Submitted:
02 June 2025
Posted:
02 June 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Cu Essentiality, Deficiency, and Toxicity
3. Cu Transport: Extracellular to Intracellular Space
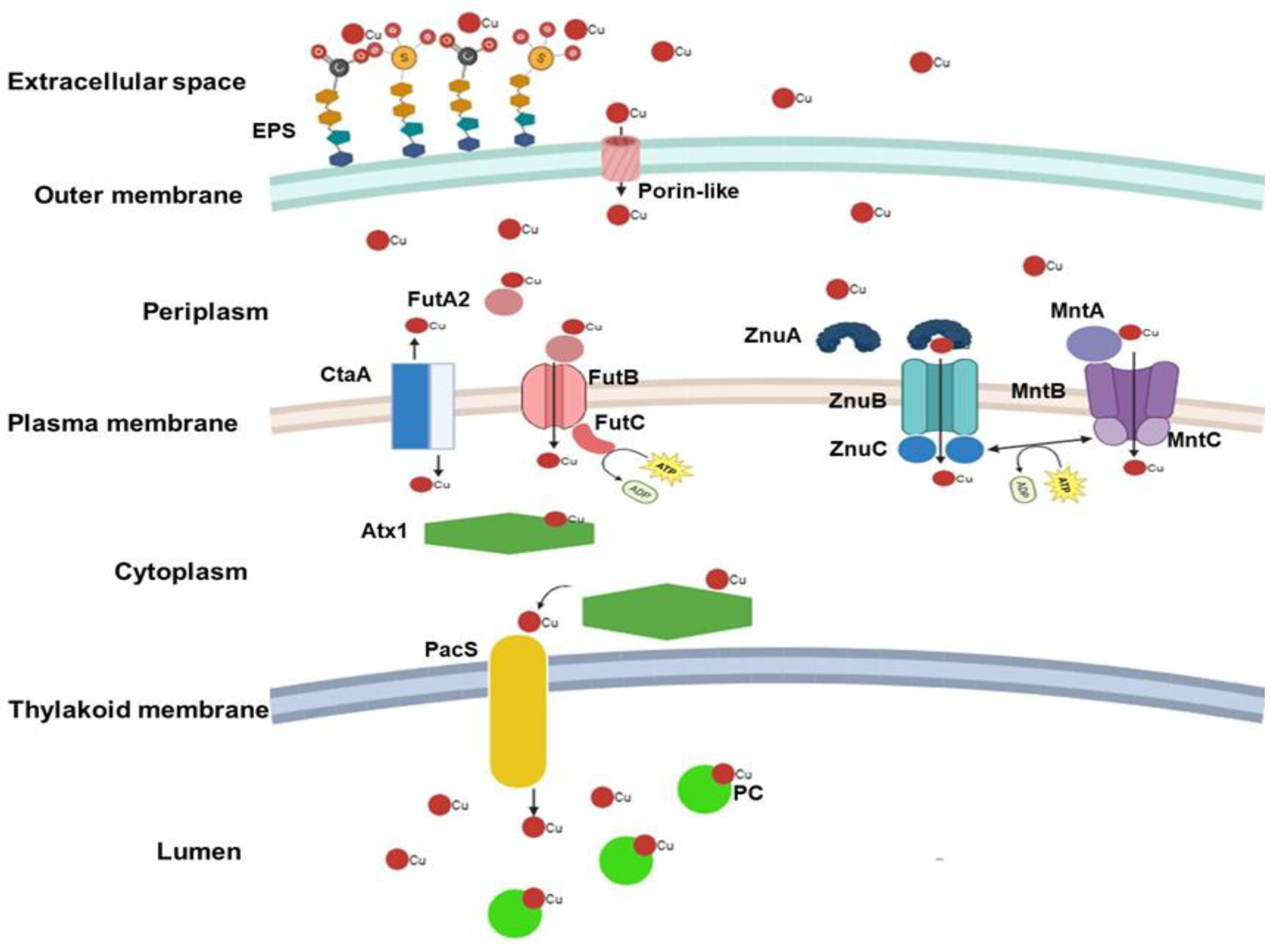
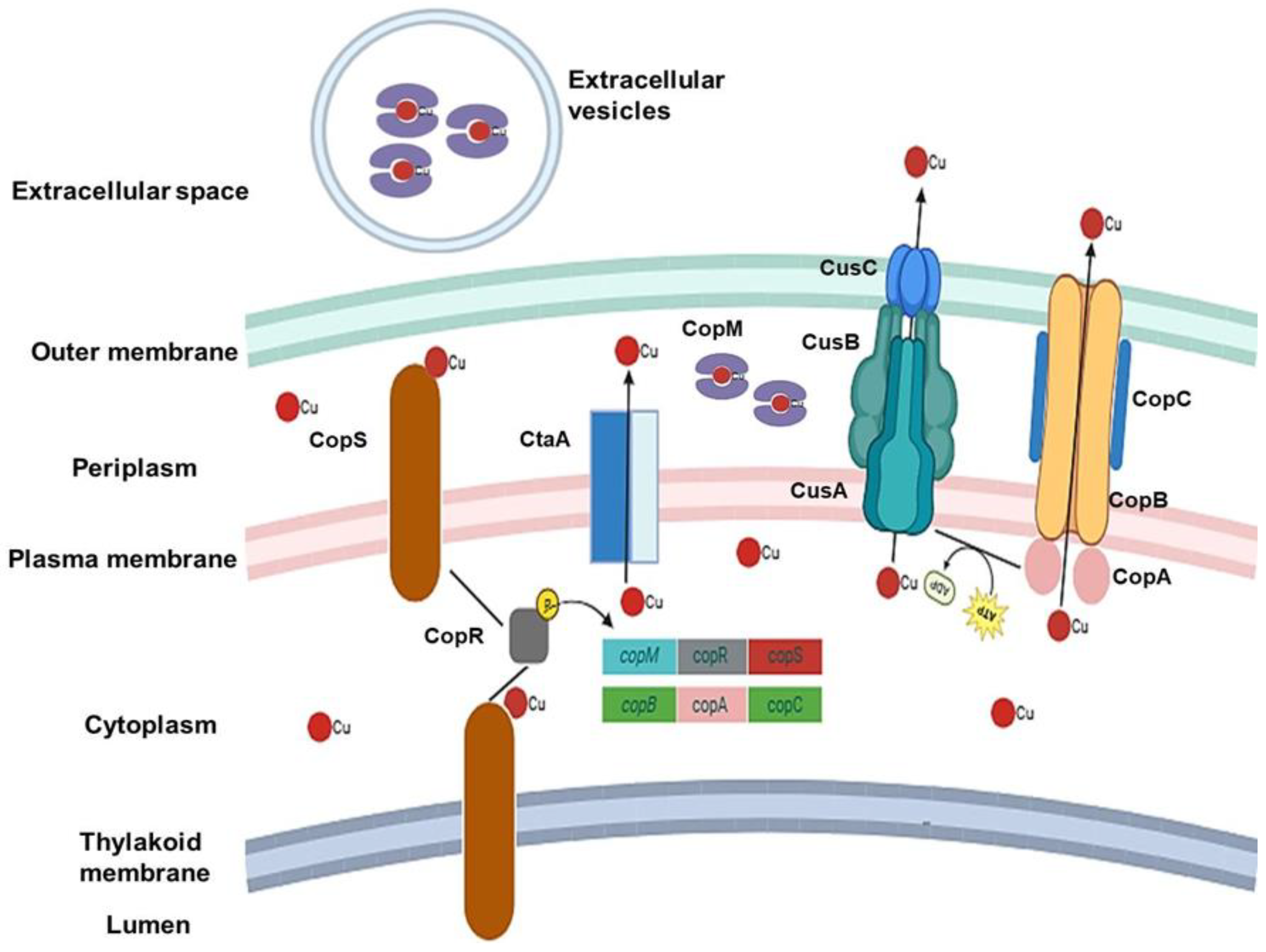
4. Response to Excess Cu
5. Exopolysaccharides (EPS) Interactions
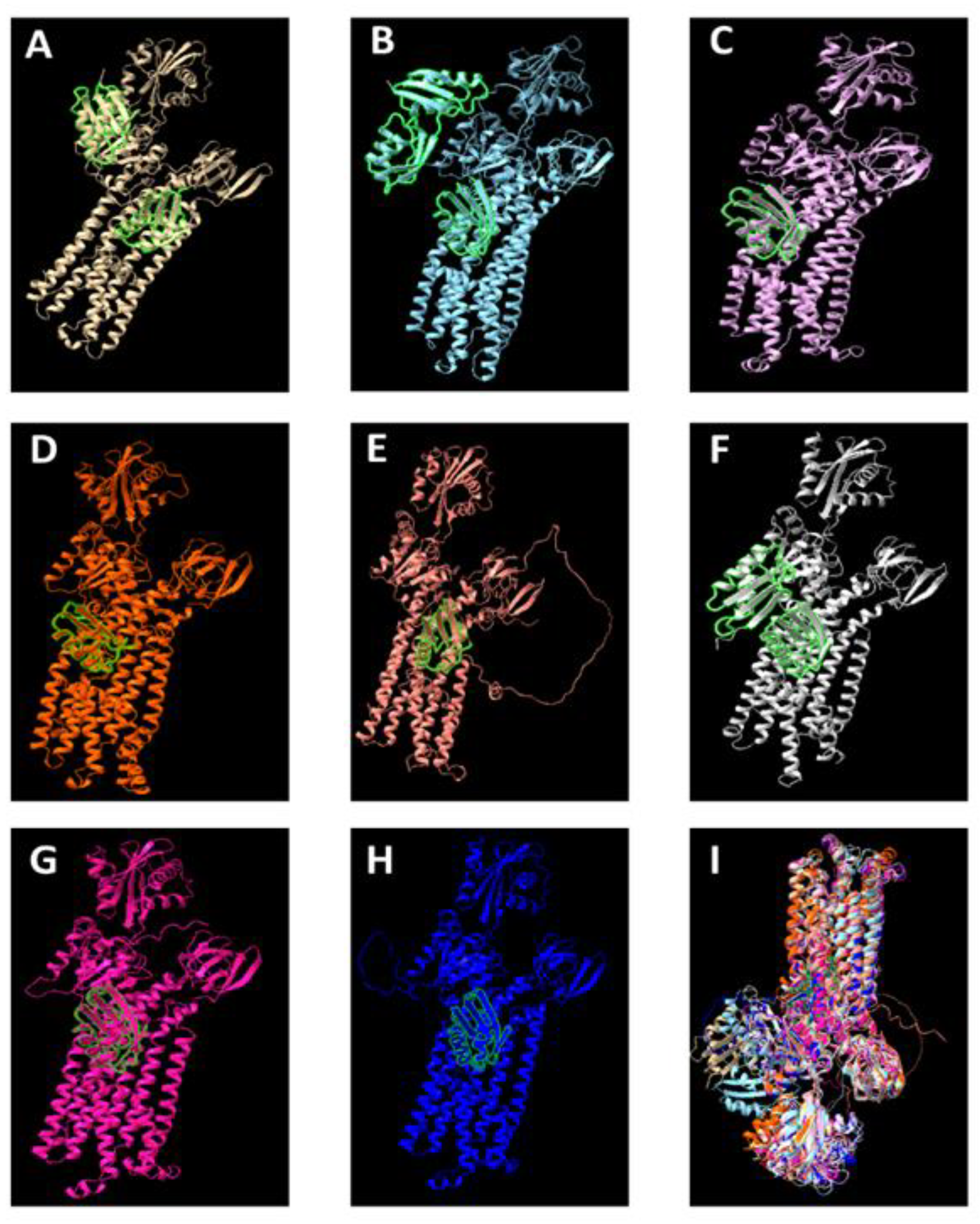
6. Phylogeny and Structure of the CopA Protein

7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Cu | copper |
| EPS | exopolysaccharides |
| Ni | nickel |
| Cd | cadmium |
| Pb | lead |
| Hg | mercury |
| Zn | zinc |
| Cr | chromium |
| Mn | manganese |
| PC | plastocyanin |
| SOD | superoxide dismutase |
| Fe-S | iron-sulfur |
| PCC | Pasteur Culture Collection |
| Cyt | c6 cytochrome c6 |
| PSI | photosystem I |
| ROS | reactive oxygen species |
| LC50 | lethal concentration 50 |
| SLH | S-layer Homologous |
| PSII | photosystem II |
| RND | Nodulation-Cell Division |
| HK | histidine kinase |
| CPS | capsular exopolysaccharides |
| RPS | released exopolysaccharides |
| NCBI | National Center for Biotechnology Information |
References
- Sinha, R.P.; Häder, D.-P. UV-protectants in cyanobacteria. Plant Sci. 2008, 174, 278–289. [Google Scholar] [CrossRef]
- De Los Ríos, A.; et al. Ultrastructural and genetic characteristics of endolithic cyanobacterial biofilms colonizing Antarctic granite rocks. FEMS Microbiol. Ecol. 2007, 59, 386–395. [Google Scholar] [CrossRef]
- Chisti, Y. Biodiesel from microalgae. Biotechnol. Adv. 2007, 25, 294–306. [Google Scholar] [CrossRef] [PubMed]
- Chittora, D.; et al. Cyanobacteria as a source of biofertilizers for sustainable agriculture. Biochem. Biophys. Rep. 2020, 22, 100737. [Google Scholar] [CrossRef] [PubMed]
- Żymańczyk-Duda, E.; et al. Versatile applications of cyanobacteria in biotechnology. Microorganisms 2022, 10, 2318. [Google Scholar] [CrossRef]
- Zhang, H.; et al. Characteristics of γ-hexachlorocyclohexane biodegradation by a nitrogen-fixing cyanobacterium, Anabaena azotica. J. Appl. Phycol. 2012, 24, 221–225. [Google Scholar] [CrossRef]
- Cepoi, L.; et al. Removal of organic pollutants from wastewater by cyanobacteria. In: Cyanobacteria for Bioremediation of Wastewaters 2016, 27–43.
- Parikh, A.; Madamwar, D. Textile dye decolorization using cyanobacteria. Biotechnol. Lett. 2005, 27, 323–326. [Google Scholar] [CrossRef]
- Tiwari, A.K.; Tiwari, B.S. Cyanotherapeutics: An emerging field for future drug discovery. Appl. Phycol. 2020, 1, 44–57. [Google Scholar] [CrossRef]
- Kalita, N.; Baruah, P.P. Cyanobacteria as a potent platform for heavy metals biosorption: Uptake, responses and removal mechanisms. J. Hazard. Mater. Adv. 2023, 100349. [Google Scholar] [CrossRef]
- Huertas, M.J.; et al. Metals in cyanobacteria: analysis of the copper, nickel, cobalt and arsenic homeostasis mechanisms. Life 2014, 4, 865–886. [Google Scholar] [CrossRef]
- Chevalier, P.; et al. Nitrogen and phosphorus removal by high latitude mat-forming cyanobacteria for potential use in tertiary wastewater treatment. J. Appl. Phycol. 2000, 12, 105–112. [Google Scholar] [CrossRef]
- Lynch, F.; et al. Screening native isolates of cyanobacteria and a green alga for integrated wastewater treatment, biomass accumulation and neutral lipid production. Algal Res. 2015, 11, 411–420. [Google Scholar] [CrossRef]
- Kabariya, J.H.; Ramani, V.M. Dairy wastewater treatment by cyanobacteria for removal of nutrients with extraction of high value compounds from biomass. Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 1527–1538. [Google Scholar] [CrossRef]
- Mukherjee, C.; et al. Parboiled rice effluent: A wastewater niche for microalgae and cyanobacteria with growth coupled to comprehensive remediation and phosphorus biofertilization. Algal Res. 2016, 19, 225–236. [Google Scholar] [CrossRef]
- Del Valle-Real, M.; et al. Wastewater from maize lime-cooking as growth media for alkaliphilic microalgae–cyanobacteria consortium to reduce chemical oxygen demand and produce biomass with high protein content. Int. J. Food Sci. Technol. 2023, 58, 6775–6783. [Google Scholar] [CrossRef]
- Nagasathya, A.; Thajuddin, N. Decolourization of paper mill effluent using hypersaline cyanobacterium. 2008.
- Blaby-Haas, C.E. Cyanobacteria provide a new paradigm in the regulation of cofactor dependence. Proceedings of the National Academy of Sciences. 2021, 118, e2100281118. [Google Scholar] [CrossRef]
- Kong, L. Copper requirement and acquisition by marine microalgae. Microorganisms. 2022, 10, 1853. [Google Scholar] [CrossRef]
- Boden, J.S.; et al. Timing the evolution of antioxidant enzymes in cyanobacteria. Nature Communications. 2021, 12, 4742. [Google Scholar] [CrossRef] [PubMed]
- Kalita, N.; Baruah, P.P. Copper removal efficacy and stress tolerance potential of Leptolyngbya sp. GUEco1015. Heliyon. 2024, 10, 8. [Google Scholar] [CrossRef]
- National Research Council et al. Copper in drinking water. 2000.
- Castielli, O.; et al. Proteomic analyses of the response of cyanobacteria to different stress conditions. FEBS letters. 2009, 583, 1753–1758. [Google Scholar] [CrossRef]
- García-Cañas, R.; et al. A protease-mediated mechanism regulates the cytochrome c 6/plastocyanin switch in Synechocystis sp. PCC 6803. Proceedings of the National Academy of Sciences. 2021, 118, e2017898118. [Google Scholar] [CrossRef]
- Bellemare, D.R.; et al. A novel copper-regulated promoter system for expression of heterologous proteins in Schizosaccharomyces pombe. Gene. 2001, 273, 191–198. [Google Scholar] [CrossRef]
- Yamamoto, K.; Ishihama, A. Characterization of copper-inducible promoters regulated by CpxA/CpxR in Escherichia coli. Bioscience, biotechnology, and biochemistry. 2006, 70, 1688–1695. [Google Scholar] [CrossRef] [PubMed]
- Rippka, R.; et al. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology. 1979, 111, 1–61. [Google Scholar] [CrossRef]
- Waterbury, J.B.; Stanier, R.Y. Isolation and growth of cyanobacteria from marine and hypersaline environments. The prokaryotes: a handbook on habitats, isolation, and identification of bacteria. Berlin, Heidelberg: Springer Berlin Heidelberg. 1981, 221-223.
- Kotai, J. Instruction of preparations of modified nutrient medium Z8 for algae. Norw. Inst. Water Res. Blind. Oslo. 1972, 32–39. [Google Scholar]
- Zarrouk, C. Contribution a l'etude d'une Cyanophycee. Influence de Divers Facteurs Physiques et Chimiques sur la croissance et la photosynthese de Spirulina mixima. Thesis. University of Paris, France. 1966.
- Nongrum, N.A.; Syiem, M.B. Effects of copper ion (Cu2+) on the physiological and biochemical activities of the cyanobacterium Nostoc ANTH. Environmental Engineering Research. 2012, 17, 63–67. [Google Scholar] [CrossRef]
- Ahad, R.I.A.; Syiem, M.B. Copper and cadmium-induced toxicity on the cyanobacterium Nostoc muscorum Meg 1: a comparative study. EurAsian Journal of BioSciences. 2018, 12, 333–345. [Google Scholar]
- Wu, Z.; et al. Response of Microcystis to copper stress–do phenotypes of Microcystis make a difference in stress tolerance? Environmental Pollution. 2007, 147, 324–330. [Google Scholar] [CrossRef]
- Wang, H.; Ebenezer, V.; Ki, J. Photosynthetic and biochemical responses of the freshwater green algae Closterium ehrenbergii Meneghini (Conjugatophyceae) exposed to the metal coppers and its implication for toxicity testing. Journal of Microbiology. 2018, 56, 426–434. [Google Scholar] [CrossRef]
- Mohy El Din, S. Effect of copper and lead on growth and some metabolic activities of cyanobacterium Spirulina platensis (Nordstedt). Egyptian Journal of Botany. 2017, 57, 445–456. [Google Scholar] [CrossRef]
- Giner-Lamia, J.; López-Maury, L.; Florencio, F.J. Global transcriptional profiles of the copper responses in the cyanobacterium Synechocystis sp. PCC 6803. PLoS One. 2014, 9, e108912. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, R.; Kenney, G.E.; Rosenzweig, A.C. Dual pathways for copper uptake by methanotrophic bacteria. Journal of biological chemistry. 2011, 286, 37313–37319. [Google Scholar] [CrossRef] [PubMed]
- Andrei, A.; et al. Cu homeostasis in bacteria: the ins and outs. Membranes. 2020, 10, 9–242. [Google Scholar] [CrossRef]
- Kowata, H.; et al. Outer membrane permeability of cyanobacterium Synechocystis sp. strain PCC 6803: studies of passive diffusion of small organic nutrients reveal the absence of classical porins and intrinsically low permeability. Journal of bacteriology. 2017, 199. [Google Scholar] [CrossRef]
- Kojima, S.; Muramoto, K.; Kusano, T. Outer membrane proteins derived from non-cyanobacterial lineage cover the peptidoglycan of Cyanophora paradoxa cyanelles and serve as a cyanelle diffusion channel. Journal of Biological Chemistry. 2016, 291, 20198–20209. [Google Scholar] [CrossRef]
- Wylie, J.L.; Worobec, E.A. The OprB porin plays a central role in carbohydrate uptake in Pseudomonas aeruginosa. Journal of bacteriology. 1995, 177, 3021–3026. [Google Scholar] [CrossRef] [PubMed]
- Schätzle, H.; et al. Comparative phenotypic analysis of Anabaena sp. PCC 7120 mutants of porinlike genes. Journal of Microbiology and Biotechnology. 2021, 31, 645. [Google Scholar] [CrossRef]
- Cardoso, D.; et al. The role of outer membrane protein (s) harboring SLH/OprB-domains in extracellular vesicles’ production in Synechocystis sp. PCC 6803. Plants. 2021, 10, 2757. [Google Scholar] [CrossRef]
- Coines, J.; et al. Glucose transport via the pseudomonad porin OprB: Implications for the design of Trojan Horse anti-infectives. Physical Chemistry Chemical Physics. 2019, 21, 8457–8463. [Google Scholar] [CrossRef]
- Pederick, V.G.; et al. ZnuA and zinc homeostasis in Pseudomonas aeruginosa. Scientific reports. 2015, 5, 13139. [Google Scholar] [CrossRef]
- Cavet, J.S.; Borrelly, G.P.M.; Robinson, N.J. Zn, Cu and Co in cyanobacteria: selective control of metal availability. FEMS Microbiology Reviews. 2003, 27, 165–181. [Google Scholar] [CrossRef]
- Banerjee, S.; et al. Structural determinants of metal specificity in the zinc transport protein ZnuA from Synechocystis 6803. Journal of molecular biology. 2003, 333, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Yatsunyk, L.A.; et al. Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli. JBIC Journal of Biological Inorganic Chemistry. 2008, 13, 271–288. [Google Scholar] [CrossRef] [PubMed]
- Patzer, S.I.; Hantke, K. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Molecular microbiology. 1998, 28, 1199–1210. [Google Scholar] [CrossRef] [PubMed]
- Hudek, L.; et al. Molecular and cellular characterisation of the zinc uptake (Znu) system of Nostoc punctiforme. FEMS microbiology ecology. 2013, 86, 149–171. [Google Scholar] [CrossRef]
- Bartsevich, V.V.; Pakrasi, H.B. Manganese transport in the cyanobacterium Synechocystis sp. PCC 6803. Journal of Biological Chemistry. 1996, 271, 26057–26061. [Google Scholar] [CrossRef]
- Al-tameemi, H.; et al. Staphylococcus aureus lacking a functional MntABC manganese import system has increased resistance to copper. Molecular microbiology. 2021, 115, 554–573. [Google Scholar] [CrossRef]
- Lim, K. HL.; et al. Metal binding specificity of the MntABC permease of Neisseria gonorrhoeae and its influence on bacterial growth and interaction with cervical epithelial cells. Infection and immunity. 2008, 76, 3569–3576. [Google Scholar] [CrossRef]
- Giner-Lamia, J.; et al. The CopRS two-component system is responsible for resistance to copper in the cyanobacterium Synechocystis sp. PCC 6803. Plant physiology. 2012, 159, 1806–1818. [Google Scholar] [CrossRef]
- Rosenzweig, A.C.; Argüello, J.M. Toward a molecular understanding of metal transport by P1B-Type ATPases. In: Current topics in membranes. Academic Press. 2012, 113-136.
- Williams, L.E.; Mills, R.F. P1B-ATPases–an ancient family of transition metal pumps with diverse functions in plants. Trends in plant science. 2005, 10, 491–502. [Google Scholar] [CrossRef]
- Salustros, Nina et al. Structural basis of ion uptake in copper-transporting P1B-type ATPases. Nature Communications. 2022, 13, 5121.
- Hederstedt, L.; Lewin, A.; Throne-Holst, M. . Heme A synthase enzyme functions dissected by mutagenesis of Bacillus subtilis CtaA. Journal of bacteriology. 2005, 187, 8361–8369. [Google Scholar] [CrossRef] [PubMed]
- Raimunda, D.; González-Guerrero, M.; Leeber, B., III; Argüello, J. The transport mechanism of bacterial Cu+-ATPases: Distinct efflux rates adapted to different function. BioMetals. 2011, 24, 467–475. [Google Scholar] [CrossRef]
- Phung, L.T.; Ajlani, G.; Haselkorn, R. P-type ATPase from the cyanobacterium Synechococcus 7942 related to the human Menkes and Wilson disease gene products. Proceedings of the National Academy of Sciences. 1994, 91, 9651–9654. [Google Scholar] [CrossRef]
- Yuan, D.S.; Dancis, A.; Klausner, R.D. Restriction of copper export in Saccharomyces cerevisiae to a late Golgi or post-Golgi compartment in the secretory pathway. Journal of Biological Chemistry. 1997, 272, 25787–25793. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Z.-H.; et al. Reciprocal effect of copper and iron regulation on the proteome of Synechocystis sp. PCC 6803. Frontiers in Bioengineering and Biotechnology. 2021, 9, 673402. [Google Scholar] [CrossRef]
- Katoh, H.; Hagino, N.; Ogawa, T. Iron-binding activity of FutA1 subunit of an ABC-type iron transporter in the cyanobacterium Synechocystis sp. strain PCC 6803. Plant and Cell Physiology. 2001, 42, 823–827. [Google Scholar] [CrossRef] [PubMed]
- Waldron, K.J.; et al. A periplasmic iron-binding protein contributes toward inward copper supply. Journal of Biological Chemistry. 2007, 282, 3837–3846. [Google Scholar] [CrossRef]
- Stevanovic, M.; et al. The components of the putative iron transport system in the cyanobacterium Anabaena sp. PCC 7120. Environmental Microbiology. 2012, 14, 1655–1670. [Google Scholar] [CrossRef] [PubMed]
- Aliaga, M.E.; et al. Redox-implications associated with the formation of complexes between copper ions and reduced or oxidized glutathione. Journal of Inorganic Biochemistry. 2016, 154, 78–88. [Google Scholar] [CrossRef]
- Rakshit, A.; et al. Cu²⁺ selective chelators relieve copper-induced oxidative stress in vivo. Chemical Science. 2018, 9, 7916–7930. [Google Scholar] [CrossRef]
- Markossian, K.A.; Kurganov, B.I. Copper chaperones, intracellular copper trafficking proteins. Function, structure, and mechanism of action. Biochemistry (Moscow). 2003, 68, 827–837. [Google Scholar] [CrossRef]
- Arnesano, F.; et al. Solution structure of the Cu(I) and apo forms of the yeast metallochaperone, Atx1. Biochemistry. 2001, 40, 1528–1539. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; et al. Transition metal transporting P-type ATPases: terminal metal-binding domains serve as sensors for autoinhibitory tails. The FEBS Journal. 2024. [Google Scholar] [CrossRef] [PubMed]
- Gounder, P.E.; et al. Metallothioneins: Diverse Protein Family to Bind Metallic Ions. Chapters. 2021. [Google Scholar]
- Bourdieunaud, J.-P.; et al. Challenging the model for induction of metallothionein gene expression. Biochimie. 2006, 88, 1787–1792. [Google Scholar] [CrossRef] [PubMed]
- Dziegiel, P.; et al. Metallothioneins: structure and functions. Metallothioneins in Normal and Cancer Cells. 2016, 3, 20. [Google Scholar]
- Robinson, N.J.; Wilson, J.R.; Turner, J.S. Expression of the type 2 metallothionein-like gene MT2 from Arabidopsis thaliana in Zn²⁺-metallothionein-deficient Synechococcus PCC 7942: putative role for MT2 in Zn²⁺ metabolism. Plant Molecular Biology. 1996, 30, 1169–1179. [Google Scholar] [CrossRef]
- Verma, S.K.; Singh, S.P. Factors regulating copper uptake in a cyanobacterium. Current Microbiology. 1990, 21, 33–37. [Google Scholar] [CrossRef]
- Munson, G.P.; et al. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. Journal of Bacteriology. 2000, 182, 5864–5871. [Google Scholar] [CrossRef]
- Rouch, D.A.; Brown, N.L. Copper-inducible transcriptional regulation at two promoters in the Escherichia coli copper resistance determinant pco. Microbiology. 1997, 143, 1191–1202. [Google Scholar] [CrossRef]
- Giner-Lamia, J.; et al. Extracellular proteins: novel key components of metal resistance in cyanobacteria? Frontiers in Microbiology. 2016, 7, 878. [Google Scholar] [CrossRef] [PubMed]
- Gittins, J.R. Cloning of a copper resistance gene cluster from the cyanobacterium Synechocystis sp. PCC 6803 by recombineering recovery. FEBS Letters. 2015, 589, 1872–1878. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; et al. Structural basis for copper/silver binding by the Synechocystis metallochaperone CopM. Biological Crystallography. 2016, 72, 997–1005. [Google Scholar]
- Yang, J.; et al. Structural basis of copper binding by a dimeric periplasmic protein forming a six-helical bundle. Journal of Inorganic Biochemistry. 2022, 229, 111728. [Google Scholar] [CrossRef] [PubMed]
- Lima, S.; et al. Extracellular vesicles as an alternative copper-secretion mechanism in bacteria. Journal of Hazardous Materials. 2022, 431, 128594. [Google Scholar] [CrossRef]
- Lawton, T.J.; et al. The CopC family: structural and bioinformatic insights into a diverse group of periplasmic copper binding proteins. Biochemistry. 2016, 55, 2278–2290. [Google Scholar] [CrossRef]
- Gautam, P.; Erill, I.; Cusick, K.D. Linking Copper-Associated Signal Transduction Systems with Their Environment in Marine Bacteria. Microorganisms. 2023, 11, 1012. [Google Scholar] [CrossRef]
- Rensing, C.; et al. CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proceedings of the National Academy of Sciences. 2000, 97, 652–656. [Google Scholar] [CrossRef]
- Gómez-Santos, N.; et al. CorE from Myxococcus xanthus is a copper-dependent RNA polymerase sigma factor. PLoS Genetics. 2011, 7, e1002106. [Google Scholar] [CrossRef]
- Cha, J.-S.; Cooksey, D.A. Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Proceedings of the National Academy of Sciences. 1991, 88, 8915–8919. [Google Scholar] [CrossRef]
- Cha, J.-S.; Cooksey, D.A. Copper hypersensitivity and uptake in Pseudomonas syringae containing cloned components of the copper resistance operon. Applied and Environmental Microbiology. 1993, 59, 1671–1674. [Google Scholar] [CrossRef] [PubMed]
- Olivan-Muro, I.; et al. Unbalancing Zur (FurB)-mediated homeostasis in Anabaena sp. PCC7120: Consequences on metal trafficking, heterocyst development and biofilm formation. Environmental Microbiology. 2023, 25, 2142–2162. [Google Scholar] [CrossRef] [PubMed]
- Behlau, F.; et al. Molecular characterization of copper resistance genes from Xanthomonas citri subsp. citri and Xanthomonas alfalfae subsp. citrumelonis. Applied and Environmental Microbiology. 2011, 77, 4089–4096. [Google Scholar] [PubMed]
- Mills, S.D.; Jasalavich, C.A.; Cooksey, D.A. A two-component regulatory system required for copper-inducible expression of the copper resistance operon of Pseudomonas syringae. Journal of Bacteriology. 1993, 175, 1656–1664. [Google Scholar] [CrossRef]
- Franke, S.; et al. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. Journal of Bacteriology. 2003, 185, 3804–3812. [Google Scholar] [CrossRef]
- Su, C.-C.; et al. Crystal structure of the CusBA heavy-metal efflux complex of Escherichia coli. Nature. 2011, 470, 558–562. [Google Scholar] [CrossRef]
- Hahn, A.; et al. The outer membrane TolC-like channel HgdD is part of tripartite resistance-nodulation-cell division (RND) efflux systems conferring multiple-drug resistance in the cyanobacterium Anabaena sp. PCC7120. Journal of Biological Chemistry. 2013, 288, 31192–31205. [Google Scholar] [CrossRef]
- Nicolaisen, K.; et al. The interplay between siderophore secretion and coupled iron and copper transport in the heterocyst-forming cyanobacterium Anabaena sp. PCC 7120. Biochimica et Biophysica Acta (BBA)-Biomembranes. 2010, 1798, 2131–2140. [Google Scholar] [CrossRef]
- Cruz, D.; et al. Exopolysaccharides from cyanobacteria: strategies for bioprocess development. Applied Sciences. 2020, 10, 3763. [Google Scholar] [CrossRef]
- Tiwari, O.N.; et al. Biosynthesis, purification and structure-property relationships of new cyanobacterial exopolysaccharides. Polymer Testing. 2020, 89, 106592. [Google Scholar] [CrossRef]
- De Philippis, R.; Vincenzini, M. Exocellular polysaccharides from cyanobacteria and their possible applications. FEMS Microbiology Reviews. 1998, 22, 151–175. [Google Scholar] [CrossRef]
- Nogueira, P.F.M.; et al. The effects of Anabaena spiroides exopolysaccharides on copper accumulation in an aquatic food chain. Aquatic Toxicology. 2009, 93, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Kuang, X.; et al. Enhancement mechanisms of copper(II) adsorption onto kaolinite by extracellular polymeric substances of Microcystis aeruginosa (cyanobacterium). International Biodeterioration & Biodegradation. 2019, 138, 8–14. [Google Scholar]
- Paperi, R.; Micheletti, E.; De Philippis, R. Optimization of copper sorbing–desorbing cycles with confined cultures of the exopolysaccharide-producing cyanobacterium Cyanospira capsulata. Journal of Applied Microbiology. 2006, 101, 1351–1356. [Google Scholar] [CrossRef] [PubMed]
- Ciani, M.; et al. Semi-continuous cultivation of EPS-producing marine cyanobacteria: A green biotechnology to remove dissolved metals obtaining metal-organic materials. New Biotechnology. 2024, 82, 33–42. [Google Scholar] [CrossRef]
- Pereira, S.; et al. Complexity of cyanobacterial exopolysaccharides: composition, structures, inducing factors and putative genes involved in their biosynthesis and assembly. FEMS Microbiology Reviews. 2009, 33, 917–941. [Google Scholar] [CrossRef]
- Pagli, C.; et al. Isolation of biocrust cyanobacteria and evaluation of Cu, Pb, and Zn immobilisation potential for soil restoration and sustainable agriculture. Science of the Total Environment. 2024, 946, 174020. [Google Scholar] [CrossRef]
- Gupta, P.; Diwan, B. Bacterial exopolysaccharide mediated heavy metal removal: a review on biosynthesis, mechanism and remediation strategies. Biotechnology Reports. 2017, 13, 58–71. [Google Scholar] [CrossRef]
- Singh, N.; et al. Thiol and exopolysaccharide production in a cyanobacterium under heavy metal stress. Process Biochemistry. 1999, 35, 63–68. [Google Scholar] [CrossRef]
- De Philippis, R.; Paperi, R.; Sili, C. Heavy metal sorption by released polysaccharides and whole cultures of two exopolysaccharide-producing cyanobacteria. Biodegradation. 2007, 18, 181–187. [Google Scholar] [CrossRef]
- Leal, P.P.; et al. Copper pollution exacerbates the effects of ocean acidification and warming on kelp microscopic early life stages. Scientific Reports. 2018, 8, 14763. [Google Scholar] [CrossRef] [PubMed]
- Comte, S.; Guibaud, G.; Baudu, M. Biosorption properties of extracellular polymeric substances (EPS) towards Cd, Cu and Pb for different pH values. Journal of Hazardous Materials. 2008, 151, 185–193. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




