Submitted:
05 August 2025
Posted:
06 August 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction to Caenorhabditis Elegans as a Model Organism
2. Overview of Machine Learning
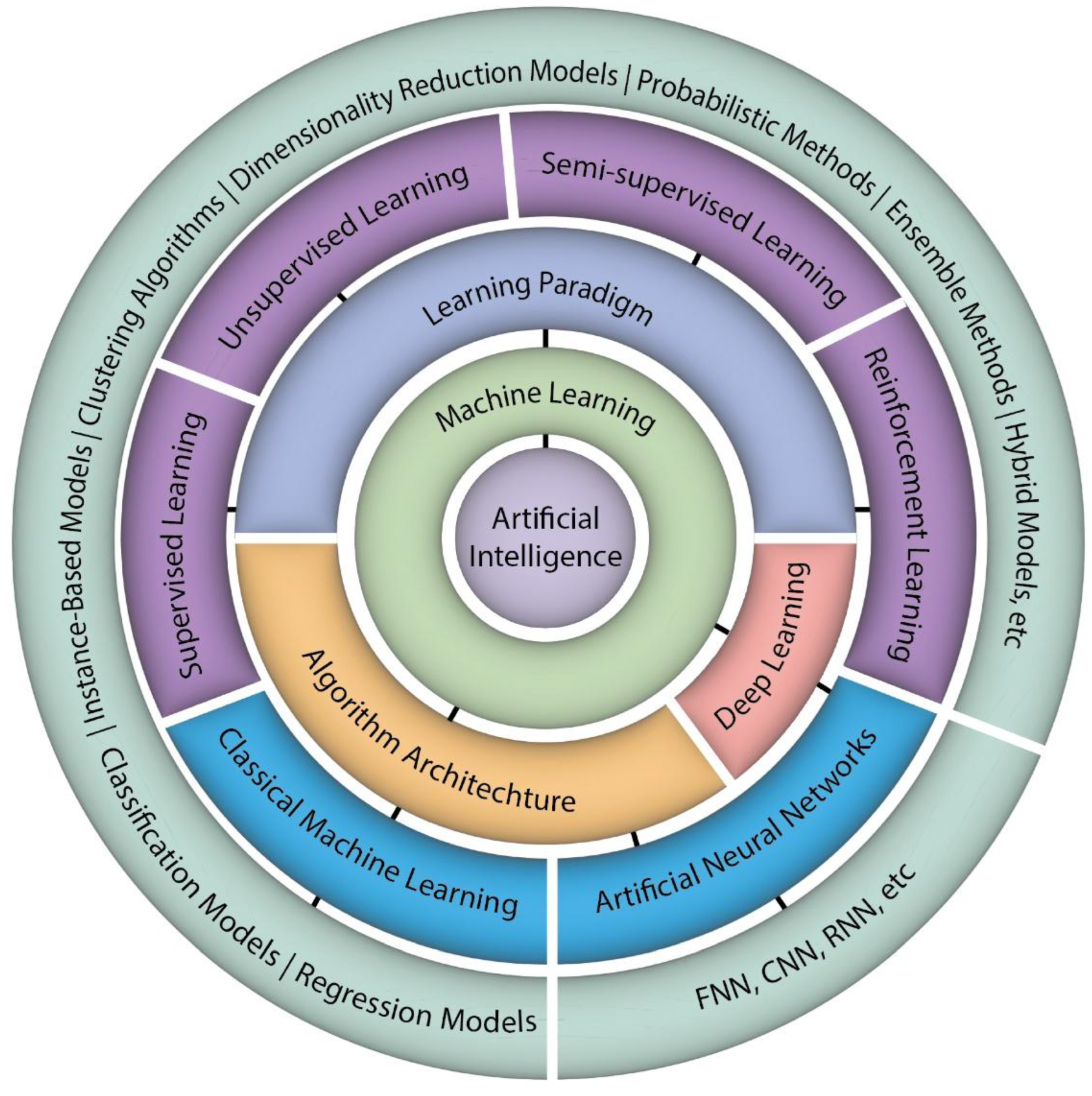
2.1. Types of Machine Learning
2.1. Types of Machine Learning Architecture
3. Machine Learning in C. elegans Developmental Research
3.1. Classification and Morphological Phenotyping of C. elegans
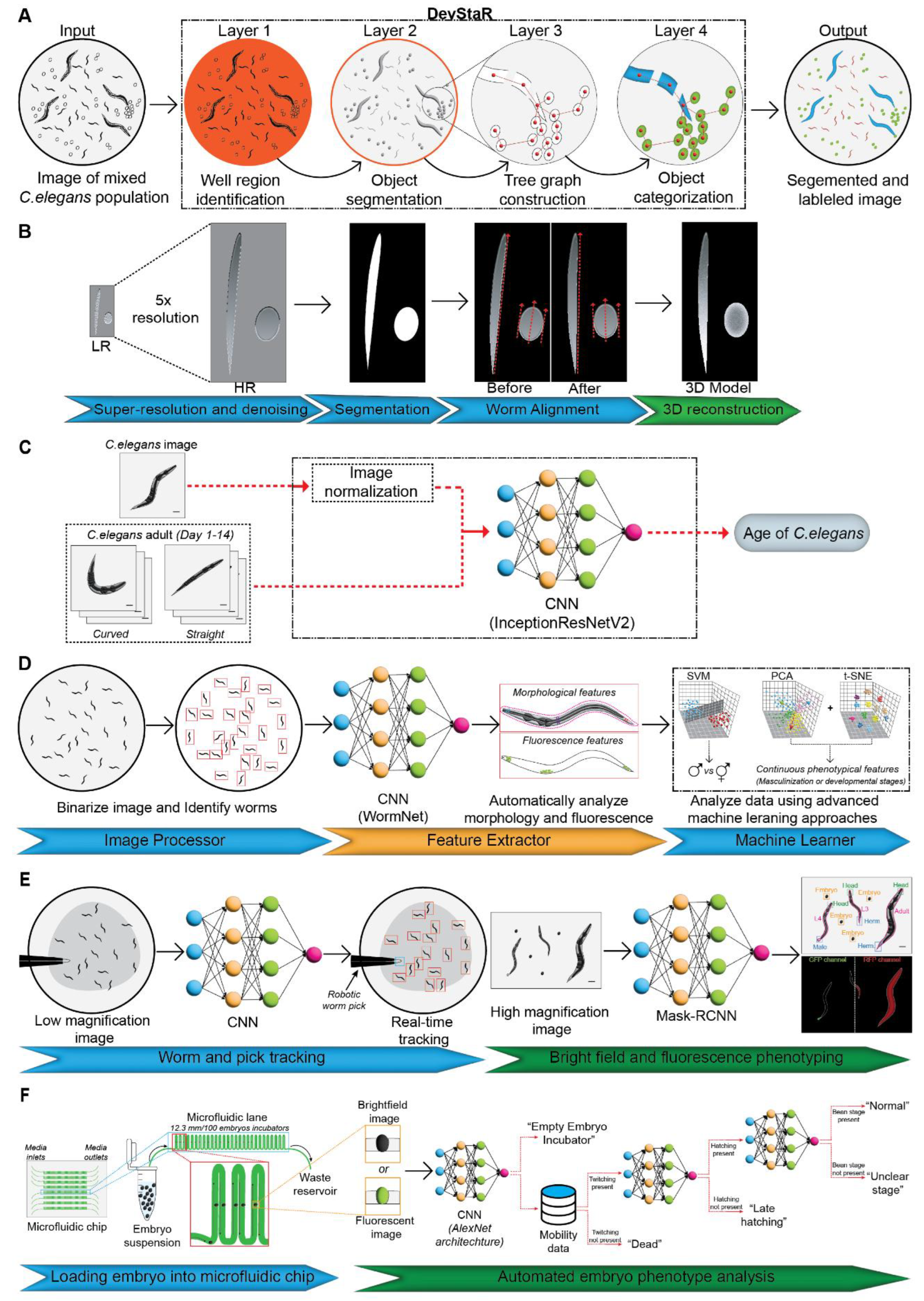
3.1.1. Classification of Developmental Stages
3.1.2. 3-dimensional Morphological Reconstruction and Phenotyping
3.1.3. Physiological Age Estimation
3.1.4. Sexual Classification
3.1.5. Real-Time Tracking and Dynamic Phenotyping
3.2. Developmental Toxicity and Tissue Analysis in C. elegans
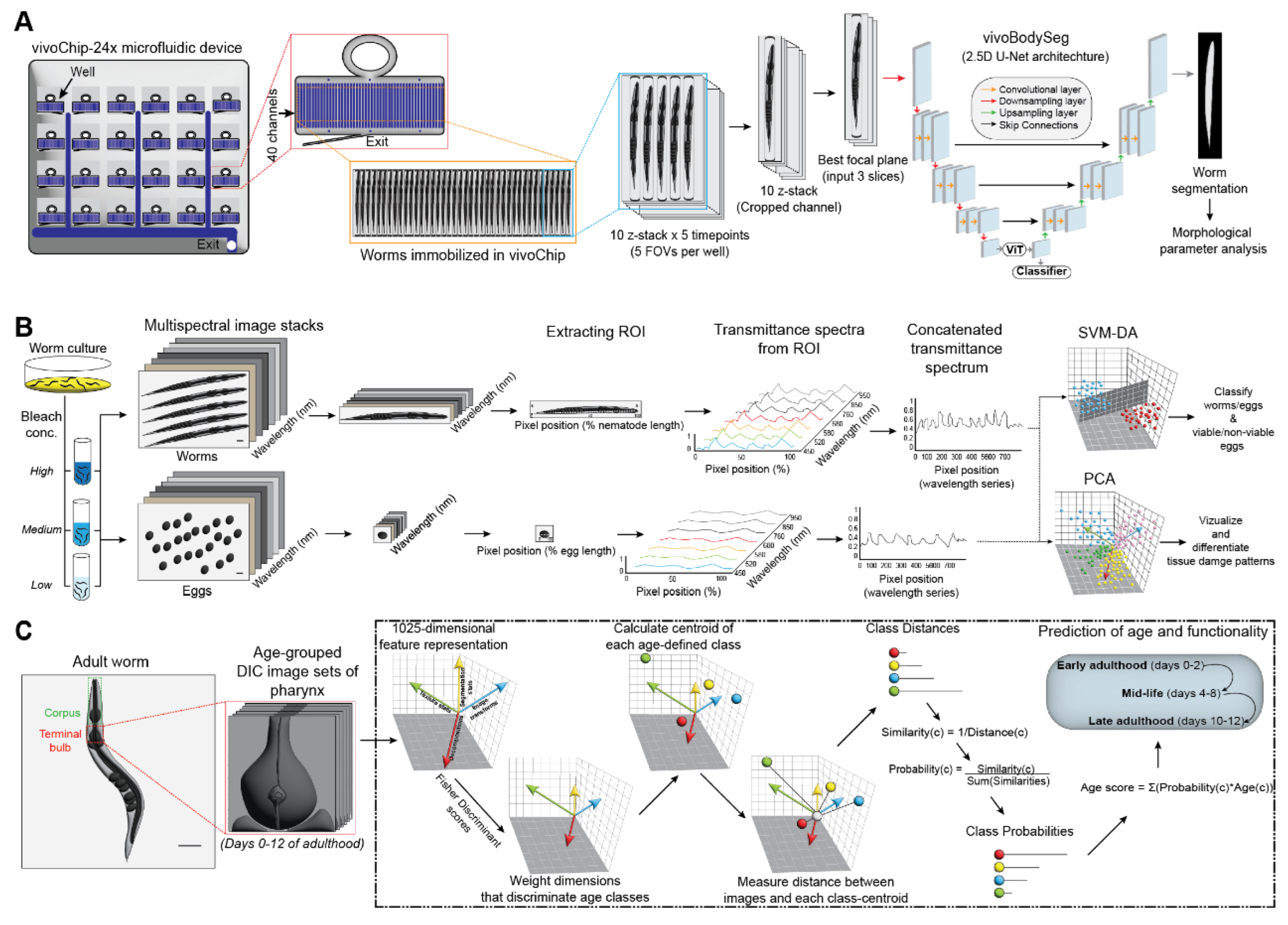
3.2.1. Developmental Toxicity Testing
3.2.1. Analysing Tissue Damage and Egg Viability
3.2.1. Tissue Morphological Transitions
3.2. Cellular Dynamics and Lineage Studies in C. elegans
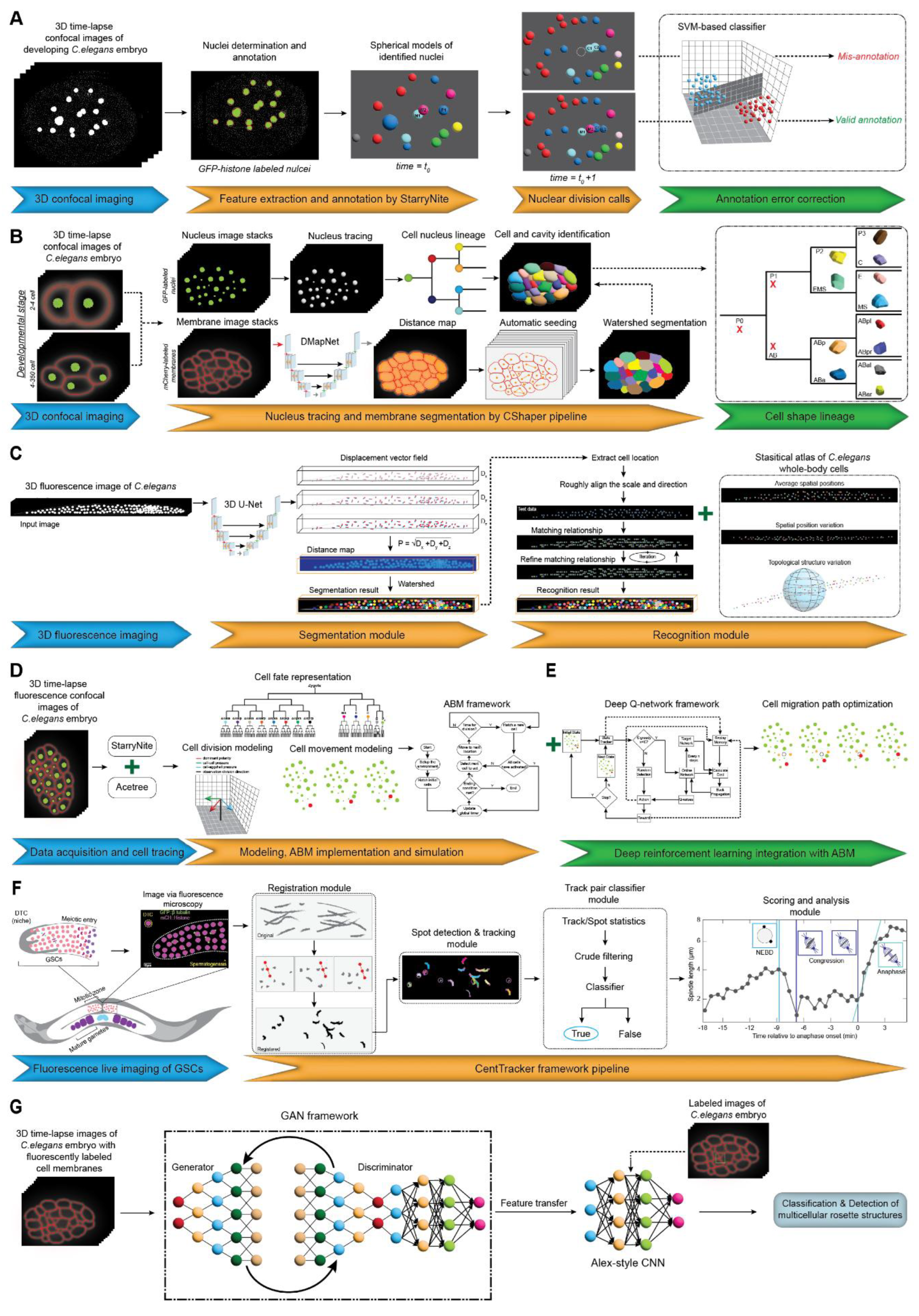
3.2.1. Cell Lineage Tracing
3.3.2. Whole-Body Cell Segmentation and Recognition
3.3.3. Modelling Cellular Dynamics in Embryogenesis
3.3.4. Tracking Germline Stem Cell Dynamics in Embryos
3.3.5. Detection and Characterization of Multicellular Structures in Embryos
4. Future Perspectives and Limitations
5. Concluding Remarks
Author Contributions
Acknowledgements
Declaration of interests
References
- Ray, A. K.; et al. A bioinformatics approach to elucidate conserved genes and pathways in C. elegans as an animal model for cardiovascular research. Sci Rep 2024, 14, 7471. [Google Scholar] [CrossRef] [PubMed]
- Valperga, G. & de Bono, M. Impairing one sensory modality enhances another by reconfiguring peptidergic signalling in Caenorhabditis elegans. Impairing one sensory modality enhances another by reconfiguring peptidergic signalling in Caenorhabditis elegans. Elife 2022, 11. [Google Scholar] [CrossRef]
- Vidal, B.; et al. An atlas of Caenorhabditis elegans chemoreceptor expression. PLoS Biol 2018, 16, e2004218. [Google Scholar] [CrossRef] [PubMed]
- Azuma, Y. , Okada, H. & Onami, S. Systematic analysis of cell morphodynamics in C. elegans early embryogenesis. Front Bioinform 2023, 3, 1082531. [Google Scholar] [CrossRef]
- Packer, J. S.; et al. A lineage-resolved molecular atlas of C. elegans embryogenesis at single-cell resolution. Science 2019, 365. [Google Scholar] [CrossRef]
- So, S. , Asakawa, M. & Sawa, H. Distinct functions of three Wnt proteins control mirror-symmetric organogenesis in the C. elegans gonad. Elife 2024, 13. [Google Scholar] [CrossRef]
- Godini, R. , Fallahi, H. & Pocock, R. The regulatory landscape of neurite development in Caenorhabditis elegans. Front Mol Neurosci 2022, 15, 974208. [Google Scholar] [CrossRef]
- Zhang, S. , Li, F., Zhou, T., Wang, G. & Li, Z. Caenorhabditis elegans as a Useful Model for Studying Aging Mutations. Front Endocrinol (Lausanne) 2020, 11, 554994. [Google Scholar] [CrossRef]
- Li, Y.; et al. A full-body transcription factor expression atlas with completely resolved cell identities in C. elegans. Nat Commun 2024, 15, 358. [Google Scholar] [CrossRef]
- Corsi, A. K. , Wightman, B. & Chalfie, M. A Transparent Window into Biology: A Primer on Caenorhabditis elegans. Genetics 2015, 200, 387–407. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, L. P. , Luke, C. J., Perlmutter, D. H., Silverman, G. A. & Pak, S. C. C. elegans in high-throughput drug discovery. Adv Drug Deliv Rev 2014, 69, 247–253. [Google Scholar] [CrossRef]
- Yuan, H.; et al. Microfluidic-Assisted Caenorhabditis elegans Sorting: Current Status and Future Prospects. Cyborg Bionic Syst 2023, 4, 0011. [Google Scholar] [CrossRef]
- An, Q. , Rahman, S., Zhou, J. & Kang, J. J. A Comprehensive Review on Machine Learning in Healthcare Industry: Classification, Restrictions, Opportunities and Challenges. Sensors (Basel) 2023, 23. [Google Scholar] [CrossRef]
- Buton, N. , Coste, F. & Le Cunff, Y. Predicting enzymatic function of protein sequences with attention. Predicting enzymatic function of protein sequences with attention. Bioinformatics 2023, 39. [Google Scholar] [CrossRef]
- Russo, E. T.; et al. DPCfam: Unsupervised protein family classification by Density Peak Clustering of large sequence datasets. PLoS Comput Biol 2022, 18, e1010610. [Google Scholar] [CrossRef]
- Mourad, R. Semi-supervised learning improves regulatory sequence prediction with unlabeled sequences. BMC Bioinformatics 2023, 24, 186. [Google Scholar] [CrossRef]
- Yang, R. , Zhang, L., Bu, F., Sun, F. & Cheng, B. AI-based prediction of protein-ligand binding affinity and discovery of potential natural product inhibitors against ERK2. BMC Chem 2024, 18, 108. [Google Scholar] [CrossRef]
- Zhang, L.; et al. A deep learning model to identify gene expression level using cobinding transcription factor signals. Brief Bioinform 2022, 23. [Google Scholar] [CrossRef]
- Wang, M. , Wei, Z., Jia, M., Chen, L. & Ji, H. Deep learning model for multi-classification of infectious diseases from unstructured electronic medical records. BMC Med Inform Decis Mak 2022, 22, 41. [Google Scholar] [CrossRef]
- Anwar, M. Y.; et al. Machine learning-based clustering identifies obesity subgroups with differential multi-omics profiles and metabolic patterns. Obesity (Silver Spring) 2024, 32, 2024–2034. [Google Scholar] [CrossRef] [PubMed]
- Ballard, J. L. , Wang, Z., Li, W., Shen, L. & Long, Q. Deep learning-based approaches for multi-omics data integration and analysis. BioData Min 2024, 17, 38. [Google Scholar] [CrossRef]
- Adam, G.; et al. Machine learning approaches to drug response prediction: challenges and recent progress. NPJ Precis Oncol 2020, 4, 19. [Google Scholar] [CrossRef]
- Guan, B. Z. , Parmigiani, G., Braun, D. & Trippa, L. Prediction of Hereditary Cancers Using Neural Networks. Ann Appl Stat 2022, 16, 495–520. [Google Scholar] [CrossRef] [PubMed]
- Poirion, O. B. , Jing, Z., Chaudhary, K., Huang, S. & Garmire, L. X. DeepProg: an ensemble of deep-learning and machine-learning models for prognosis prediction using multi-omics data. Genome Med 2021, 13, 112. [Google Scholar] [CrossRef]
- Firat Atay, F.; et al. A hybrid machine learning model combining association rule mining and classification algorithms to predict differentiated thyroid cancer recurrence. Front Med (Lausanne) 2024, 11, 1461372. [Google Scholar] [CrossRef] [PubMed]
- Choi, R. Y. , Coyner, A. S., Kalpathy-Cramer, J., Chiang, M. F. & Campbell, J. P. Introduction to Machine Learning, Neural Networks, and Deep Learning. Transl Vis Sci Technol 2020, 9, 14. [Google Scholar] [CrossRef]
- Ravindran, U. & Gunavathi, C. Deep learning assisted cancer disease prediction from gene expression data using WT-GAN. BMC Med Inform Decis Mak 2024, 24, 311. [Google Scholar] [CrossRef]
- Shimasaki, K.; et al. Deep learning-based segmentation of subcellular organelles in high-resolution phase-contrast images. Cell Struct Funct 2024, 49, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Kandathil, S. M. , Lau, A. M. & Jones, D. T. Machine learning methods for predicting protein structure from single sequences. Curr Opin Struct Biol 2023, 81, 102627. [Google Scholar] [CrossRef]
- Cao, R.; et al. ProLanGO: Protein Function Prediction Using Neural Machine Translation Based on a Recurrent Neural Network. Molecules 2017, 22. [Google Scholar] [CrossRef]
- White, A. G.; et al. Rapid and accurate developmental stage recognition of C. elegans from high-throughput image data. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit 2010, 2010, 3089–3096. [Google Scholar] [CrossRef]
- Pan, P.; et al. High-Resolution Imaging and Morphological Phenotyping of C. elegans through Stable Robotic Sample Rotation and Artificial Intelligence-Based 3-Dimensional Reconstruction. Research (Wash D C) 2024, 7, 0513. [Google Scholar] [CrossRef]
- Lin, J. L.; et al. Using Convolutional Neural Networks to Measure the Physiological Age of Caenorhabditis elegans. IEEE/ACM Trans Comput Biol Bioinform 2021, 18, 2724–2732. [Google Scholar] [CrossRef] [PubMed]
- Moore, B. T. , Jordan, J. M. & Baugh, L. R. WormSizer: high-throughput analysis of nematode size and shape. PLoS One 2013, 8, e57142. [Google Scholar] [CrossRef]
- Schindelin, J.; et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Jung, S. K. , Aleman-Meza, B., Riepe, C. & Zhong, W. QuantWorm: a comprehensive software package for Caenorhabditis elegans phenotypic assays. PLoS One 2014, 9, e84830. [Google Scholar] [CrossRef]
- Wahlby, C.; et al. An image analysis toolbox for high-throughput C. elegans assays. Nat Methods 2012, 9, 714–716. [Google Scholar] [CrossRef]
- Hakim, A.; et al. WorMachine: machine learning-based phenotypic analysis tool for worms. BMC Biol 2018, 16, 8. [Google Scholar] [CrossRef]
- Li, Z.; et al. A robotic system for automated genetic manipulation and analysis of Caenorhabditis elegans. PNAS Nexus 2023, 2, pgad197. [Google Scholar] [CrossRef]
- Baris Atakan, H. , Alkanat, T., Cornaglia, M., Trouillon, R. & Gijs, M. A. M. Automated phenotyping of Caenorhabditis elegans embryos with a high-throughput-screening microfluidic platform. Microsyst Nanoeng 2020, 6, 24. [Google Scholar] [CrossRef]
- Boyd, W. A. , Smith, M. V., Kissling, G. E. & Freedman, J. H. Medium- and high-throughput screening of neurotoxicants using C. elegans. Neurotoxicol Teratol 2010, 32, 68–73. [Google Scholar] [CrossRef]
- Hunt, P. R. The C. elegans model in toxicity testing. J Appl Toxicol 2017, 37, 50–59. [Google Scholar] [CrossRef]
- Yoon, S.; et al. Microfluidics in High-Throughput Drug Screening: Organ-on-a-Chip and C. elegans-Based Innovations. Biosensors (Basel) 2024, 14. [Google Scholar] [CrossRef]
- DuPlissis, A.; et al. Machine learning-based analysis of microfluidic device immobilized C. elegans for automated developmental toxicity testing. Sci Rep 2025, 15, 15. [Google Scholar] [CrossRef] [PubMed]
- Chow, D. J. X.; et al. Quantifying DNA damage following light sheet and confocal imaging of the mammalian embryo. Sci Rep 2024, 14, 20760. [Google Scholar] [CrossRef] [PubMed]
- Tian, W. , Chen, R. & Chen, L. Computational Super-Resolution: An Odyssey in Harnessing Priors to Enhance Optical Microscopy Resolution. Anal Chem 2025, 97, 4763–4792. [Google Scholar] [CrossRef] [PubMed]
- Nigamatzyanova, L. & Fakhrullin, R. Dark-field hyperspectral microscopy for label-free microplastics and nanoplastics detection and identification in vivo: A Caenorhabditis elegans study. Environ Pollut 2021, 271, 116337. [Google Scholar] [CrossRef] [PubMed]
- Verdu, S. , Fuentes, C., Barat, J. M. & Grau, R. Characterisation of chemical damage on tissue structures by multispectral imaging and machine learning procedures: Alkaline hypochlorite effect in C. elegans. Comput Biol Med 2022, 145, 105477. [Google Scholar] [CrossRef]
- Dybiec, J. , Szlagor, M., Mlynarska, E., Rysz, J. & Franczyk, B. Structural and Functional Changes in Aging Kidneys. Structural and Functional Changes in Aging Kidneys. Int J Mol Sci 2022, 23. [Google Scholar] [CrossRef]
- Johnston, J. , Iser, W. B., Chow, D. K., Goldberg, I. G. & Wolkow, C. A. Quantitative image analysis reveals distinct structural transitions during aging in Caenorhabditis elegans tissues. PLoS One 2008, 3, e2821. [Google Scholar] [CrossRef]
- Bao, Z.; et al. Automated cell lineage tracing in Caenorhabditis elegans. Proc Natl Acad Sci U S A 2006, 103, 2707–2712. [Google Scholar] [CrossRef] [PubMed]
- Aydin, Z. , Murray, J. I., Waterston, R. H. & Noble, W. S. Using machine learning to speed up manual image annotation: application to a 3D imaging protocol for measuring single cell gene expression in the developing C. elegans embryo. BMC Bioinformatics 2010, 11, 84. [Google Scholar] [CrossRef]
- Cao, J.; et al. Establishment of a morphological atlas of the Caenorhabditis elegans embryo using deep-learning-based 4D segmentation. Nat Commun 2020, 11, 6254. [Google Scholar] [CrossRef]
- Li, Y.; et al. Automated segmentation and recognition of C. elegans whole-body cells. Bioinformatics 2024, 40. [Google Scholar] [CrossRef] [PubMed]
- Setty, Y. Multi-scale computational modeling of developmental biology. Bioinformatics 2012, 28, 2022–2028. [Google Scholar] [CrossRef]
- Wang, Z.; et al. An Observation-Driven Agent-Based Modeling and Analysis Framework for C. elegans Embryogenesis. PLoS One 2016, 11, e0166551. [Google Scholar] [CrossRef] [PubMed]
- Wang, D. , Wang, Z., Zhao, X., Xu, Y. & Bao, Z. An Observation Data Driven Simulation and Analysis Framework for Early Stage C. elegans Embryogenesis. J Biomed Sci Eng 2018, 11, 225–234. [Google Scholar] [CrossRef]
- Wang, Z.; et al. Deep reinforcement learning of cell movement in the early stage of C.elegans embryogenesis. Bioinformatics 2018, 34, 3169–3177. [Google Scholar] [CrossRef]
- Zellag, R. M.; et al. CentTracker: a trainable, machine-learning-based tool for large-scale analyses of Caenorhabditis elegans germline stem cell mitosis. Mol Biol Cell 2021, 32, 915–930. [Google Scholar] [CrossRef]
- Wang, D.; et al. Cellular structure image classification with small targeted training samples. IEEE Access 2019, 7, 148967–148974. [Google Scholar] [CrossRef]
- Kore, M. , Acharya, D., Sharma, L., Vembar, S. S. & Sundriyal, S. Development and experimental validation of a machine learning model for the prediction of new antimalarials. BMC Chem 2025, 19, 28. [Google Scholar] [CrossRef]
- Godec, P.; et al. Democratized image analytics by visual programming through integration of deep models and small-scale machine learning. Nat Commun 2019, 10, 4551. [Google Scholar] [CrossRef]
- Rajeev, P. A.; et al. Advancing e-waste classification with customizable YOLO based deep learning models. Sci Rep 2025, 15, 18151. [Google Scholar] [CrossRef]
- Dineva, K. & Atanasova, T. Health Status Classification for Cows Using Machine Learning and Data Management on AWS Cloud. Health Status Classification for Cows Using Machine Learning and Data Management on AWS Cloud. Animals (Basel) 2023, 13. [Google Scholar] [CrossRef]
- Mittal, P. A comprehensive survey of deep learning-based lightweight object detection models for edge devices. Artificial Intelligence Review 2024, 57. [Google Scholar] [CrossRef]
- Garcia Garvi, A. , Puchalt, J. C., Layana Castro, P. E., Navarro Moya, F. & Sanchez-Salmeron, A. J. Towards Lifespan Automation for Caenorhabditis elegans Based on Deep Learning: Analysing Convolutional and Recurrent Neural Networks for Dead or Live Classification. Sensors (Basel) 2021, 21. [Google Scholar] [CrossRef]
- Restif, C.; et al. CeleST: computer vision software for quantitative analysis of C. elegans swim behavior reveals novel features of locomotion. PLoS Comput Biol 2014, 10, e1003702. [Google Scholar] [CrossRef] [PubMed]
- Van Camp, B. T. , Zapata, Q. N. & Curran, S. P. WormRACER: Robust Analysis by Computer-Enhanced Recording. GeroScience 2025, 47, 5377–5387. [Google Scholar] [CrossRef]
- Garcia-Garvi, A. , Layana-Castro, P. E. & Sanchez-Salmeron, A. J. Analysis of a C. elegans lifespan prediction method based on a bimodal neural network and uncertainty estimation. Comput Struct Biotechnol J 2023, 21, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A. & Kirkegaard, J. B. Fast detection of slender bodies in high density microscopy data. Commun Biol 2023, 6, 754. [Google Scholar] [CrossRef]
- Yang, S. R. , Liaw, M., Wei, A. C. & Chen, C. H. Deep learning models link local cellular features with whole-animal growth dynamics in zebrafish. Life Sci Alliance 2025, 8. [Google Scholar] [CrossRef]
- Melkani, Y. , Pant, A., Guo, Y. & Melkani, G. C. Automated assessment of cardiac dynamics in aging and dilated cardiomyopathy Drosophila models using machine learning. Commun Biol 2024, 7, 702. [Google Scholar] [CrossRef]
- Aljovic, A.; et al. A deep learning-based toolbox for Automated Limb Motion Analysis (ALMA) in murine models of neurological disorders. Commun Biol 2022, 5, 131. [Google Scholar] [CrossRef] [PubMed]
| Sl. No | Phenotype | Input data | Machine learning model | Pros | Cons | Reference |
| 1 | Developmental stage classification (eggs, larvae, adult) | High-resolution image datasets (brightfield microscopy) | SVM | High precision for adults, reduces human errors | Low precision for eggs and larvae | [31] |
| 2 | 3D worm body structure, key morphological traits | Stacked 2D confocal or widefield microscopy images | Customized machine learning pipeline with noise reduction and segmentation | Accurate 3D reconstructions, applicable to drug screens | Limited real-time dynamic phenotyping | [32] |
| 3 | Physiological age estimation | Brightfield images of worms across 14-day lifespan | CNN (InceptionResNetV2) | Granular day-level age prediction | Potential bias due to manual preprocessing | [33] |
| 4 | Sex determination (male, hermaphrodite) | High-contrast fluorescence and morphological images | SVM with PCA and t-SNE for dimensionality reduction | High sexual classification accuracy | Memory constraints for large image files | [38] |
| 5 | Dynamic phenotypic changes during development | Brightfield and fluorescence microscopy images | CNN and Mask-RCNN | Reduces manual interventions | Limited for worms with extreme morphologies | [39] |
| 6 | Embryonic developmental stages, motility, and viability states | Brightfield and fluorescent image patches of embryos | AlexNet-based CNN with standard image processing | Rapid phenotyping of embryos, suitable for large-scale screenings, reduces manual interventions | Requires high-performance GPUs and is sensitive to labelled data quality and quantity | [40] |
| 7 | Morphological and developmental changes due to toxins | High-resolution brightfield and fluorescence images | 2.5D U-Net for segmentation | Low variability, high reproducibility | Requires high-performance GPUs and memory | [44] |
| 8 | Tissue damage, egg viability under stress conditions | Multispectral images (450-950 nm) of worms and eggs | PCA, SVM-DA (Discriminant Analysis) | Non-invasive imaging with high specificity | Sophisticated imaging systems needed | [48] |
| 9 | Pharynx structure changes across lifespan | DIC microscopy images of pharynx tissue | Pattern recognition-based machine learning algorithm | Quantitative insights into structural aging | Limited to pharynx tissue | [50] |
| 10 | Cell lineage development, nuclear divisions | 3D confocal microscopy images of embryos | SVM classifier integrated with StarryNite software | Reduces errors and manual curation time | Does not address false negatives | [52] |
| 11 | Cell shape, volume, surface area, nucleus position, and spatial organization | 3D time-lapse confocal microscopy images of embryos (4-cell to 350-cell stages) | DMapNet deep learning model (distance map-based segmentation) | Generates comprehensive 3D morphological atlas, high accuracy in densely packed cellular environments | Requires significant computational resources and lacks a user-friendly visualization platform | [53] |
| 12 | Whole-body cell identification and segmentation | 3D fluorescence microscopy images | DVF-based deep learning model | Adaptable to other animal models | Requires extensive statistical priors | [54] |
| 13 | Cell migration, division, fate determination | Time-lapse 3D confocal microscopy images | ABM combined with reinforcement learning | Provides cellular behavior insights | High computational requirements | [57,58] |
| 14 | Germline stem cell division dynamics | Live imaging of germline stem cells | Random forest-based track pair classifier | Spatial clustering analysis of GSCs | Performance drops in noisy datasets | [59] |
| 15 | Detection of multicellular rosette structures | 3D live images with fluorescently labeled cell membranes | GAN-based deep learning model with feature transfer | Efficient classification with small datasets | Performance depends on high-performance GPUs | [60] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





