1. Introduction
The emergence of the novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), responsible for the coronavirus disease 2019 (COVID-19) pandemic, has brought unprecedented challenges to global healthcare systems [
1]. While much attention has rightfully been directed towards understanding the clinical manifestations and epidemiology of COVID-19 in the general population [2, 3,4], there is a pressing need to elucidate its effects on vulnerable populations, particularly pregnant individuals and their developing fetuses.
Pregnancy represents a unique immunological state characterized by profound physiological changes that modulate maternal immune responses to accommodate the semi-allogeneic fetus [
5,
6]. Emerging evidence suggests that pregnant individuals may experience altered immune responses, potentially rendering them more susceptible to severe outcomes upon SARS-CoV-2 infection [
7,
8]. Moreover, concerns have been raised regarding the potential vertical transmission of the virus from mother to fetus, highlighting the importance of understanding the impact of maternal infection on placental health and fetal development [
9,
10,
11].
Epigenetic mechanisms, including histone modifications, play crucial roles in regulating gene expression and chromatin structure, thereby influencing various physiological processes during pregnancy [
12,
13]. Among these modifications, histone methylation at lysine 4 (H3K4) and lysine 9 (H3K9) have garnered significant attention for their involvement in placental development, trophoblast differentiation, and maternal-fetal communication [
14,
15]. As we already investigated H3K4 and H3K9 in patients with praeclampsia, we were able to demonstrate that levels of H3K4me3 and H3K9ac are significantly reduced in placentas from individuals with preeclampsia compared to normotensive controls, suggesting a potential link between epigenetic alterations and the pathophysiology of this pregnancy complication [
16,
17].
Inspired by these findings, our study aims to explore the impact of maternal SARS-CoV-2 infection on histone modifications, particularly H3K4me3 and H3K9ac, in placental tissues. Given the similarities between the pathophysiological features of preeclampsia and COVID-19-related pregnancy complications, such as endothelial dysfunction and inflammation, we hypothesize that maternal SARS-CoV-2 infection may induce epigenetic changes reminiscent of those observed in preeclampsia.
Despite the growing body of literature elucidating the role of histone modifications in normal pregnancy, our understanding of their dysregulation in the context of maternal SARS-CoV-2 infection remains limited.
This study aims to provide a comprehensive overview of the current understanding of H3K4 and H3K9 histone modifications in pregnancies affected by SARS-CoV-2. By immunohistochemistry we seek to elucidate the potential mechanisms underlying epigenetic alterations in response to maternal viral infection and their implications for pregnancy outcomes. Furthermore, we will discuss the therapeutic potential of targeting histone-modifying enzymes as a novel approach to mitigate the adverse effects of COVID-19 on maternal-fetal health.
In conclusion, unraveling the complex interplay between SARS-CoV-2 infection and histone modifications in pregnancy holds promise for identifying novel biomarkers, therapeutic targets, and preventive strategies to improve maternal and neonatal outcomes in the face of this ongoing global health crisis.
2. Materials and Methods
2.1. Sample – Placenta Tissue
This study collected placenta samples at the department of obstetrics and gynecology at Hospital Großhadern of the LMU. The cohort of patients was stratified into four distinct groups, encompassing two control groups: one group of women who had received SARS-CoV-2 vaccination(n=13) and another group of women who had not (n=17). Notably, no distinction was made among the available vaccines. The two groups of interest consisted of women who got infected with SARS-CoV-2 during gestation (n=19) and those who tested positive for the virus upon parturition (n=18). Promptly postpartum, following explicit patient consent, the placenta samples were collected. The specimens were divided, with one portion subject to fixation in 4% formalin. Subsequent processing involved paraffin embedding of the fixed samples for utilization in immunohistochemistry and immunofluorescence.
2.2. Immunohistochemical Staining (IHC)
Employing established methodologies as previously described by (Meister et al., 2020) immunohistochemical staining was carried out. Briefly, following multiple washing steps and antigen retrieval via citric acid solution, a blocking agent was used to prevent unspecific staining (Detection system ZytoChem Plus HRP polymer system (mouse/rabbit), Zytomed, nr. POLHRP-100). Next, incubation with primary antibodies (Anti-H3K9ac, Order no. Ab32129 Rabbit monoclonal, and Anti-H3K4me3, Order no. Ab8580 Rabbit polyclonal) followed for a duration of 16 hours overnight. Post-incubation, a post-blocking step was carried out, followed by visualization of the staining using liquid chromogen 3,3′-Diaminobenzidine (Liquid DAB+ Substrate Chromogen System (Dako; Best.Nr. K3468) with an individual incubation period for each antibody. Lastly, counterstaining followed by putting the slides into Mayer’s hemalaun.
2.3. Staining Evaluation
2.3.1. Staining Evaluation via IRS
The semi-quantative immunoreactive score (IRS) served as the evaluation tool, integrating the staining intensity and the proportion of positively stained cells. Intensity gradation (0 = no reaction, 1 = weak intensity, 2 = moderate intensity, 3 = high intensity) was multiplied by the percentage of positively stained cells (0 = none, 1 = less than 10%, 2 = 10-50%, 3 = 51-80%, 4 = more than 81%), thereby resulting in the IRS. Three scores were obtained for each slide – one for the fetal part of the placenta, the syncytiotrophoblasts, and two for the maternal part of the placenta, the decidua. Within the decidua the extravillous trophoblasts (EVT) and decidual stromal cells were evaluated.
2.3.2. Staining Evaluation via QuPath
To objectify the staining results the program QuPath (v0.5.1) was used. To distinguish between the fetal and maternal part of the placenta, the syncytiotrophoblasts and decidua were evaluated separately.
2.4. Double-Staining Immunofluorescence
The immunofluorescence staining followed established protocols as described by (Meister et al., 2020). Initially the slides underwent deparaffinization in Roticlear, followed by rehydration in a descending alcohol series. Incubation overnight followed a blocking step to prevent unspecific staining. The primary antibodies (H3K4me3, H3K9ac) were mixed with CD163, a marker for macrophages. The next day, the slides were incubated with secondary antibodies functioning as the fluorochromes (AlexaFluorTM Plus 555, Donkey Anti-rabbit, Thermofisher A32794, CyTM5 AffiniPureTM, Donkey Anti-mouse, Jackson Immuno Research, Order-No 715-175-151). For nucleus staining 4’,6’-Diamino-2-phenylindole (DAPI) was used.
2.5. Statistics
Statistical analysis was undertaken using the SPSS software (Version 29; IBM company, Chicago, IL). Non parametric tests were used for statistical analysis, due to the not normally distributed variables. The Mann-Whitney-U-test was used for independent samples and the Wilcoxon-signed-rank-test for paired samples. The results are given as mean value ± standard deviation. The Spearman-Rho correlation test was used to examine the correlations. The correlation coefficient r indicates the strength of the correlation (r < 0.3 weak relation, r > 0.3 medium relation, r > 0.5 strong relation). For the western blot analysis the unpaired student’s T-test was used to examine differences between the two groups. The significance level for all tests was assumed at p < 0.05.
3. Results
3.1. Down-Regulation of H3K4me3 and H3K9ac in COVID Placentas
3.1.1. Trimethylated Histone H3K4me3 Reduced in COVID Placentas
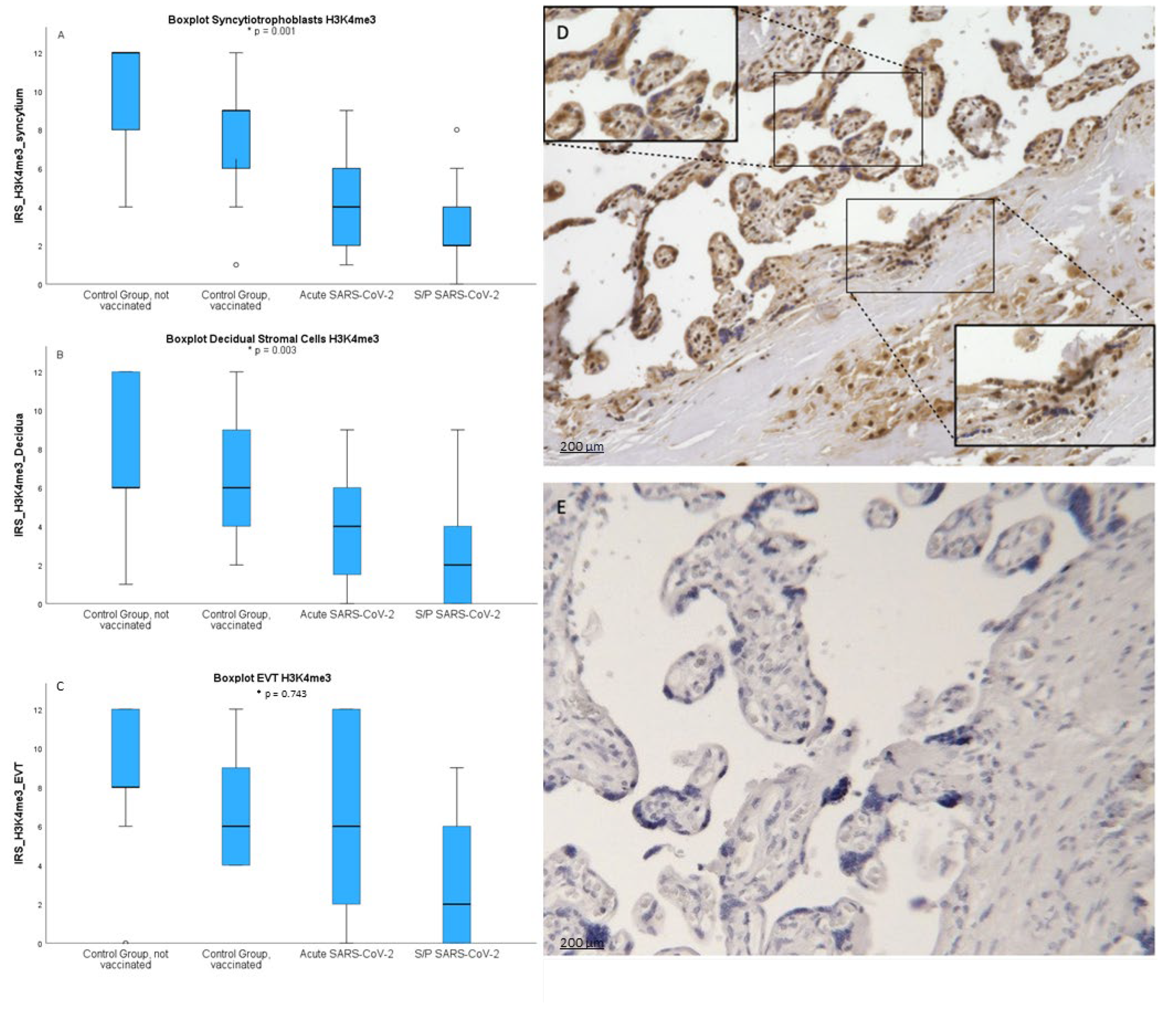
After staining four groups of placenta sections with anti- H3K4me3 antibody we have determined IRS score for each sample. As shown in
Figure 1 obtained results show broad distribution and individual variability within groups. Nevertheless, SCT of infected patiens showed significantly lower levels of the H3K4me3 mark (mean=2.95 +/- 1.9; p=0.001). IRS in SCT was highest in not vaccinated healthy women (mean=9.29 +/- 3.31) followed by vaccinated (mean=7.62 +/- 3.45). Similar results could be shown in the decidua resembling the maternal side. Here IRS was lowest in placentas with an Sars-CoV-2 infection during pregnancy (p=0.003).
In the EVT there was no significant downregulation of trimethylated histone H3K4me3 after a Sars-CoV-2 infection (p=0.743).
To objectify the results via IHC IRS we conducted examinations via Qupath applying automated detection and measurement of signal and calculation of H score. Here we were able to show similar results. The trimethylated histone H3K4me3 was lowest in the SCT of COVID placentas (p=0.001) and with no significant difference in the decidua (p=0.179).
3.1.2. Acetylated Histone H3K9ac Reduced in COVID Placentas
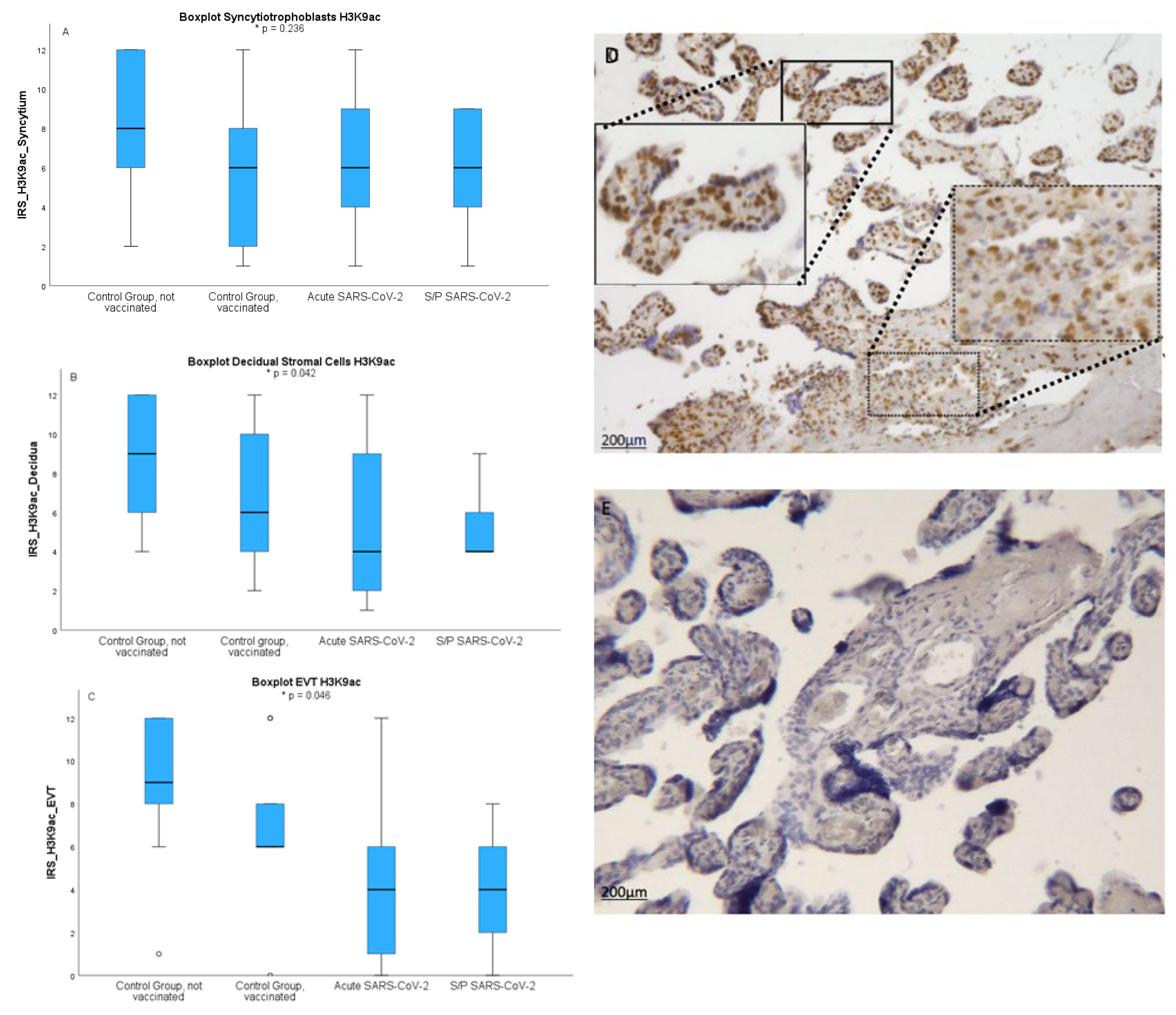
Next we wanted to asses the pattern of occurrence of H3K9ac, another epigenetic marker involved in gene regulation. After staining four groups of placenta sections with anti- H3K9ac antibody we have determined IRS score for each sample. As shown in
Figure 2 obtained results show broad distribution and individual variability within groups. In H3K9ac the results were not as clear. Here in IHC IRS in SCT was highest in not vaccinated healthy controls (8.18+/-3.33) followed by an acute infection (6.47+/-3.84) infection during pregnancy (6.00+/-2.65) and vaccination in healthy women (5.78+/-3.35; p=0.236) shown in
Figure 2A. In the decidua IRS of H3K9ac as H3K4me3 was highest in healthy not vaccinated women followed by not vaccinated controls, acute infection and infection during pregnancy (p=0.042) shown in
Figure 2B. The gestational age was no confounder.
In the EVT there was also a significant downregulation of acetylated histone H3K9ac after a Sars-CoV-2 infection (p=0.046). To objectify the results via IHC IRS we conducted examinations via Qupath. Here we were able to show similar results. With IRS lowest in healthy vaccinated placentas in the SCT (p=0.007) and after a Sars-Cov-2 infection in the decidua (p=0.042). The values between male and female did not show any significant difference in the staining both histone modifications neither in the decidua nor in the syncytiotrophoblast. The analysis of correlations between histone acetylation and maternal age and gestational age showed no significant results.
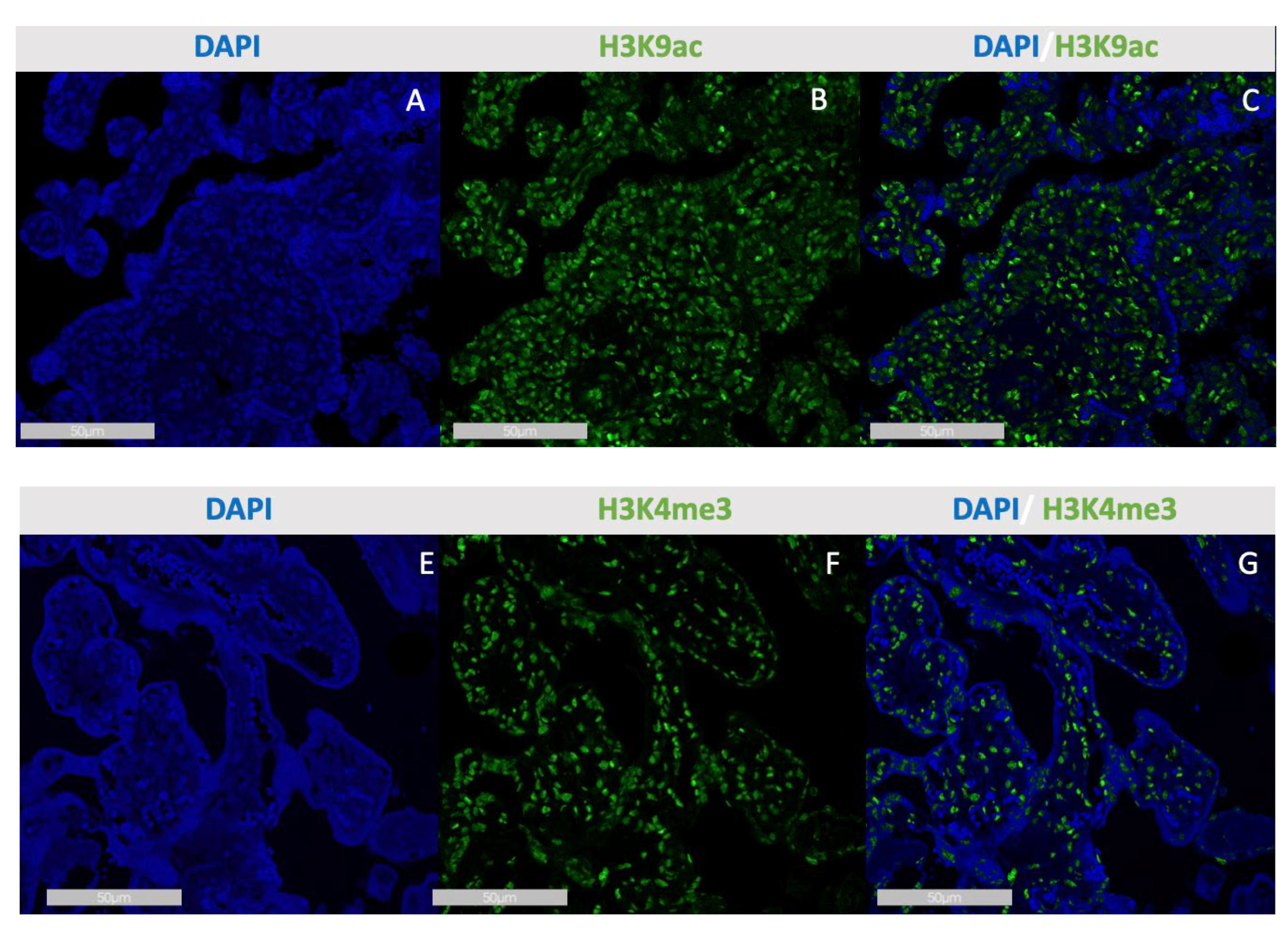
3.2. Double-Staining Immunofluorescence
In the double staining we were able to reproduce the results that we showed in IHC. Trimethylated histone H3K4me3 was downregulated after an infection with Sars-CoV-2 (p=0.038).
Acetylated histone H3K9ac was also downregulated after an infection with Sars-CoV-2 (p=0.003), CD 163 positive macrophages were increased during infection (p=0.012)
Figure 3.
Immunofluorescence staining of H3K9ac (A-C) and H3K4me3 (E-G) in the SCT of Sars-Cov-2placentas. Single immunofluorescence of DAPI in Sars-Cov-2 placenta (A and E), single immunofluorescence of H3K9ac in Sars-Cov-2 placenta (B), merge of H3K9ac and DAPI in Sars-Cov-2 placenta (C), immunofluorescence of H3K4me3 in Sars-Cov-2 placenta (F), merge of H3K4me3 and DAPI in Sars-Cov-2 placenta (G).
Figure 3.
Immunofluorescence staining of H3K9ac (A-C) and H3K4me3 (E-G) in the SCT of Sars-Cov-2placentas. Single immunofluorescence of DAPI in Sars-Cov-2 placenta (A and E), single immunofluorescence of H3K9ac in Sars-Cov-2 placenta (B), merge of H3K9ac and DAPI in Sars-Cov-2 placenta (C), immunofluorescence of H3K4me3 in Sars-Cov-2 placenta (F), merge of H3K4me3 and DAPI in Sars-Cov-2 placenta (G).
4. Discussion
Protein modifications of histones are known for the exhibit strong effect on the regulation of transcription [
18]. In a previous study we were able to show an influence of preeclampsia on histone modifications trimethylated histone H3K4me3 and the acetylated H3K9ac [
16] which both were reduced in the study group compared to healthy controls and therefore associated with a reduction in transcription access [
16]. Histones facilitate the dense packing of a large amount of DNA, while their N-terminal tails remain flexible [
19]. These N-terminal tails of histone proteins can undergo post-translational modifications by enzymes, adding chemical modifications that alter the DNA packaging structure and either allow or prevent gene transcription [
20]. When the configuration is open (“decondensed”) transcription factors are enabled to access binding sites, while a closed (“condensed”) configuration blocks these sites, thus regulating gene transcription [
21].
Various histone modifications have been implicated in mechanisms similar to those occurring in Sars-Cov-2 Infection [
16]. Additionally, histone modifications have been shown to affect the expression of specific growth factors. Vascular endothelial growth factor (VEGF) exhibits abnormal trimethylation of H3K9 in its promoter region [
22], while changes in histone acetylation of H3 and H4 in the promoter region due to hypoxia impact placental growth factor (PlGF) [
23]. Importantly, histone modifications regulate factors essential for trophoblast invasion and migration, which are defective in preeclampsia and potentially in Sars-Cov-2 Infection [
24,
25,
26]. This becomes relevant in trophoblast invasion and migration and results in insufficient placentation and higher risks for IUGR, elevated blood pressure and preterm birth [
27]. Now we wanted to investigate the role of histone modifications in placentas of women with or after an infection with Sars-CoV-2. Our data show that trimethylated histone H3K4me3 and the acetylated H3K9ac after a Sars-CoV-2 infection during pregnancy are reduced in comparison to control placentas. More precisely when compared to an infection during pregnancy to an infection during birth trimethylated histone H3K4me3 and the acetylated H3K9ac are more affected during pregnancy. In a next step we also focused on the vaccination. Here we were able to show that methylated H3K4 me3 as well as acetylated H3K9ac were more affected through the Sars-Cov-2 infection, followed by women who got vaccinated. Despite the observed differences in histone modifications, we found no significant variation in staining between placentas of male and female foetuses for either histone modification, indicating that sex does not influence these specific epigenetic changes. Correlation analyses between histone acetylation and maternal epidemiological data yielded no significant results, suggesting that the observed changes may be driven more by direct viral interaction rather than external factors. Acetylation of histones, particularly H3K9ac, is generally associated with transcriptional activation and increased gene accessibility [
28]. The observed decrease in H3K9 acetylation suggests a shift towards a more repressive chromatin state in the placenta post-infection. This reduction could be attributed to several factors linked to the viral infection. SARS-CoV-2 has been shown to induce a pro-inflammatory response, leading to the upregulation of histone deacetylases (HDACs) as part of the cellular response to stress and inflammation [
29,
30]. Increased HDAC activity can result in the removal of acetyl groups from histones, thereby silencing genes crucial for placental health and function [
31]. Similarly, the downregulation of trimethylated H3K4 (H3K4me3) is particularly noteworthy, as this modification is commonly associated with active transcription and is found enriched at promoter regions of actively expressed genes [
32]. The loss of H3K4me3 in the placenta could indicate a broader repression of gene expression, potentially affecting trophoblast function, nutrient transport, and immune modulation. This is especially concerning in the context of pregnancy, where the placenta plays a critical role in maintaining a favorable environment for fetal development. The interplay between viral infection and epigenetic modifications could lead to significant consequences for placental biology. The downregulation of these histone marks may hinder the placenta’s ability to adapt to the demands of a changing environment, particularly under inflammatory conditions induced by the virus. This could result in impaired trophoblast proliferation and differentiation, ultimately affecting placental structural integrity and function [
33]. Moreover, the altered epigenetic landscape may have long-term implications for fetal development. The genes affected by the loss of H3K9ac and H3K4me3 are likely involved in critical processes such as cell signaling, immune response, and metabolic regulation. As a result, the downregulation of these histone modifications may contribute to adverse pregnancy outcomes, including preterm birth, low birth weight, or even fetal developmental issues.
5. Conclusion
In conclusion, the downregulation of acetylated H3K9 and methylated H3K4 in the placenta following SARS-CoV-2 infection underscores the intricate relationship between viral infections and epigenetic regulation. Future studies should focus on identifying specific genes impacted by these modifications and elucidating the functional consequences on placental health and fetal development. Understanding these mechanisms will be crucial for developing therapeutic strategies to mitigate the effects of viral infections on pregnancy outcomes.
Author Contributions
Franziska Ganster: Conceptualization, Methodology, Writing - original draft. Friederike Kallenber: Investigation. Anna Titiva: Investigation. Laura Hahn: Validation, Visualization. Susanne Beyer: Formal analysis. Mirjana Kessler: Validation. Thomas Kolben: Validation. Sven Mahner: Writing - review & editing. Sarah Meister: Conceptualization, Supervision.
Funding
no funding was received for this study.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board (or Ethics Committee) of the Ethikkomission ber der LMU München (protocol code 18-700 on March 25th 2019)
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Conflicts of Interest
Sv.M: Research support, advisory board, honoraria and travel expenses from AbbVie, AstraZeneca, Clovis, Eisai, GlaxoSmithKline, Medac, MSD, Novartis, Olympus, PharmaMar, Roche, Sensor Kinesis, Teva, Tesaro; TK: holds stock of Roche, relative employed at Roche. This study is part of the doctoral thesis of Friedrike Kallenberg.
Abbreviations
The following abbreviations are used in this manuscript:
IHC Immunohistochemical Staining
IRS semi-quantative immunoreactive score
SCT syncytiotrophoblast
EVT extravillous trophoblast
H3K4 histone methylation at lysine 4
H3K9 histone acethylation at lysine 9
References
- da Silva RGL, Chammas R, Novaes HMD. Rethinking approaches of science, technology, and innovation in healthcare during the COVID-19 pandemic: the challenge of translating knowledge infrastructures to public needs. Health Res Policy Syst. 2021 Jul 21;19(1):104. [CrossRef] [PubMed] [PubMed Central]
- Ochani R, Asad A, Yasmin F, Shaikh S, Khalid H, Batra S, Sohail MR, Mahmood SF, Ochani R, Hussham Arshad M, Kumar A, Surani S. COVID-19 pandemic: from origins to outcomes. A comprehensive review of viral pathogenesis, clinical manifestations, diagnostic evaluation, and management. Infez Med. 2021 Mar 1;29(1):20-36. [PubMed]
- Atzrodt CL, Maknojia I, McCarthy RDP, Oldfield TM, Po J, Ta KTL, Stepp HE, Clements TP. A Guide to COVID-19: a global pandemic caused by the novel coronavirus SARS-CoV-2. FEBS J. 2020 Sep;287(17):3633-3650. [CrossRef] [PubMed] [PubMed Central]
- Hassan M, Zalkifal M, Wahab A, Afzal S, Rafique S, Shahid M, Khan MA, Ahmed N, Idrees M, Shahid AA. Novel Coronavirus: A Review from Origin to Current Status of Therapeutic Strategies. Crit Rev Eukaryot Gene Expr. 2021;31(3):21-34. [CrossRef] [PubMed]
- Morelli AE, Sadovsky Y. Extracellular vesicles and immune response during pregnancy: A balancing act. Immunol Rev. 2022 Jul;308(1):105-122. [CrossRef] [PubMed] [PubMed Central]
- Abu-Raya B, Michalski C, Sadarangani M, Lavoie PM. Maternal Immunological Adaptation During Normal Pregnancy. Front Immunol. 2020 Oct 7;11:575197. [CrossRef] [PubMed] [PubMed Central]
- Chen G, Liao Q, Ai J, Yang B, Bai H, Chen J, Liu F, Cao Y, Liu H, Li K. Immune Response to COVID-19 During Pregnancy. Front Immunol. 2021 May 3;12:675476. [CrossRef] [PubMed] [PubMed Central]
- Juttukonda LJ, Wachman EM, Boateng J, Jain M, Benarroch Y, Taglauer ES. Decidual immune response following COVID-19 during pregnancy varies by timing of maternal SARS-CoV-2 infection. J Reprod Immunol. 2022 Jun;151:103501. [CrossRef] [PubMed] [PubMed Central]
- Oliveira KF, Oliveira JF, Wernet M, Paschoini MC, Ruiz MT. Vertical transmission and COVID-19: a scoping review. Rev Bras Enferm. 2021 May 21;74(suppl 1):e20200849. English, Portuguese. [CrossRef] [PubMed]
- Thapa B, Acharya S, Karki S. Vertical Transmission of COVID-19: A Case Report and Review of Literature. J Nepal Health Res Counc. 2021 Apr 23;19(1):203-205. [CrossRef] [PubMed]
- Al-Kuraishy HM, Al-Gareeb AI, Albezrah NKA, Bahaa HA, El-Bouseary MM, Alexiou A, Al-Ziyadi SH, Batiha GE. Pregnancy and COVID-19: high or low risk of vertical transmission. Clin Exp Med. 2023 Aug;23(4):957-967. [CrossRef] [PubMed] [PubMed Central]
- Socha MW, Flis W, Wartęga M. Epigenetic Genome Modifications during Pregnancy: The Impact of Essential Nutritional Supplements on DNA Methylation. Nutrients. 2024 Feb 28;16(5):678. [CrossRef] [PubMed] [PubMed Central]
- Stonawski V, Frey S, Golub Y, Moll GH, Heinrich H, Eichler A. Affektive Belastungen der Mutter in der Schwangerschaft und assoziierte epigenetische Veränderungen beim Kind: Eine Übersicht [Epigenetic modifications in children associated with maternal emotional stress during pregnancy]. Z Kinder Jugendpsychiatr Psychother. 2018 Mar;46(2):155-167. German. [CrossRef] [PubMed]
- Lund PJ, Lehman SM, Garcia BA. Quantitative analysis of global protein lysine methylation by mass spectrometry. Methods Enzymol. 2019;626:475-498. [CrossRef] [PubMed] [PubMed Central]
- Karmodiya K, Krebs AR, Oulad-Abdelghani M, Kimura H, Tora L. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics. 2012 Aug 24;13:424. [CrossRef] [PubMed] [PubMed Central]
- Meister S, Hahn L, Beyer S, Kuhn C, Jegen M, von Schönfeldt V, Corradini S, Schulz C, Kolben TM, Hester A, Appelt T, Mahner S, Jeschke U, Kolben T. Epigenetic modification via H3K4me3 and H3K9ac in human placenta is reduced in preeclampsia. J Reprod Immunol. 2021 Jun;145:103287. [CrossRef] [PubMed]
- Meister S, Hahn L, Beyer S, Paul C, Mitter S, Kuhn C, von Schönfeldt V, Corradini S, Sudan K, Schulz C, Kolben TM, Mahner S, Jeschke U, Kolben T. Regulation of Epigenetic Modifications in the Placenta during Preeclampsia: PPARγ Influences H3K4me3 and H3K9ac in Extravillous Trophoblast Cells. Int J Mol Sci. 2021 Nov 18;22(22):12469. [CrossRef] [PubMed] [PubMed Central]
- Patsouras MD, Vlachoyiannopoulos PG. Evidence of epigenetic alterations in thrombosis and coagulation: A systematic review. J Autoimmun. 2019 Nov;104:102347. [CrossRef] [PubMed]
- Iwasaki W, Miya Y, Horikoshi N, Osakabe A, Taguchi H, Tachiwana H, Shibata T, Kagawa W, Kurumizaka H. Contribution of histone N-terminal tails to the structure and stability of nucleosomes. FEBS Open Bio. 2013 Aug 22;3:363-9. . Erratum in: FEBS Open Bio. 2018 Aug 23;8(9):1567. 10.1002/2211-5463.12508. [CrossRef] [PubMed] [PubMed Central]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007 Nov;14(11):1025-1040. [CrossRef] [PubMed] [PubMed Central]
- Yu Y, Zeng P, Xiong J, Liu Z, Berger SL, Merlino G. Epigenetic drugs can stimulate metastasis through enhanced expression of the pro-metastatic Ezrin gene. PLoS One. 2010 Sep 13;5(9):e12710. [CrossRef] [PubMed] [PubMed Central]
- Rahat B, Najar RA, Hamid A, Bagga R, Kaur J. The role of aberrant methylation of trophoblastic stem cell origin in the pathogenesis and diagnosis of placental disorders. Prenat Diagn. 2017 Feb;37(2):133-143. [CrossRef] [PubMed]
- Tudisco L, Della Ragione F, Tarallo V, Apicella I, D’Esposito M, Matarazzo MR, De Falco S. Epigenetic control of hypoxia inducible factor-1α-dependent expression of placental growth factor in hypoxic conditions. Epigenetics. 2014 Apr;9(4):600-10. [CrossRef] [PubMed] [PubMed Central]
- Apicella C, Ruano CSM, Méhats C, Miralles F, Vaiman D. The Role of Epigenetics in Placental Development and the Etiology of Preeclampsia. Int J Mol Sci. 2019 Jun 11;20(11):2837. [CrossRef] [PubMed] [PubMed Central]
- Kamrani A, Alipourfard I, Ahmadi-Khiavi H, Yousefi M, Rostamzadeh D, Izadi M, Ahmadi M. The role of epigenetic changes in preeclampsia. Biofactors. 2019 Sep;45(5):712-724. [CrossRef] [PubMed]
- Staff AC, Dechend R, Pijnenborg R. Learning from the placenta: acute atherosis and vascular remodeling in preeclampsia-novel aspects for atherosclerosis and future cardiovascular health. Hypertension. 2010 Dec;56(6):1026-34. [CrossRef] [PubMed]
- Moser G, Windsperger K, Pollheimer J, de Sousa Lopes SC, Huppertz B. Human trophoblast invasion: new and unexpected routes and functions. Histochem Cell Biol. 2018 Oct;150(4):361-370. [CrossRef] [PubMed] [PubMed Central]
- Shvedunova M, Akhtar A. Modulation of cellular processes by histone and non-histone protein acetylation. Nat Rev Mol Cell Biol. 2022 May;23(5):329-349. [CrossRef] [PubMed]
- Gandhi S, Mitterhoff R, Rapoport R, Farago M, Greenberg A, Hodge L, Eden S, Benner C, Goren A, Simon I. Mitotic H3K9ac is controlled by phase-specific activity of HDAC2, HDAC3, and SIRT1. Life Sci Alliance. 2022 Aug 18;5(10):e202201433. [CrossRef] [PubMed] [PubMed Central]
- Sen R, Garbati M, Bryant K, Lu Y. Epigenetic mechanisms influencing COVID-19. Genome. 2021 Apr;64(4):372-385. [CrossRef] [PubMed]
- Shen Y, Wei W, Zhou DX. Histone Acetylation Enzymes Coordinate Metabolism and Gene Expression. Trends Plant Sci. 2015 Oct;20(10):614-621. [CrossRef] [PubMed]
- Wang H, Fan Z, Shliaha PV, Miele M, Hendrickson RC, Jiang X, Helin K. H3K4me3 regulates RNA polymerase II promoter-proximal pause-release. Nature. 2023 Mar;615(7951):339-348. . Epub 2023 Mar 1. Erratum in: Nature. 2023 Apr;616(7956):E7. 10.1038/s41586-023-05958-0. Erratum in: Nature. 2023 Nov;623(7987):E8. 10.1038/s41586-023-06778-y. [CrossRef] [PubMed] [PubMed Central]
- Gallardo V, González M, Toledo F, Sobrevia L. Role of heme oxygenase 1 and human chorionic gonadotropin in pregnancy associated diseases. Biochim Biophys Acta Mol Basis Dis. 2020 Feb 1;1866(2):165522. [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).