1. Introduction
Inteins are mobile genetic elements invading highly conserved genes throughout all domains of life and viruses. An intein invades its host gene at the DNA level, similar to an intron [
1]. Unlike introns which are spliced out at the RNA level, inteins splice themselves out at the protein level using an autocatalytic self-splicing reaction enabled by the intein’s self-splicing domain. During protein splicing, the intein is able to seamlessly ligate the two halves of its host protein back together, allowing the host protein to function [
2]. This natural capacity of inteins to engage in seamless protein splicing has made them invaluable tools in the development of protein engineering technology [
3]. Recent large-scale intein characterization studies have revealed increasingly diverse intein architectures, such as varied architectures in the inteins of phages [
4]. Such novel intein architectures hold the potential for new technological applications, emphasizing the importance of continued mass intein-characterization efforts.
Along with their novel biochemical capabilities, inteins also engage in unorthodox evolutionary behaviors. In addition to the self-splicing domain, full inteins contain a central homing endonuclease domain which bestows them with the ability to be inherited at super-Mendelian frequencies through the process of homing [
5]. When in the presence of an uninvaded copy of the intein’s host gene, the homing endonuclease domain will make a double-strand DNA break at the intein insertion site. Then, through homologous recombination-based DNA repair, the intein-containing copy of the gene can be used as a template leading the intein DNA sequence to be pasted into the previously uninvaded copy. As a result, homing allows inteins to rapidly proliferate through a population in spite of their fitness cost to the host [
6]. Per the Goddard-Burt life cycle of a homing endonuclease driven selfish genetic element such as an intein, once the element reaches saturation in a population and there are no more empty target sites, the homing endonuclease is no longer under selective pressure to be maintained [
5]. Once the homing endonuclease of an intein has severely decayed beyond function, it is referred to as a mini intein. In the Goddard-Burt model, the mini intein is eventually lost from the population. Recent models have expanded on the Goddard-Burt model to suggest the co-existence of the three states (intein-free, full intein-containing, and mini intein-containing) as opposed to a synchronized progression through the states [
7,
8], but to date no evidence of such co-existence of all three states in a single population has been shown [
9].
There remains an incredible wealth of public sequence data to be explored for new intein architectures and evolutionary behaviors, particularly in archaea. Archaea contain inteins in a wide range of genes [
10], but the majority of intensive archaeal intein characterization efforts have been focused on a few select groups such as haloarchaea. In addition to being a group ripe for the exploration of intein architectures and evolutionary dynamics, archaea offer a unique area of intein exploration in which both architecture and evolution can be studied: genes invaded by multiple inteins simultaneously [
11,
12]. The archaeal gene
mcm, which encodes the MCM subunit of replicative DNA helicase [
13,
14], contains five intein insertion sites named MCM-a through e respectively. Inteins at sites MCM-a through d have been the subject of intein insertion site recognition and self-splicing investigations, particularly in haloarchaea [
12,
15]. Insertion site MCM-e is not invaded in any haloarchaea analyzed to date, but the site is active in some groups of non-haloarchaea. The MCM-e intein published to the intein database InBase 2.0 [
16] in 2012 was from
Thermococcus litoralis, with the insertion site name CDC21-e [
17]. An analysis of the MCM inteins at sites MCM-a through d in haloarchaeal MCM homologs from the order Haloferacales revealed a wide array of intein invasion statuses (empty, single, double, triple, and quadruple), mini and full inteins at the same insertion sites in different homologs, and sporadic distribution of the four inteins across the host protein phylogeny [
12]. The diversity of MCM inteins in this order alone begs the question of whether such patterns will hold when a similar analysis is performed on other archaeal lineages, and whether such diversity can also be found in a single group of geographically overlapping populations of archaea as opposed to a mass sampling of sequences from a wide array of timepoints and geographic locations.
To address these questions of intein architectural diversity, distribution patterns, and evolutionary dynamics at the population level, we gathered 4,243 complete archaeal MCM homologs from NCBI’s non-redundant protein sequence database. To obtain as accurate a description of the MCM inteins across all archaea as possible with available data, an iterative search approach was used to thoroughly sample all available groups of archaea. A combination of sequence alignment and predicted structure-based analyses were used to characterize the inteins at all sites, through which six new archaeal MCM intein insertion sites were discovered. These new insertion sites all fall within the same catalytic ATPase domain of MCM as the known five (MCM-a through e). The sites are not active in all groups, with Nanobdellati (DPANN) being the only group to have at least one intein at all 11 MCM intein insertion sites. Our structural analyses revealed three sites within the MCM inteins where insertions resembling DNA-binding domains are found. These insertions vary in presence and size, adding a second facet to the inteins’ architectural diversity beyond the status of their homing endonucleases (mini or full). Within this dataset were 26 haloarchaeal sequences from the Atacama Desert all sampled in June of 2013 from the same three locations as part of a metagenomic study by Finstad et al. [
18]. This single group of geographically overlapping archaeal populations had greatly diverse MCM intein compositions, including no, mini, and full inteins at the same site in different individuals. Such a mixture of alleles strongly supports the co-existence model of intein persistence and captures the varied histories of inteins found at the different sites of a multi-intein gene.
2. Materials and Methods
Retrieving and curating amino acid sequence collection of archaeal MCM homologs. Using the MCM extein (host protein only, inteins removed in silico) sequence of
Haloferax mediterranei (Protein Accession: WP_004058379.1) as the query sequence, PSI-BLAST [
19] searches were performed against NCBI’s non-redundant protein sequence database with maximum 500 target sequences and an e-value cutoff of 0.0001. No more than five iterations were allowed, and the resulting matches to be used for the subsequent iteration were manually pruned to remove partial MCM sequences (less than 600aa) and any non-MCM sequences. Each search was restricted to a different taxonomic group, such that effective sampling could be performed even for highly sequenced groups. After combining the smaller subsets of matches into taxonomically relevant groups (i.e., combining the four orders of Haloarchaea into a single Haloarchaea subgroup), 16 subgroups were formed: Haloarchaea (taxid 183963), Methanomicrobia (taxid 224756), Methanoliparia (taxid 2545688) Archaeoglobi (taxid 183980), Methanonatronarchaeia (taxid 171536), Thermoplasmatota (taxid 2283796), Nanohaloarchaea (taxid 1051663), Nanobdellati (DPANN) (taxid 1783276), Theionarchaea (taxid 1980645), Methanofastidiosa (taxid 1705400), Thermococci (taxid 183968), Hadarchaeota (taxid 3055124), Thermoproteati (TACK) (taxid 1783275), Promethearchaeati (Asgard) (taxid 1935183), and Hydrothermarchaeota (taxid 1935019).
Combined sequence and structure-based approach for characterizing the architectures of all inteins at each insertion site. For each of the 16 sets of MCM homologs, the sequences were initially aligned using MUSCLE [
20] in SeaView [
21] with slight manual adjustments to clarify intein versus extein (host protein) boundaries. For more complex cases such as Nanobdellati (DPANN) where all 11 intein insertion sites are active, and to varying degrees, no tried alignment algorithms (MUSCLE, clustalo [
22], and MAFFT [
23]) were able to properly align the sequences. For these cases, more extensive manual adjustments were required to establish the intein-host protein boundaries. These alignments were never directly used for phylogenetic reconstruction, and rather used to establish boundaries between host protein and intein sequences which were then extracted and re-aligned algorithmically for further analyses. For each intein insertion site within each taxonomic group sampled, the largest intein at the site was extracted, de-aligned, and used as input for AlphaFold3 [
24]. Through this process, three sites within the inteins which occasionally contained insertions were identified: Insert Site 1 at the end of the N-terminal portion of the self-splicing domain, Insert Site 2 at the start of the C-terminal portion of the self-splicing domain, and Insert Site 3 ~12aa after Insert Site 2. Guided by the predicted structure of the largest intein, the homing endonuclease LAGLIDADG motif blocks and any within-intein insertions (Insertions 1, 2, and/or 3) were marked as selectable Sites in the sequence alignment in SeaView. By defining these Sites, each intein could be characterized based on its homing endonuclease and insertion architecture. The insertions were categorized as either small (20aa-60aa) or large (greater than 60aa) to capture the size variation observed between insertions at the same sites in different inteins. The minimum cutoff of 20aa was chosen based on the minimum length of a helix-turn-helix DNA-binding domain [
25]. The sequence alignments for each group with declared Sites are provided as .mase files (viewable in SeaView) in
Supplemental Data 1. The NCBI Protein Accession numbers are provided in the annotation line of every sequence. The inteins at each site were extracted into joined files, where Sites indicating Inserts 1, 2, and 3 were established in SeaView (.mase files available in
Supplemental Data 2).
Unrooted amino acid sequence phylogenies. All phylogenies generated for this work should be considered unrooted, and are arbitrarily rooted when presented as such. For construction, the respective alignments were used as input for IQ-TREE2 [
26], allowing ModelFinder [
27] to identify the best fit model, and with 1000 replicates of ultrafast boostrapping [
28]. The selected models are provided in the figure legends for each respective phylogeny. Treefiles were visualized using FigTree v.1.4.4 and Inkscape v.1.2.2.
3. Results
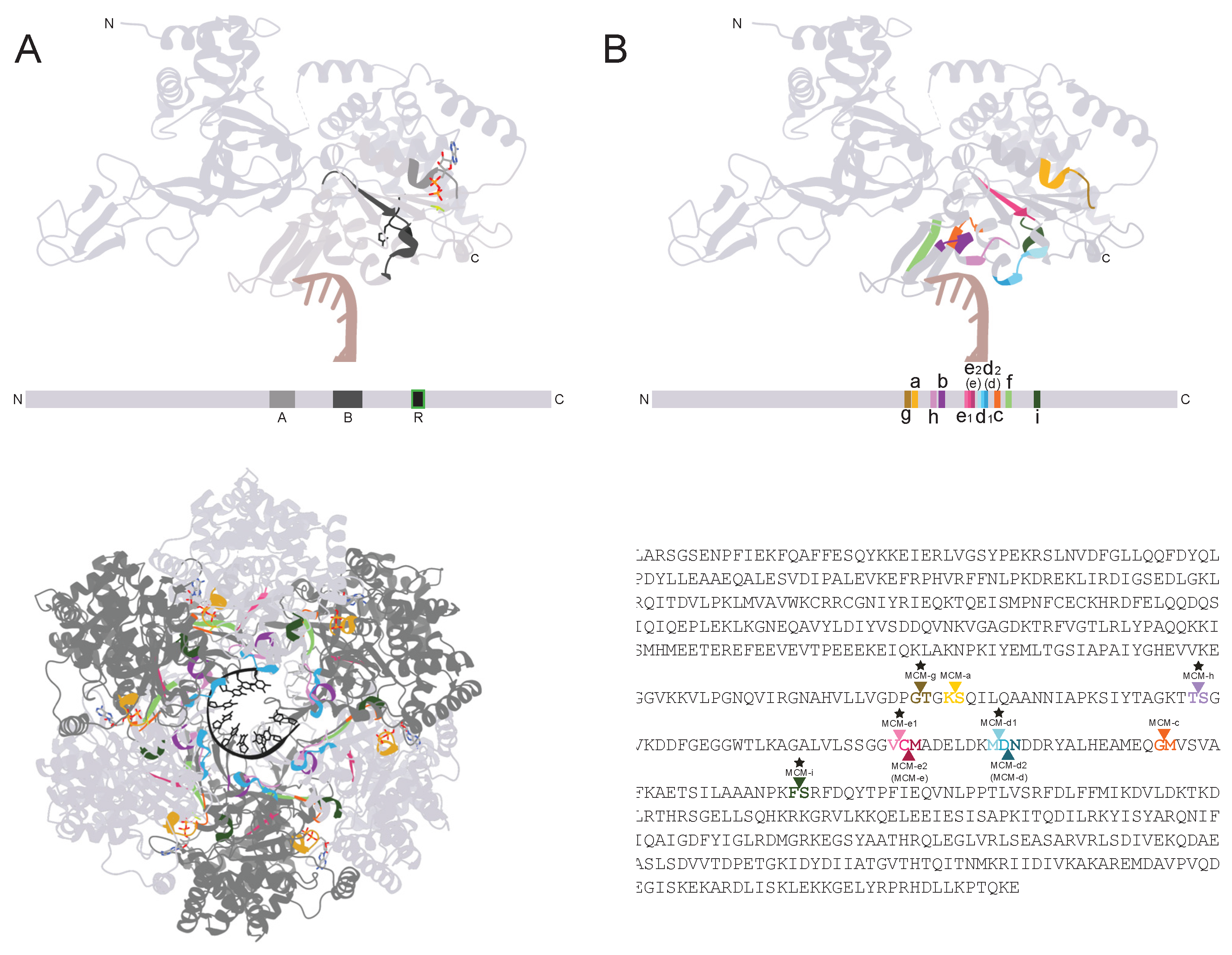
Analysis of MCM homologs across archaea reveals six new active MCM insertion sites. To investigate the abundance, structural features, and distribution of archaeal MCM inteins, 4,243 archaeal MCM homologs from NCBI’s non-redundant protein sequence database were systematically collected. The domain Archaea was divided into subgroups following NCBI’s Taxonomy Browser classifications, with more heavily sampled groups such as Stenosarchaea broken down into smaller groups for more thorough sampling. With thorough manual inspection of the sequence alignments generated for each subgroup, 11 distinct MCM intein insertion sites were identified (
Figure 1). To our best knowledge, the only previously reported archaeal MCM intein insertion sites were MCM-a, b, c, d, and e. The new sites are all located in the same catalytic region as the known five (
Figure 1A-C), owing to inteins’ propensity to invade highly conserved regions [
29]. The insertion sites cluster around especially important motifs for ATP binding by MCM subunits: the Walker A, Walker B, and arginine finger motifs [
30]. Following the naming convention used for these intein insertion sites thus far, with slight alteration due to the very close proximity of two new sites to two pre-existing sites, we refer to these new sites as MCM-f, MCM-g, MCM-h, MCM-i, and MCM-d1 and MCM-e1. MCM-f through i are named in order of discovery and not their position in the linear sequence (
Figure 1D), as this has been followed for naming the previously known sites. MCM-d1 and e1 are distinctly different from but very close (1 residue upstream) to MCM-d and e, thus we felt it beneficial to stray slightly from the traditional naming convention to reflect this. In this work, we refer to the original MCM-d and e as MCM-d2 and e2 due to them being one residue downstream of MCM-d1 and e1 respectively. Due to lack of sequence variation in the three MCM-d1 inteins, using phylogenetic reconstruction to further cement them as inteins of a separate insertion site than the MCM-d2 (d) inteins was not possible using an alignment of the MCM-d1 and d2 (d) inteins. However, there was sufficient variation among the inteins found at MCM-e1, allowing all MCM-e1 and MCM-e2 (e) inteins to be extracted, re-aligned, and used for phylogenetic reconstruction (
Figure S1). In the resulting phylogeny, the MCM-e1 inteins group together as opposed to grouping with the MCM-e2 inteins from their respective archaeal groups (Nanobdellati (DPANN) and Promethearchaeati (Asgard)), providing further support for the MCM-e1 inteins being distinct from MCM-e2 (e).
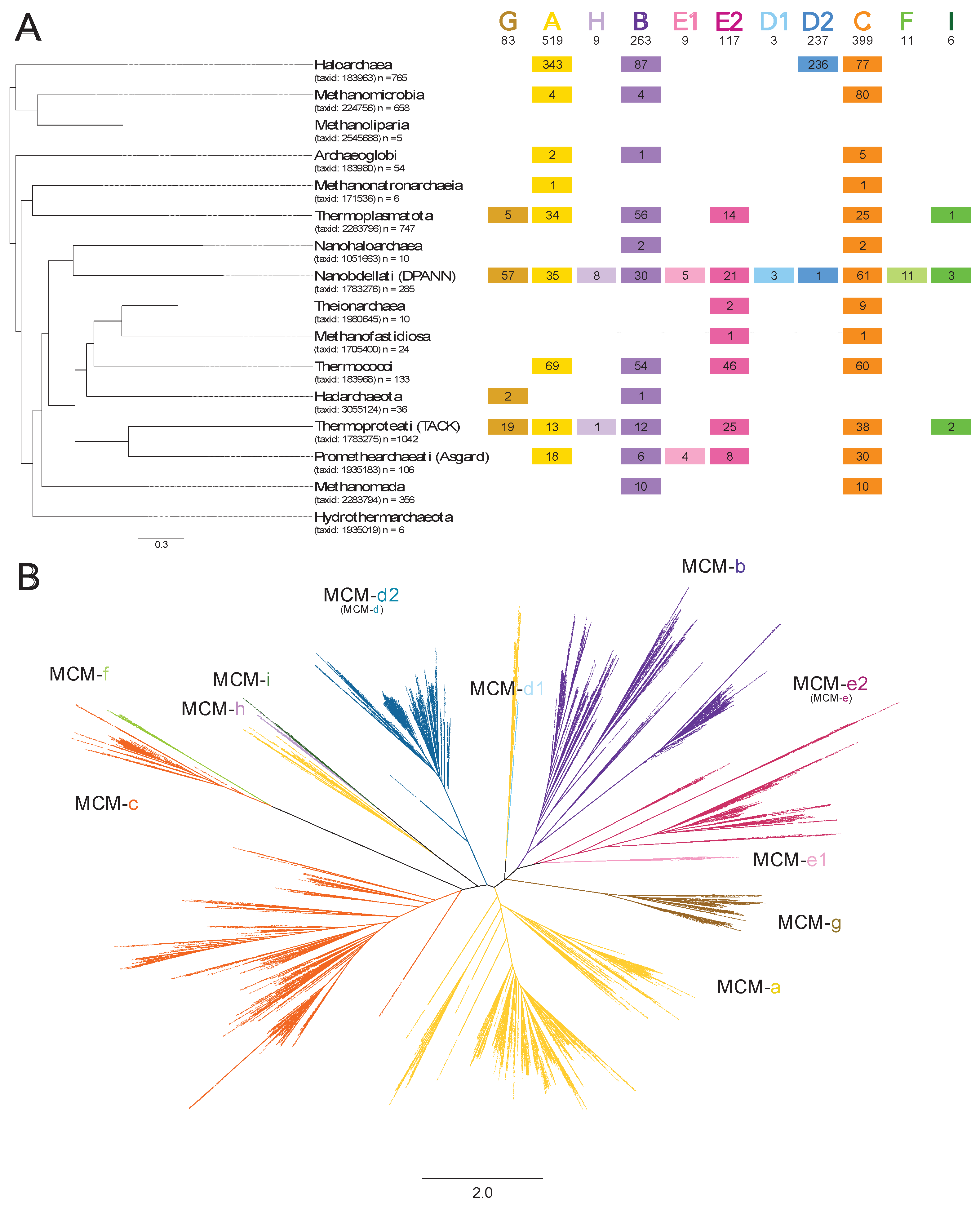
Varied invasion activity levels and distinct evolutionary histories at each MCM intein insertion site. After establishing the positions of all MCM intein insertion sites, the insertion activity levels for each site across each archaeal group were assessed (
Figure 2A). Out of the 16 subgroups, 14 had at least one active MCM intein insertion site. The only group in which all 11 sites are active, meaning at least one homolog from the group contains an intein at that site, is Nanobdellati (DPANN). Nanobdellati (DPANN) is also the only group with an active MCM intein insertion site which is inactive in all other groups (MCM-f). In contrast to MCM-f whose activity is seemingly limited to Nanobdellati (DPANN), in all intein-containing groups except for Hadarchaeota, at least one homolog had an intein at MCM-c. All new MCM intein insertion sites are less populated with inteins than the previously known sites. Similarly, instances of multi-intein invasions more frequently involved with previously known sites, with all quadruple invasions involving sites MCM-a, b, and c, with the fourth occupied site either being MCM-d2 (d) or e2 (e) (
Table S1). In total, ~73.5% of homologs (3125) had no inteins, ~17% (709) had one intein, ~7% (305) had two inteins, ~2% (79) had three inteins, and ~0.5% (25) had four inteins (
Table 1). While having 11 intein insertion sites and accounting for an intein status of empty, mini, or full at each site yields 177,147 theoretically possible MCM intein combinations, only 105 were observed. Out of those 105, 37 of the arrangements were observed in only one homolog each. All observed combinations of MCM inteins and their occurrences are available in
Table S1. In addition to assessing intein invasion levels at each site, phylogenetic analysis was performed to assess grouping patterns of the MCM inteins (
Figure 2B). Inteins at sites MCM-g, b, e1, d1, d2 (d), and f are monophyletic. The MCM-e2 (e) inteins all group together, with the MCM-e1 intein group emerging from them, adding further support to the differentiation between MCM-e1 and e2 (e) inteins despite their insertion sites being a single residue apart. This analysis also strengthens confidence in the differentiation between MCM-d1 and d2 (d) inteins, as the MCM-d1 inteins exhibit evolutionary distance from the MCM-d2 (d) inteins. The MCM-d1 inteins emerge from a group of MCM-a inteins. An additional small group of MCM-a inteins from which the MCM-h and i intein groups emerge is observed, and the majority of MCM-a inteins group together. The MCM-c inteins all group together, with the MCM-f inteins emerging from them. Over all, the inteins group strongly by insertion site as opposed to archaeal group.
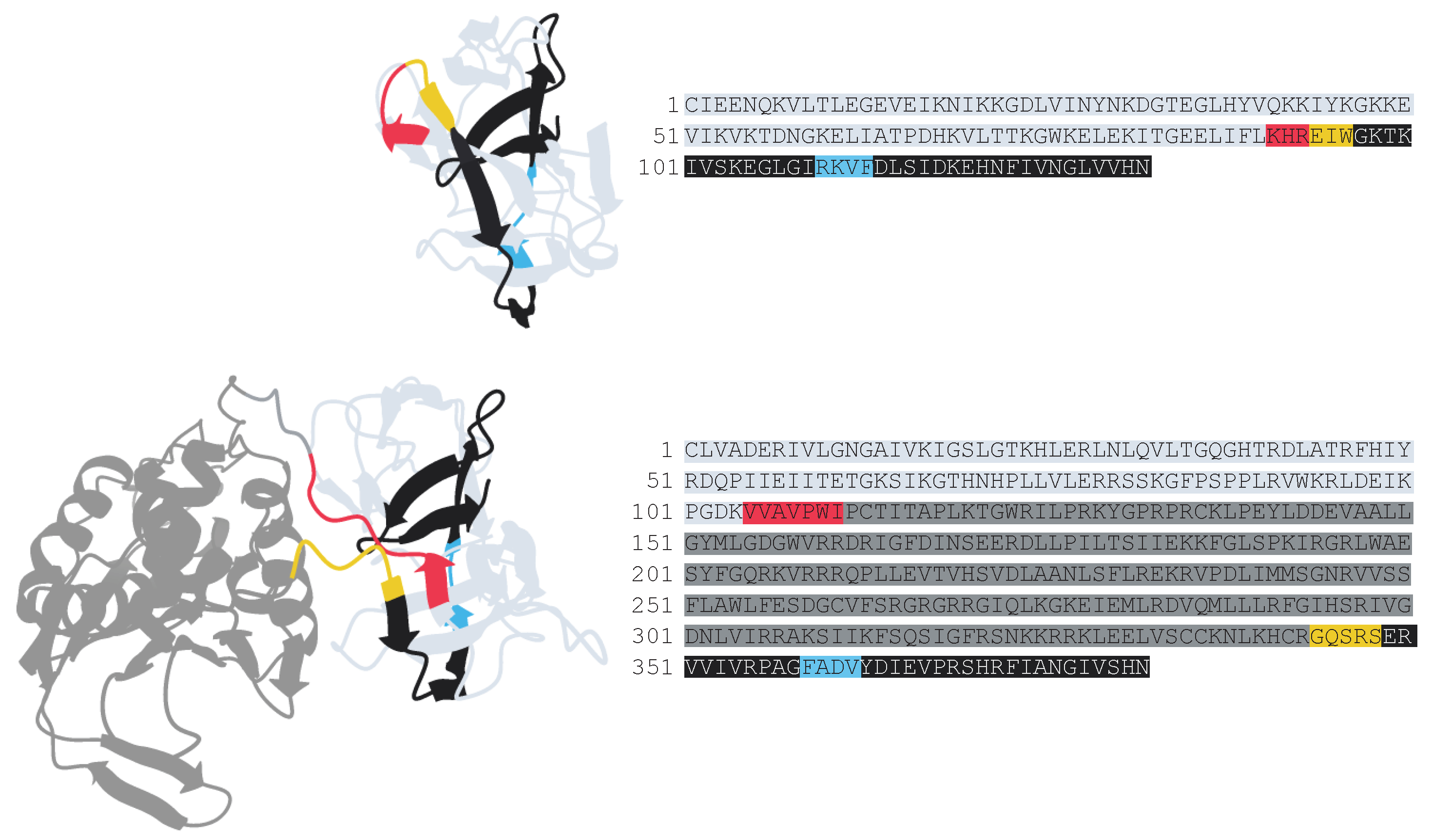
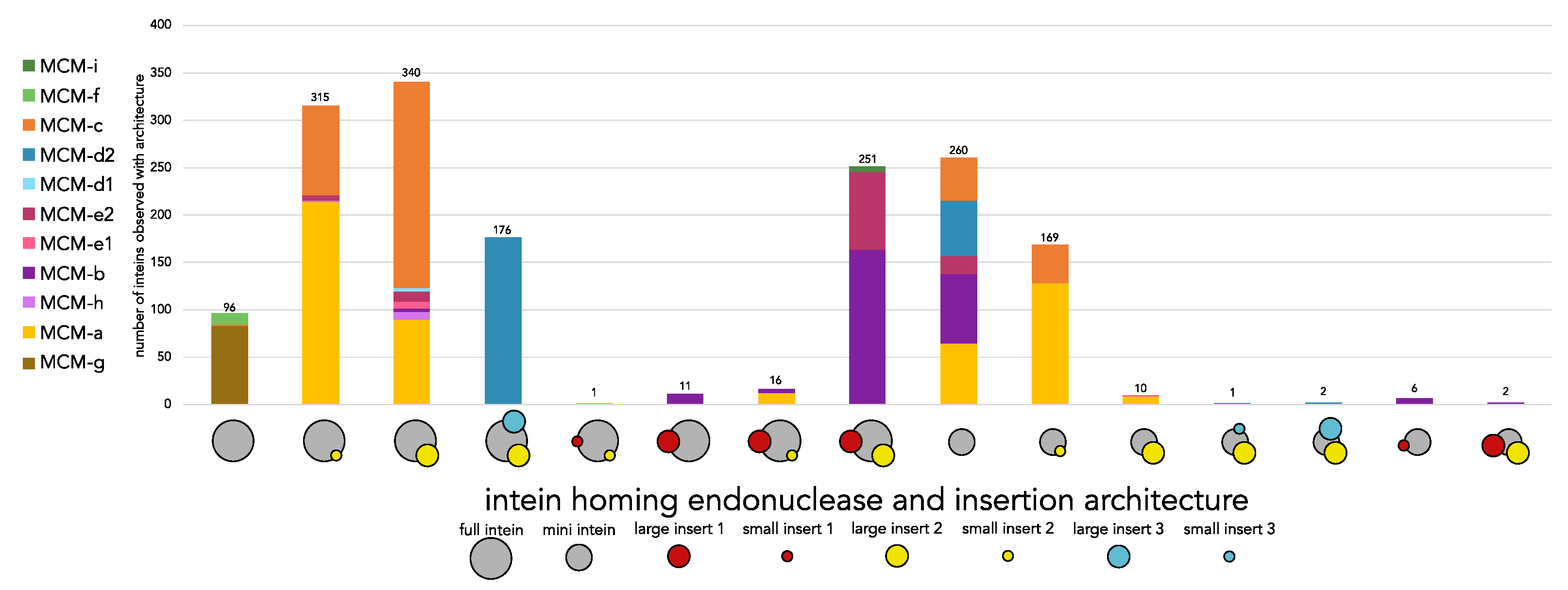
Decaying versus full homing endonucleases and insertions within inteins at three distinct sites generate architectural diversity. For each MCM intein insertion site in each of the 16 groups of homologs, all inteins were categorized as either mini (no detectable homing endonuclease domain) or full (detectable homing endonuclease domain with both LAGLIDADG motifs). Mini inteins were only identified at sites MCM-a, b, c, d2, e1, and e2. For those sites, there were between 1.5 and 8 times more full inteins found than mini inteins (
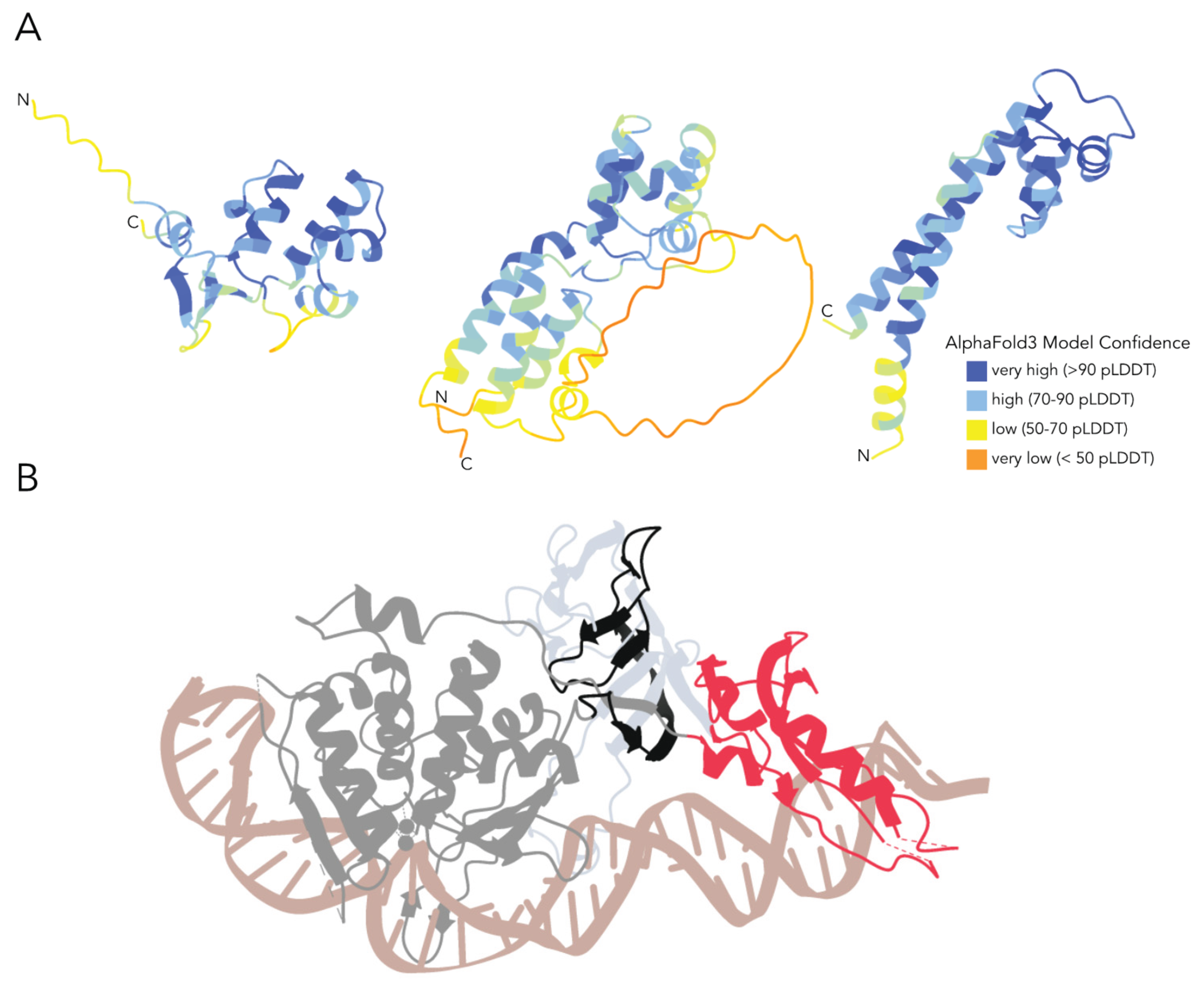
Figure S3). Using a combined sequence and predicted-structure based approach to define the domains of the inteins found at each site, inteins with additional domains beyond a homing endonuclease and self-splicing domain were identified. These inserted domains were identified in both full and mini inteins. Across all 1,656 inteins investigated, three distinct sub-insertion sites within the intein were identified: Insertion 1 located at the end of the N-terminal portion of the self-splicing domain; Insertion 2 at the beginning of the C-terminal portion of the self-splicing domain; Insertion 3 located ~12aa downstream of Insertion 2, placing it just after a conserved beta-strand in the C-terminal portion of the self-splicing domain [
32,
33] (
Figure 3). Accounting for both the intein’s homing endonuclease and sub-insertion status (mini or full intein; no, small, or large Insert 1; no, small, or large Insert 2; no, small, or large Insert 3) a total of 17 architectural variants were identified. The distribution of these architectural variants across the MCM intein insertion sites was assessed (
Figure 4). Certain MCM intein insertion sites exhibited little variation in the architecture of their inteins, such as MCM-g which contained only full inteins with no insertions. This homogeneity is not due to limited distribution, as MCM-g inteins are present across several archaeal groups: Thermoplasmatota, Nanobdellati (DPANN), Hadarchaeota, and Thermoproteati (TACK) (
Figure 2). In contrast, site MCM-b contained seven architectural variants. Insertion 3 was only identified in inteins located at site MCM-d2 (d).
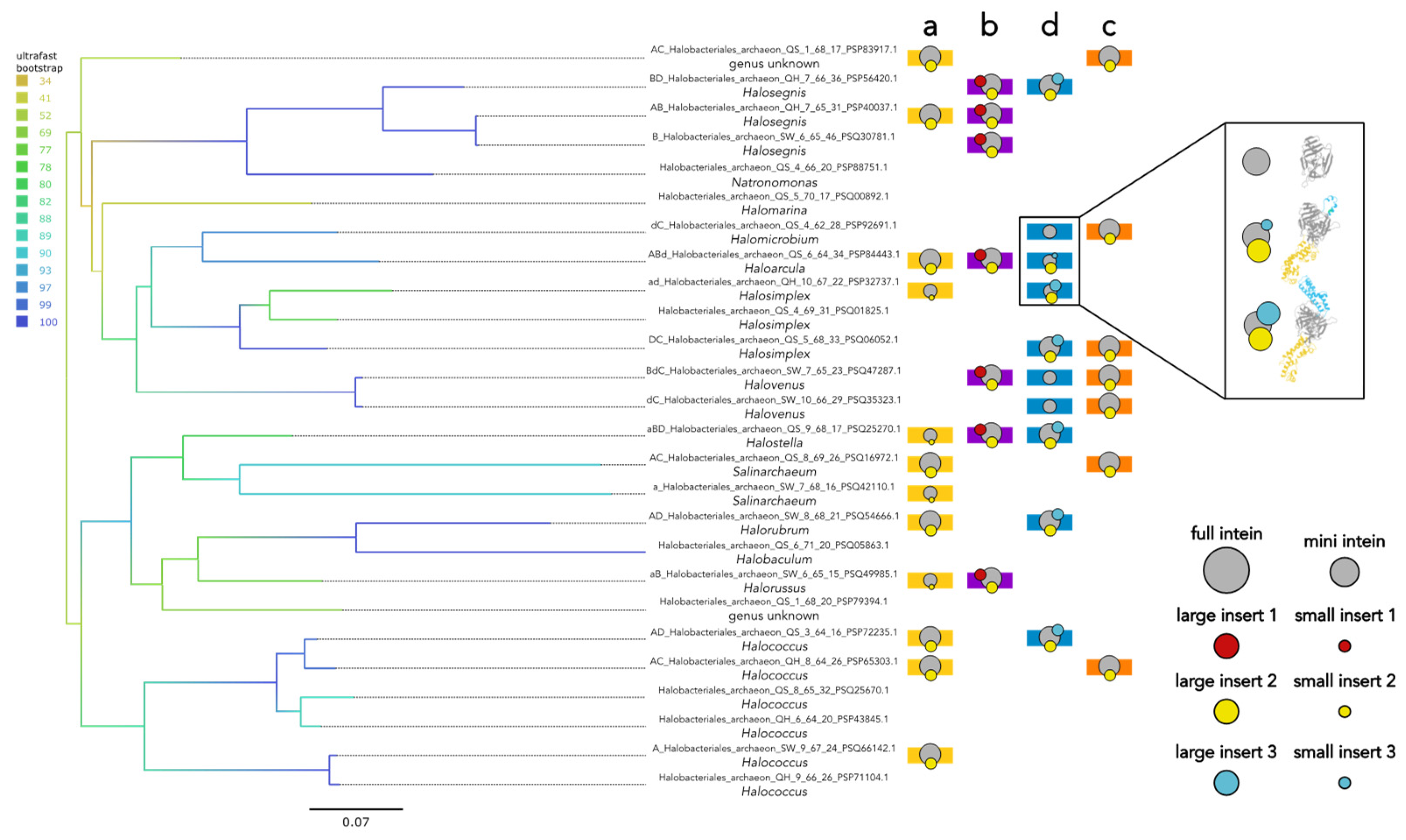
Geographically overlapping populations in Atacama Desert have a wide range of MCM intein architectures and invasion statuses including co-existing empty, mini intein, and full intein alleles. To investigate models of intein persistence which involve co-existence of intein-free, full-intein, and mini-intein alleles [
9], the haloarchaeal Atacama Desert sequences generated during the halite metagenome-based project of Finstad et al. [
18] were utilized. All samples were collected from three regions in the Atacama Desert in Chile during June of 2013. From their sequence data, we identified 26 complete haloarchaeal MCM homologs. While the sequences are classified only as Halobacteriales archaea through the Finstad et al. project, we were able to provide more certainty on the genus-level identities of 24/26 archaea by comparing to sequences in our dataset of haloarchaea with known genus-level identities (
Figure S4). By these classifications, these archaeal populations span the genera
Salinarchaeum, Natronomonas, Halovenus, Halostella, Halosimplex, Halosegnis, Halorussus, Halorubrum, Halomicrobium, Halomarina, Halococcus, Halobaculum, and
Haloarcula. Mapping the intein presence and architectures of these sequences onto a phylogeny of the MCM host proteins reveals a mixture of vertical inheritance and horizontal transfer, and varied intein architectures at a single site in closely related individuals (
Figure 5). All degrees of MCM intein invasion except quadruple (empty, single, double, and triple) are observed in the population, as well as mini and full inteins at sites MCM-a and d. The Atacama Desert sequences provide concrete evidence for the co-existence of empty, mini-intein, and full-intein alleles in geographically overlapping populations of archaea from a single time period (June 2013), with each insertion site exhibiting different degrees of balance between the three alleles owing to their unique evolutionary histories.
4. Discussion
Archaeal MCM is a powerful system for the continued exploration of multi-intein gene dynamics. Our work presents six previously unknown MCM intein insertion sites, and provides extensive characterization of the frequencies and architectures of inteins found at each site across archaea. We find the maximum degree for invasion of the
mcm gene to be four inteins, despite many archaeal groups having more than four active MCM intein insertion sites. Similar caps on intein invasion have been observed for multi-intein genes such as the archaeal gene
polB with up to three inteins simultaneously invading investigated copies of the gene [
11] and a bacterial ribonucleotide reductase gene with up to four inteins [
34]. Interestingly, the previous work in archaeal
polB revealed
Haloquadratum walsbyi to harbor the highest degree of intein invasion (triple), which is also the case for several strains of
Haloquadratum walsbyi analyzed in this study (invaded at sites MCM-a, b, c, and d) making this a species of interest for future intein fitness cost investigations. However, the highest degree of invasion is still ultimately the rarest configuration in both
polB and
mcm. The additional sites presented in this work offer new avenues for exploring biochemical and molecular dynamics between inteins co-inhabiting a gene. In addition to the new insertion sites, the range of intein architectures discovered opens avenues for further investigation of the biochemical versatility of inteins with additional, potentially DNA-binding, domains.
Potential origin and role of the within-intein insertions. Archaea utilize several small DNA-binding proteins for transcriptional regulation, with some even interacting directly with MCM [
35]. The core domain responsible for the DNA-binding abilities of these proteins is a helix-turn-helix domain [
25], to which the sub-insertions found within many of the MCM inteins presented in this work bear strong resemblance in predicted structures (
Figure 6). Thus, the pool of small DNA-binding helix-turn-helix proteins encoded in archaeal genomes could potentially be the source of some of the MCM intein sub-insertions. Within inteins, such additional domains have been observed in a region analogous to Insertion 1 in this work, at the end of the N-terminal portion of the self-splicing domain. In the crystal structure of the yeast vacuolar ATPase intein PI-
SceI (Protein Databank (PDB) entry 1LWS [
36]), such an insertion is present. Work preceding the solving of PDB 1LWS implicated this region in DNA recognition and binding [
37,
38], with the crystal structure confirming direct interaction between this region and the DNA target sequence [
36]. Thus, it is possible that the insertions within the MCM inteins are involved in the homing process, potentially in stabilizing the binding of the intein to its target DNA.
Atacama Desert archaea provide support for co-existence model of intein persistence. Several models have been proposed to explain the life cycles of inteins in populations, with more recent proposals expanding on the Goddard-Burt homing cycle to suggest the co-existence of the three alleles (intein-free, full intein-containing, and mini intein-containing) in populations as a means of intein persistence [
5,
9]. Intein alleles of geographically overlapping populations at a single timepoint had not yet been assessed, and the Atacama Desert samples investigated in this work provide for the first time a clear picture of the MCM intein dynamics in a group of geographically overlapping archaeal populations. In these archaea, there is a balance of empty, mini, and full alleles for sites MCM-a and d, and empty and full alleles for sites MCM-b and c (the other seven sites are inactive, which is true of all haloarchaea analyzed). Thus, the MCM inteins in these populations operate in a manner more in line with a co-existence model for intein persistence [
7,
8,
9], rather than the synchronized Goddard-Burt life cycle [
5]. Future investigations of intein dynamics within geographically overlapping populations will continue to shed light on the frequencies of co-existence versus synchronized progression modes for intein persistence in natural populations.
Author Contributions
Conceptualization, D.A. and J.P.G.; methodology, D.A., G.F.S., and J.P.G.; software, D.A., G.F.S., and J.P.G.; validation, D.A., G.F.S., and J.P.G.; formal analysis, D.A. and G.F.S.; investigation, D.A., G.F.S., and J.P.G.; resources, D.A., G.F.S., and J.P.G.; data curation, D.A. and G.F.S.; writing—original draft preparation, D.A., G.F.S., and J.P.G.; writing—review and editing, D.A., G.F.S., and J.P.G.; visualization, D.A.; supervision, D.A., G.F.S., and J.P.G.; project administration, J.P.G.; funding acquisition, J.P.G. All authors have read and agreed to the published version of the manuscript.