Submitted:
02 June 2025
Posted:
05 June 2025
You are already at the latest version
Abstract
Keywords:
Introduction
Materials and Methods
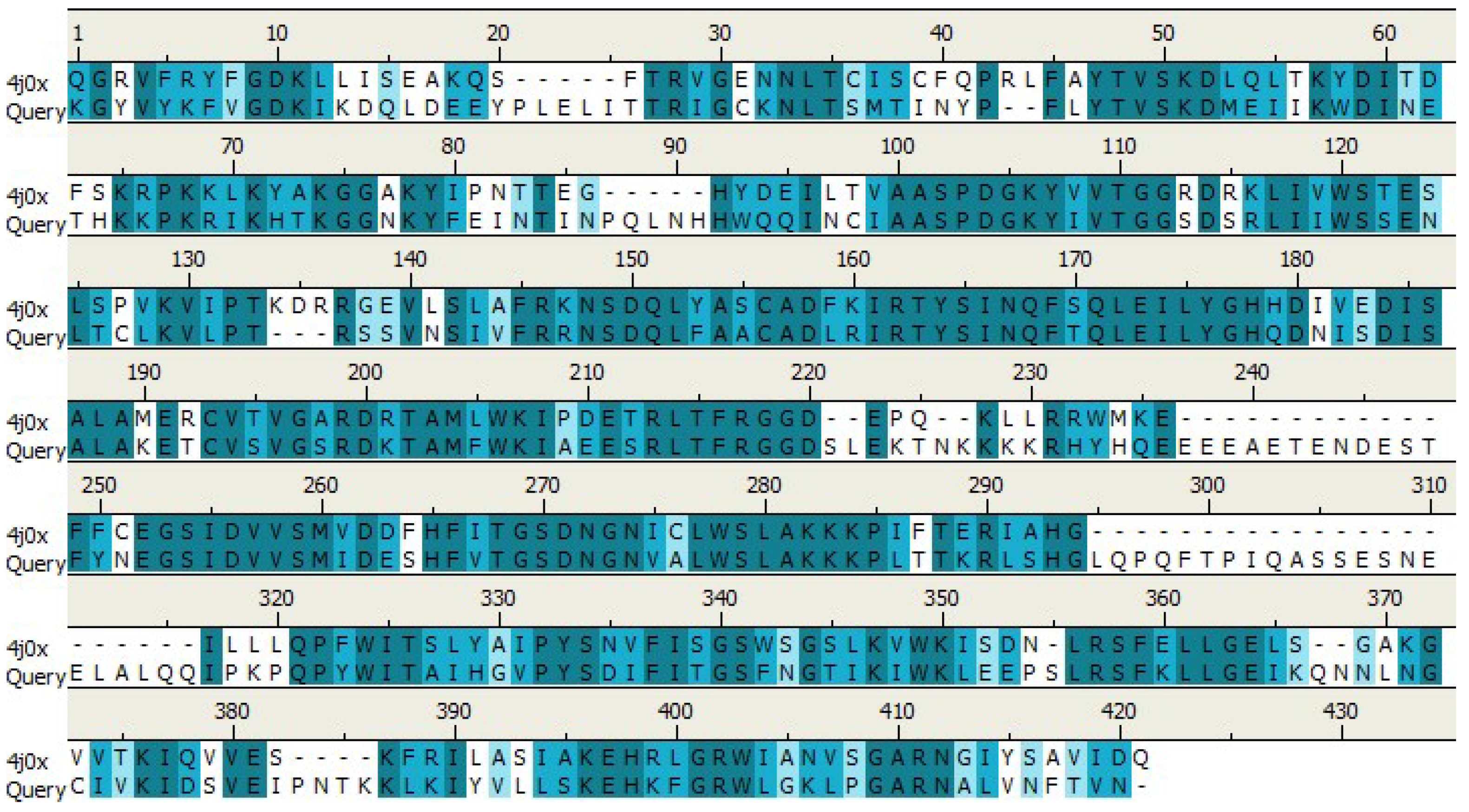
Retrieve and analysis of protein sequence
Modeling of 3D Structure
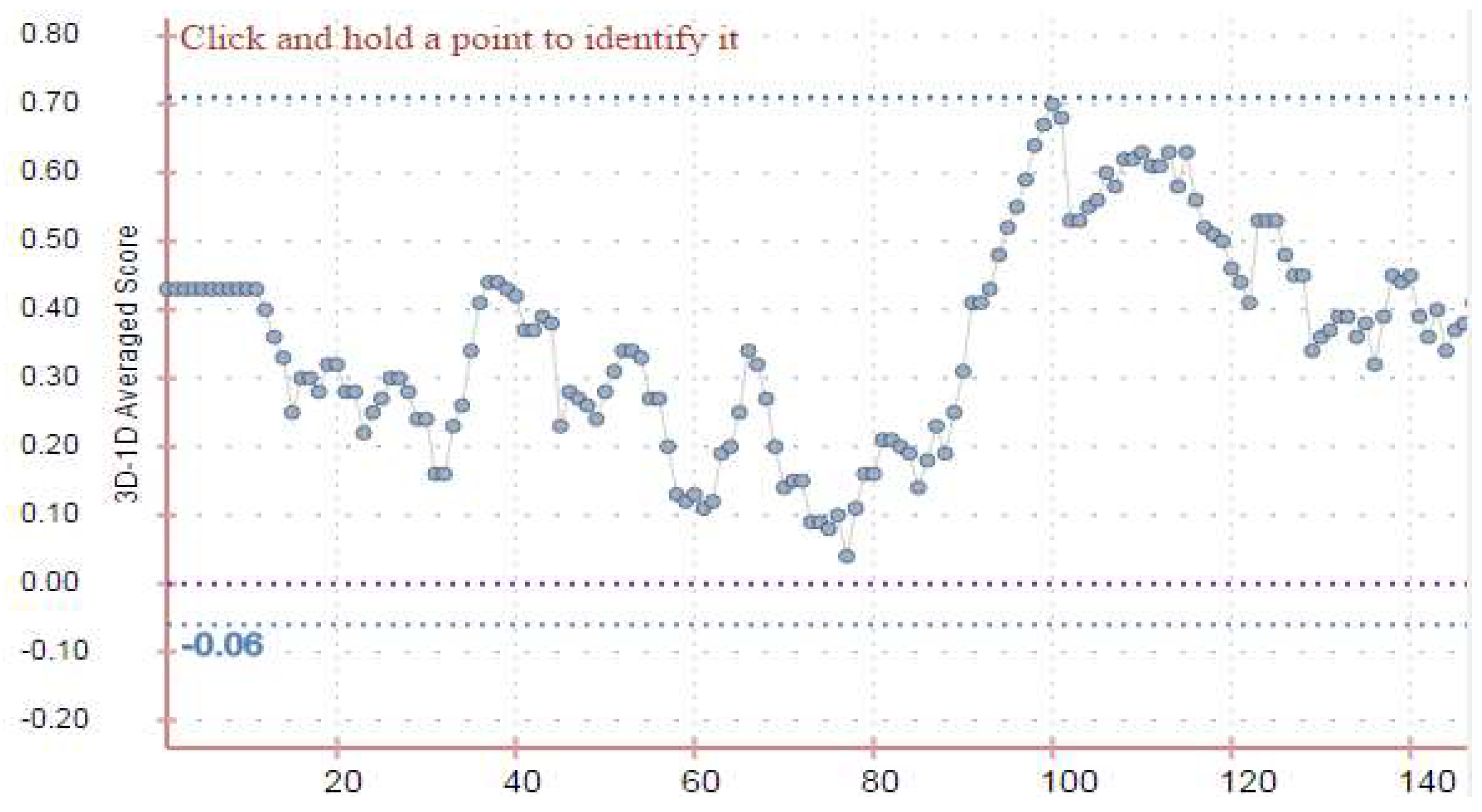
Model Evaluation:
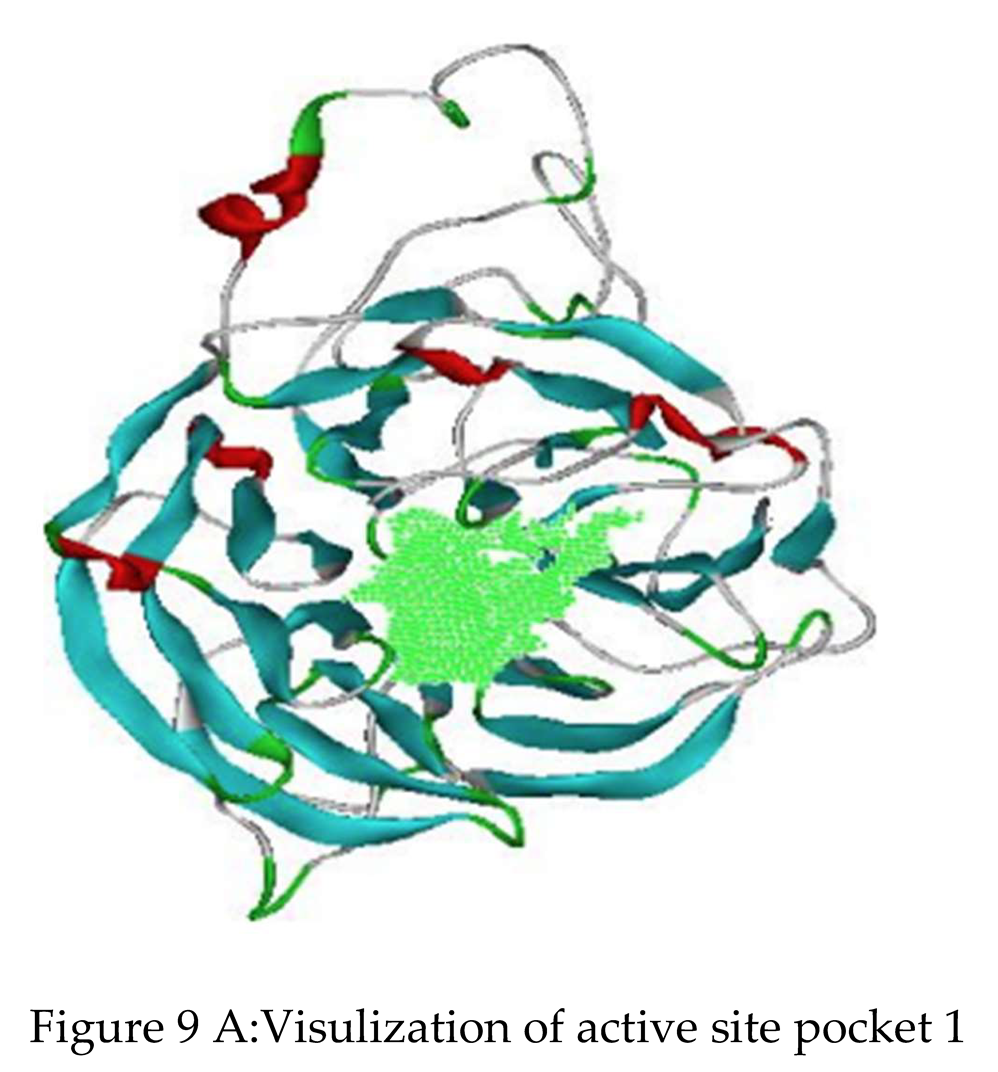
Identification of Active Site
Generation of Ligand data set and their optimization
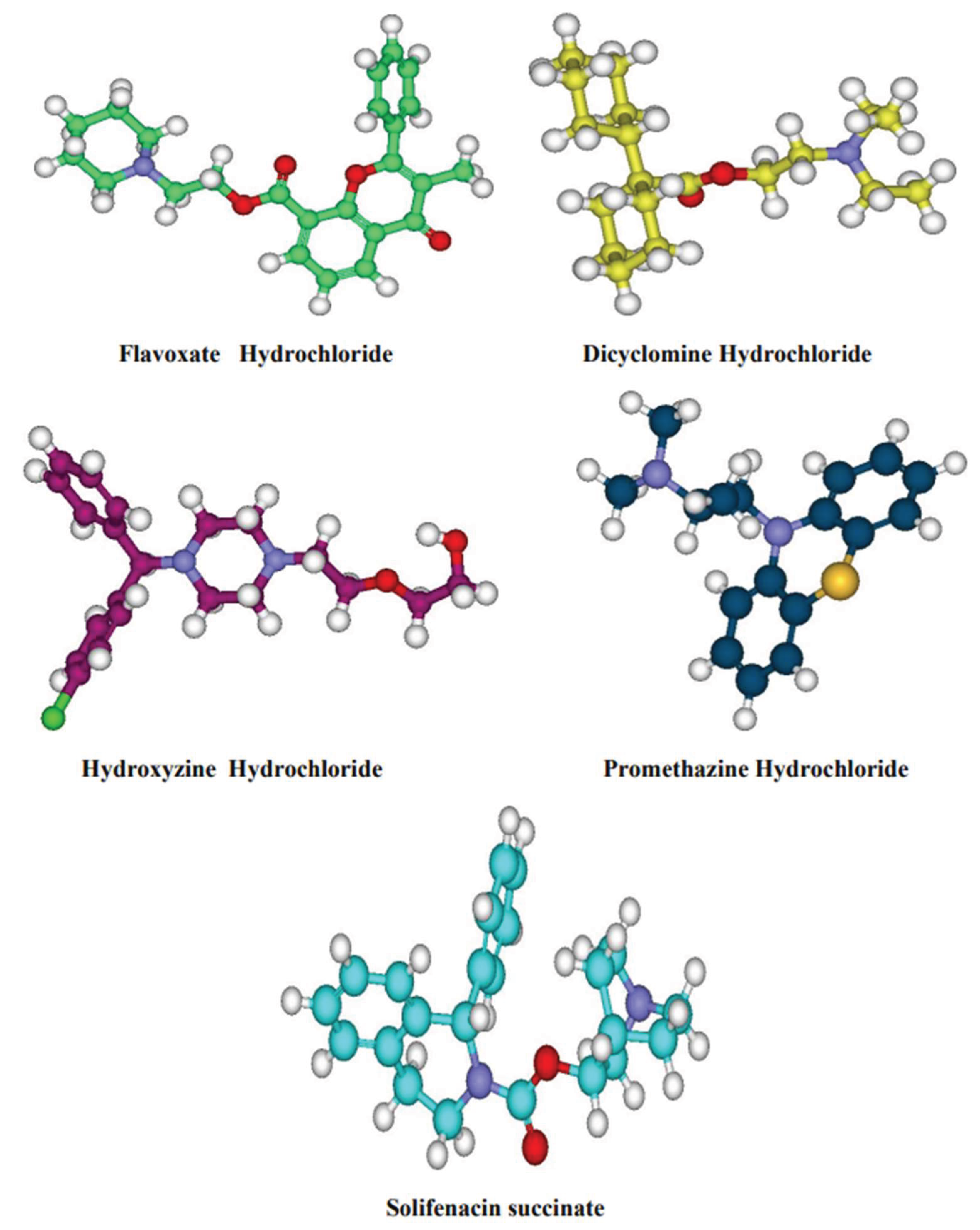
Molecular docking
Results and discussion
Retrieve and analysis of sequence
3. D Structure modeling
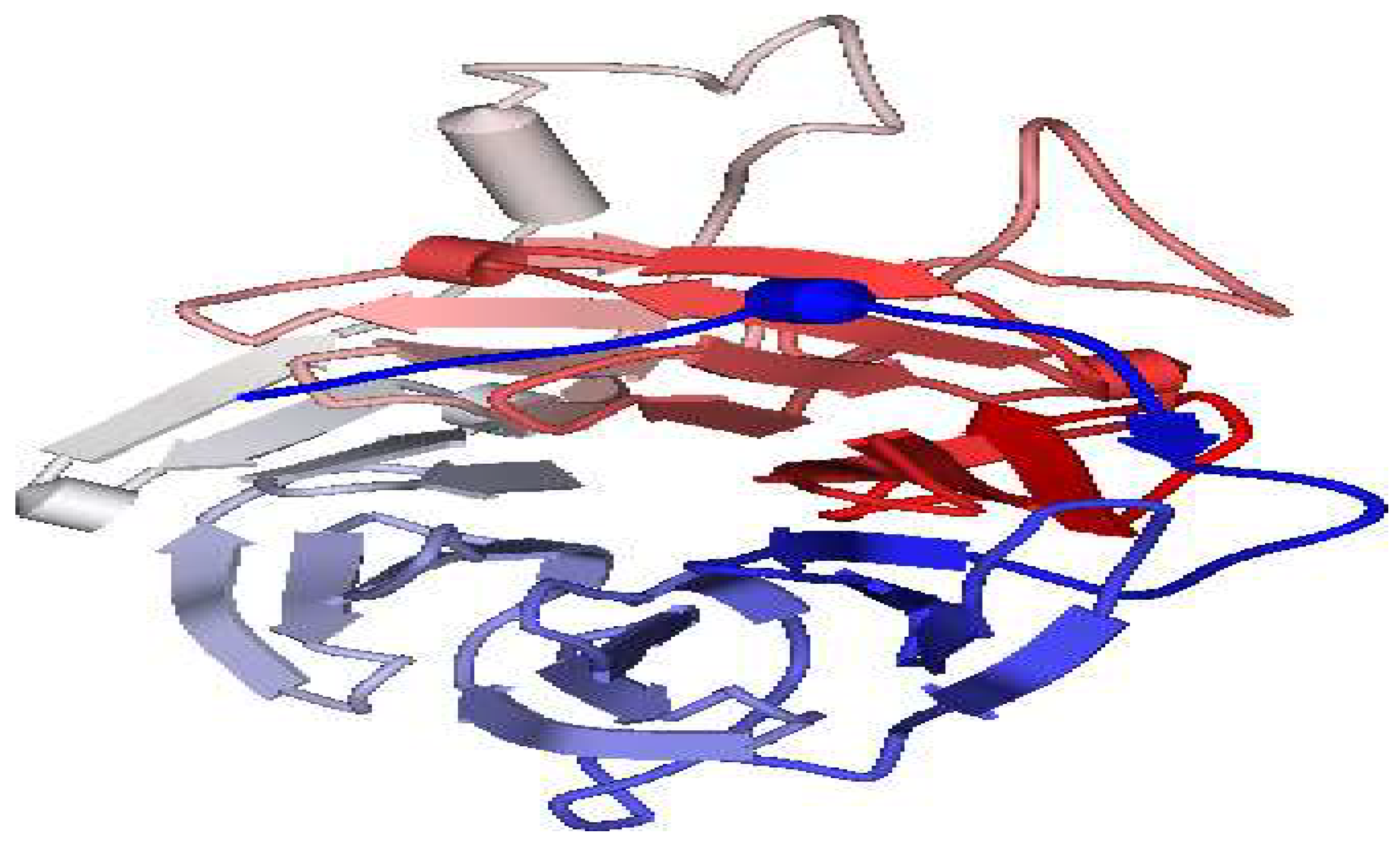
Model Validation
Identification of Active Site
Molecular Docking
Conclusion
Acknowledgement
Author contributions
Competing interest
References
- F.M Garibotto, A.D.Garro, M.F.Masman, A.M.Rodriguez, P.G.Luiten, M.M.Raimondi, S.A.Zacchino, C.Somlai, B.Penke, R.D.Enriz, New small-size peptides possessing antifungal activity. Bioorg. Med. Chem., 2010;18: 158–167.
- Narang R, Narasimhan B, Sharma S, Sriram D, Yogeeswari P, Clercq ED, Balzarini J, Nicotinic acid benzylidene hydrazides: Synthesis, antitubercular, antiviral, antimicrobial evaluation and QSAR studies. Med Chem Res, 2012; 21:1557-1576.
- Kankate, R.S. et al., Design, synthesis and antifungal evaluation of novel benzimidazole tertiary amine type of fluconazole analogues. Arabian Journal of Chemistry (2015). [CrossRef]
- Brown, G.D.; Denning, D.W.; Levitz, S.M. Tackling human fungal infections. Science 2012, 336. [Google Scholar] [CrossRef] [PubMed]
- D.Barrett, From natural products to clinically useful antifungals. Biochim. Biophys. Acta. 2002;1587: 224–233.
- Yapar, N. Epidemiology and risk factors for invasive candidiasis. Ther Clin Risk Manag 10,95–105 (2014).
- Salgado, Paula S., et al. "Structural basis for the broad specificity to host-cell ligands by the pathogenic fungus Candida albicans." Proceedings of the National Academy of Sciences 108.38 (2011): 15775-15779.
- Ge SH, Wan Z, Li J, Xu J, Li RY, Bai FY (2010) Correlation between azole susceptibilities, genotypes, and ERG11 mutations in Candida albicans isolates associated with vulvovaginal candidiasis in China. Antimicrob Agents Chemother 54(8):3126–3131.
- Lum, Kah Yean, et al. "Activity of novel synthetic peptides against Candida albicans." Scientific reports 5 (2015).
- Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2009–2010. Infect Control Hosp Epidemiol. 2013;34:1–14.
- Pfaller, MA. Antifungal drug resistance: mechanisms, epidemiology and consequences for treatment. Am J Med. 2012;125:3–13.
- Chin VK, Lee TY, Rusliza B, Chong PP. Dissecting Candida albicans Infection from the Perspective of C. albicans Virulence and Omics Approaches on Host–Pathogen Interaction: A Review. Woo PCY, ed. International Journal of Molecular Sciences. 2016;17(10):1643. [CrossRef]
- Xu, Kehan, et al. "Design, synthesis, and antifungal activities of novel triazole derivatives containing the benzyl group." Drug design, development and therapy 9 (2015): 1459.
- Granneman, Sander, et al. "Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs." Proceedings of the National Academy of Sciences 106.24 (2009): 9613-9618.
- Enjalbert B, Moran GP, Vaughan C, Yeomans T, MacCallum DM, et al. (2009) Genome-wide gene expression ƉƌŽĮůŝnŐ and a forward ŐĞnĞtic screen show that ĚŝīĞƌĞntiĂů expression of the sodium ion transporter Ena21 contributes to the ĚŝīĞƌĞntiĂů tolerance of Candida albicans and Candida dubliniensis to ŽƐmŽtic stress. Mol Microbiol 72: 216-228.
- Aoyama T, Nakayama H, Ueno K, Inukai T, Tanabe K, et al. (2014) Genome-wide survey of ƚƌĂnƐcƌŝƉtiŽnĂů ŝnŝtiĂtiŽn in the pathogenic fungus, Candida glabrata. Genes Cells 9: 478-503.
- Zhang, L., Lin, J., & Ye, K. (2013). Structural and functional analysis of the U3 snoRNA binding protein Rrp9 . RNA, 19(5), 701–711. [CrossRef]

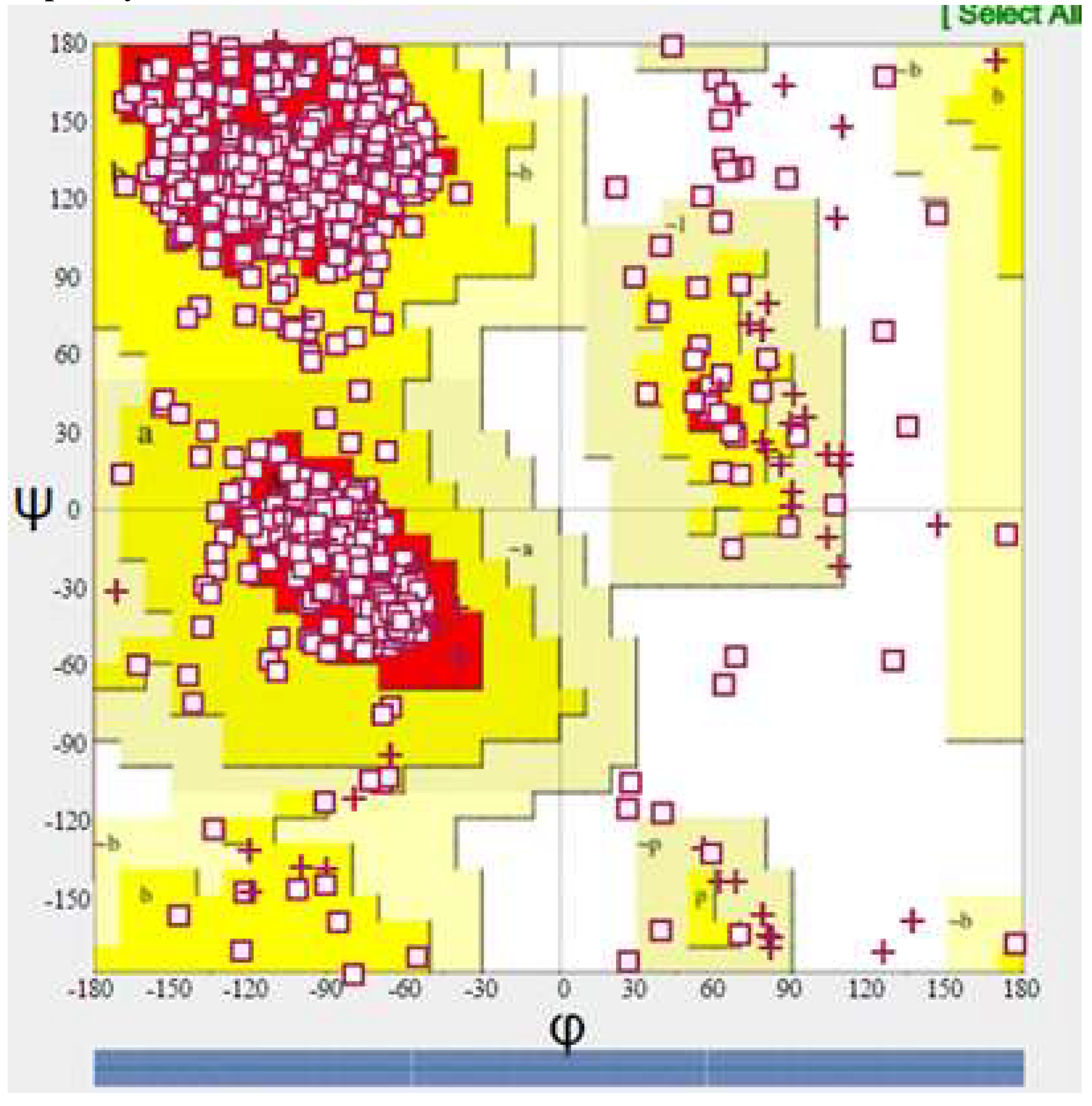
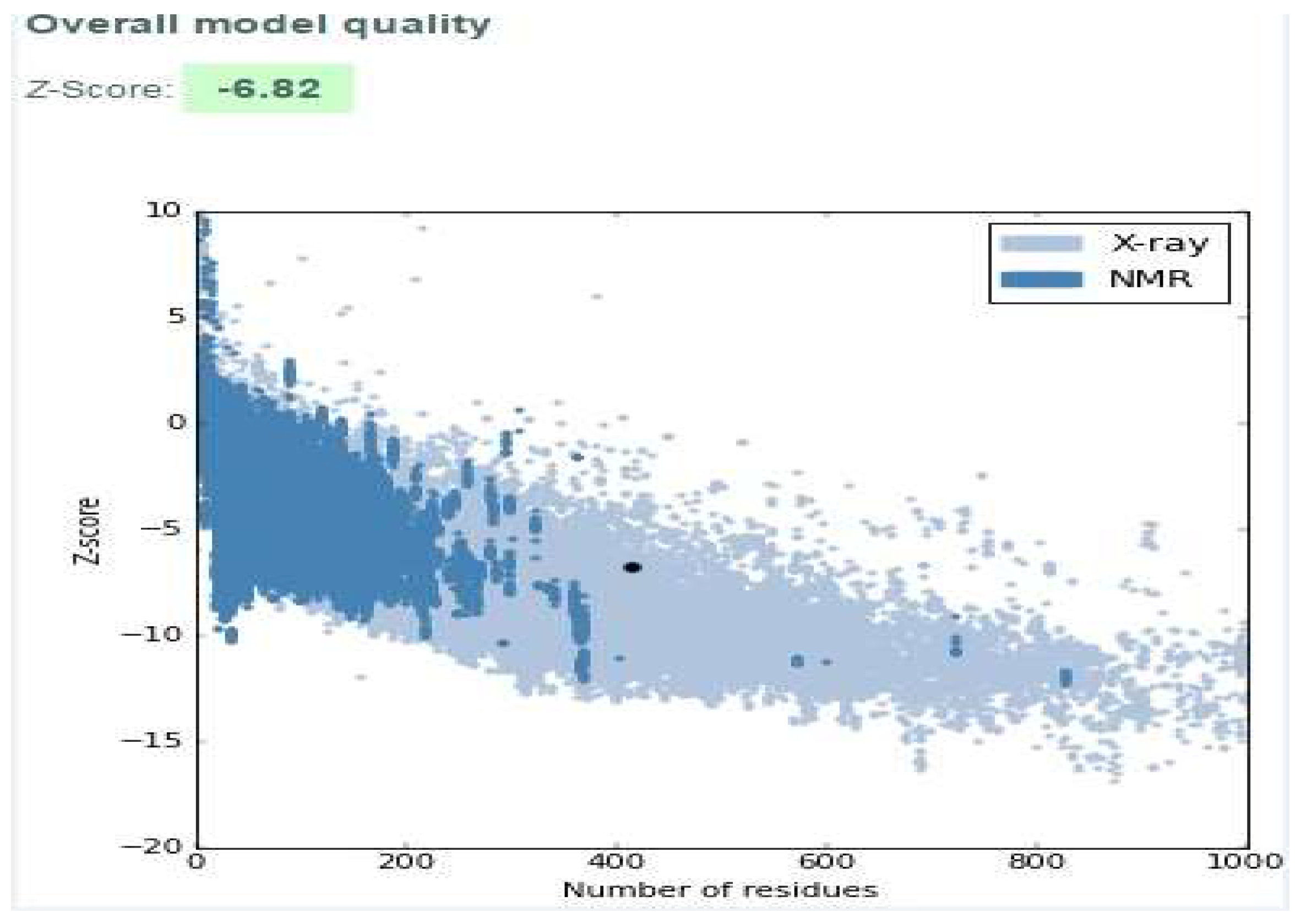

| Parameter | Value | |
| Amino Acid Length | 549 | |
| Molecular Weight (M.wt.) | 62414.05 | |
| PI | 6.59 | |
| Total number of negatively charged residues (Asp + Glu) | 78 | |
| Total number of positively charged residues (Arg + Lys) | 75 | |
| Instability index (II) | 44.74 | |
| Aliphatic index (AI) | 76.03 | |
| Half-life | Mammalian reticulocytes | 30 hours |
| Yeast | >20 hours | |
| E. coli | >10 hours | |
| GRAVY | -0.753 | |
| Parameters | Number of amino acids | Amino acids (%) |
| Alpha helix (Hh) | 148 | 26.96 |
| 310 helix (Gg) | 0 | 0.00 |
| Pi helix (Ii) | 0 | 0.00 |
| Beta bridge (Bb) | 0 | 0.00 |
| Extended strand (Ee) | 141 | 25.68 |
| Beta turn (Tt) | 46 | 7.47 |
| Bend region (Ss) | 0 | 0.00 |
| Random coil (Cc) | 219 | 39.89 |
| Ambigous states | 0 | 0.00 |
| Other states | 0 | 0.00 |
| Name | Fitness score | S(hb_ext) | S(Vdw_ext) | S(hb_int) | S(Int) |
| Dicyclomine hydrochloride | 42.14 | 0.00 | 37.83 | 0.00 | -9.88 |
| Hydroxyine hydrochloride | 54.55 | 0.00 | 44.39 | 0.00 | -6.49 |
| Promethazine Hydrochloride |
45.91 | 0.00 | 36.84 | 0.00 | -4.75 |
| Solifenacin succinate | 62.17 | 0.00 | 44.26 | 0.00 | 1.32 |
| Flavoxate Hydrochloride | 57.67 | 0.00 | 48.75 | 0.00 | -9.36 |
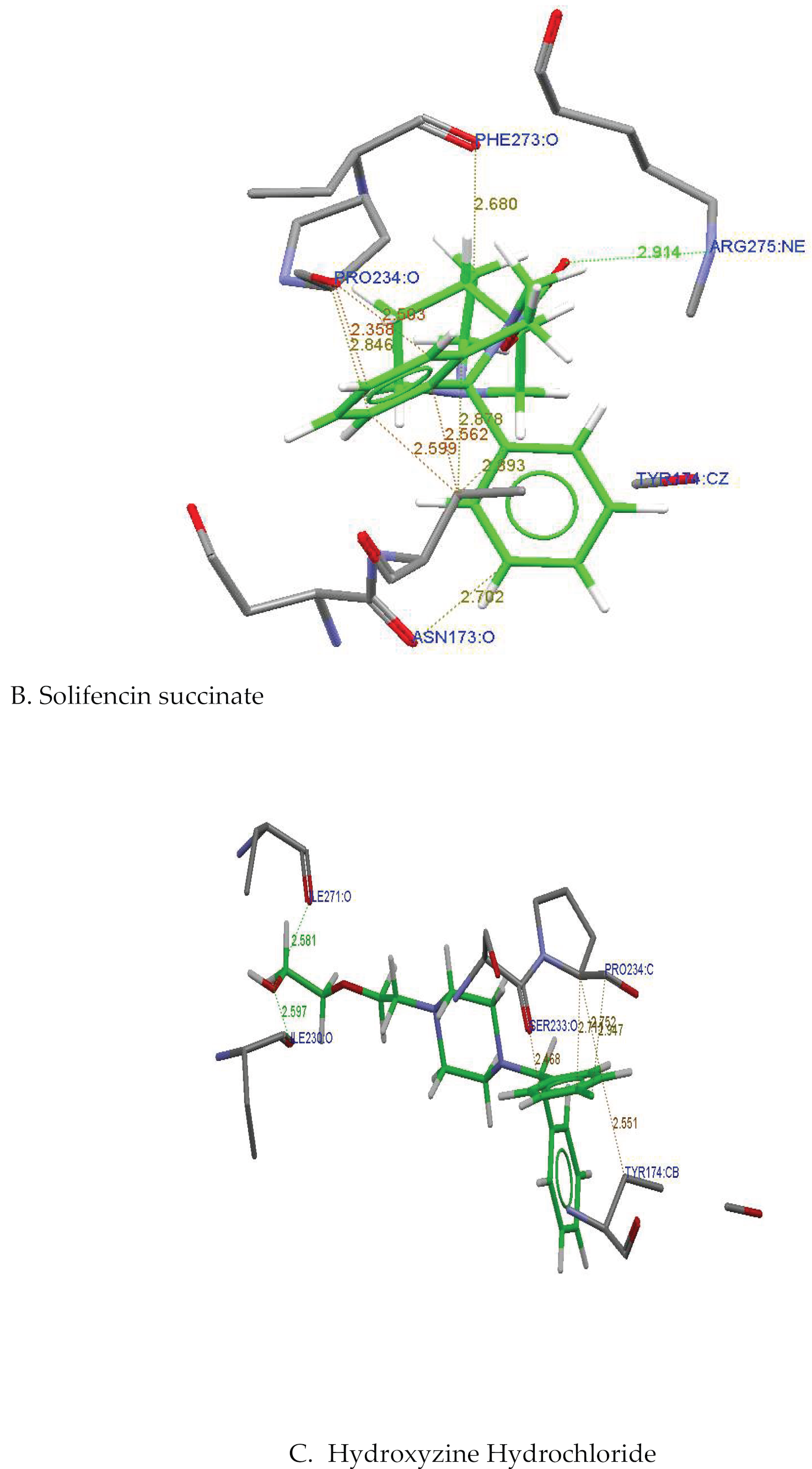
| Ligand name | Interacting aminoacids | Interacting atoms | H-distance |
| Solifenacin Succinate | Pro234,Arg275, tyr174,Asn173, Phe273 |
Arg275:NE----O2: Solifenacin Succinate Phe273:O---C8: Solifenacin Succinate Pro234:CA---C24: Solifenacin Succinate Asn173:O---C17: Solifenacin Succinate |
2.914 2.503 2.356 2.702 |
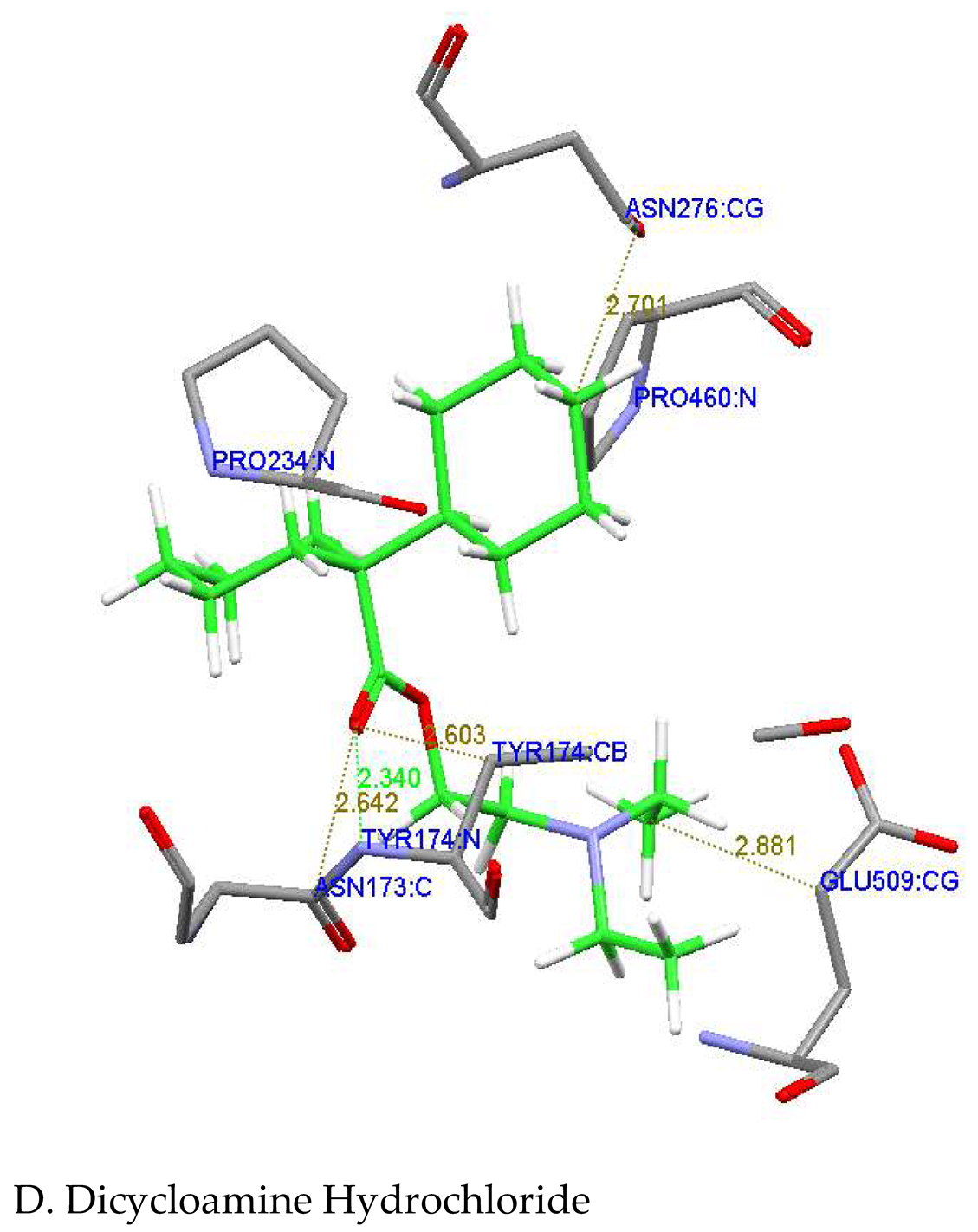
| Dicycloamine |
Glu509,Asn173, Tyr174,Asn276, Pro460,Pro234 |
Tyr174:N---O2:Dicycloamine TYR174:CB---C8: Dicycloamine Asn173:C..O2: Dicycloamine Glu509:CG...C21: Dicycloamin |
2.340 2.603 2.642 2.881 |
| Hydroxyzine hydrochloride |
ILE271,ILE230, Pro234,Ser232, Tyr174,Asp506 Ser233 |
Ile271:O---OH: Hdroxygene Hydrochloride Ile230:O—OH:Hydroxygene Hydrochloride Ser233:O—C21: Hydroxygene Hydrochloride Pro234:C---C17: Hydroxygene Hydrochloride |
2.581 2.597 2.468 2.752 |
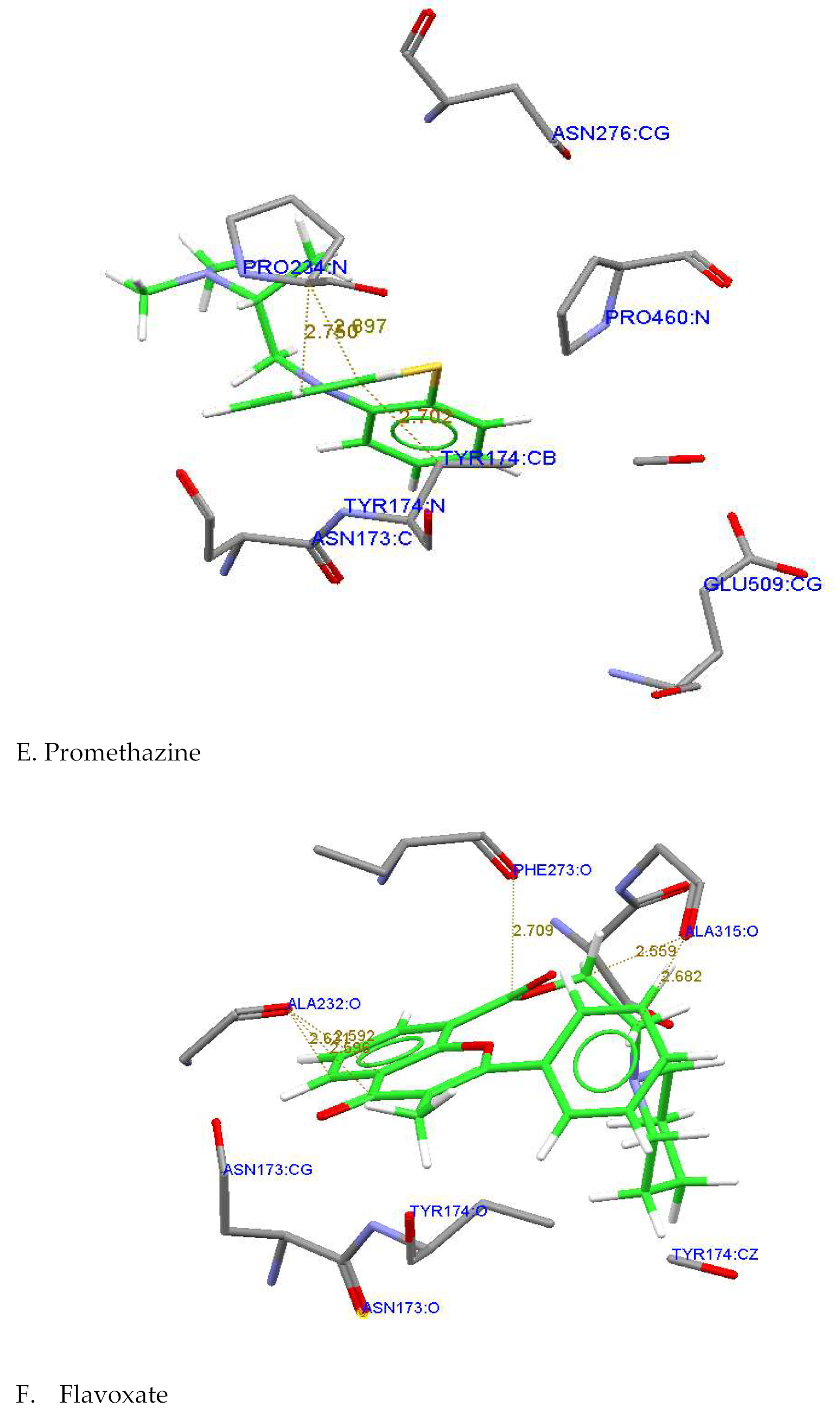
| Promethazine Hydrochloride |
Pro234,Phe273 Ser233,Ala232 Ty174,Arg275 |
Tyr174:CB---C13:Promethazine Hydrochloride Pro:234:N....C13 : Promethazine Hydrochloride |
2.730 2.897 |
| Flavoxate |
Pro234,Phe273 Ser233,Ala232 Ty174,Arg275 |
Phe273:O---O3:Flavoxate Ala232:O---C21: Flavoxate Ala315:O---C11: Flavoxate Ala232;O---O4: Flavoxate |
2.709 2.621 2.559 2.596 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




