Submitted:
30 May 2025
Posted:
02 June 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Efficient and Accurate Bypass of 8-oxoA in Reactions in the Presence of Mg2+
2.2. Mn2+ Ions Decrease Accuracy of PrimPol on DNA Substrates with 8-Oxopurines
2.3. The Effect of DNA Sequence Context on A and 8-oxoA Bypass
3. Discussion
4. Materials and Methods
4.1. DNA Templates and Enzymes
| Oligonucleotide | Sequence 5`-3` |
|---|---|
| Pr18-Cy5 | Сy5-AGGGCAGAGTATTCTTCT |
| Pr18 | AGGGCAGAGTATTCTTCT |
| TemplateXA | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTATACCGCAGGCAXAGAAGAATACTCTGCCCT |
| TemplateXG | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTATACCGCAGGCGXAGAAGAATACTCTGCCCT |
| TemplateXT | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTATACCGCAGGCTXAGAAGAATACTCTGCCCT |
| TemplateXC | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTATACCGCAGGCCXAGAAGAATACTCTGCCCT |
4.2. DNA Polymerase Reactions for the Primer Extension Assay
4.3. Steady-State Kinetics Analysis of dNMP Incorporation
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 8-oxoA | 8-oxoadenine |
| 8-oxoG | 8-oxoguanine |
References
- García-Gómez, S.; Reyes, A.; Martínez-Jiménez, M.I.; Chocrón, S.; Mourón, S.; Terrados, G.; Powell, C.; Salido, E.; Méndez, J.; Holt, I.J.; et al. PrimPol, an Archaic Primase/Polymerase Operating in Human Cells. Mol. Cell 2013, 52, 541–553. [Google Scholar] [CrossRef]
- Bianchi, J.; Rudd, S.G.; Jozwiakowski, S.K.; Bailey, L.J.; Soura, V.; Taylor, E.; Stevanovic, I.; Green, A.J.; Stracker, T.H.; Lindsay, H.D.; et al. Primpol Bypasses UV Photoproducts during Eukaryotic Chromosomal DNA Replication. Mol. Cell 2013, 52, 566–573. [Google Scholar] [CrossRef]
- Mourón, S.; Rodriguez-Acebes, S.; Martínez-Jiménez, M.I.; García-Gómez, S.; Chocrón, S.; Blanco, L.; Méndez, J. Repriming of DNA Synthesis at Stalled Replication Forks by Human PrimPol. Nat. Struct. Mol. Biol. 2013, 20, 1383–1389. [Google Scholar] [CrossRef]
- Piberger, A.L.; Bowry, A.; Kelly, R.; Walker, A.K.; Gonzalez, D.; Bailey, L.J.; Doherty, A.J.; Mendez, J.; Morris, J.R.; Bryant, H.E.; et al. PrimPol-Dependent Single-Stranded Gap Formation Mediates Homologous Recombination at Bulky DNA Adducts. Nat. Commun. 2020, 11, 5863. [Google Scholar] [CrossRef]
- González-Acosta, D.; Blanco-Romero, E.; Ubieto-Capella, P.; Mutreja, K.; Míguez, S.; Llanos, S.; García, F.; Muñoz, J.; Blanco, L.; Lopes, M.; et al. PrimPol-mediated Repriming Facilitates Replication Traverse of DNA Interstrand Crosslinks. EMBO J. 2021, 40, e106355. [Google Scholar] [CrossRef]
- Schiavone, D.; Jozwiakowski, S.K.; Romanello, M.; Guilbaud, G.; Guilliam, T.A.; Bailey, L.J.; Sale, J.E.; Doherty, A.J. PrimPol Is Required for Replicative Tolerance of G Quadruplexes in Vertebrate Cells. Mol. Cell 2016, 61, 161–169. [Google Scholar] [CrossRef]
- Li, T.; Tang, L.; Kou, H.; Wang, F. Primpol Competes with Rad51 to Resolve G-Quadruplexinduced Replication Stress via Its Interaction with Rpa. Acta Biochim. Biophys. Sin. (Shanghai). 2023, 55, 498–507. [Google Scholar] [CrossRef]
- Butler, T.J.; Estep, K.N.; Sommers, J.A.; Maul, R.W.; Moore, A.Z.; Bandinelli, S.; Cucca, F.; Tuke, M.A.; Wood, A.R.; Bharti, S.K.; et al. Mitochondrial Genetic Variation Is Enriched in G-Quadruplex Regions That Stall DNA Synthesis in Vitro. Hum Mol Genet. 2020, 29, 1292–1309. [Google Scholar] [CrossRef]
- Šviković, S.; Crisp, A.; Tan-Wong, S.M.; Guilliam, T.A.; Doherty, A.J.; Proudfoot, N.J.; Guilbaud, G.; Sale, J.E. R-loop Formation during S Phase Is Restricted by PrimPol-mediated Repriming. EMBO J. 2019, 38, e99793. [Google Scholar] [CrossRef]
- Kobayashi, K.; Guilliam, T.A.; Tsuda, M.; Yamamoto, J.; Bailey, L.J.; Iwai, S.; Takeda, S.; Doherty, A.J.; Hirota, K. Repriming by PrimPol Is Critical for DNA Replication Restart Downstream of Lesions and Chain-Terminating Nucleosides. Cell Cycle 2016, 15, 1997–2008. [Google Scholar] [CrossRef]
- Bailey, L.J.; Bianchi, J.; Doherty, A.J. PrimPol Is Required for the Maintenance of Efficient Nuclear and Mitochondrial DNA Replication in Human Cells. Nucleic Acids Res. 2019, 47, 4026–4038. [Google Scholar] [CrossRef]
- Makarova, A.V.; Boldinova, E.O.; Belousova, E.A.; Lavrik, O.I. In Vitro Lesion Bypass by Human PrimPol. DNA Repair (Amst). 2018, 70, 18–24. [Google Scholar] [CrossRef]
- Boldinova, E.O.; Ghodke, P.P.; Sudhakar, S.; Mishra, V.K.; Manukyan, A.A.; Miropolskaya, N.; Pradeepkumar, P.I.; Makarova, A. V Translesion Synthesis across the N2-Ethyl-Deoxyguanosine Adduct by Human PrimPol. ACS Chem. Biol. 2022, 17, 3238–3250. [Google Scholar] [CrossRef]
- Yudkina, A.V.; Barmatov, A.E.; Bulgakov, N.A.; Boldinova, E.O.; Shilkin, E.S.; Makarova, A.V.; Zharkov, D.O. Bypass of Abasic Site–Peptide Cross-Links by Human Repair and Translesion DNA Polymerases. Int. J. Mol. Sci. 2023, 24, 10877. [Google Scholar] [CrossRef]
- Boldinova, E.O.; Yudkina, A.V.; Shilkin, E.S.; Gagarinskaya, D.I.; Baranovskiy, A.G.; Tahirov, T.H.; Zharkov, D.O.; Makarova, A.V. Translesion Activity of PrimPol on DNA with Cisplatin and DNA-Protein Cross-Links. Sci. Rep. 2021, 11, 17588. [Google Scholar] [CrossRef]
- Stojkovic, G.; Makarova, A.V.; Wanrooij, P.H.; Forslund, J.; Burgers, P.M.J.; Wanrooij, S. Oxidative DNA Damage Stalls the Human Mitochondrial Replisome. Sci. Rep. 2016, 6, 28942. [Google Scholar] [CrossRef]
- Helbock, H.J.; Beckman, K.B.; Shigenaga, M.K.; Walter, P.B.; Woodall, A.A.; Yeo, H.C.; Ames, B.N. DNA Oxidation Matters: The HPLC-Electrochemical Detection Assay of 8-Oxo-Deoxyguanosine and 8-Oxo-Guanine. Proc. Natl. Acad. Sci. U. S. A. 1998, 95, 288–293. [Google Scholar] [CrossRef]
- Møller, P.; Cooke, M.S.; Collins, A.; Olinski, R.; Rozalski, R.; Loft, S. Harmonising Measurements of 8-Oxo-7,8-Dihydro-2′-Deoxyguanosine in Cellular DNA and Urine. Free Radic. Res. 2012, 46, 541–553. [Google Scholar] [CrossRef]
- Fuciarelli, A.F.; Wegher, B.J.; Gajewski, E.; Dizdaroglu, M.; Blakely, W.F. Quantitative Measurement of Radiation-Induced Base Products in DNA Using Gas Chromatography-Mass Spectrometry. Radiat. Res. 1989, 119, 219–231. [Google Scholar] [CrossRef]
- Tuo, J.; Jaruga, P.; Rodriguez, H.; Dizdaroglu, M.; Bohr, V.A. The Cockayne Syndrome Group B Gene Product Is Involved in Cellular Repair of 8-Hydroxyadenine in DNA. J. Biol. Chem. 2002, 277, 30832–30837. [Google Scholar] [CrossRef]
- Leonard, G.A.; Brown, T.; Guy, A.; Téoule, R.; Hunter, W.N. Conformation of Guanine-8-Oxoadenine Base Pairs in the Crystal Structure of d(CGCGAATT(08A)GCG). Biochemistry 1992, 31, 8415–8420. [Google Scholar] [CrossRef]
- Yudkina, A.V.; Shilkin, E.S.; Endutkin, A.V.; Makarova, A.V.; Zharkov, D.O. Reading and Misreading 8-Oxoguanine, a Paradigmatic Ambiguous Nucleobase. Crystals 2019, 9, 269. [Google Scholar] [CrossRef]
- Moriya, M. Single-Stranded Shuttle Phagemid for Mutagenesis Studies in Mammalian Cells: 8-Oxoguanine in DNA Induces Targeted G·C → T·A Transversions in Simian Kidney Cells. Proc. Natl. Acad. Sci. U. S. A. 1993, 90, 1122–1126. [Google Scholar] [CrossRef]
- Wood, M.L.; Esteve, A.; Morningstar, M.L.; Kuziemko, G.M.; Essigmann, J.M. Genetic Effects of Oxidative DNA Damage: Comparative Mutagenesis of 7,8-Dihydro-8-Oxoguanine and 7,8-Dihydro-8-Oxoadenine in Escherichia Coli. Nucleic Acids Res. 1992, 20, 6023–6032. [Google Scholar] [CrossRef]
- Kamiya, H.; Miura, H.; Murata-kamiya, N.; Ishikawa, H.; Sakaguchi, T.; Inoue, H.; Sasaki, T.; Masutanl, C.; Hanaoka, F.; Nishimura, S.; et al. 8-Hydroxyadenine (7, 8-Dihydro-8-Oxoadenine) Induces Misincorporation in in Vitro DNA Synthesis and Mutations in NIH 3t3 Cells. Nucleic Acids Res. 1995, 23, 2893–2899. [Google Scholar] [CrossRef]
- Kamiya, H.; Murata-kamiya, N.; Koizume, S.; Inoue, H.; Nishimura, S.; Ohtsuka, E. 8-Hydroxyguanine (7,8-Dihydro-8-Oxoguanine) in Hot Spots of the c-Ha-Ras Gene: Effects of Sequence Contexts on Mutation Spectra. Carcinogenesis 1995, 16, 883–889. [Google Scholar] [CrossRef]
- Tan, X.; Grollman, A.P.; Shibutani, S. Comparison of the Mutagenic Properties of 8-Oxo-7,8-Dihydro-2’-Deoxyadenosine and 8-Oxo-7,8-Dihydro-2’-Deoxyguanosine DNA Lesions in Mammalian Cells. Carcinogenesis 1999, 20, 2287–2292. [Google Scholar] [CrossRef]
- Martínez-Jiménez, M.I.; García-Gómez, S.; Bebenek, K.; Sastre-Moreno, G.; Calvo, P.A.; Díaz-Talavera, A.; Kunkel, T.A.; Blanco, L. Alternative Solutions and New Scenarios for Translesion DNA Synthesis by Human PrimPol. DNA Repair (Amst). 2015, 29, 127–138. [Google Scholar] [CrossRef]
- Shibutani, S.; Bodepudi, V.; Johnson, F.; Grollman, A.P. Translesional Synthesis on DNA Templates Containing 8-Oxo-7,8-Dihydrodeoxyadenosine. Biochemistry 1993, 32, 4615–4621. [Google Scholar] [CrossRef]
- Koag, M.C.; Jung, H.; Lee, S. Mutagenic Replication of the Major Oxidative Adenine Lesion 7,8-Dihydro-8-Oxoadenine by Human DNA Polymerases. J. Am. Chem. Soc. 2019, 141, 4584–4596. [Google Scholar] [CrossRef]
- Torregrosa-Muñumer, R.; Forslund, J.; Goffart, S.; Pfeiffer, A.; Stojkovic, G.; Carvalho, G.; Al-Furoukh, N.; Blanco, L.; Wanrooij, S.; Pohjoismäki, J.L.O. PrimPol Is Required for Replication Reinitiation after MtDNA Damage. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 11398–11403. [Google Scholar] [CrossRef]
- Guilliam, T.A.; Jozwiakowski, S.K.; Ehlinger, A.; Barnes, R.P.; Rudd, S.G.; Bailey, L.J.; Skehel, J.M.; Eckert, K.A.; Chazin, W.J.; Doherty, A.J. Human PrimPol Is a Highly Error-Prone Polymerase Regulated by Single-Stranded DNA Binding Proteins. Nucleic Acids Res. 2015, 43, 1056–1068. [Google Scholar] [CrossRef]
- Yung, C.; Suzuki, T.; Okugawa, Y.; Kawakami, A.; Loakes, D.; Negishi, K.; Negishi, T. Nucleotide Incorporation against 7,8-Dihydro-8-Oxoguanine Is Influenced by Neighboring Base Sequences in TLS DNA Polymerase Reaction. In Proceedings of the Nucleic acids symposium series; 2007; pp. 49–50. [Google Scholar]
- Zafar, M.K.; Ketkar, A.; Lodeiro, M.F.; Cameron, C.E.; Eoff, R.L. Kinetic Analysis of Human PrimPol DNA Polymerase Activity Reveals a Generally Error-Prone Enzyme Capable of Accurately Bypassing 7,8-Dihydro-8-Oxo-2′-Deoxyguanosine. Biochemistry 2014, 53, 6584–6594. [Google Scholar] [CrossRef]
- Rechkoblit, O.; Johnson, R.E.; Gupta, Y.K.; Prakash, L.; Prakash, S.; Aggarwal, A.K. Structural Basis of DNA Synthesis Opposite 8-Oxoguanine by Human PrimPol Primase-Polymerase. Nat. Commun. 2021, 12, 4020. [Google Scholar] [CrossRef]
- Gromova, A.S.; Boldinova, E.O.; Kim, D.V.; Chuprov-Netochin, R.N.; Leonov, S.V.; Pustovalova, M.V.; Zharkov, D.O.; Makarova, A.V. Response of PRIMPOL-Knockout Human Lung Adenocarcinoma A549 Cells to Genotoxic Stress. Biochem. 2023, 88, 1933–1943. [Google Scholar] [CrossRef]
- Andrs, M.; Stoy, H.; Boleslavska, B.; Chappidi, N.; Kanagaraj, R.; Nascakova, Z.; Menon, S.; Rao, S.; Oravetzova, A.; Dobrovolna, J.; et al. Excessive Reactive Oxygen Species Induce Transcription-Dependent Replication Stress. Nat. Commun. 2023, 14. [Google Scholar] [CrossRef]
- Gyüre, Z.; Póti, Á.; Németh, E.; Szikriszt, B.; Lózsa, R.; Krawczyk, M.; Richardson, A.L.; Szüts, D. Spontaneous Mutagenesis in Human Cells Is Controlled by REV1-Polymerase ζ and PRIMPOL. Cell Rep. 2023, 42, 112887. [Google Scholar] [CrossRef]
- Petushkov, I.V.; Aralov, A.V.; Ivanov, I.A.; Baranov, M.S.; Zatsepin, T.S.; Kulbachinskiy, A.V. Effect of 8-Oxo-1,N6-Ethenoadenine Derivatives on the Activity of RNA Polymerases from SARS-CoV-2 and Escherichia Coli. Biochemistry. (Mosc). 2024, 89, 2263–2273. [Google Scholar] [CrossRef]
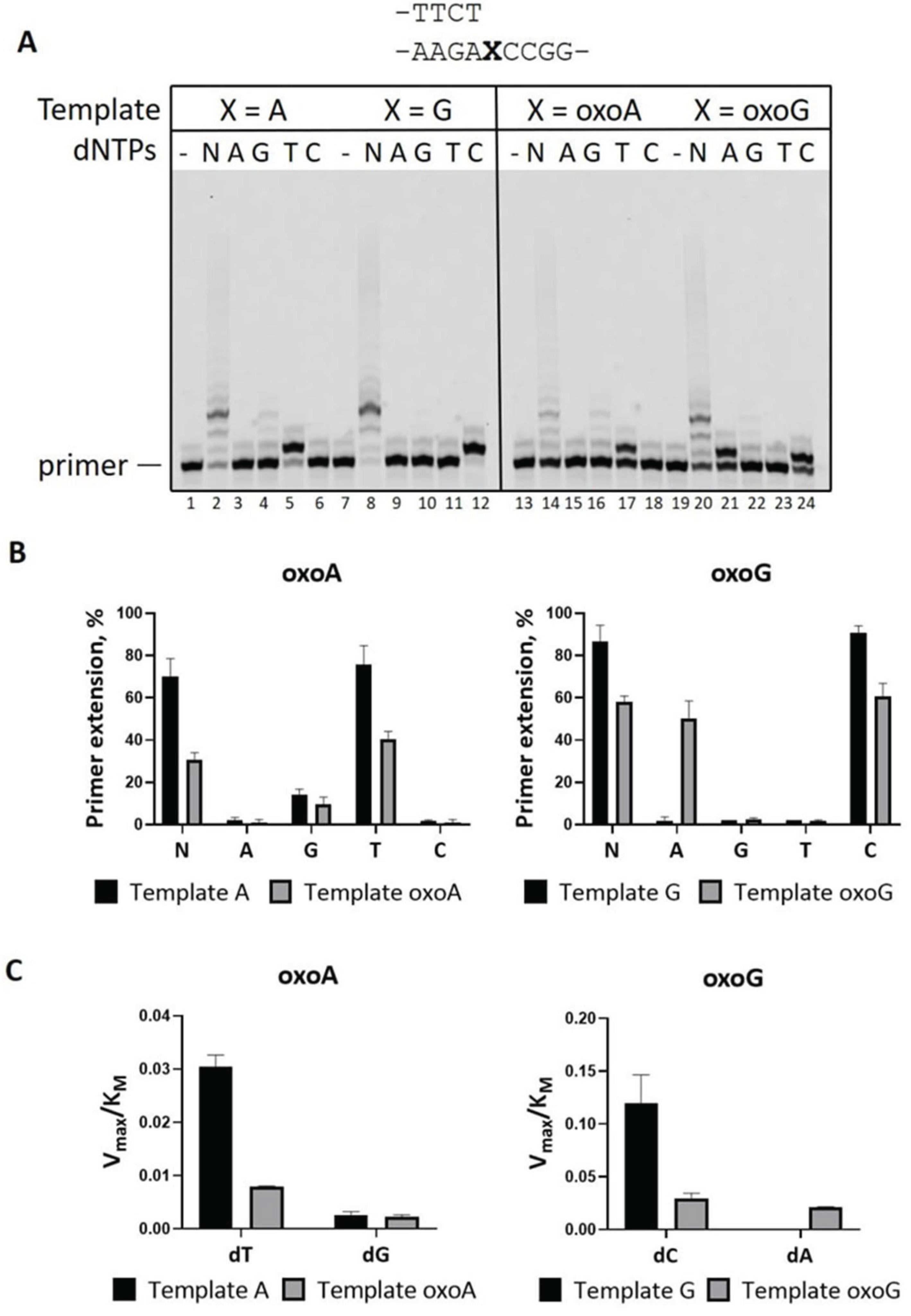
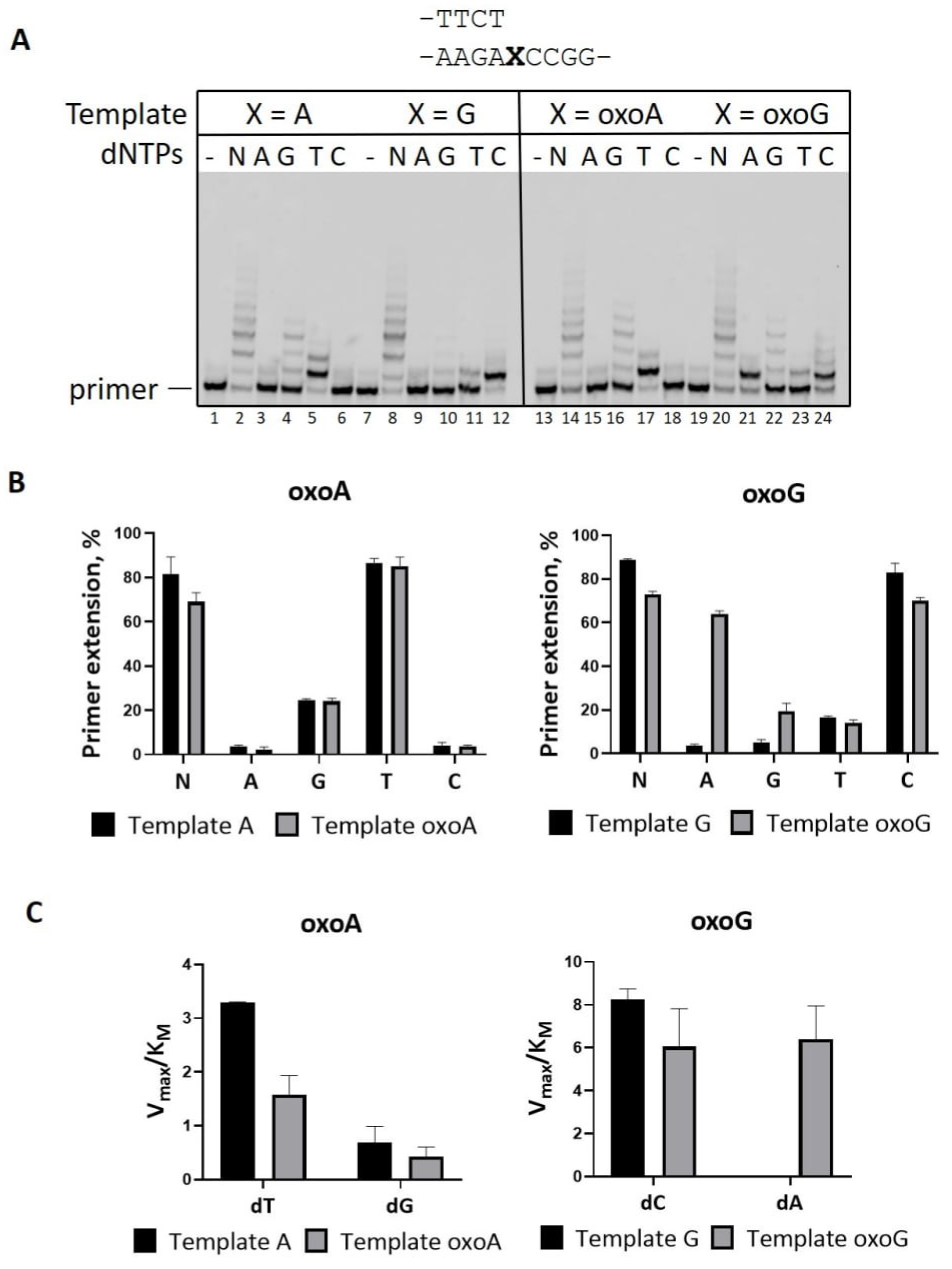
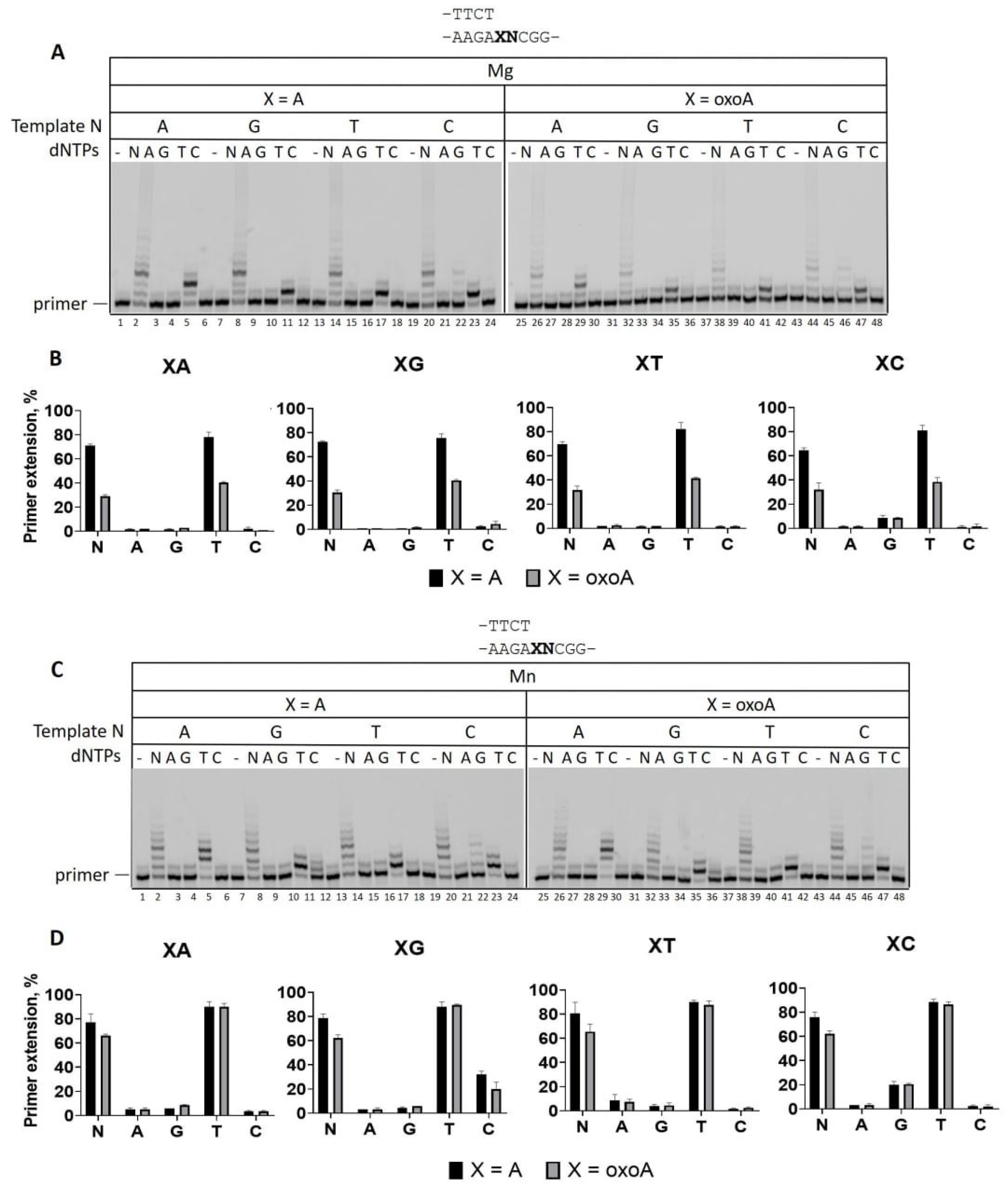
| Template | dNMP | Vmax, % per min | KM, μM | Vmax/KM | Finc |
|---|---|---|---|---|---|
| Mg2+ | |||||
| Template A | dTradio | 12.9 ± 0.7 | 420 ± 37 | 0.031 ±0.003 | |
| dT | 12.2 ± 0.7 | 400 ± 42 | 0.031 ± 0.002 | 1 | |
| dG | 0.4 ± 0.004 | 184 ± 46 | 0.003 ± 0.0005 | 0.09 | |
| Template oxoA | dTradio | 3.6 ±0.3 | 610 ± 11 | 0.006 ±0.001 | |
| dT | 5.3 ± 0.1 | 677 ± 16 | 0.008 ± 0.0001 | 1 | |
| dG | 0.4 ± 0.005 | 182 ± 33 | 0.002 ± 0.0001 | 0.25 | |
| Template G | dC | 9 ± 0.9 | 73 ± 5 | 0.123 ± 0.019 | |
| dA | ND | ||||
| Template oxoG | dC | 3.8 ± 0.3 | 128 ± 3 | 0.029 ± 0.004 | 1 |
| dA | 2.9 ± 0.3 | 137 ± 9 | 0.021 ± 0.0005 | 0.72 | |
| Mn2+ | |||||
| Template A | dTradio | 50 ± 1.5 | 11 ± 1 | 4.5 ± 0.3 | |
| dT | 40 ± 0.9 | 12.2 ± 0.3 | 3.3 ± 0.01 | 1 | |
| dG | 2.7 ± 0.5 | 4 ± 0.3 | 0.7 ± 0.2 | 0.2 | |
| Template oxoA | dTradio | 28 ± 1.4 | 10 ± 0.5 | 2.7 ± 0.1 | |
| dT | 22.5 ± 0.02 | 14.6 ± 1.9 | 1.6 ± 0.2 | 1 | |
| dG | 2.4 ± 0.2 | 3.9 ± 0.8 | 0.4 ± 0.1 | 0.25 | |
| Template G | dC | 41.5 ± 0.1 | 5.1 ± 0.2 | 8.2 ± 0.3 | |
| dA | ND | ||||
| Template oxoG | dC | 23.5 ± 1 | 4 ± 0.5 | 6 ± 1 | 1 |
| dA | 15.9 ± 0.6 | 2.5 ± 0.3 | 6.4 ± 0.9 | 1.1 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





