et al. IDseq—An open source cloud-based pipeline and analysis service for metagenomic pathogen detection and monitoring. GigaScience 2020, 9, giaa111. [Google Scholar] [CrossRef]
- Dobin, A.; et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Niu, B.; Fu, L.; Sun, S.; Li, W. Artificial and natural duplicates in pyrosequencing reads of metagenomic data. BMC Bioinformatics 2010, 11, 187. [Google Scholar] [CrossRef]
- Wu, T.D.; Nacu, S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 2010, 26, 873–881. [Google Scholar] [CrossRef]
- Zhao, Y.; Tang, H.; Ye, Y. RAPSearch2: a fast and memory-efficient protein similarity search tool for next-generation sequencing data. Bioinformatics 2012, 28, 125–126. [Google Scholar] [CrossRef]
- Chen, I.-M. A.; et al. The IMG/M data management and analysis system v.7: content updates and new features. Nucleic Acids Res. 2023, 51, D723–D732. [Google Scholar] [CrossRef]
- Lomsadze, A.; Gemayel, K.; Tang, S.; Borodovsky, M. Modeling leaderless transcription and atypical genes results in more accurate gene prediction in prokaryotes. Genome Res. 2018, 28, 1079–1089. [Google Scholar] [CrossRef]
- Mistry, J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Haft, D.H.; et al. TIGRFAMs and Genome Properties in 2013. Nucleic Acids Res. 2013, 41, D387–D395. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Möller, S.; Croning, M.D.R.; Apweiler, R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 2001, 17, 646–653. [Google Scholar] [CrossRef]
- McCulloch, J.A.; et al. JAMS - A framework for the taxonomic and functional exploration of microbiological genomic data. 2023. [Google Scholar] [CrossRef]
- Zdobnov, E.M.; Apweiler, R. InterProScan – an integration platform for the signature-recognition methods in InterPro. Bioinformatics 2001, 17, 847–848. [Google Scholar] [CrossRef]
- Chivian, D.; et al. Metagenome-assembled genome extraction and analysis from microbiomes using KBase. Nat. Protoc. 2023, 18, 208–238. [Google Scholar] [CrossRef]
- Zheng, J.; et al. dbCAN3: automated carbohydrate-active enzyme and substrate annotation. Nucleic Acids Res. 2023, 51, W115–W121. [Google Scholar] [CrossRef]
- Shaffer, M.; et al. DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res. 2020, 48, 8883–8900. [Google Scholar] [CrossRef] [PubMed]
- Song, H.S.; et al. OMEGGA: A Computationally Efficient Omics-Guided Global Gapfilling Algorithm for Phenotype-Consistent Metabolic Network Reconstruction. U.S. Department of Energy Genomic Science Program, (2023).
- Faria, J.P.; et al. ModelSEED v2: High-throughput genome-scale metabolic model reconstruction with enhanced energy biosynthesis pathway prediction. 2023. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2 – Approximately Maximum-Likelihood Trees for Large Alignments. PLOS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Edgar, R.C. Muscle5: High-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nat. Commun. 2022, 13, 6968. [Google Scholar] [CrossRef]
- Churcheward, B.; Millet, M.; Bihouée, A.; Fertin, G.; Chaffron, S. MAGNETO: An Automated Workflow for Genome-Resolved Metagenomics. mSystems, 2022; 7. [Google Scholar]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef]
- Benoit, G.; et al. Multiple comparative metagenomics using multiset k-mer counting. PeerJ Comput. Sci. 2016, 2, e94. [Google Scholar] [CrossRef]
- Steinegger, M.; Söding, J. Clustering huge protein sequence sets in linear time. Nat. Commun. 2018, 9, 2542. [Google Scholar] [CrossRef] [PubMed]
- Sunagawa, S.; et al. Metagenomic species profiling using universal phylogenetic marker genes. Nat. Methods 2013, 10, 1196–1199. [Google Scholar] [CrossRef]
- Murovec, B.; Deutsch, L.; Stres, B. Computational Framework for High-Quality Production and Large-Scale Evolutionary Analysis of Metagenome Assembled Genomes. Mol. Biol. Evol. 2020, 37, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Wu, Y.-W. ezTree: an automated pipeline for identifying phylogenetic marker genes and inferring evolutionary relationships among uncultivated prokaryotic draft genomes. BMC Genomics 2018, 19, 921. [Google Scholar] [CrossRef] [PubMed]
- Maurice, N.; Lemaitre, C.; Vicedomini, R.; Frioux, C. Mapler: a pipeline for assessing assembly quality in taxonomically rich metagenomes sequenced with HiFi reads. Bioinformatics 2025, 41, btaf334. [Google Scholar] [CrossRef]
- Benoit, G.; et al. High-quality metagenome assembly from long accurate reads with metaMDBG. Nat. Biotechnol. 2024, 42, 1378–1383. [Google Scholar] [CrossRef] [PubMed]
- Mapleson, D.; Garcia Accinelli, G.; Kettleborough, G.; Wright, J.; Clavijo, B.J. KAT: a K-mer analysis toolkit to quality control NGS datasets and genome assemblies. Bioinformatics 2017, 33, 574–576. [Google Scholar] [CrossRef] [PubMed]
- Zorrilla, F.; Buric, F.; Patil, K.R.; Zelezniak, A. metaGEM: reconstruction of genome scale metabolic models directly from metagenomes. Nucleic Acids Res. 2021, 49, e126. [Google Scholar] [CrossRef] [PubMed]
- Machado, D.; Andrejev, S.; Tramontano, M.; Patil, K.R. Fast automated reconstruction of genome-scale metabolic models for microbial species and communities. Nucleic Acids Res. 2018, 46, 7542–7553. [Google Scholar] [CrossRef]
- Zelezniak, A.; et al. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc. Natl. Acad. Sci. 2015, 112, 6449–6454. [Google Scholar] [CrossRef]
- Lieven, C.; et al. MEMOTE for standardized genome-scale metabolic model testing. Nat. Biotechnol. 2020, 38, 272–276. [Google Scholar] [CrossRef]
- Emiola, A.; Oh, J. High throughput in situ metagenomic measurement of bacterial replication at ultra-low sequencing coverage. Nat. Commun. 2018, 9, 4956. [Google Scholar] [CrossRef]
- Shaban, B.; et al. MetaGenePipe: An Automated, Portable Pipeline for Contig-based Functional and Taxonomic Analysis. The Journal of Open Source Software 2023, 8, 4851. [Google Scholar] [CrossRef]
- Poux, S.; et al. On expert curation and scalability: UniProtKB/Swiss-Prot as a case study. Bioinformatics 2017, 33, 3454–3460. [Google Scholar] [CrossRef]
- Eddy, S.R. A Probabilistic Model of Local Sequence Alignment That Simplifies Statistical Significance Estimation. PLOS Comput. Biol. 2008, 4, e1000069. [Google Scholar] [CrossRef]
- Aramaki, T.; et al. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 2020, 36, 2251–2252. [Google Scholar] [CrossRef]
- Kieser, S.; Brown, J.; Zdobnov, E.M.; Trajkovski, M.; McCue, L.A. ATLAS: A Snakemake workflow for assembly, annotation, and genomic binning of metagenome sequence data. BMC Bioinformatics 2020, 21, 1–8. [Google Scholar] [CrossRef]
- Belmann, P.; et al. Metagenomics-Toolkit: the flexible and efficient cloud-based metagenomics workflow featuring machine learning-enabled resource allocation. NAR Genomics Bioinforma. 2025, 7, lqaf093. [Google Scholar] [CrossRef]
- Kokot, M.; Długosz, M.; Deorowicz, S. KMC 3: counting and manipulating k-mer statistics. Bioinformatics 2017, 33, 2759–2761. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-R, L.M.; Gunturu, S.; Tiedje, J.M.; Cole, J.R.; Konstantinidis, K.T. Nonpareil 3: Fast Estimation of Metagenomic Coverage and Sequence Diversity. mSystems 2018, 3, 10.1128–msystems.00039. [Google Scholar] [CrossRef]
- Mallawaarachchi, V.; Lin, Y. MetaCoAG: Binning Metagenomic Contigs via Composition, Coverage and Assembly Graphs. in Research in Computational Molecular Biology (ed. Pe’er, I.) 70–85 (2022).
- Wang, Z.; Huang, P.; You, R.; Sun, F.; Zhu, S. MetaBinner: a high-performance and stand-alone ensemble binning method to recover individual genomes from complex microbial communities. Genome Biol. 2023, 24, 1–18. [Google Scholar] [CrossRef]
- Mirdita, M.; Steinegger, M.; Breitwieser, F.; Söding, J.; Levy Karin, E. Fast and sensitive taxonomic assignment to metagenomic contigs. Bioinformatics 2021, 37, 3029–3031. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; et al. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef]
- Zimmermann, J.; Kaleta, C.; Waschina, S. gapseq: informed prediction of bacterial metabolic pathways and reconstruction of accurate metabolic models. Genome Biol. 2021, 22, 81. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal. Methods 2015, 8, 12–24. [Google Scholar] [CrossRef]
- Wittler, R. Alignment- and reference-free phylogenomics with colored de Bruijn graphs. Algorithms Mol. Biol. 2020, 15, 4. [Google Scholar] [CrossRef]
- Salazar, V.W.; et al. Metaphor—A workflow for streamlined assembly and binning of metagenomes. GigaScience 2023, 12, 1–12. [Google Scholar] [CrossRef]
- Mainguy, J.; et al. metagWGS, a comprehensive workflow to analyze metagenomic data using Illumina or PacBio HiFi reads. 2024. [Google Scholar] [CrossRef]
- Joshi NA & Fass, JN. Source code for: Sickle-A sliding-window, adaptive, quality-based trimming tool for FastQ files. 2014. Available online: https://github.com/najoshi/sickle.
- Mainguy, J.; Hoede, C. Binette: a fast and accurate bin refinement tool to construct high quality Metagenome Assembled Genomes. J. Open Source Softw. 2024, 9, 6782. [Google Scholar] [CrossRef]
- Kumar, S.; Jones, M.; Koutsovoulos, G.; Clarke, M.; Blaxter, M. Blobology: exploring raw genome data for contaminants, symbionts and parasites using taxon-annotated GC-coverage plots. Front. Genet. 2013; 4. [Google Scholar]
- Hildebrand, F.; et al. Dispersal strategies shape persistence and evolution of human gut bacteria. Cell Host Microbe 2021, 29, 1167–1176.e9. [Google Scholar] [CrossRef]
- Hildebrand, F.; Tadeo, R.; Voigt, A.Y.; Bork, P.; Raes, J. LotuS: an efficient and user-friendly OTU processing pipeline. Microbiome 2014, 2, 30. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-C.; et al. MetaDecoder: a novel method for clustering metagenomic contigs. Microbiome 2022, 10, 46. [Google Scholar] [CrossRef] [PubMed]
- Drula, E.; et al. The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef]
- Milanese, A.; et al. Microbial abundance, activity and population genomic profiling with mOTUs2. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Blanco-Míguez, A.; et al. Extending and improving metagenomic taxonomic profiling with uncharacterized species using MetaPhlAn 4. Nat. Biotechnol. 2023; 1–12. [Google Scholar]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. 2012. [Google Scholar] [CrossRef]
- Cokelaer, T.; Desvillechabrol, D.; Legendre, R.; Cardon, M. ‘Sequana’: a Set of Snakemake NGS pipelines. J. Open Source Softw. 2017, 2, 352. [Google Scholar] [CrossRef]
- Richardson, L.; et al. MGnify: the microbiome sequence data analysis resource in 2023. Nucleic Acids Res. 2023, 51, D753–D759. [Google Scholar] [CrossRef]
- Cock, P.J.A.; et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef] [PubMed]
- Suzek, B.E.; et al. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 2015, 31, 926–932. [Google Scholar] [CrossRef]
- Rho, M.; Tang, H.; Ye, Y. FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res. 2010, 38, e191. [Google Scholar] [CrossRef]
- Blin, K.; et al. antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Ziemski, M.; et al. MOSHPIT: accessible, reproducible metagenome data science on the QIIME 2 framework. 2025. [Google Scholar] [CrossRef]
- Damme, R. van et al. Metagenomics workflow for hybrid assembly, differential coverage binning, metatranscriptomics and pathway analysis (MUFFIN). PLOS Comput. Biol. 2021, 17, 1–13. [Google Scholar]
- Parks, D.H.; et al. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 2022, 50, D785–D794. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Grabherr, M.G.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Liu, L.; Yang, Y.; Deng, Y.; Zhang, T. Nanopore long-read-only metagenomics enables complete and high-quality genome reconstruction from mock and complex metagenomes. Microbiome 2022, 10, 209. [Google Scholar] [CrossRef]
- Vaser, R.; Sović, I.; Nagarajan, N.; Šikić, M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017, 27, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Oxford Nanopore Technologies Ltd. Source code for: medaka-Sequence correction provided by ONT Research. 2025. Available online: https://github.com/nanoporetech/medaka.
- The UniProt Consortium. UniProt: the Universal Protein Knowledgebase in 2025. Nucleic Acids Res. 2025, 53, D609–D617. [Google Scholar] [CrossRef] [PubMed]
- Krakau, S.; Straub, D.; Gourlé, H.; Gabernet, G.; Nahnsen, S. nf-core/mag: a best-practice pipeline for metagenome hybrid assembly and binning. NAR Genomics Bioinforma. 2022; 4. [Google Scholar]
- Schubert, M.; Lindgreen, S.; Orlando, L. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res. Notes 2016, 9, 88. [Google Scholar] [CrossRef] [PubMed]
- Levy Karin, E.; Mirdita, M.; Söding, J. MetaEuk—sensitive, high-throughput gene discovery, and annotation for large-scale eukaryotic metagenomics. Microbiome 2020, 8, 48. [Google Scholar] [CrossRef]
- Borry, M.; Hübner, A.; Rohrlach, A.B.; Warinner, C. PyDamage: automated ancient damage identification and estimation for contigs in ancient DNA de novo assembly. PeerJ 2021, 9, e11845. [Google Scholar] [CrossRef]
- Camargo, A.P.; et al. Identification of mobile genetic elements with geNomad. Nat. Biotechnol. 2024, 42, 1303–1312. [Google Scholar] [CrossRef]
- Karlicki, M.; Antonowicz, S.; Karnkowska, A. Tiara: deep learning-based classification system for eukaryotic sequences. Bioinformatics 2022, 38, 344–350. [Google Scholar] [CrossRef]
- Almeida, F.M. de, Campos, T.A. de & Pappas, G.J. Scalable and versatile container-based pipelines for de novo genome assembly and bacterial annotation. F1000Research 2023, 12, 1205. [Google Scholar]
- Seemann, T. Source code for: Shovill-Assemble bacterial isolate genomes from Illumina paired-end reads. 2020. Available online: https://github.com/tseemann/shovill.
- Haghshenas, E.; Asghari, H.; Stoye, J.; Chauve, C.; Hach, F. HASLR: Fast Hybrid Assembly of Long Reads. iScience 2020, 23, 101389. [Google Scholar] [CrossRef]
- Vaser, R.; Šikić, M. Time- and memory-efficient genome assembly with Raven. Nat. Comput. Sci. 2021, 1, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Shafin, K.; et al. Nanopore sequencing and the Shasta toolkit enable efficient de novo assembly of eleven human genomes. Nat. Biotechnol. 2020, 38, 1044–1053. [Google Scholar] [CrossRef]
- Ruan, J.; Li, H. Fast and accurate long-read assembly with wtdbg2. Nat. Methods 2020, 17, 155–158. [Google Scholar] [CrossRef]
- Graham, E.D.; Heidelberg, J.F.; Tully, B.J. Potential for primary productivity in a globally-distributed bacterial phototroph. ISME J. 2018, 12, 1861–1866. [Google Scholar] [CrossRef]
- Seemann, T. Source code for: Barrnap-Bacterial ribosomal RNA predictor. 2018. Available online: https://github.com/tseemann/shovill.
- Feldgarden, M.; et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef]
- Starikova, E.V.; et al. Phigaro: high-throughput prophage sequence annotation. Bioinformatics 2020, 36, 3882–3884. [Google Scholar] [CrossRef]
- Dong, W.; et al. An expanded database and analytical toolkit for identifying bacterial virulence factors and their associations with chronic diseases. Nat. Commun. 2024, 15, 8084. [Google Scholar] [CrossRef]
- Carattoli, A.; et al. In Silico Detection and Typing of Plasmids using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Maiden, M.C. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 2010, 11, 595. [Google Scholar] [CrossRef]
- Schwengers, O.; et al. Platon: identification and characterization of bacterial plasmid contigs in short-read draft assemblies exploiting protein sequence-based replicon distribution scores. Microb. Genomics 2020, 6, e000398. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
- Arango-Argoty, G.A.; et al. ARGminer: a web platform for the crowdsourcing-based curation of antibiotic resistance genes. Bioinformatics 2020, 36, 2966–2973. [Google Scholar] [CrossRef]
- Florensa, A.F.; Kaas, R.S.; Clausen, P.T.L.C.; Aytan-Aktug, D.; Aarestrup, F.M. ResFinder – an open online resource for identification of antimicrobial resistance genes in next-generation sequencing data and prediction of phenotypes from genotypes. Microb. Genomics 2022, 8, 000748. [Google Scholar] [CrossRef]
- Narayanasamy, S.; et al. IMP: a pipeline for reproducible reference-independent integrated metagenomic and metatranscriptomic analyses. Genome Biol. 2016, 17, 260. [Google Scholar] [CrossRef] [PubMed]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef]
- Schudoma, C. Source code for: gff_quantifier. 2023. Available online: https://github.com/cschu/gff_quantifier.
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Tadrent, N.; et al. SnakeMAGs: a simple, efficient, flexible and scalable workflow to reconstruct prokaryotic genomes from metagenomes. F1000Research 2023, 11, 1522. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.S.B.; et al. SPIRE: a Searchable, Planetary-scale mIcrobiome REsource. Nucleic Acids Res. 2024, 52, D777–D783. [Google Scholar] [CrossRef]
- Coelho, L.P.; et al. NG-meta-profiler: fast processing of metagenomes using NGLess, a domain-specific language. Microbiome 2019, 7, 84. [Google Scholar] [CrossRef]
- Seemann, T. Source code for: ABRicate-Mass screening of contigs for antimicrobial and virulence genes. 2020. Available online: https://github.com/tseemann/abricate.
- Doster, E.; et al. MEGARes 2.0: a database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic Acids Res. 2020, 48, D561–D569. [Google Scholar] [CrossRef]
- Santos-Júnior, C.D.; Pan, S.; Zhao, X.-M.; Coelho, L.P. Macrel: antimicrobial peptide screening in genomes and metagenomes. PeerJ 2020, 8, e10555. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D.; et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef]
- Tamames, J.; Puente-Sánchez, F. SqueezeMeta, a highly portable, fully automatic metagenomic analysis pipeline. Front. Microbiol. 2019, 10, 3349. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Parks, D.H. Source code for: CompareM-A toolbox for comparative genomics. 2020. Available online: https://github.com/donovan-h-parks/CompareM.
- Marçais, G.; et al. MUMmer4: A fast and versatile genome alignment system. PLOS Comput. Biol. 2018, 14, e1005944. [Google Scholar] [CrossRef]
- Puente-Sánchez, F.; García-García, N.; Tamames, J. SQMtools: automated processing and visual analysis of ’omics data with R and anvi’o. BMC Bioinformatics 2020, 21, 358. [Google Scholar] [CrossRef]
- Sjöqvist, C.; Delgado, L.F.; Alneberg, J.; Andersson, A.F. Ecologically coherent population structure of uncultivated bacterioplankton. ISME J. 2021, 15, 3034–3049. [Google Scholar] [CrossRef]
- Clarke, E.L.; et al. Sunbeam: An extensible pipeline for analyzing metagenomic sequencing experiments. Microbiome 2019, 7, 1–13. [Google Scholar] [CrossRef]
- Espinoza, J.L.; et al. Unveiling the microbial realm with VEBA 2.0: a modular bioinformatics suite for end-to-end genome-resolved prokaryotic, (micro)eukaryotic and viral multi-omics from either short- or long-read sequencing. Nucleic Acids Res. 2024, 52, e63. [Google Scholar] [CrossRef]
- Bushmanova, E.; Antipov, D.; Lapidus, A.; Prjibelski, A.D. rnaSPAdes: a de novo transcriptome assembler and its application to RNA-Seq data. GigaScience 2019, 8, giz100. [Google Scholar] [CrossRef]
- Nayfach, S.; et al. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat. Biotechnol. 2021, 39, 578–585. [Google Scholar] [CrossRef]
- Ren, J.; Ahlgren, N.A.; Lu, Y.Y.; Fuhrman, J.A.; Sun, F. VirFinder: a novel k-mer based tool for identifying viral sequences from assembled metagenomic data. Microbiome 2017, 5, 69. [Google Scholar] [CrossRef]
- Zdouc, M.M.; et al. MIBiG 4.0: advancing biosynthetic gene cluster curation through global collaboration. Nucleic Acids Res. 2025, 53, D678–D690. [Google Scholar] [CrossRef]
- Eberhardt, R.Y.; et al. AntiFam: a tool to help identify spurious ORFs in protein annotation. Database 2012, 2012, bas003. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Perez, C.A.; Conrad, R.E.; Konstantinidis, K.T. MicrobeAnnotator: a user-friendly, comprehensive functional annotation pipeline for microbial genomes. BMC Bioinformatics 2021, 22, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Yu, Y.W. Rapid species-level metagenome profiling and containment estimation with sylph. Nat. Biotechnol. 2025, 43, 1348–1359. [Google Scholar] [CrossRef]
- Weber, N.; et al. Nephele: a cloud platform for simplified, standardized and reproducible microbiome data analysis. Bioinformatics 2018, 34, 1411–1413. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Doak, T.G. A Parsimony Approach to Biological Pathway Reconstruction/Inference for Genomes and Metagenomes. PLOS Comput. Biol. 2009, 5, e1000465. [Google Scholar] [CrossRef]
- Goussarov, G.; et al. Benchmarking short-, long- and hybrid-read assemblers for metagenome sequencing of complex microbial communities. Microbiology 2024, 170, 001469. [Google Scholar] [CrossRef]
- Meyer, F.; et al. Tutorial: assessing metagenomics software with the CAMI benchmarking toolkit. Nat. Protoc. 2021, 16, 1785–1801. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Y.; Fuhrman, J.A.; Sun, F.; Zhu, S. Assessment of metagenomic assemblers based on hybrid reads of real and simulated metagenomic sequences. Brief. Bioinform. 2020, 21, 777–790. [Google Scholar] [CrossRef]
- Rozov, R.; Goldshlager, G.; Halperin, E.; Shamir, R. Faucet: streaming de novo assembly graph construction. Bioinformatics 2018, 34, 147–154. [Google Scholar] [CrossRef]
- Brown, C.L.; et al. Critical evaluation of short, long, and hybrid assembly for contextual analysis of antibiotic resistance genes in complex environmental metagenomes. Sci. Rep. 2021, 11, 3753. [Google Scholar] [CrossRef] [PubMed]
- Herazo-Álvarez, J.; Mora, M.; Cuadros-Orellana, S.; Vilches-Ponce, K.; Hernández-García, R. A review of neural networks for metagenomic binning. Brief. Bioinform. 2025, 26, bbaf065. [Google Scholar] [CrossRef] [PubMed]
- Cansdale, A.; Chong, J.P.J. MAGqual: a stand-alone pipeline to assess the quality of metagenome-assembled genomes. Microbiome 2024, 12, 1–10. [Google Scholar] [CrossRef]
- Imelfort, M.; et al. GroopM: an automated tool for the recovery of population genomes from related metagenomes. PeerJ 2014, 2, e603. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; et al. Evaluating metagenomics tools for genome binning with real metagenomic datasets and CAMI datasets. BMC Bioinformatics 2020, 21, 1–15. [Google Scholar] [CrossRef]
- Yepes-García, J.; Falquet, L. Metagenome quality metrics and taxonomical annotation visualization through the integration of MAGFlow and BIgMAG. F1000Research 2024, 13, 640. [Google Scholar] [CrossRef]
- Simion, P.; et al. A Large and Consistent Phylogenomic Dataset Supports Sponges as the Sister Group to All Other Animals. Curr. Biol. 2017, 27, 958–967. [Google Scholar] [CrossRef]
- Edwin, N.R.; Fitzpatrick, A.H.; Brennan, F.; Abram, F.; O’Sullivan, O. An in-depth evaluation of metagenomic classifiers for soil microbiomes. Environ. Microbiome 2024, 19, 19. [Google Scholar] [CrossRef]
- Timilsina, M.; Chundru, D.; Pradhan, A.K.; Blaustein, R.A.; Ghanem, M. Benchmarking Metagenomic Pipelines for the Detection of Foodborne Pathogens in Simulated Microbial Communities. J. Food Prot. 2025, 88, 100583. [Google Scholar] [CrossRef] [PubMed]
- Irankhah, L.; Khorsand, B.; Naghibzadeh, M.; Savadi, A. Analyzing the performance of short-read classification tools on metagenomic samples toward proper diagnosis of diseases. J. Bioinform. Comput. Biol. 2024, 22, 2450012. [Google Scholar] [CrossRef]
- Van Uffelen, A.; et al. Benchmarking bacterial taxonomic classification using nanopore metagenomics data of several mock communities. Sci. Data 2024, 11, 864. [Google Scholar] [CrossRef]
- Liang, Q.; Bible, P.W.; Liu, Y.; Zou, B.; Wei, L. DeepMicrobes: taxonomic classification for metagenomics with deep learning. NAR Genomics Bioinforma. 2020, 2, lqaa009. [Google Scholar] [CrossRef]
- Pusadkar, V.; Azad, R.K. Benchmarking Metagenomic Classifiers on Simulated Ancient and Modern Metagenomic Data. Microorganisms 2023, 11, 2478. [Google Scholar] [CrossRef]
- Marić, J.; Križanović, K.; Riondet, S.; Nagarajan, N.; Šikić, M. Comparative analysis of metagenomic classifiers for long-read sequencing datasets. BMC Bioinformatics 2024, 25, 15. [Google Scholar] [CrossRef]
- Lin, B.; Luo, X.; Liu, Y.; Jin, X. A comprehensive review and comparison of existing computational methods for protein function prediction. Brief. Bioinform. 2024, 25, bbae289. [Google Scholar] [CrossRef] [PubMed]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef]
- Rawlings, N.D.; et al. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef] [PubMed]
- Zeller, M.; Huson, D.H. Comparison of functional classification systems. NAR Genomics Bioinforma. 2022, 4, lqac090. [Google Scholar] [CrossRef]
- Xu, Y.; et al. A new massively parallel nanoball sequencing platform for whole exome research. BMC Bioinformatics 2019, 20, 153. [Google Scholar] [CrossRef]
- Liang, H.; et al. Efficiently constructing complete genomes with CycloneSEQ to fill gaps in bacterial draft assemblies. Gigabyte 2025, 2025, gigabyte154–0. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-M.; et al. Comparative analysis of 7 short-read sequencing platforms using the Korean Reference Genome: MGI and Illumina sequencing benchmark for whole-genome sequencing. GigaScience 2021, 10, giab014. [Google Scholar] [CrossRef]
- Vosloo, S.; et al. Evaluating de Novo Assembly and Binning Strategies for Time Series Drinking Water Metagenomes. Microbiol. Spectr. 2021; 9. [Google Scholar]
- Lynn, H.M.; Gordon, J.I. Sequential co-assembly reduces computational resources and errors in metagenome-assembled genomes. Cell Rep. Methods, 2025; 5. [Google Scholar]
- Goldfarb, T.; et al. NCBI RefSeq: reference sequence standards through 25 years of curation and annotation. Nucleic Acids Res. 2025, 53, D243–D257. [Google Scholar] [CrossRef] [PubMed]
- Arkin, A.P.; et al. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef]
- Achudhan, A.B.; Kannan, P.; Gupta, A.; Saleena, L.M. A Review of Web-Based Metagenomics Platforms for Analysing Next-Generation Sequence Data. Biochem. Genet. 2024, 62, 621–632. [Google Scholar] [CrossRef]
- Wratten, L.; Wilm, A.; Göke, J. Reproducible, scalable, and shareable analysis pipelines with bioinformatics workflow managers. Nat. Methods 2021, 18, 1161–1168. [Google Scholar] [CrossRef]
- Köster, J.; et al. Sustainable data analysis with Snakemake. F1000Research 2021, 10, 33. [Google Scholar] [PubMed]
- Tommaso, P.D.; et al. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 2017, 35, 316–319. [Google Scholar] [CrossRef]
- OpenWDL. Source code for: Specification for the Workflow Description Language (WDL). 2025. Available online: https://github.com/openwdl/wdl.
- Ewels, P.A.; et al. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 2020, 38, 276–278. [Google Scholar] [CrossRef]
- Roach, M.J.; et al. Ten simple rules and a template for creating workflows-as-applications. PLOS Comput. Biol. 2022, 18, e1010705. [Google Scholar] [CrossRef] [PubMed]
- Reiter, T.; et al. Streamlining data-intensive biology with workflow systems. GigaScience, 2021; 10. [Google Scholar]
- Kadri, S.; Sboner, A.; Sigaras, A.; Roy, S. Containers in Bioinformatics: Applications, Practical Considerations, and Best Practices in Molecular Pathology. J. Mol. Diagn. 2022, 24, 442–454. [Google Scholar] [CrossRef]
- Badia, R.M.; et al. COMP Superscalar, an interoperable programming framework. SoftwareX, 4.
- Espinoza, J.L. Source code for: Genopype-Architecture for creating bash pipelines, in particular, for bioinformatics. 2025. Available online: https://github.com/jolespin/genopype.
| Pipeline/Platform | Category | Short reads | Long reads* | Hybrid Assembly | Multiple samples |
Co-assembly and/or Co-binning** | Bin refinement | Infrastructure *** |
Interface∇ | Workflow manager | Software execution | Special features | Last update° | Number of citations° | License⏶ | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Ancient DNA [44] | Special | Yes | No | No | No | No | Yes | Local, HPC | CLI | Local | Ancient DNA identification | 2024 | 0 | Not specified | |
| 2 | Anvi'o [50] | Short-read centered | Yes | No | No | Yes | Yes | Yes | Local, HPC | CLI/GUI | Conda | Visualization module | 2025 | 678 | GNU GPL v3 | |
| 3 | Aviary [60] | Hybrid | Yes | Yes | Yes | Yes | No | Yes | Local, HPC, CC | CLI | Snakemake | Conda | Genotype recovery | 2025 | NA | GNU GPL v3 |
| 4 | BugBuster [70] | Short-read centered | Yes | No | No | Yes | No | Yes | Local, HPC, CC | CLI | Nextflow | Docker | Taxonomic profiling, antimicrobial resistance gene prediction | 2025 | 0 | Not specified |
| 5 | BV-BRC [79] | Web-based | Yes | No | No | Yes | No | No | External | GUI | External | Taxonomic profiling, Viral MAGs | 2024 | 783 | MIT License | |
| 6 | DATMA [87] | Short-read centered | Yes | No | No | No | No | No | Local, HPC | CLI | COMP Superscalar [300] | Local | Reads first grouped (binning) and assembled in batches | 2020 | 4 | GNU GPL v3 |
| 7 | EasyMetagenome [94] | Short-read centered | Yes | No | No | Yes | Yes | Yes | Local, HPC | CLI | Conda | Taxonomic profiling | 2024 | 14 | GNU GPL v3 | |
| 8 | EasyNanoMeta [99] | Long-read focused | No | Yes (ONT) | Yes | Yes | No | No | Local, HPC | CLI | Conda, Singularity | Taxonomic profiling | 2024 | 0 | GNU GPL v3 | |
| 9 | Eukfinder [106] | Special | Yes | Yes | No | No | No | No | Local, HPC | CLI | Conda | Eukaryotic MAGs | 2025 | 1 | MIT License | |
| 10 | EURYALE (MEDUSA) [110,111] | Short-read centered | Yes | No | No | Yes | No | No | Local, HPC, CC | CLI | Nextflow | Conda, Singularity, Docker | 2024 | 7 | MIT License | |
| 11 | Galaxy [113] | Web-based | Yes | Yes | Yes | No | No | Yes | External | GUI | External | Taxonomic profiling | 2024 | 1168 | Academic Free License v3 | |
| 12 | GEN-ERA [116] | Dual | Yes | Yes (ONT) | No | Yes | No | No | Local, HPC, CC | CLI | Nextflow | Singularity | Metabolic modeling | 2024 | 7 | GNU GPL v3 |
| 13 | HiFi-MAG [130] | Long-read focused | No | Yes (PacBio) | No | Yes | No | Yes | Local, HPC, CC | CLI | Snakemake | Conda | 2025 | 8 | BSD-3-Clause-Clear | |
| 14 | IDseq [131] | Web-based | Yes | Yes (ONT) | No | No | No | No | External | GUI | External | Viral MAGs | 2025 | 347 | MIT License | |
| 15 | IMG/M [136] | Web-based | NA | NA | NA | No | No | No | External | GUI | External | Eukaryotic MAGs | 2025 | 268 | IMG Expert Review Submission Agreement | |
| 16 | JAMS [142] | Short-read centered | Yes | No | No | No | No | No | Local, HPC | CLI | Conda | Direct sample comparison | 2025 | 7 | GNU GPL v3 | |
| 17 | KBase [144] | Web-based | Yes | Yes | Yes | Yes | Yes | Yes | External | GUI | External | Taxonomic profiling, metabolic modeling | 2024 | 63 | MIT License | |
| 18 | MAGNETO [152] | Short-read centered | Yes | No | No | Yes | Yes | No | Local, HPC, CC | CLI | Snakemake | Conda | Taxonomic profiling | 2025 | 13 | GNU GPL v3 |
| 19 | MAGO [157] | Short-read centered | Yes | No | No | No | No | Yes | Local, HPC | CLI | Singularity,Docker | Phylogenetic tree generation, pangenome analysis | 2020 | 21 | Creative Commons BY 4.0 | |
| 20 | Mapler [160] | Long-read focused | No | Yes (PacBio) | No | Yes | No | No | Local, HPC, CC | CLI | Snakemake | Conda | Visualization module | 2025 | 0 | GNU AGPL v3 |
| 21 | MetaGEM [163] | Short-read centered | Yes | No | No | Yes | No | Yes | Local, HPC, CC | CLI | Snakemake | Conda | Eukaryotic MAGs, Metabolic modeling | 2023 | 99 | MIT License |
| 22 | MetaGenePipe [168] | Short-read centered | Yes | No | No | Yes | Yes | No | Local, HPC, CC | CLI | Workflow Definition Language (WDL) [295] | Singularity | 2023 | 1 | Apache License 2.0 |
|
| 23 | Metagenome-Atlas [172] | Short-read centered | Yes | No | No | Yes | No | Yes | Local, HPC, CC | CLI | Snakemake | Conda | 2024 | 159 | BSD-3-Clause-Clear | |
| 24 |
Metagenomics- Toolkit [173] |
Dual | Yes | Yes (ONT) | No | Yes | No | Yes | Local, HPC, CC | CLI | Nextflow | Docker | Plasmid assembly, metabolic modeling, controlled resource allocation |
2025 | 0 | GNU AGPL v3 |
| 25 | Metaphor [183] | Short-read centered | Yes | No | No | Yes | Yes | Yes | Local, HPC, CC | CLI | Snakemake | Conda | Visualization module | 2024 | 13 | MIT License |
| 26 | metagWGS [184] | Dual | Yes | Yes (PacBio) | No | Yes | Yes | Yes | Local, HPC, CC | CLI | Nextflow | Singularity | Taxonomic profiling | 2025 | 2 | GNU GPL v3 |
| 27 | MetaWRAP [74] | Short-read centered | Yes | No | No | Yes | Yes | Yes | Local, HPC | CLI | Conda, Docker | Taxonomic profiling | 2020 | 1917 | MIT License | |
| 28 | MG-TK [188] | Dual | Yes | No | No | Yes | Yes | No | Local, HPC | CLI | Conda | Taxonomic profiling, strain delineation | 2025 | 99 | GNU GPL v2 | |
| 29 | MGnify [197] | Web-based | Yes | Yes | Yes | Yes | Yes | No | External | GUI | External | Taxonomic profiling | 2025 | 286 | Apache License 2.0 | |
| 30 | MOSHPIT [202] | Short-read centered | Yes | No | No | Yes | No | Yes | Local, HPC | CLI | Conda | Taxonomic profiling | 2025 | 1 | BSD-3-Clause-Clear | |
| 31 | MUFFIN [203] | Hybrid pipelines | No | Yes (ONT) | Yes | Yes | No | Yes | Local, HPC, CC | CLI | Nextflow | Conda, Docker, Singularity |
Metatranscriptome support | 2022 | 34 | GNU GPL v3 |
| 32 | NanoPhase [207] | Long-read focused | No | Yes (ONT) | Yes | No | No | Yes | Local, HPC | CLI | Conda | 2023 | 73 | MIT License | ||
| 33 | nf-core/mag [211] | Hybrid | Yes | No | Yes | Yes | Yes | Yes | Local, HPC, CC | CLI | Nextflow | Conda, Docker, Singularity, Others |
Taxonomic profiling, ancient DNA identification | 2025 | 57 | MIT License |
| 34 |
ngs-preprocess MpGAp Bacannot [217] |
Hybrid | Yes | Yes | Yes | Yes | No | No | Local, HPC, CC | CLI | Nextflow | Conda, Docker, Singularity | Antimicrobial resistance gene prediction, virulence factor annotation, plasmid assembly |
2025 | 2 | GNU GPL v3 |
| 35 | nIMP3 [234] | Short-read centered | Yes | No | No | Yes | No | No | Local, HPC, CC | CLI | Nextflow | Docker, Singularity | Metatranscriptome support, taxonomic profiling | 2024 | 150 | MIT License |
| 36 | SnakeMAGs [238] | Short-read centered | Yes | No | No | Yes | No | No | Local, HPC, CC | CLI | Snakemake | Conda | 2024 | 6 | CeCILL Free Software License Agreement v2.1 | |
| 37 | SPIRE [239] | Short-read centered | Yes | No | No | Yes | No | No | Local, HPC, CC | CLI | Nextflow | Antimicrobial resistance gene prediction, virulence factor annotation | 2025 | 41 | MIT License | |
| 38 | SqueezeMeta [245] | Hybrid | Yes | Yes | Yes | Yes | Yes | Yes | Local, HPC | CLI | Conda | Taxonomic profiling, metatranscriptome support, visualization module | 2025 | 400 | GNU GPL v3 | |
| 39 | Sunbeam [251] | Short-read centered | Yes | No | No | Yes | No | No | Local, HPC | CLI | Snakemake | Conda, Docker | Taxonomic profiling | 2025 | 184 | GNU GPL v3 |
| 40 | VEBA [252] | Dual | Yes | Yes (ONT or PacBio) | No | Yes | Yes and pseudo- coassembly |
Yes | Local, HPC | CLI | GenoPype [301] | Conda, Docker | Eukaryotic or Viral MAGs, antimicrobial resistance gene prediction, virulence factor annotation | 2025 | 23 | GNU AGPL v3 |
| 41 | WGSA2+/LoRA [260] | Web-based | Yes | Yes (ONT or PacBio) | No | Yes | No | No | External, CC | GUI | AWS environment | External | Visualization module, metatranscriptome support, antimicrobial resistance gene prediction |
2025 | 138 | CC0 1.0 Universal |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





