Submitted:
16 May 2025
Posted:
16 May 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
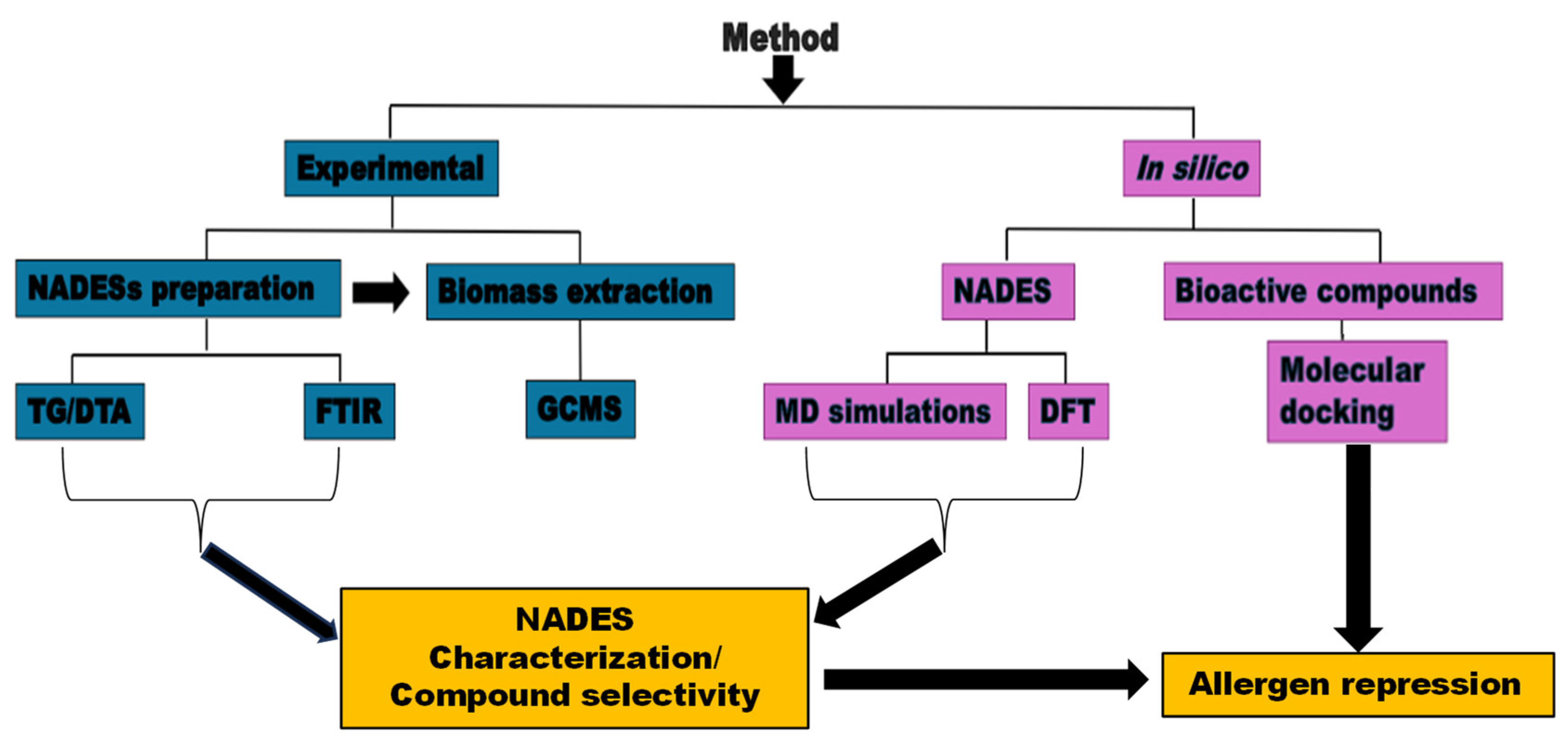
2.1. Experimental
2.1.1. Chemicals/ Reagents
2.1.2. Synthesis of the Natural Deep Eutectic Solvents
2.1.3. NADES Characterization
2.1.4. Ultrasonic Assisted Extraction Principle
2.1.5. Determination of Components of the Extract
2.2. In Silico Methods
2.2.1. Density Functional Theory Calculations
2.2.2. Molecular Dynamics Simulations
2.2.3. Molecular Docking Simulation
2.3. Statistical Analysis
3. Results and Discussion
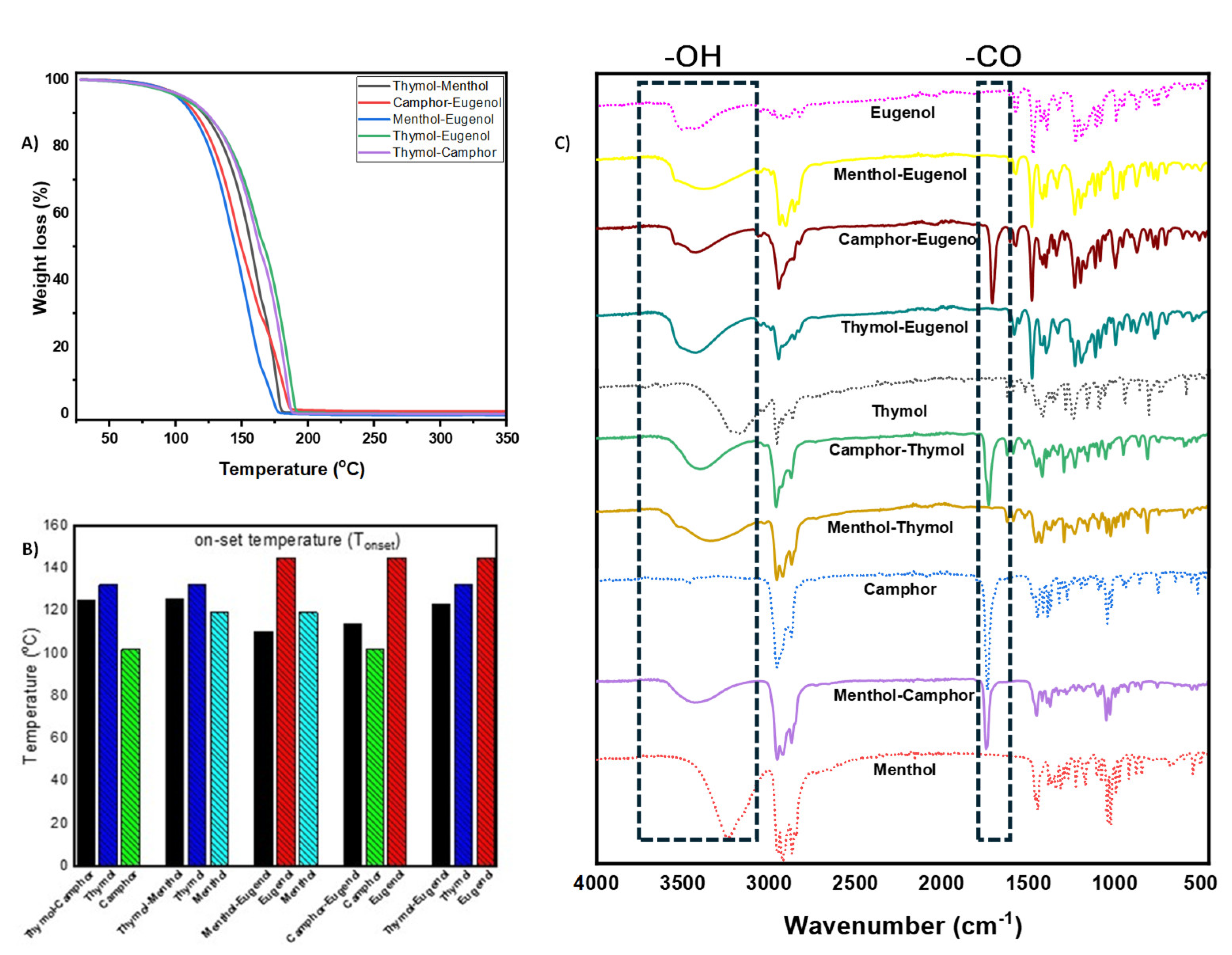
3.1. NADES Synthesis and Characterization
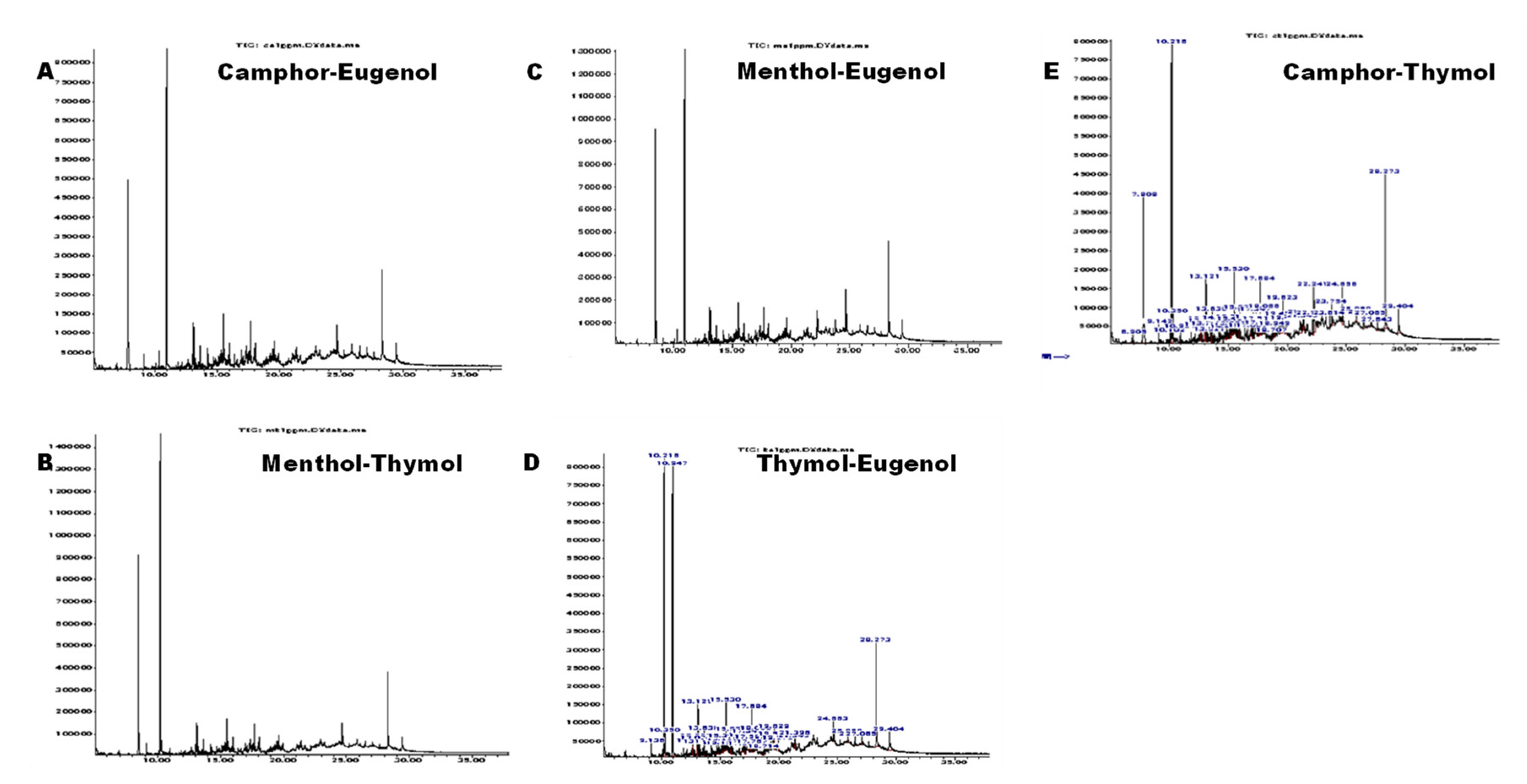
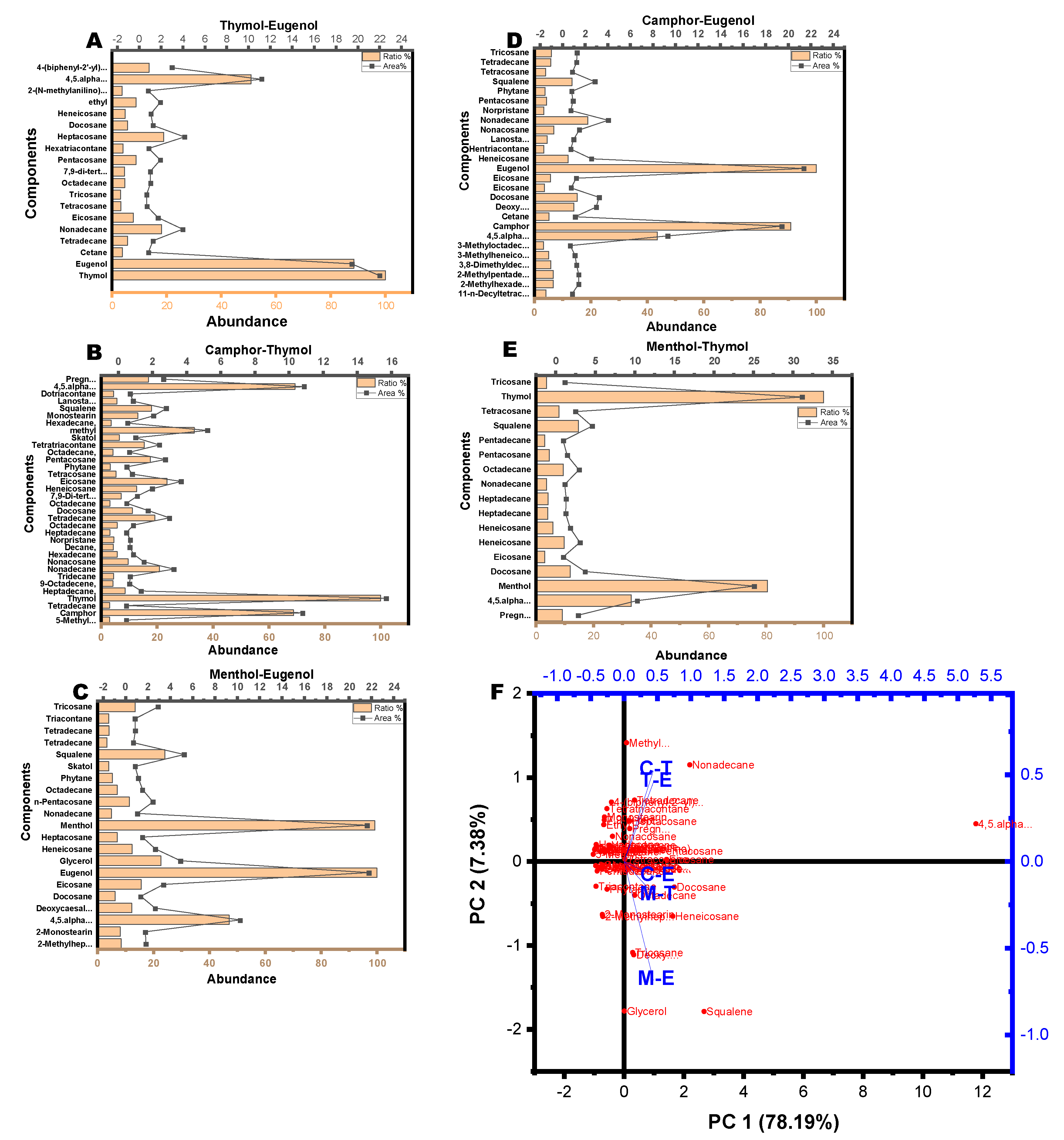
3.2. Phytochemical Composition
3.3. In Silico Simulation
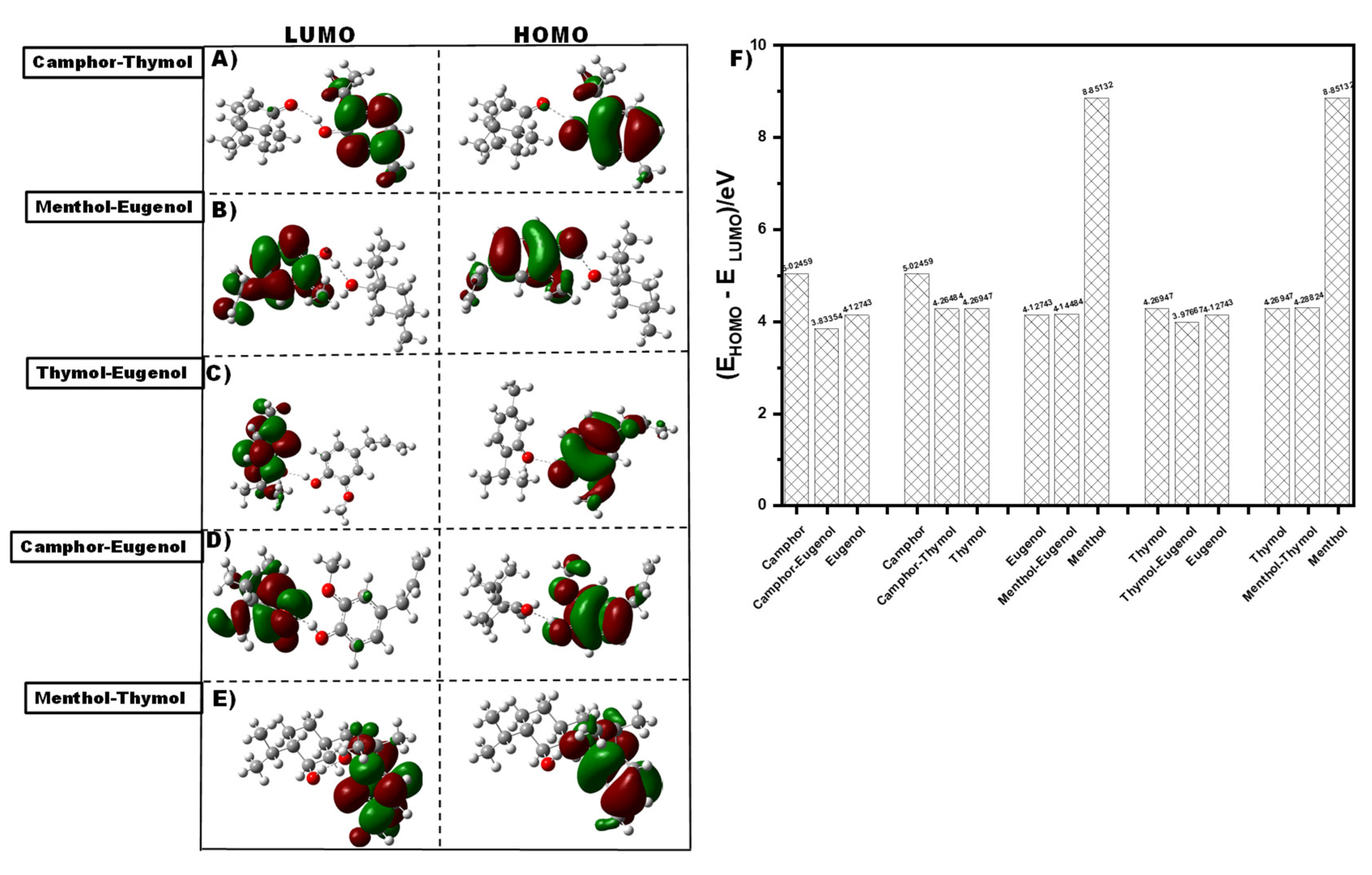
3.3.1. Density Functional Theory (DFT)
Frontier Molecular orbital Analysis
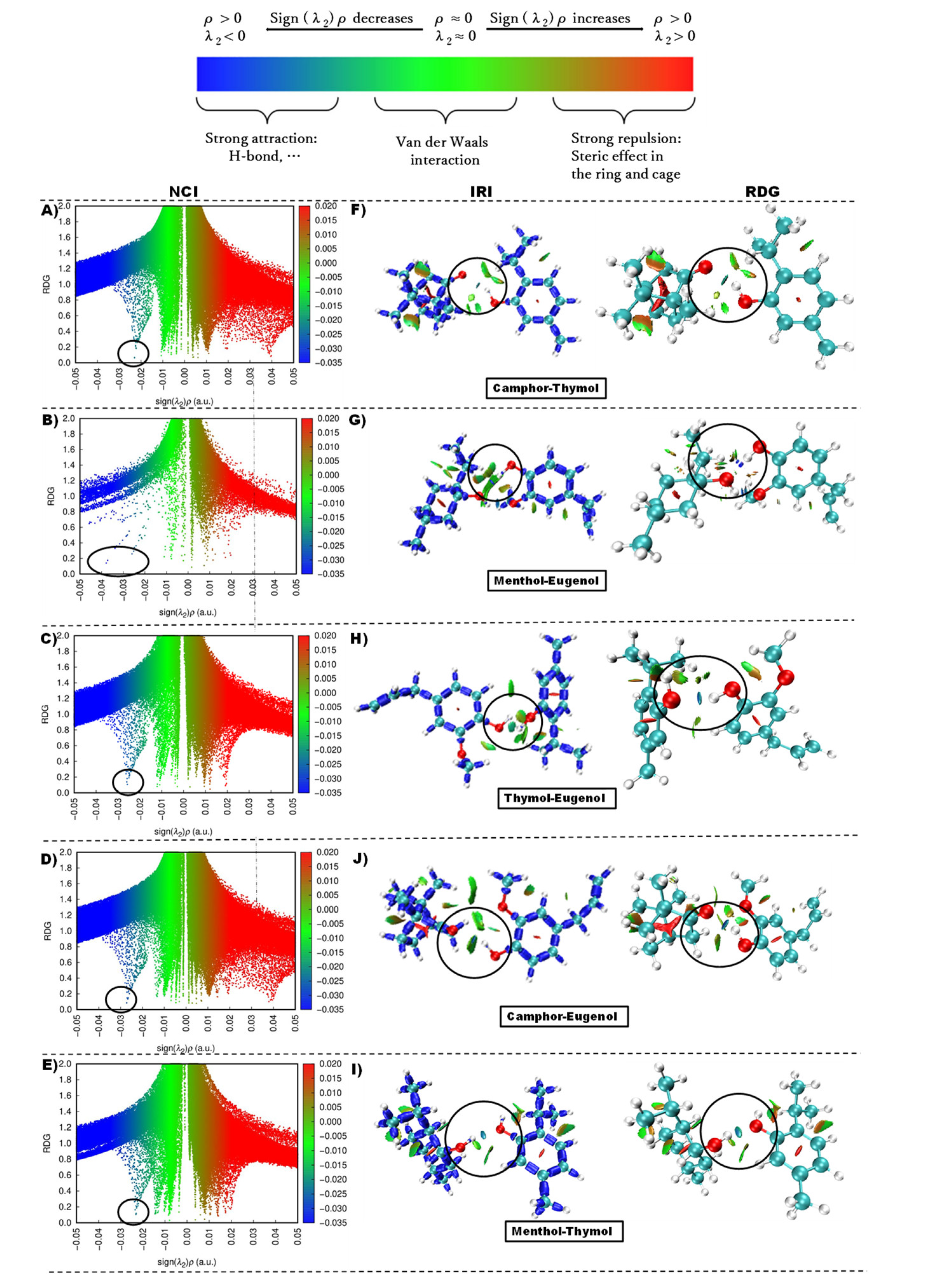
- Investigation of hydrogen bond interactions in the NADES system (QTAIM, NCI, and IRI analysis)
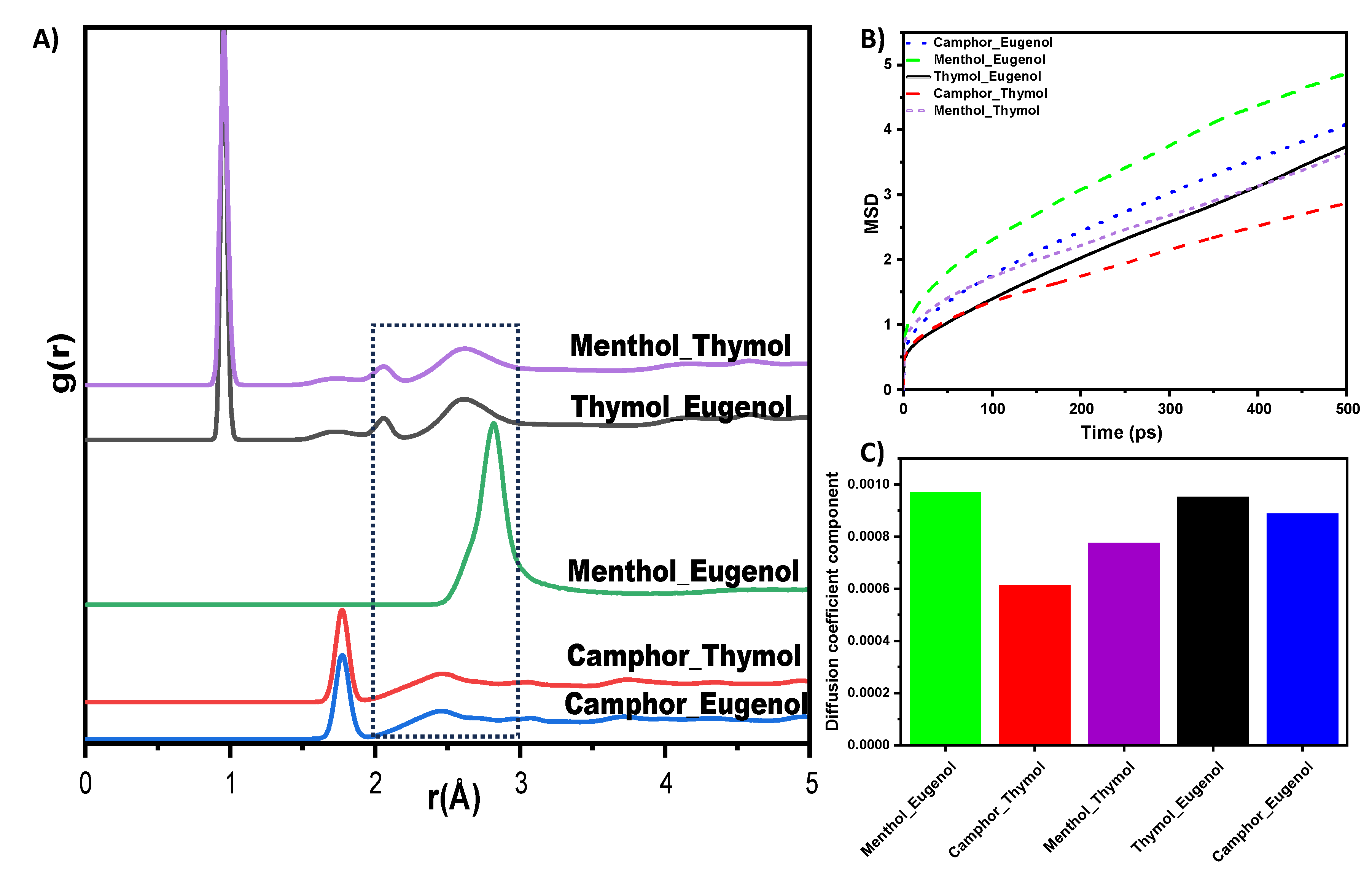
3.3.2. Molecular Dynamics Simulations of the NADES
Mean Square Displacement (MSD) and Diffusion Coefficient
Radial Distribution Functions (RDFs)
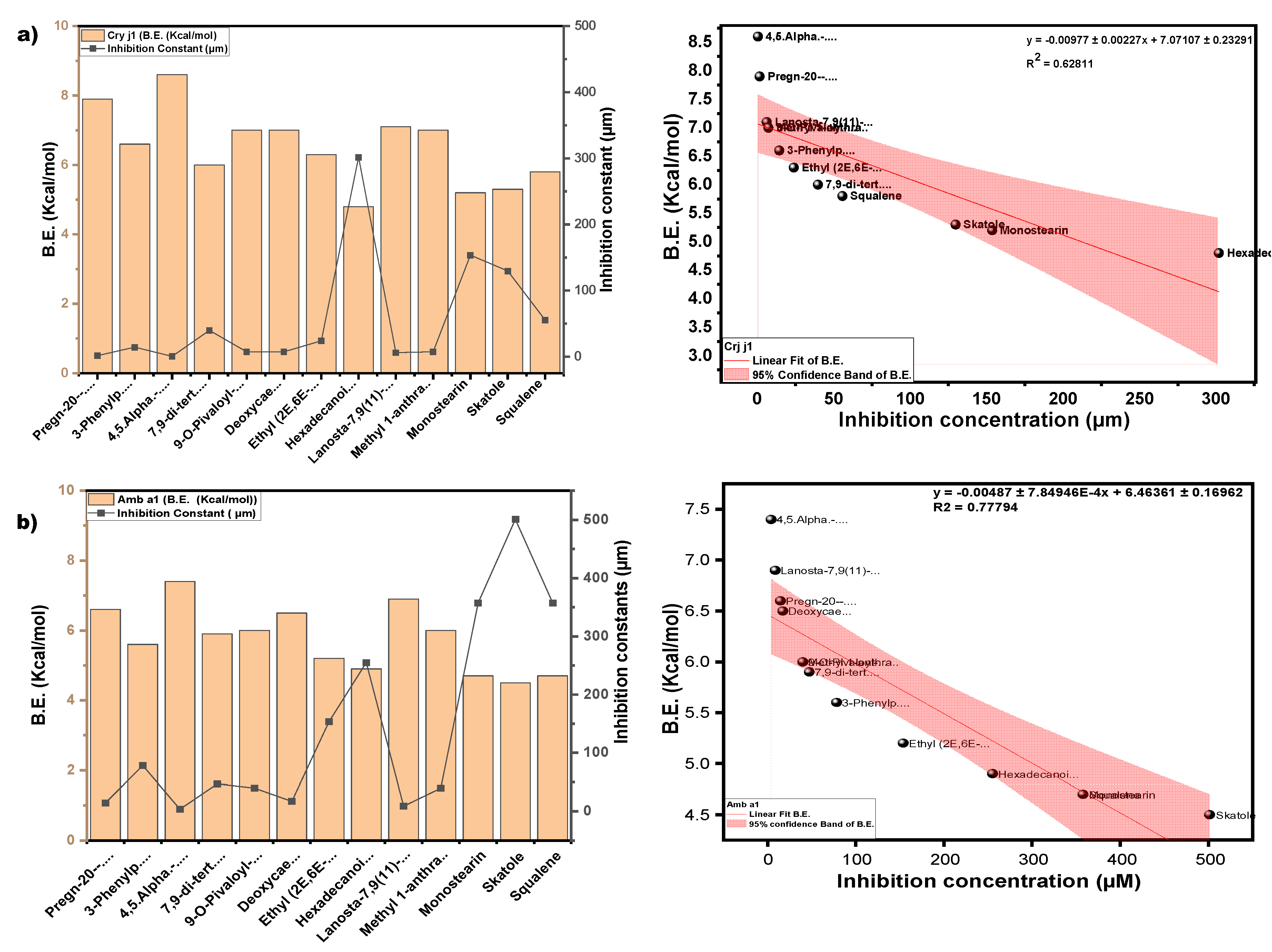
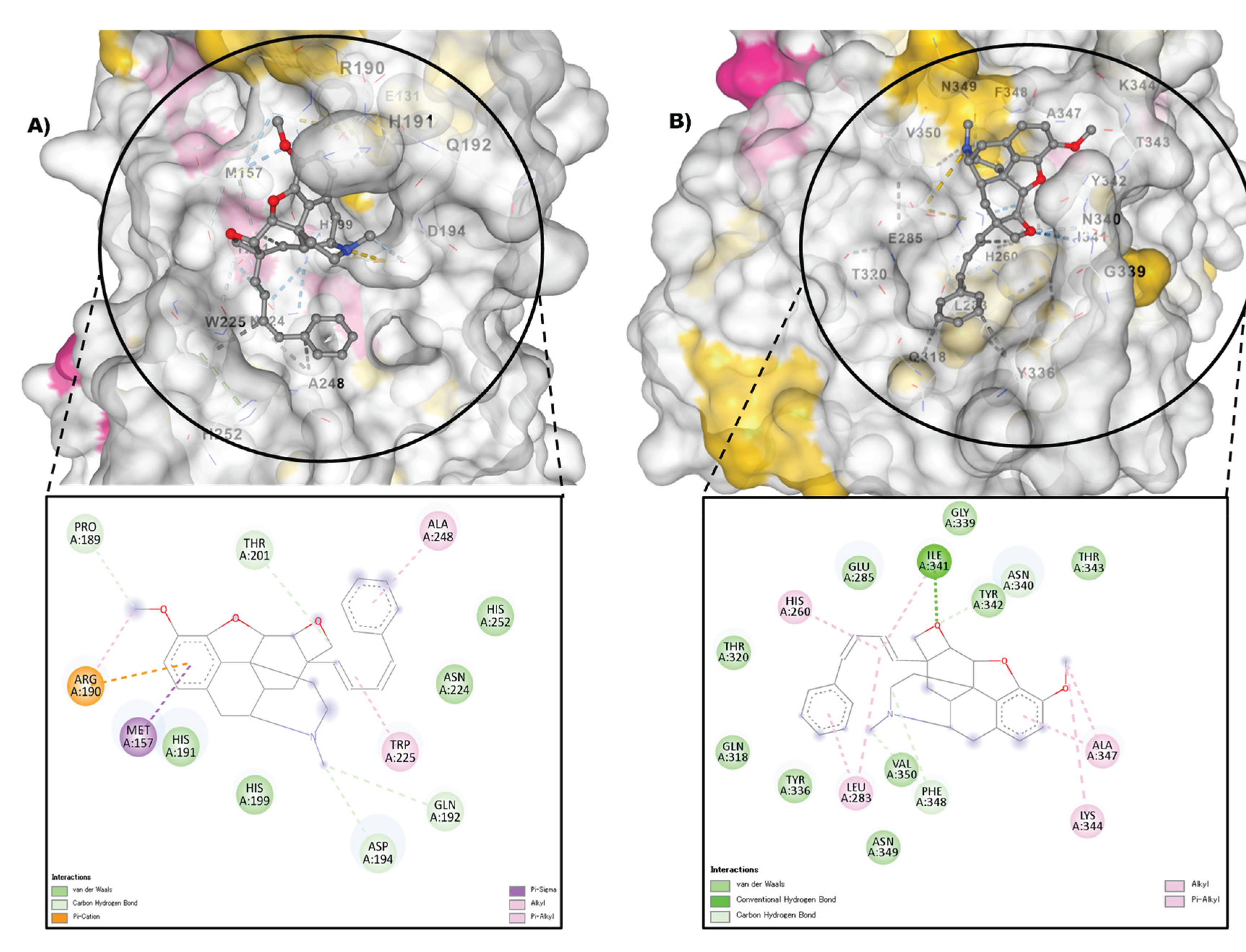
3.3.3. Molecular Docking Approach: Allergen repressing Potential of the Bioactive Compounds
| s/n | Compound | Structure | Pub chem id | Amb a 1 (B.E) Kcal/mol |
Cry j 1 (B.E) Kcal/mol |
Amb a 1 IC (µm) |
Cry j 1 (I.C) µm |
Cry j 1 Bonding: Interaction site | Amb a 1 Bonding: Interaction site |
| 1 | Pregn-20-en-3-ol,20-methyl-,(3β,5α)- |  |
22296144 | -6.6 | -7.9 | 14.44 | 1.61 | ALK: LEU 283 VDW: GLN 318, TYR 336, ILE 341, HIS 260, ASN 340, TYR 342, ALA 347, PHE 348, GLU 285, THR 320 |
CHB: ALA 368, LYS 77, ALA 78 VDW: GLY 74, GLY 369, PHE 75, GLY 229, THR 228, SER 204, ASP 205 |
| 2 | 3-Phenylpropionic acid,5-methoxy-2-[3,4-dimethoxyphenyl]- |  |
628993 | -5.6 | -6.6 | 78.18 | 14.44 | CVB: THR 320, SER 284, TYR 336 CHB: GLU 285 π-T: TYR 336 ALK/π-ALKLYS 335, LEU 283, ILE 341, TYR 342 VDW: ASN 340, GLN 318, GLU 285, SER 319 |
CHB: ASP 194, HIS 191, GLU 131 π-S: TRP 225, MET 157 π-π-T: HIS 199 ALK/π-ALK: ALA 248, ARG 190 VDW: ASN 224, THR 201, ASP 196, GLY 195, THR 155, SER 193 |
| 3 | 4,5.Alpha.-Epoxy-3-Methoxy-17-Methyl7.Alpha.-(4-Phenyl-1,3-Butadienyl)-6.Beta.,7.Beta.-(Oxymethylene)Morphinan |  |
- | -7.4 | -8.6 | 3.74 | 0.49 | CHB: GLY 339, GLY 338 CVB: THR 320 π-CAT: LYS 335 ALK/π-ALK:ILE 341, VAL 350, HIS 260 VDW: THR 342, PHE 348, GLY 285, LEU 283, GLN 318, TYR 336, ASN 340 |
CHB: ASP 194, GLN 192, THR 201 π-ALK: TRP 225, ALA 248 π-CAT: MET 157 VDW: HIS 191, HIS 199, ASN 224, HIS 253 |
| 4 | 7,9-di-tert-butyl-1-oxaspiro[4.5]deca-6,9-diene-2,8-dione |  |
545303 | -5.9 | -6.0 | 47.10 | 39.79 | CVB: ILE 341, TYR 342 ALK: LEU 283 VDW: HIS 260, GLU 285, THR 320, ASN 340, GLY 339, GLY 338, TYR 336 |
CVB: THR 201 π-ALK: ALA 248, TRP 225 VDW: ASN 158, MET 157, HIS 199, ASN 224, ASP 218, ASP 194, ASP 196, LEU 245 |
| 5 | 9-O-Pivaloyl-N-acetylcolchinol | 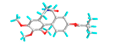 |
634928 | -6.0 | -7.0 | 39.79 | 7.35 | CVB: GLU 285 CHB: PHE 348 π-π-T: HIS 260, VAL 350, ILE 341, LEU 283 VDW: TYR 342, ASN 340, TYR 336, GLN 318, LYS 335, GLN 321, THR 320 |
CVB: THR 201 π-π: TRP 225 π-ALK: ALA 248 |
| 6 | Deoxycaesaldekarin C | 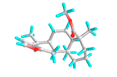 |
572829 | -6.5 | -7.0 | 17.10 | 7.35 | CVB: THR 320, TYR 342 ALK/π-ALK: ALA 347, LEU 283, ILE 341 VDW: GLU 285, GLN 318, TYR 336, ASN 340 |
ALK: ALA 78, LYS 77, ALA 368 π-AN: ASP 205 VDW: GLY 74, GLY 369, THR 228, HIS 259, GLY 229, PHE 75, GLU 365 |
| 7 | Ethyl (2E,6E,10E)-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraenoate | 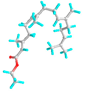 |
5366011 | -5.2 | -6.3 | 153.61 | 23.97 | CVB: LYS 132, GLN 169 ALK/π-ALK: HIS 167, PHE 130, VAL 134, PHE 161, ALA 108, ILE 157, ARG 133 VDW: ASN 158, GLU 164, PRO 165, TYR 106, VAL 166, ARG 177 |
CHB: LYS 77, GLN 309 π-ALK: ALA 78, PHE 75, LEU 362, ARG 341, PRO 360 VDW: GLY 74, ALA 368, GLU 365, GLY 228, ASP 204, HIS 80 |
| 8 | Hexadecanoic acid, 1-(hydroxymethyl)-1,2-ethanediyl | 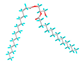 |
99931 | -4.9 | -4.8 | 254.94 | 301.84 | CVB: ILE 341, TYR 342 ALK/π-ALK: LEU 283, TYR 336 VDW: LEU 283, TYR 336 GLN 318, ALA 347, VAL 350, THR 320, GLY 339, ASN 340, GLN 285, PHE 348 |
CVB: GLN 309 (ester and OH) ALK: LEU 362, LEU 307, PRO 360, ARG 341 VDW: GLN 284, ALA 373, MET 371, GLN 372, MET 370, ASP 359 |
| 9 | Lanosta-7,9(11)-diene-3,18,20-triol | 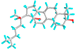 |
91695604 | -6.9 | -7.1 | 8.70 | 6.21 | CVB: PHE 348 UDD: PHE 348 ALK/π-ALK: LEU 283, TYR 336 VDW: GLN 318, SER 284, GLU 285, THR 320, GLY 338, ILE 341, GLY 339, ASN 340, TYR 342, ASN 349, ALA 347 |
CHB: SER 204 ALK: LYS 257 AC-AC: ASP 205 VDW: HIS 259, GLU 365, THR 228, GLY 229, PHE 75, ALA 78, GLY 369, ALA 368 |
| 10 | Methyl 1-anthraquinonesulfenate | 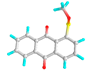 |
349031192 | -6.0 | -7.0 | 39.79 | 7.35 | CVB: ILE 341 ALK/π-ALK: LEU 283, ILE 341 π-HB: THR 320 VDW: GLN 318, SER 284, HIS 260, GLU 285, TYR 342, ASN 340, GLY 339, TYR 336 |
CHB: GLN 309, LEU 362 π-S: LEU 307 π-ALK: ARG 341, PRO 360 VDW: ASP 359, GLN 343, MET 370, ASN 367, VAL 361 |
| 11 | Monostearin | 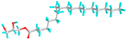 |
24699 | -4.7 | -5.2 | 357.37 | 153.61 | CVB: GLU 285, VAL 350 CHB: HIS 260 VDW: ASN 340, GLY 339, TYR 342, GLN 321, GLN 318, THR 320, ASN 349 ALK/π-ALK: ILE 341, LEU 283, TYR 336 |
CVB: THR 155, ASP 194, HIS 199 ALK/π-ALK: MET 157, TRP 225 VDW: GLY 195, GLY 369, GLU 365, THR 228 |
| 12 | Skatole | 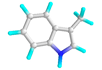 |
6736 | -4.5 | -5.3 | 500.95 | 129.74 | π-CAT: LYS 132 VDW: ARG 133, GLU 164 π-S: ILE 157 π-π-T: PHE 161 ALK/π-ALK: VAL 166, TYR 106 |
CHB: ASP 205 π-ALK: ALA 78, ALA 368, PHE 75, HIS 259 VDW: GLY 369, GLY 229, THR 228, GLU 365 |
| 13 | Squalene |  |
638072 | -4.7 | -5.8 | 357.37 | 55.77 | ALK/π-ALK: TYR 336, LEU 283, ILE 341, TYR 342, ALA 347 VDW: GLN 318, GLY 339, THR 320, PHE 348, VAL 350, ASN 340, GLU 285, HIS 260 |
ALK: LEU 307, PRO 360, MET 370, LEU 362, ARG 341, ALA 373 VDW: MET 371, GLN 372, GLN 309, ASP 359, GLN 343 |
4. Conclusion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bader R F W 1990 In Atoms in Molecules: A Quantum Theory (Oxford: Clarendon Press); (b) Bader R F W 1985 Acc. Chem. Res. 18 9; (c) Bader R F W 1991 Chem. Rev. 91 893.
- Bader, R F W; Johnson, S; Tang, H T; Popelier, P L A. Phys. Chem. 1996; 100, 15398.
- Bader, R. F.; Essén, H. The characterization of atomic interactions. The Journal of chemical physics 1984, 80(5), 1943–1960. [Google Scholar] [CrossRef]
- Bitwell, C.; Indra, S. S.; Luke, C.; Kakoma, M. K. A review of modern and conventional extraction techniques and their applications for extracting phytochemicals from plants. Scientific African 2023, 19, e01585. [Google Scholar] [CrossRef]
- Bordas-Le Floch, V.; Groeme, R.; Chabre, H.; Baron-Bodo, V.; Nony, E.; Mascarell, L.; Moingeon, P. New insights into ragweed pollen allergens. Current allergy and asthma reports 2015, 15(11), 63. [Google Scholar] [CrossRef] [PubMed]
- Boudesocque-Delaye, L.; Ardeza, I. M.; Verger, A.; Grard, R.; Théry-Koné, I.; Perse, X.; Munnier, E. Natural Deep Eutectic Solvents as a Novel Bio-Based Matrix for Ready-to-Use Natural Antioxidants-Enriched Ingredients: Extraction and Formulation Optimization. Cosmetics 2024, 11(1), 17. [Google Scholar] [CrossRef]
- Bubalo, M. C.; Vidović, S.; Redovniković, I. R.; Jokić, S. New perspective in extraction of plant biologically active compounds by green solvents. Food and Bioproducts Processing 2018, 109, 52–73. [Google Scholar] [CrossRef]
- Choi, Y. H.; van Spronsen, J.; Dai, Y.; Verberne, M.; Hollmann, F.; Arends, I. W.; Verpoorte, R. Are natural deep eutectic solvents the missing link in understanding cellular metabolism and physiology? Plant physiology 2011, 156(4), 1701–1705. [Google Scholar] [CrossRef]
- Dai, Y.; Van Spronsen, J.; Witkamp, G. J.; Verpoorte, R.; Choi, Y. H. Natural deep eutectic solvents as new potential media for green technology. Analytica chimica acta 2013, 766, 61–68. [Google Scholar] [CrossRef]
- Dávila, M. J.; Alcalde, R.; Atilhan, M.; Aparicio, S.; PρT measurements and derived properties of liquid 1-alkanols; Hoover, W.G. Canonical dynamics: equilibrium phase-space distributions. The Journal of Chemical Thermodynamics Phys. Rev. A 2012, 47 31, 241–259 1695. [Google Scholar] [CrossRef]
- Emamian, S.; Lu, T.; Kruse, H.; Emamian, H. Exploring nature and predicting strength of hydrogen bonds: a correlation analysis between atoms-in-molecules descriptors, binding energies, and energy components of symmetry-adapted perturbation theory. Journal of computational chemistry 2019, 40(32), 2868–2881. [Google Scholar] [CrossRef]
- Enyoh, C. E.; Wang, Q.; Wang, W.; Chowdhury, T.; Rabin, M. H.; Islam, R.; Xiao, K. Sorption of per-and polyfluoroalkyl substances (PFAS) using Polyethylene (PE) microplastics as adsorbent: Grand canonical Monte Carlo and molecular dynamics (GCMC-MD) studies. International Journal of Environmental Analytical Chemistry 2024, 104(12), 2719–2735. [Google Scholar] [CrossRef]
- Enyoh, C.E.; Wang, Qingyue; Wang, Weiqian; Chowdhury, Tanzin; Rabin, Mominul Haque; Islam; Md Rezwanul, Yue; Guo, Yichun; Lin; Xiao, Kai. Sorption of per-and polyfluoroalkyl substances (PFAS) using polyethylene (PE) microplastics as adsorbent: grand canonical Monte Carlo and molecular dynamics (GCMC-MD) studies. Int. J. Environ. Anal. Chem. 2022b. [Google Scholar] [CrossRef]
- Enyoh, CE; Wang, Q.; Ovuoraye, PE; Maduka, TO. Toxicity evaluation of microplastics to aquatic organisms through molecular simulations and fractional factorial designs. Chemosphere 2022, 308, 136342. [Google Scholar] [CrossRef] [PubMed]
- Escobar, A.; Perez, M.; Romanelli, G.; Blustein, G. Thymol bioactivity: A review focusing on practical applications. Arabian Journal of Chemistry 2020, 13(12), 9243–9269. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M. L.; Darden, T.; Lee, H.; Pedersen, L. G. A smooth particle mesh Ewald method. The Journal of chemical physics 1995, 103(19), 8577–8593. [Google Scholar] [CrossRef]
- Fan, C.; Liu, Y.; Sebbah, T.; Cao, X. A theoretical study on terpene-based natural deep eutectic solvent: relationship between viscosity and hydrogen-bonding interactions. Global Challenges 2021, 5(3), 2000103. [Google Scholar] [CrossRef]
- Fan, C.; Liu, Y.; Sebbah, T.; Cao, X. A theoretical study on terpene-based natural deep eutectic solvent: relationship between viscosity and hydrogen-bonding interactions. Global Challenges 2021, 5(3), 2000103. [Google Scholar] [CrossRef]
- Farooq, M. Q.; Odugbesi, G. A.; Abbasi, N. M.; Anderson, J. L. Elucidating the role of hydrogen bond donor and acceptor on solvation in deep eutectic solvents formed by ammonium/phosphonium salts and carboxylic acids. ACS Sustainable Chemistry & Engineering 2020, 8(49), 18286–18296. [Google Scholar]
- Fattahi, N.; Shamsipur, M.; Nematifar, Z.; Babajani, N.; Moradi, M.; Soltani, S.; Akbari, S. Novel deep eutectic solvent-based liquid phase microextraction for the extraction of estrogenic compounds from environmental samples. RSC advances 2022, 12(23), 14467–14476. [Google Scholar] [CrossRef]
- Fourmentin, S.; Gomes, M. C.; Lichtfouse, E. (Eds.) Deep eutectic solvents for medicine, gas solubilization and extraction of natural substances; Springer Nature, 2020; Vol. 56. [Google Scholar]
- Frisch, M. J.; Trucks, G. W.; Schlegel, H. B.; Scuseria, G. E.; Robb, M. A.; Cheeseman, J. R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G. A.; Nakatsuji, H.; Caricato, M.; Li, X.; Hratchian, H. P.; Izmaylov, A. F.; Bloino, J.; Zheng, G.; Sonnenberg, J. L.; Hada, M.; Fox, D. J. Gaussian 09, Revision D.01; Gaussian, Inc, 2013. [Google Scholar]
- Fujimura, T.; Fujinami, K.; Ishikawa, R.; Tateno, M.; Tahara, Y.; Okumura, Y.; Ohta, H.; Miyazaki, H.; Taniguchi, M. Recombinant Fusion Allergens, Cry j 1 and Cry j 2 from Japanese Cedar Pollen, Conjugated with Polyethylene Glycol Potentiate the Attenuation of Cry j 1-Specific IgE Production in Cry j 1-Sensitized Mice and Japanese Cedar Pollen Allergen-Sensitized Monkeys. International archives of allergy and immunology 2015, 168(1), 32–43. [Google Scholar] [CrossRef]
- Fuster, F.; Sevin, A.; Silvi, B. Topological analysis of the electron localization function (ELF) applied to the electrophilic aromatic substitution E.C. Ihmels, Jr Gmehling, Densities of toluene, carbon dioxide, carbonyl sulfide, and hydrogen sulfide over a wide temperature and pressure range in the sub-and supercritical state. The Journal of Physical Chemistry A Ind. Eng. Chem. Res. 2000, 104((4) 40), 852-858 4470–4477. [Google Scholar]
- García-Roldán, A.; Piriou, L.; Jauregi, P. Natural deep eutectic solvents as a green extraction of polyphenols from spent coffee ground with enhanced bioactivities. Frontiers in Plant Science 2023, 13, 1072592. [Google Scholar] [CrossRef] [PubMed]
- Goswami, A.; Rahman, S. N. R.; Sree, A.; Shunmugaperumal, T. Solubility of cinnarizine in natural deep eutectic solvent (camphor+ menthol) and correlation with different solubility models. Fluid Phase Equilibria 2024, 578, 114008. [Google Scholar] [CrossRef]
- Hanwell, M. D.; Curtis, D. E.; Lonie, D. C.; Vandermeersch, T.; Zurek, E.; Hutchison, G. R. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform R.F.W. Bader, Atoms in molecules. Journal of cheminformatics Acc. Chem. Res. 2012, 4 18, 1-17 9–15. [Google Scholar]
- Huang, M. M.; Yiin, C. L.; Lock, S. S. M.; Chin, B. L. F.; Othman, I.; Chan, Y. H. Natural deep eutectic solvents (NADES) for sustainable extraction of bioactive compounds from medicinal plants: Recent advances, challenges, and future directions. Journal of Molecular Liquids 2025, 127202. [Google Scholar] [CrossRef]
- Ikpa, C. C. B.; Maduka, T. O. D. Antimicrobial Properties of Methanol Extract of Dacryodes edulis Seed and Determination of Phytochemical Composition Using FTIR and GCMS. Chemistry Africa 2020, 3(4), 927–935. [Google Scholar] [CrossRef]
- Iman, M.; et al. “Computational prediction of drug binding energies and pharmacological properties.”. Journal of Molecular Modeling 2015, 21(6), 1–12. [Google Scholar]
- Ishaq, M.; Gilani, M. A.; Ahmad, F.; Afzal, Z. M.; Arshad, I.; Bilad, M. R.; Khan, A. L. Theoretical and experimental investigation of CO2 capture through choline chloride based supported deep eutectic liquid membranes. Journal of Molecular Liquids 2021, 335, 116234. [Google Scholar] [CrossRef]
- Jangir, A. K.; Mandviwala, H.; Patel, P.; Sharma, S.; Kuperkar, K. Acumen into the effect of alcohols on choline chloride: L-lactic acid-based natural deep eutectic solvent (NADES): A spectral investigation unified with theoretical and thermophysical characterization. Journal of Molecular Liquids 2020, 317, 113923. [Google Scholar] [CrossRef]
- Javeed, A.; Sarfraz, M.; Bhutta, N. K.; Han, B. Alkaloids as natural anti-allergy agents: A mini review. Allergy Medicine 2024, 100014. [Google Scholar] [CrossRef]
- Johnson, E. R.; Keinan, S.; Mori-Sánchez, P.; Contreras-García, J.; Cohen, A. J.; Yang, W. Revealing noncovalent interactions. Journal of the American Chemical Society 2010, 132(18), 6498–6506. [Google Scholar] [CrossRef]
- Kalyniukova, A.; Holuša, J.; Musiolek, D.; Sedlakova-Kadukova, J.; Płotka-Wasylka, J.; Andruch, V. Application of deep eutectic solvents for separation and determination of bioactive compounds in medicinal plants. Industrial Crops and Products 2021, 172, 114047. [Google Scholar] [CrossRef]
- Kamatou, G. P.; Vermaak, I.; Viljoen, A. M.; Lawrence, B. M. Menthol: A simple monoterpene with remarkable biological properties. Phytochemistry 2013, 96, 15–25. [Google Scholar] [CrossRef]
- Khan, P.; Jamshaid, M.; Tabassum, S.; Perveen, S.; Mahmood, T.; Ayub, K.; Gilani, M. A. Exploring the interaction of ionic liquids with Al12N12 and Al12P12 nanocages for better electrode-electrolyte materials in super capacitors. Journal of Molecular Liquids 2021, 344, 117828. [Google Scholar] [CrossRef]
- Kianpoor, A.; Sadeghi, R. Novel hydrophobic acetanilide-based deep eutectic solvents. Preparation, properties, their applications in liquid–liquid extraction. Chemical Engineering Science 2024, 288, 119866. [Google Scholar] [CrossRef]
- Kim, C. H.; Lee, T.; Oh, I.; Nam, K. W.; Kim, K. H.; Oh, K. B.; Mar, W. Mast cell stabilizing effect of (−)-Elema-1, 3, 11 (13)-trien-12-ol and Thujopsene from Thujopsis dolabrata is mediated by Down-regulation of Interleukin-4 secretion in antigen-induced RBL-2H3 cells. Biological and Pharmaceutical Bulletin 2013, 36(3), 339–345. [Google Scholar] [CrossRef]
- Klix, C. L.; Maret, G.; Keim, P. Discontinuous shear modulus determines the glass transition temperature. Physical Review X 2015, 5(4), 041033. [Google Scholar] [CrossRef]
- Kumar, P. S. V.; Raghavendra, V.; Subramanian, V. Bader’s theory of atoms in molecules (AIM) and its applications to chemical bonding. Journal of Chemical Sciences 2016, 128, 1527–1536. [Google Scholar] [CrossRef]
- Kusano, S.; Kukimoto-Niino, M.; Satta, Y.; Ohsawa, N.; Uchikubo-Kamo, T.; Wakiyama, M.; Ikeda, M.; Terada, T.; Yamamoto, K.; Nishimura, Y.; Shirouzu, M.; Sasazuki, T.; Yokoyama, S. Structural basis for the specific recognition of the major antigenic peptide from the Japanese cedar pollen allergen Cry j 1 by HLA-DP5. Journal of molecular biology 2014, 426(17), 3016–3027. [Google Scholar] [CrossRef]
- Kusano, S.; Ueda, S.; Oryoji, D.; Toyoumi, A.; Hashimoto-Tane, A.; Kishi, H.; Yokoyama, S. Contributions of the N-terminal flanking residues of an antigenic peptide from the Japanese cedar pollen allergen Cry j 1 to the T-cell activation by HLA-DP5. International immunology 2023, 35(9), 447–458. [Google Scholar] [CrossRef]
- Lashgari, A.; Ghamami, S.; Govindarajan, M.; Salgado-Morán, G.; Montes Romero, P.; Gerli Candia, L. A theoretical quantum study of the electronic properties of mentoxy dichloro phosphorous (C10H19OPCl2). Journal of the Chilean Chemical Society 2018, 63(1), 3887–3897. [Google Scholar] [CrossRef]
- Lin, Y.; Xiao, K.; Wang, Q.; Lu, S.; Wang, W.; Seguchi, A. Research on Repressing Allergen Cry j 1 Released from Japanese Cedar Pollen Using Todomatsu Oil. Atmosphere 2023, 14(6), 991. [Google Scholar] [CrossRef]
- Lin, Y.; Xiao, K.; Wang, W.; Lu, S. ) Study on Lowering the Group 1 Protease Allergens from House Dust Mites by Exposing toTodomatsu Oil Atmosphere. Atmosphere 2023b, 14, 548. [Google Scholar] [CrossRef]
- Liu, Y.; Friesen, J. B.; McAlpine, J. B.; Lankin, D. C.; Chen, S. N.; Pauli, G. F. Natural deep eutectic solvents: properties, applications, and perspectives. Journal of natural products 2018, 81(3), 679–690. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, A. R.; Scheurer, S.; Vieths, S. Food allergens: molecular and immunological aspects, allergen databases and cross-reactivity. Chemical immunology and allergy 2015, 101, 18–29. [Google Scholar] [CrossRef]
- Lu, T.; Chen, Q. Interaction region indicator: a simple real space function clearly revealing both chemical bonds and weak interactions. Chemistry-Methods 2021, 1(5), 231–239. [Google Scholar] [CrossRef]
- Lu, T.; Chen, F. Multiwfn: A multifunctional wavefunction analyzer. Journal of computational chemistry 2012, 33(5), 580–592. [Google Scholar] [CrossRef]
- Mahdy; AHS; Zayed; SE; Abo-Bakr, AM; Hassan, EA. Camphor: Synthesis, reactions and uses as a potential moiety in the development of complexes and organocatalysts. Tetrahedron 2022, 121, 132913. [Google Scholar] [CrossRef]
- Martins, M. A. R.; Pinho, S. P.; Coutinho, J. A. P. Insights into the nature of eutectic and deep eutectic mixtures. Journal of Solution Chemistry 2018. [Google Scholar] [CrossRef]
- Martins, M. A.; Silva, L. P.; Schaeffer, N.; Abranches, D. O.; Maximo, G. J.; Pinho, S. P.; Coutinho, J. A. Greener terpene–terpene eutectic mixtures as hydrophobic solvents. ACS sustainable chemistry & engineering 2019, 7(20), 17414–17423. [Google Scholar]
- Martins, M. A.; Silva, L. P.; Schaeffer, N.; Abranches, D. O.; Maximo, G. J.; Pinho, S. P.; Coutinho, J. A. Greener terpene–terpene eutectic mixtures as hydrophobic solvents. ACS sustainable chemistry & engineering 2019, 7(20), 17414–17423. [Google Scholar]
- Naseem, Z.; Shehzad, R. A.; Jabeen, S.; Tahir, S.; Mushtaq, F.; Zahid, M.; Iqbal, J. Quantum chemical investigation of choline chloride-based deep eutectic solvents. Chemical Physics 2023, 571, 111936. [Google Scholar] [CrossRef]
- Naseem, Z.; Shehzad, RA; Jabeen, S.; Tahir, S.; Mushtaq, F.; Zahid, M.; Iqbal, J. Quantum chemical investigation of choline chloride-based deep eutectic solvents. Chemical Physics 2023, 571, 111936. [Google Scholar] [CrossRef]
- Nejad, S. M.; Özgüneş, H.; Başaran, N. Pharmacological and toxicological properties of eugenol. Turkish journal of pharmaceutical sciences 2017, 14(2). [Google Scholar]
- Nisar, M. F.; Khadim, M.; Rafiq, M.; Chen, J.; Yang, Y.; Wan, C. C. Pharmacological properties and health benefits of eugenol: A comprehensive review. Oxidative medicine and cellular longevity 2021, 2021(1), 2497354. [Google Scholar] [CrossRef] [PubMed]
- Nortjie, E.; Basitere, M.; Moyo, D.; Nyamukamba, P. Extraction methods, quantitative and qualitative phytochemical screening of medicinal plants for antimicrobial textiles: a review. Plants 2022, 11(15). [Google Scholar] [CrossRef] [PubMed]
- Omar, K. A.; Sadeghi, R. Database of deep eutectic solvents and their physical properties: A review. Journal of Molecular Liquids 2023, 384, 121899. [Google Scholar] [CrossRef]
- Oppong, E.; Flink, N.; Cato, A. C. Molecular mechanisms of glucocorticoid action in mast cells. Molecular and cellular endocrinology 2013, 380(1-2), 119–126. [Google Scholar] [CrossRef]
- Osiecka, D.; Vakh, C.; Makoś-Chełstowska, P.; Kubica, P. Plant-based meat substitute analysis using microextraction with deep eutectic solvent followed by LC-MS/MS to determine acrylamide, 5-hydroxymethylfurfural and furaneol. Analytical and Bioanalytical Chemistry 2024, 416(5), 1117–1126. [Google Scholar] [CrossRef]
- Padilla, N.; Delso, I.; Bergua, F.; Lafuente, C.; Artal, M. Characterization of camphor: Thymol or dl-menthol eutectic mixtures; Structure, thermophysical properties, and lidocaine solubility. Journal of Molecular Liquids 2024, 405, 125069. [Google Scholar] [CrossRef]
- Pádua, AA; Costa Gomes, MF; Canongia Lopes, JN. Molecular solutes in ionic liquids: a structural perspective. Accounts of chemical research 2007, 40(11), 1087–1096. [Google Scholar] [CrossRef]
- Paiva, A.; Craveiro, R.; Aroso, I.; Martins, M.; Reis, R. L.; Duarte, A. R. C. Natural deep eutectic solvents–solvents for the 21st century. ACS Sustainable Chemistry & Engineering 2014, 2(5), 1063–1071. [Google Scholar]
- Parham, S.; Kharazi, A. Z.; Bakhsheshi-Rad, H. R.; Nur, H.; Ismail, A. F.; Sharif, S.; Berto, F. Antioxidant, antimicrobial and antiviral properties of herbal materials. Antioxidants 2020, 9(12), 1309. [Google Scholar] [CrossRef] [PubMed]
- Parr, R. G.; Szentpály, L. V.; Liu, S. Electrophilicity index. Journal of the American Chemical Society 1999, 121(9), 1922–1924. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. Journal of Applied physics 1981, 52(12), 7182–7190. [Google Scholar] [CrossRef]
- Parsaee, Z.; Mohammadi, K.; Ghahramaninezhad, M.; Hosseinzadeh, B. A novel nano-sized binuclear nickel (II) Schiff base complex as a precursor for NiO nanoparticles: synthesis, characterization, DFT study and antibacterial activity. New Journal of Chemistry 2016, 40(12), 10569–10583. [Google Scholar] [CrossRef]
- Perkins, S. L.; Painter, P.; Colina, C. M. Experimental and computational studies of choline chloride-based deep eutectic solvents. Journal of Chemical & Engineering Data 2014, 59(11), 3652–3662. [Google Scholar]
- Popelier, P. L. A.; Aicken, F. M.; O’Brien, S. E. Atoms in molecules; Prentice Hall; Manchester, 2000; Vol. 188. [Google Scholar]
- Quesada-Moreno, M. M.; Fatima, M.; Medel, R.; Pérez, C.; Schnell, M. Sniffing out camphor: the fine balance between hydrogen bonding and London dispersion in the chirality recognition with α-fenchol. Physical Chemistry Chemical Physics 2022, 24(21), 12849–12859. [Google Scholar] [CrossRef]
- Rafati, S.; Ebrahimi, N.; Sadeghi, R. New family of type V natural hydrophobic deep eutectic solvents based on thymol-acetamide/acetanilide: Characteristics, intermolecular interactions and applications in liquid–liquid extraction. Separation and Purification Technology 2025, 359, 130583. [Google Scholar] [CrossRef]
- Rodríguez, Y. F.; Benito, C.; Aparicio, S.; Trenzado, J. L. Unveiling the high-pressure behavior of thymol+ carvone NADES: A combined experimental-computational approach. The Journal of Supercritical Fluids 2025, 215, 106408. [Google Scholar] [CrossRef]
- Rodríguez-Llorente, D.; Cañada-Barcala, A.; Álvarez-Torrellas, S.; Águeda, V. I.; García, J.; Larriba, M. A review of the use of eutectic solvents, terpenes and terpenoids in liquid–liquid extraction processes. Processes 2020, 8(10), 1220. [Google Scholar] [CrossRef]
- Rozas, S.; Zamora, L.; Benito, C.; Atilhan, M.; Aparicio, S. A study on monoterpenoid-based natural deep eutectic solvents. Green Chemical Engineering 2023, 4(1), 99–114. [Google Scholar] [CrossRef]
- Santos, C. D.; Cabot, J. C. Persistent effects after camphor ingestion: a case report and literature review. The Journal of Emergency Medicine 2015, 48(3), 298–304. [Google Scholar] [CrossRef] [PubMed]
- Smith, E. L.; Abbott, A. P.; Ryder, K. S. Deep eutectic solvents (DESs) and their applications. Chemical reviews 2014, 114(21), 11060–11082. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.; Bhardwaj, N.; Jain, S. K.; Metya, A. K.; Parambil, J. V. Unveiling the potential of acetic acid-based hydrophobic natural deep eutectic solvents for phytochemical extraction. Journal of Molecular Liquids 2024, 408, 125314. [Google Scholar] [CrossRef]
- Su, H. G.; Peng, X. R.; Shi, Q. Q.; Huang, Y. J.; Zhou, L.; Qiu, M. H. Lanostane triterpenoids with anti-inflammatory activities from Ganoderma lucidum. Phytochemistry 2020, 173, 112256. [Google Scholar] [CrossRef]
- Sun, X.; Luo, H.; Dai, S. Ionic liquids-based extraction: a promising strategy for the advanced nuclear fuel cycle. Chemical reviews 2012, 112(4), 2100–2128. [Google Scholar] [CrossRef]
- Thakur, M.; Pathania, D. Environmental fate of organic pollutants and effect on human health. In Abatement of Environmental Pollutants; Elsevier, 2020; pp. 245–262. [Google Scholar]
- Ullah, A.; Munir, S.; Badshah, S. L.; Khan, N.; Ghani, L.; Poulson, B. G.; Jaremko, M. Important flavonoids and their role as a therapeutic agent. Molecules 2020, 25(22), 5243. [Google Scholar] [CrossRef]
- Van Osch, D. J.; Dietz, C. H.; Van Spronsen, J.; Kroon, M. C.; Gallucci, F.; van Sint Annaland, M.; Tuinier, R. A search for natural hydrophobic deep eutectic solvents based on natural components. ACS Sustainable Chemistry & Engineering 2019, 7(3), 2933–2942. [Google Scholar]
- Wopfner, N.; Jahn-Schmid, B.; Schmidt, G.; Christ, T.; Hubinger, G.; Briza, P.; Schwarzenbacher, R. The alpha and beta subchain of Amb a 1, the major ragweed-pollen allergen divergent reactivity at the IgE and T-cell level. Molecular immunology 2009, 46(10), 2090–2097. [Google Scholar] [CrossRef]
- Wu, K.; Zhang, H.; Lou, X.; Wu, X.; Wang, Y.; Zhao, K.; Xia, X. Analysis of NADES and its water tailoring effects constructed from inulin and L-proline based on structure, physicochemical and antifreeze properties. International Journal of Biological Macromolecules 2024, 277, 134049. [Google Scholar] [CrossRef]
- Yao, J.; Xiao, L.; Li, C.; Wang, B.; Chen, Y.; Yan, X.; Cui, Z. Exploration of the multiscale interaction mechanism between natural deep eutectic solvents and silybin by QC calculation and MD simulation. Journal of Molecular Liquids 2022, 363, 119768. [Google Scholar] [CrossRef]
- Zahid, S.; Rasool, A.; Ans, M.; Yaseen, M.; Iqbal, J. Quantum chemical approach of donor− π–acceptor based arylborane–arylamine macrocycles with outstanding photovoltaic properties toward high-performance organic solar cells. Energy & Fuels 2021, 35(18), 15018–15032. [Google Scholar]
- Zbîrcea, L. E.; Buzan, M. R.; Grijincu, M.; Babaev, E.; Stolz, F.; Valenta, R.; Păunescu, V.; Panaitescu, C.; Chen, K. W. Relationship between IgE Levels Specific for Ragweed Pollen Extract, Amb a 1 and Cross-Reactive Allergen Molecules. International journal of molecular sciences 2023, 24(4), 4040. [Google Scholar] [CrossRef]
- Zielińska-Błajet, M.; Pietrusiak, P.; Feder-Kubis, J. Selected monocyclic monoterpenes and their derivatives as effective anticancer therapeutic agents. International journal of molecular sciences 2021, 22(9), 4763. [Google Scholar] [CrossRef] [PubMed]

| s/n | Chemical | MF | M/gmol-1 | Purity | Phase at RT | Structure |
| 1 | Camphor | C10H16O | 152.23 | >0.98 | Solid | 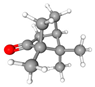 |
| 2 | Thymol | C10H14O | 150.22 | >0.98 | Solid | 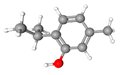 |
| 3 | Eugenol | C10H12O2 | 164.20 | >0.98 | Liquid | 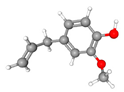 |
| 4 | Menthol | C10H20O | 156.27 | >0.98 | Solid |  |
| NaDES | T onset (°C) | Tp (°C) | DTA Peak (°C) |
| Thymol-Camphor | 125.2 | 174.31 | 180 |
| Thymol-Eugenol | 123.4 | 185.12 | 190 |
| Menthol-Eugenol | 110.2 | 155.35 | 160 |
| Camphor-Eugenol | 114.0 | 178 | 180 |
| Thymol-Menthol | 125.5 | 176.88 | 180 |
| Thymol | 132.5 | 181.8 | 180 |
| Camphor | 102.0 | 165.4 | 160 |
| Eugenol | 145.0 | 206.8 | 200 |
| Menthol | 119.3 | 167 | 160 |
| Phytochemical Family | Compounds |
| Alkane related compounds | 1-Cyano-1,1-dideuterio hexadecane (II) 2-Methylhexadecane (III) 2-Methylpentadecane (IV) 3,8-Dimethyldecane (V) 3-Methylheneicosane (VI) 3-Methyloctadecane (VII) 5-Methylundecane, (VIII) Cetane (IX) Decane, 3,8-dimethyl- (X) Docosane (XI) Eicosane (XI) Heneicosane (XII) Heptadecane (XIII) Heptadecane, 8-methyl- (XIV) 8-methyl- Heptacosane (XV) Hentriacontane (XVI) Hexadecane (XVII) Hexadecane, 7,9-dimethyl- (XIII) Hexatriacontane (XIX) Nonadecane (XX) Nonacosane (XXI) Norpristane (XXII) Octadecane (XXIII) Octadecane, 2-methyl- (XXIV) Pentacosane (XXV) Pentadecane (XXVI) Phytane (XXVII) Tetracosane (XXVIII) Tetradecane (XXIX) Tricosane (XXX) Triacontane |
| Polycyclic Aromatic Compound derivative | 4-(biphenyl-2′-yl)-7-chloro-1,2-dihydronaphthalene |
| Alkaloids | 4,5α-Epoxy-3-methoxy-17-methyl-7α-(4-phenyl-1,3-butadienyl)-6β,7β-(oxymethylene) morphinan) (II) Skatole |
| Terpenes and Terpenoids | Ethyl (2E,6E,10E)-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraenoate (II) Deoxycaesaldekarin C (III) Lanosta-7,9(11)-diene-3,18,20-triol (IV) Squalene |
| Fatty Acids and Esters | Hexadecanoic acid, 1-(hydroxymethyl)-1,2-ethanediyl ester (II) Monostearin (III) 7,9-di-tert-butyl-1-oxaspiro[4.5]deca-6,9-diene-2,8-dione |
| Phenolic Compounds | Methyl 1-anthraquinonesulfenate (II) 3-Phenylpropionic acid, 5-methoxy-2-[3,4-dimethoxyphenyl]- |
| Steroids | Pregn-20-en-3-ol, 20-methyl-, (3β,5α)- |
| Components | E gap | I (eV) | A (eV) | µ(eV) | η (eV) | S (eV) | Χ (eV) | Ѡ(eV) |
| Cam-Eug | 3.83 | 8.64 | 4.81 | -6.72 | 1.92 | 0.96 | 6.72 | 11.80 |
| Cam-Thy | 4.26 | 9.20 | 4.94 | -7.07 | 2.13 | 1.07 | 7.07 | 11.72 |
| Men-Eug | 4.14 | 8.99 | 4.84 | -6.91 | 2.07 | 1.04 | 6.91 | 11.53 |
| Thy-Eug | 3.98 | 9.00 | 5.02 | -7.01 | 1.99 | 0.99 | 7.01 | 12.35 |
| Men-Thy | 4.29 | 9.24 | 4.95 | -7.10 | 2.14 | 1.07 | 7.10 | 11.74 |
| Camphor | 5.02 | 9.76 | 4.74 | -7.25 | 2.51 | 1.26 | 7.25 | 10.46 |
| eugenol | 4.13 | 8.93 | 1.68 | -5.30 | 3.62 | 1.81 | 5.30 | 3.89 |
| menthol | 8.85 | 10.54 | 4.80 | -7.67 | 2.87 | 1.43 | 7.67 | 10.25 |
| thymol | 4.27 | 9.21 | 4.94 | -7.07 | 2.13 | 1.07 | 7.07 | 11.72 |
| NADES | (ρe) (a.u.) | (Δ2ρe) (a.u.) | CVB |
| Camphor_Thymol | 0.023 | 0.078 | 0.031 |
| Eugenol_Camphor | 0.027 | 0.086 | 0.009 |
| Eugenol_Thymol | 0.026 | 0.082 | 0.015 |
| Thymol_Menthol | 0.023 | 0.074 | 0.010 |
| Menthol_Eugenol | 0.039 | 0.117 | -0.051 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





