Submitted:
05 June 2025
Posted:
06 June 2025
You are already at the latest version
Abstract
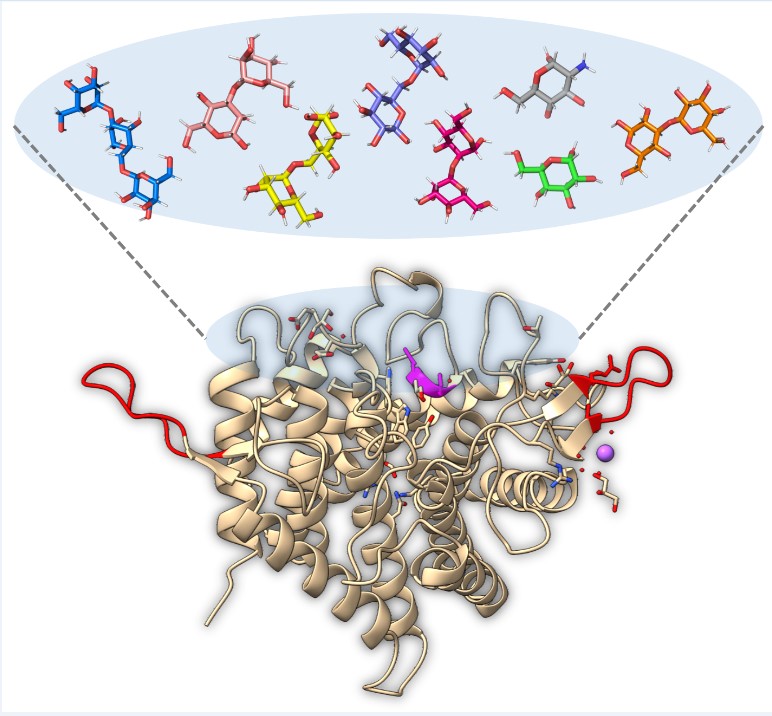
Keywords:
1. Introduction
2. Results and Discussion
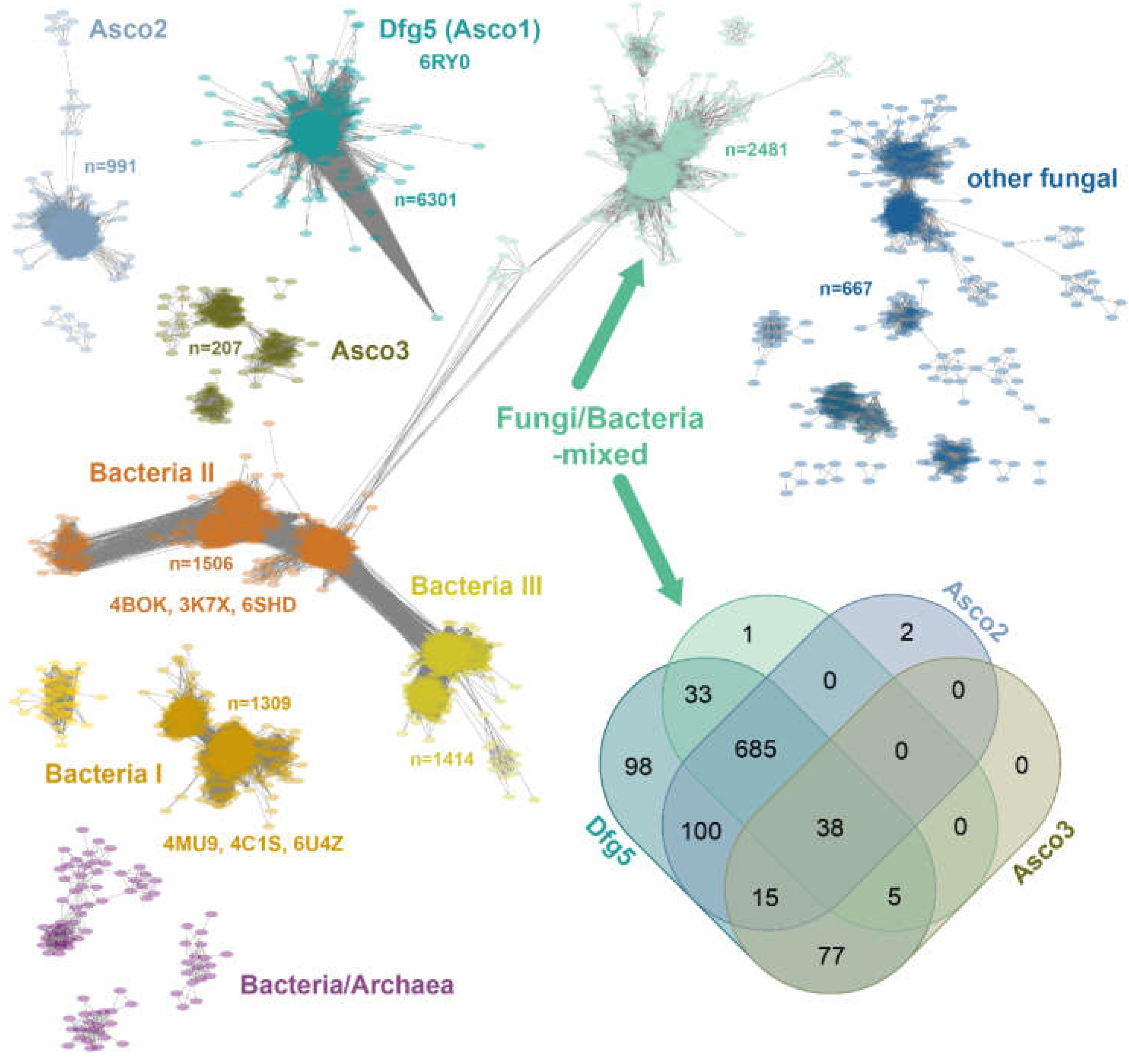
2.1. Sequence Similarity Network of the GH76 Family
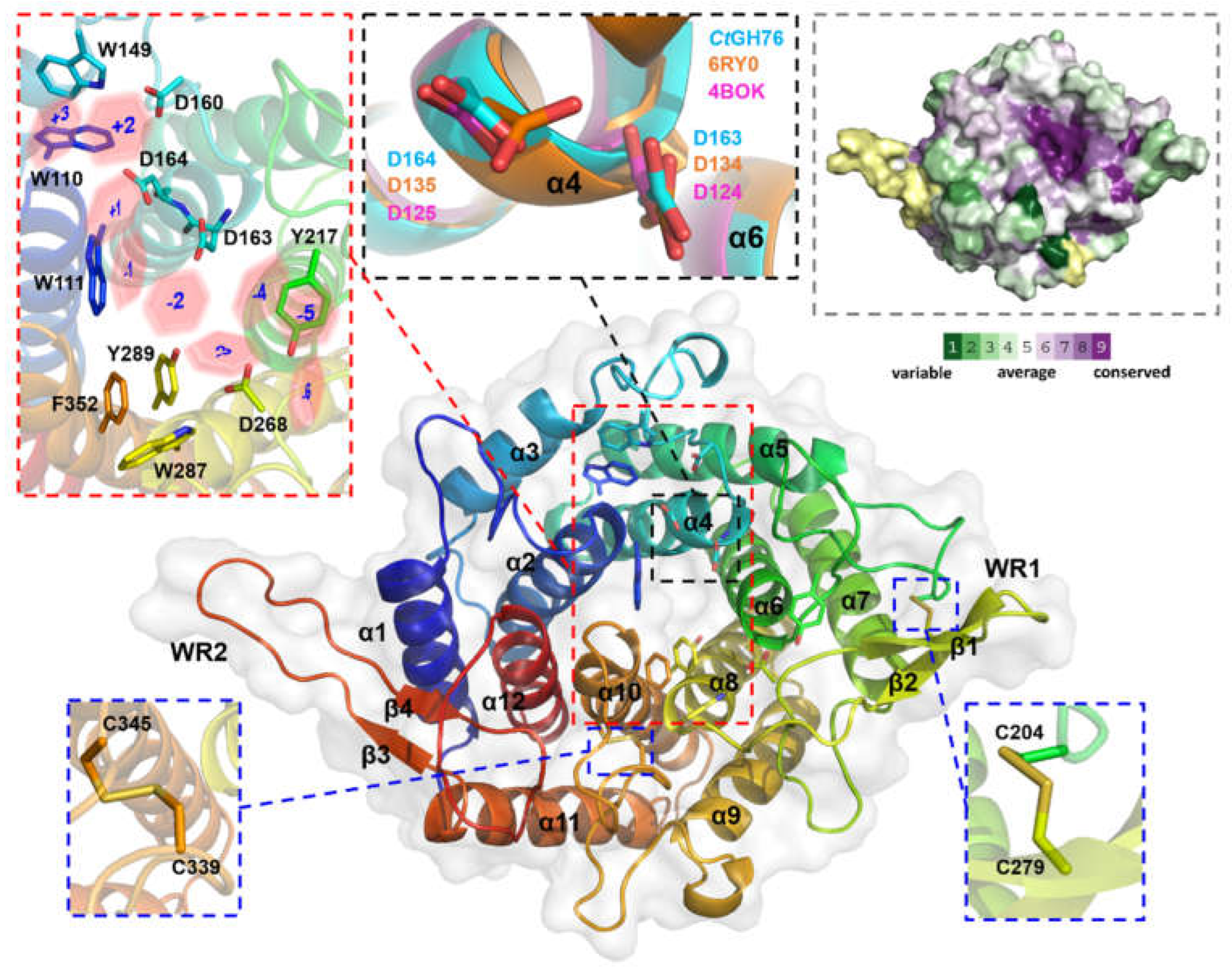
2.2. Overall Structure of CtGH76 and Its Active Site
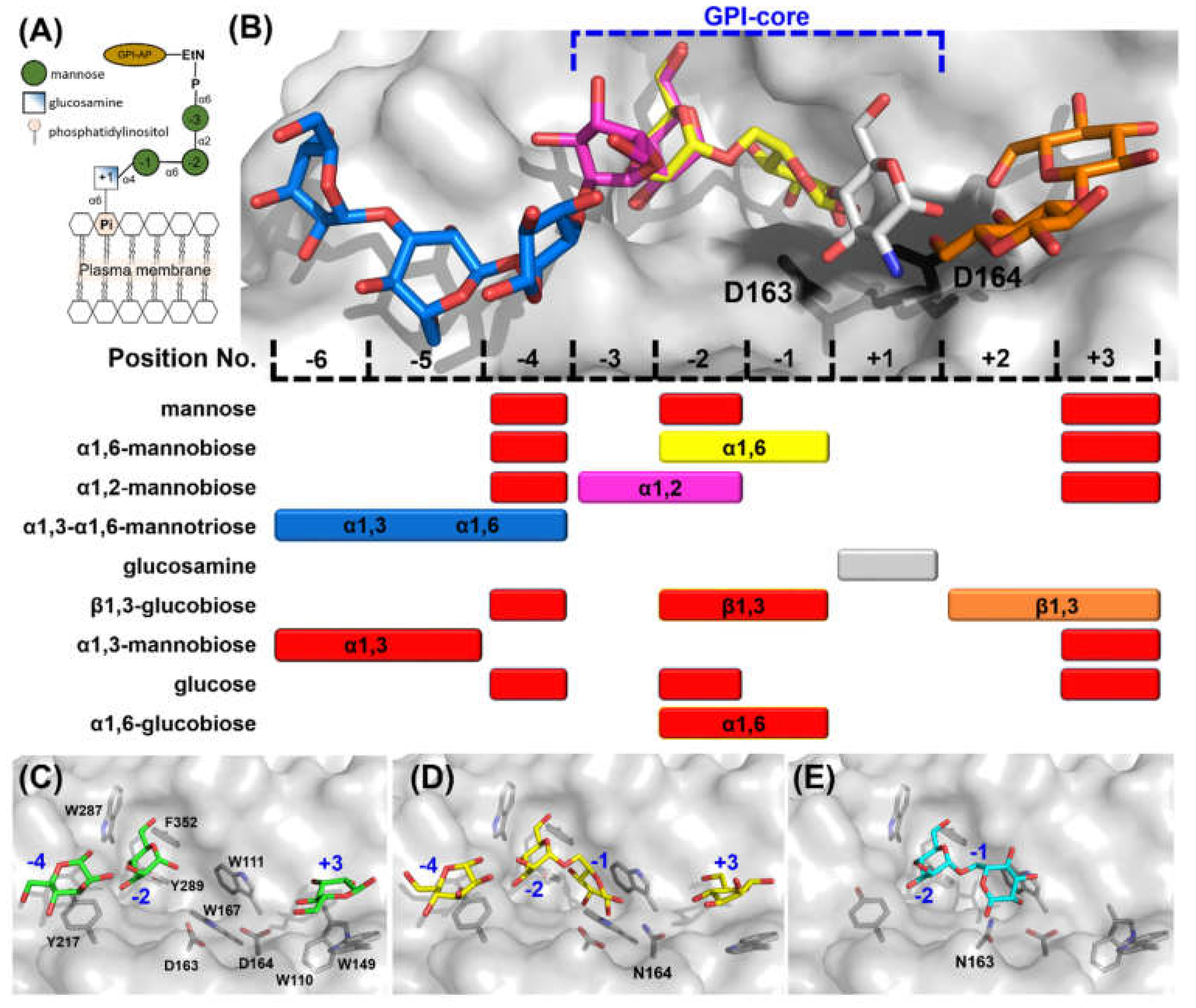
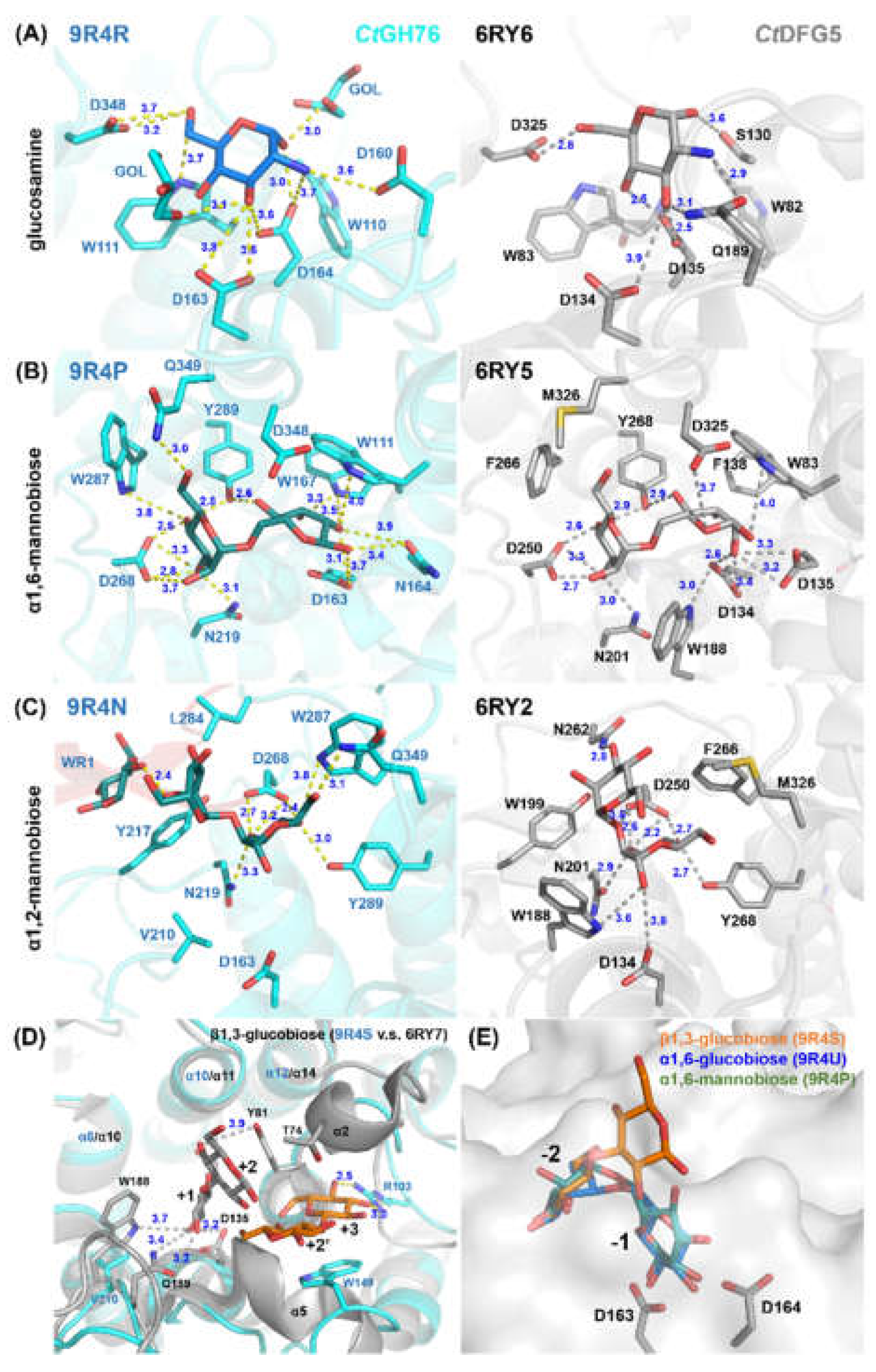
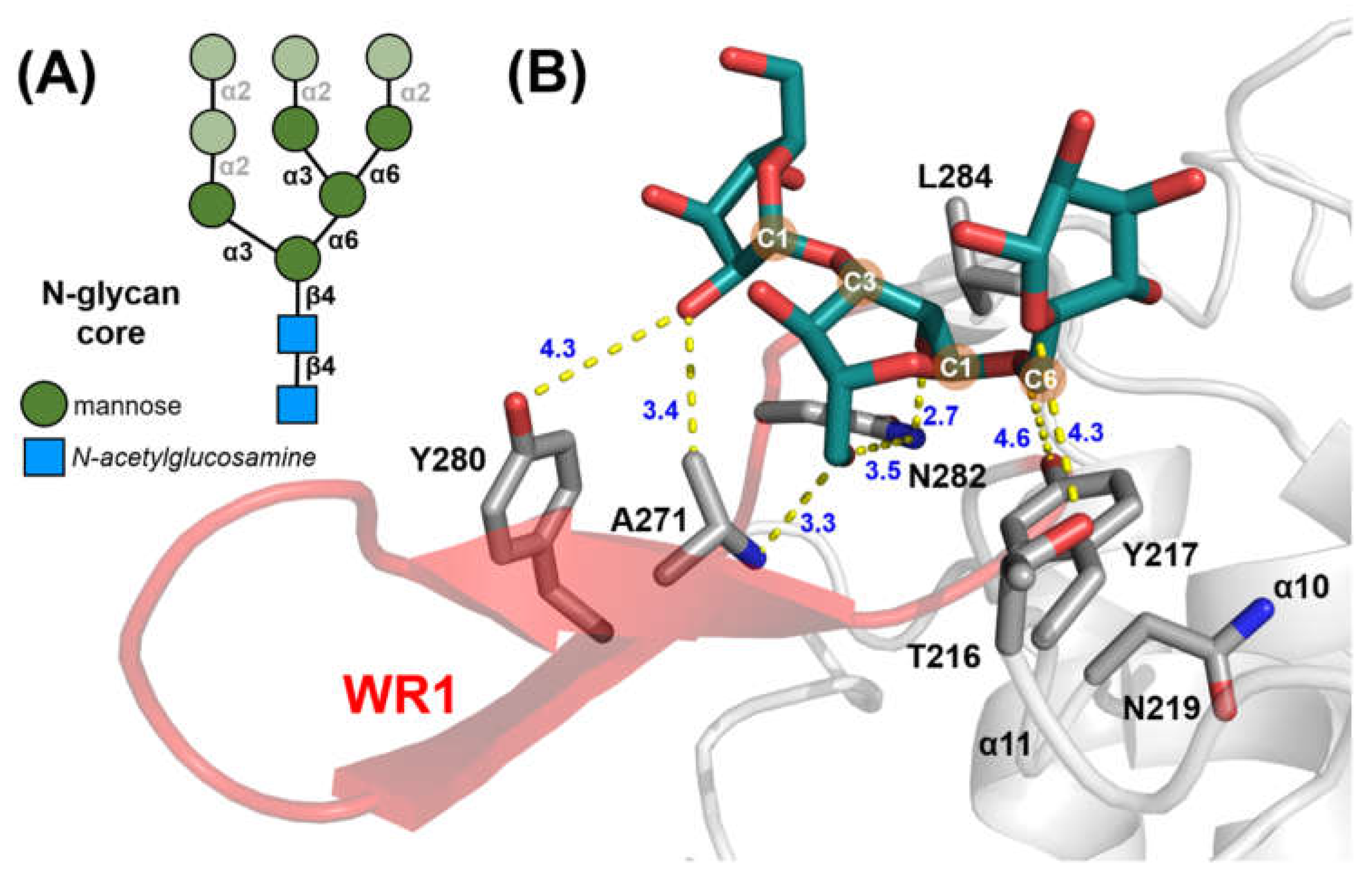
2.3. Glycan Fragment Mapping Reveals a Recognition Mode for GPI-Core Glycans
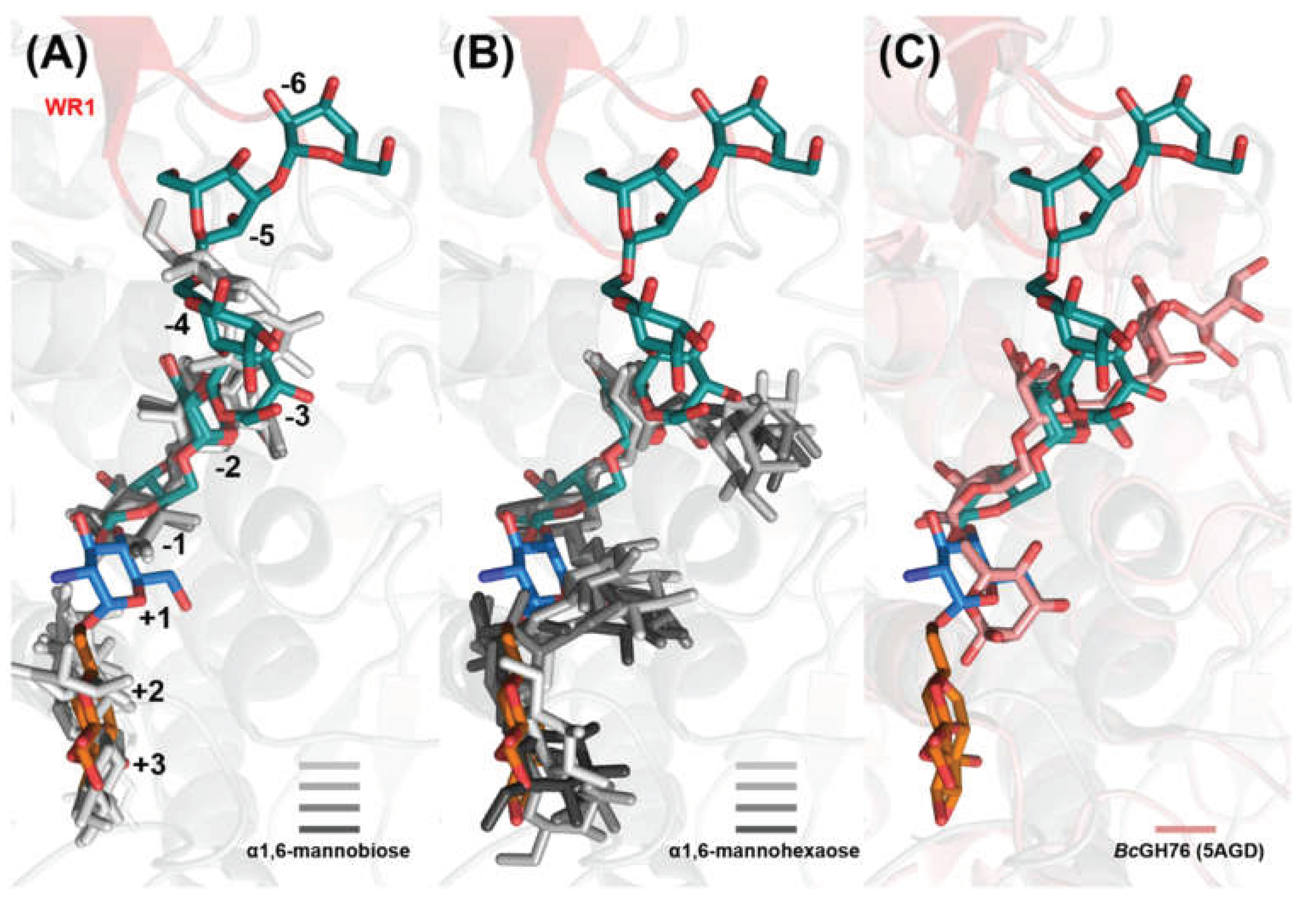
2.4. Glycan Fragment Mapping Shows the Prolonged Glycan Binding Canyon of CtGH76
2.5. Comparison with Alphafold 3-Mediated CtGH76-Glycan Complex Models
2.6. Structural Repertoire Within the F/B-Mixed Subfamily
3. Conclusion
4. Materials and Methods
4.1. Sequence Similarity Network Analysis of the CtGH76
4.2. Cloning, Overexpression and Purification of CtGH76
4.3. Crystallization of CtGH76 and Its Mutants
4.4. Glycan Fragment Screening Using Crystal Soaking
4.5. Data Processing and Structure Determination
4.6. Conservation Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bowman, S.M.; Free, S.J. The structure and synthesis of the fungal cell wall. BioEssays 2006, 28, 799–808. [Google Scholar] [CrossRef]
- Gow, N.A.R., J.-P. Latge, and C.A. Munro, The Fungal Cell Wall: Structure, Biosynthesis, and Function, in The Fungal Kingdom. 2017. p. 267-292.
- Fang, W.; Navratilova, I.; Tejero, T.; Castro-López, J.; van Aalten, D.M.F.; Merino, P.; Delso, I.; Hurtado-Guerrero, R.; Valero-Gonzalez, J.; Gomollón-Bel, F. Inhibitors against Fungal Cell Wall Remodeling Enzymes. ChemMedChem 2018, 13, 128–132. [Google Scholar] [CrossRef]
- Bernard, M.; J.-P. Latgé, Aspergillus fumigatus cell wall: composition and biosynthesis. Medical Mycology 2001, 39, 9–17. [CrossRef]
- Klis, F.M.; Verkleij, A.J.; Vliegenthart, J.F.; Humbel, B.M.; Grün, C.H.; Hochstenbach, F.; Sietsma, J.H.; Kamerling, J.P. The structure of cell wall -glucan from fission yeast. Glycobiology 2005, 15, 245–257. [Google Scholar] [CrossRef]
- Roberts, R.L.; Cabib, E.; Bowers, B. Vectorial synthesis of a polysaccharide by isolated plasma membranes. Proc. Natl. Acad. Sci. USA 1983, 80, 3318–3321. [Google Scholar] [CrossRef]
- Cabib, E.; Braatz, J.; Shematek, E. Biosynthesis of the yeast cell wall. I. Preparation and properties of beta-(1 leads to 3)glucan synthetase. J. Biol. Chem. 1980, 255, 888–894. [Google Scholar] [CrossRef]
- Kapteyn, J.C.; Van Den, E.H.; Klis, F.M. The contribution of cell wall proteins to the organization of the yeast cell wall. Biochim. Biophys. Acta 1999, 1426, 373–383. [Google Scholar] [CrossRef]
- Lesage, G.; Bussey, H. Cell Wall Assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2006, 70, 317–343. [Google Scholar] [CrossRef]
- Hu, H.; Ouyang, H.; Jin, C.; Zhou, H.; Luo, Y.; Li, H. Glycosylphosphatidylinositol (GPI) anchor is required in Aspergillus fumigatus for morphogenesis and virulence. Mol. Microbiol. 2007, 64, 1014–1027. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2013, 42, D490–D495. [Google Scholar] [CrossRef]
- Latgé, J.-P.; Shepherd, S.; Schüttelkopf, A.W.; Ibrahim, A.F.M.; Mouyna, I.; Fontaine, T.; van Aalten, D.M.F.; Hurtado-Guerrero, R. Molecular Mechanisms of Yeast Cell Wall Glucan Remodeling. J. Biol. Chem. 2009, 284, 8461–8469. [Google Scholar] [CrossRef]
- Free, S.J.; Ao, J.; Patel, P.; Kar, B. Neurospora crassa family GH72 glucanosyltransferases function to crosslink cell wall glycoprotein N-linked galactomannan to cell wall lichenin. Fungal Genet. Biol. 2019, 123, 60–69. [Google Scholar] [CrossRef]
- Maddi, A.; Free, S.J.; Ao, J.; Chinnici, J.L. The N-Linked Outer Chain Mannans and the Dfg5p and Dcw1p Endo-α-1,6-Mannanases Are Needed for Incorporation of Candida albicans Glycoproteins into the Cell Wall. Eukaryot. Cell 2015, 14, 792–803. [Google Scholar] [CrossRef]
- Thompson, A.J. , et al., Structure of the GH76 α-mannanase homolog, BT2949, from the gut symbiont Bacteroides thetaiotaomicron. Acta Crystallogr D Biol Crystallogr, 2015. 71(Pt 2): p. 408-15.
- Mösch, H.-U.; Vogt, M.S.; Essen, L.-O. Diversity of GPI-anchored fungal adhesins. Biol. Chem. 2020, 401, 1389–1405. [Google Scholar] [CrossRef]
- Schmitz, G.F.; Silva, D.V.; Mösch, H.-U.; Vogt, M.S.; Essen, L.-O. Structural base for the transfer of GPI-anchored glycoproteins into fungal cell walls. Proc. Natl. Acad. Sci. USA 2020, 117, 22061–22067. [Google Scholar] [CrossRef]
- Tian, W. and J. Skolnick, How well is enzyme function conserved as a function of pairwise sequence identity? J Mol Biol, 2003. 333(4): p. 863-82.
- Rost, B. Twilight zone of protein sequence alignments. Protein Eng. Des. Sel. 1999, 12, 85–94. [Google Scholar] [CrossRef]
- Ziemer, C.J.; Vervecken, W.; Rogowski, A.; Peña, M.J.; Zhu, Y.; Martens, E.C.; Pudlo, N.A.; Suits, M.D.; Hamilton, B.S.; McLean, R.; et al. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature 2015, 517, 165–169. [Google Scholar] [CrossRef]
- Abbott, D.W.; Hoang, K.L.M.; Pardo-Vargas, A.; Solanki, V.; Danglad-Flores, J.; Seeberger, P.H.; Klassen, L.; Crawford, C.J.; Krüger, K.; Teeling, H.; et al. Glycoside hydrolase from the GH76 family indicates that marine Salegentibacter sp. Hel_I_6 consumes alpha-mannan from fungi. ISME J. 2022, 16, 1818–1830. [Google Scholar] [CrossRef]
- Thompson, A.J. , et al., Evidence for a boat conformation at the transition state of GH76 α-1,6-mannanases--key enzymes in bacterial and fungal mannoprotein metabolism. Angew Chem Int Ed Engl, 2015. 54(18): p. 5378-82.
- Gurcha, S.S.; Banzhaf, M.; Prados-Rosales, R.; Mize, T.; Salgueiro, V.C.; Anso, I.; Sullivan, R.; Benedict, S.T.; Besra, G.S.; Scott, N.E.; et al. The mycobacterial glycoside hydrolase LamH enables capsular arabinomannan release and stimulates growth. Nat. Commun. 2024, 15, 5740. [Google Scholar] [CrossRef]
- Oliveros, J.C. , Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. 2024: https://bioinfogp.cnb.csic.es/tools/venny/.
- Groenewald, M.; Chavez, C.M.; Rokas, A.; Hittinger, C.T.; Hulfachor, A.B.; Kpurubu, G.; Huerta, R. The cell morphological diversity of Saccharomycotina yeasts. FEMS Yeast Res. 2024, 24. [Google Scholar] [CrossRef]
- Hallgren, J. , et al., DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. bioRxiv, 2022: p. 2022.04.08.487609.
- Eisenhaber, F.; Wildpaner, M.; Schneider, G.; Eisenhaber, B. A Sensitive Predictor for Potential GPI Lipid Modification Sites in Fungal Protein Sequences and its Application to Genome-wide Studies for Aspergillus nidulans, Candida albicans Neurospora crassa, Saccharomyces cerevisiae and Schizosaccharomyces pombe. J. Mol. Biol. 2004, 337, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Geng, W.; Head-Gordon, T.; Li, P.; Holst, M.J.; Jurrus, E.; Gohara, D.W.; Chen, J.; Konecny, R.; Engel, D.; Felberg, L.E.; et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018, 27, 112–128. [Google Scholar] [CrossRef]
- Maddi, A.; Free, S.J.; Fu, C. The Neurospora crassa dfg5 and dcw1 Genes Encode α-1,6-Mannanases That Function in the Incorporation of Glycoproteins into the Cell Wall. PLOS ONE 2012, 7, e38872. [Google Scholar] [CrossRef] [PubMed]
- Lipowsky, R.; Wehle, M.; Santer, M.; Seeberger, P.H.; Vilotijevic, I.; Silva, D.V. Mechanical Compressibility of the Glycosylphosphatidylinositol (GPI) Anchor Backbone Governed by Independent Glycosidic Linkages. J. Am. Chem. Soc. 2012, 134, 18964–18972. [Google Scholar] [CrossRef]
- Ding, X.; Li, W.; Pan, Y.; Zhang, Y.; Shao, M.; Ran, Y.; Ji, Q.; Kang, P. Soil saprophytic fungi could be used as an important ecological indicator for land management in desert steppe. Ecol. Indic. 2023, 150. [Google Scholar] [CrossRef]
- Mouyna, I.; Fontaine, T.; Henry, C.; Malosse, C.; Moyrand, F.; Li, J.; Janbon, G.; Latgé, J.-P.; Chamot-Rooke, J. Glycosylphosphatidylinositol Anchors from Galactomannan and GPI-Anchored Protein Are Synthesized by Distinct Pathways in Aspergillus fumigatus. J. Fungi 2018, 4, 19. [Google Scholar] [CrossRef]
- Fontaine, T. , et al., Molecular organization of the alkali-insoluble fraction of Aspergillus fumigatus cell wall. J Biol Chem, 2000. 275(36): p. 27594-607.
- Singh, S.; Low, C.M.R.; Bapst, V.; Fuchs, F.B.; Potapenko, A.; Dunger, J.; Arvaniti, E.; Zielinski, M.; Jain, R.; Thillaisundaram, A.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- krinjar, P. , et al., HAVE PROTEIN-LIGAND CO-FOLDING METHODS MOVED BEYOND MEMORISATION? bioRxiv, 2025.
- Djokic, M.; Kumar, M.; Schumbera, E.; Valsecchi, C.I.K.; Polat, F.; Gibson, T.J.; Schäfer, C.; Luck, K.; Lee, C.Y.; Schueler-Furman, O.; et al. Systematic discovery of protein interaction interfaces using AlphaFold and experimental validation. Mol. Syst. Biol. 2024, 20, 75–97. [Google Scholar] [CrossRef]
- Trouillas, F.P.; Úrbez-Torres, J.R.; Gubler, W.D. Diversity of diatrypaceous fungi associated with grapevine canker diseases in California. Mycologia 2010, 102, 319–336. [Google Scholar] [CrossRef]
- Ortiz-Basurto, R.I.; Montalvo-González, E.; Villagrán, Z.; Gómez-Rodríguez, H.; Ríos-García, U.; Anaya-Esparza, L.M.; Pérez-Moreno, J.; Martínez-Reyes, M. Huitlacoche (Ustilago maydis), an Iconic Mexican Fungal Resource: Biocultural Importance, Nutritional Content, Bioactive Compounds, and Potential Biotechnological Applications. Molecules 2023, 28, 4415. [Google Scholar] [CrossRef]
- Wölfle, T.; Conz, C.; Fitzke, E.; Gumiero, A.; Schermann, G.; Gesé, G.V.; Weyer, F.A.; Kappes, J.; Zhang, Y.; Lapouge, K.; et al. Interaction of the cotranslational Hsp70 Ssb with ribosomal proteins and rRNA depends on its lid domain. Nat. Commun. 2016, 7, 13563. [Google Scholar] [CrossRef]
- Lan, D.; Durrani, R.; Huan, W.; Zhao, Z.; Khan, F.I.; Wang, Y. The Lid Domain in Lipases: Structural and Functional Determinant of Enzymatic Properties. Front. Bioeng. Biotechnol. 2017, 5, 16. [Google Scholar] [CrossRef]
- Birren, B.W.; Grabherr, M.; Untereiner, W.A.; Ma, L.-J.; Cuomo, C.A. Draft Genome Sequence of the Cellulolytic Fungus Chaetomium globosum. Genome Announc. 2015, 3, e00021–15. [Google Scholar] [CrossRef]
- Kämper, J.; Kahmann, R.; Bölker, M.; Ma, L.-J.; Brefort, T.; Saville, B.J.; Banuett, F.; Kronstad, J.W.; Gold, S.E.; Müller, O.; et al. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 2006, 444, 97–101. [Google Scholar] [CrossRef]
- Guthke, R.; Linde, J.; Walther, G.; Horn, F.; Mattern, D.J.; Brakhage, A.A.; Valiante, V. Draft Genome Sequence of the Fungus Penicillium brasilianum MG11. Genome Announc. 2015, 3, e00724–15. [Google Scholar] [CrossRef]
- Veelders, M. and L.O. Essen, Complex gadolinium-oxo clusters formed along concave protein surfaces. Chembiochem 2012, 13, 2187–2190. [Google Scholar] [CrossRef]
- Kabsch, W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. Sect. D Struct. Biol. 2010, 66 Pt 2, 133–144. [Google Scholar] [CrossRef]
- Winn, M.D. , et al., Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 2011, 67 Pt 4, 235–242. [Google Scholar] [CrossRef]
- Echols, N.; Headd, J.J.; Zwart, P.H.; Urzhumtsev, A.; Grosse-Kunstleve, R.W.; Mustyakimov, M.; Adams, P.D.; Terwilliger, T.C.; Afonine, P.V.; Moriarty, N.W. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Sect. D Struct. Biol. 2012, 68 Pt 4, 352–367. [Google Scholar] [CrossRef]
- Emsley, P. , et al. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 2010, 66 Pt 4, 486–501. [Google Scholar] [CrossRef]
- Schrödinger, L. and W. Delano, PyMOL. 2020.
- Martz, E.; Abadi, S.; Chay, O.; Pupko, T.; Ashkenazy, H.; Mayrose, I.; Ben-Tal, N. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016, 44, W344–W350. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




