Submitted:
08 May 2025
Posted:
09 May 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
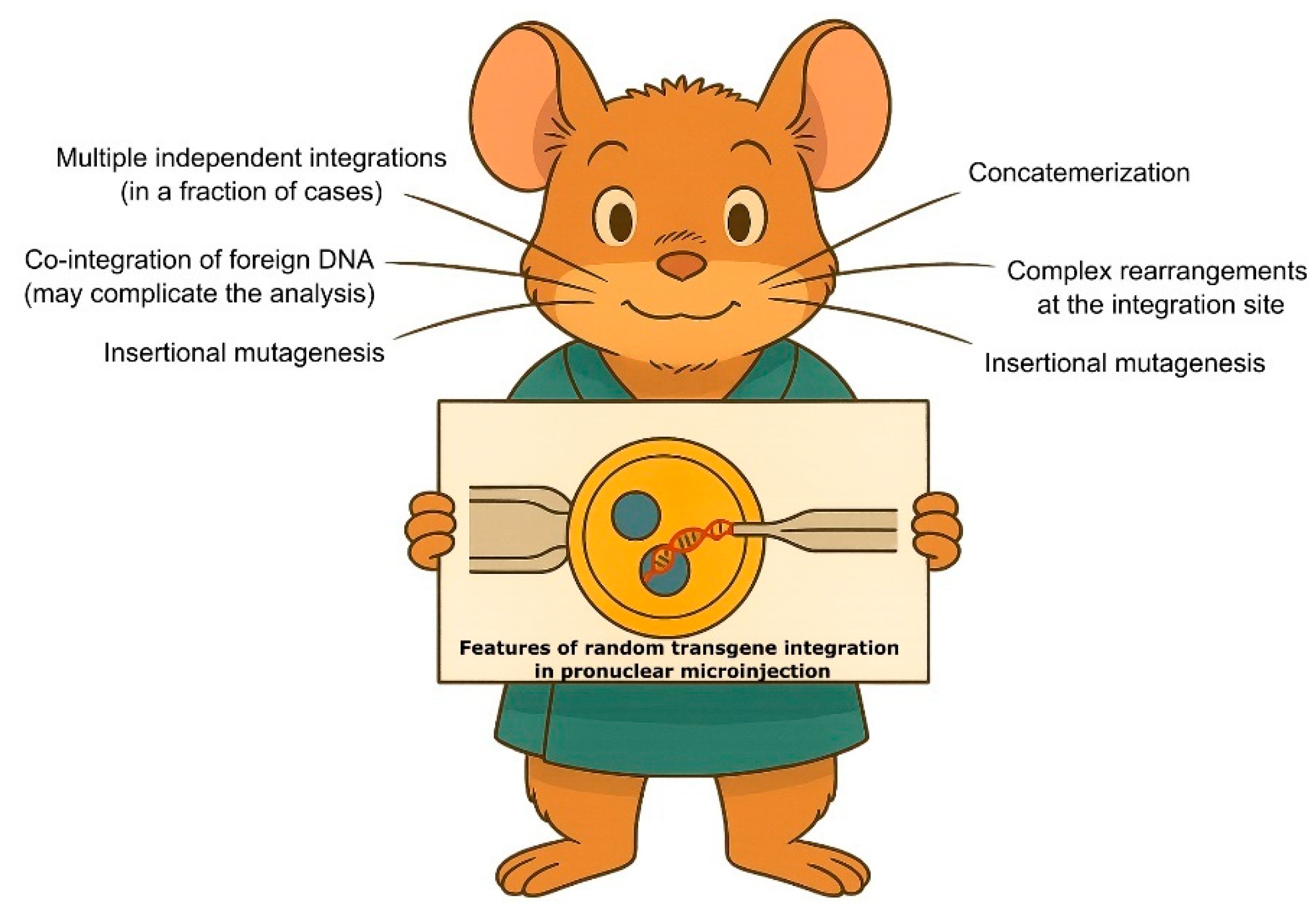
1.1. Features of Random Transgenic Insertion in Animals
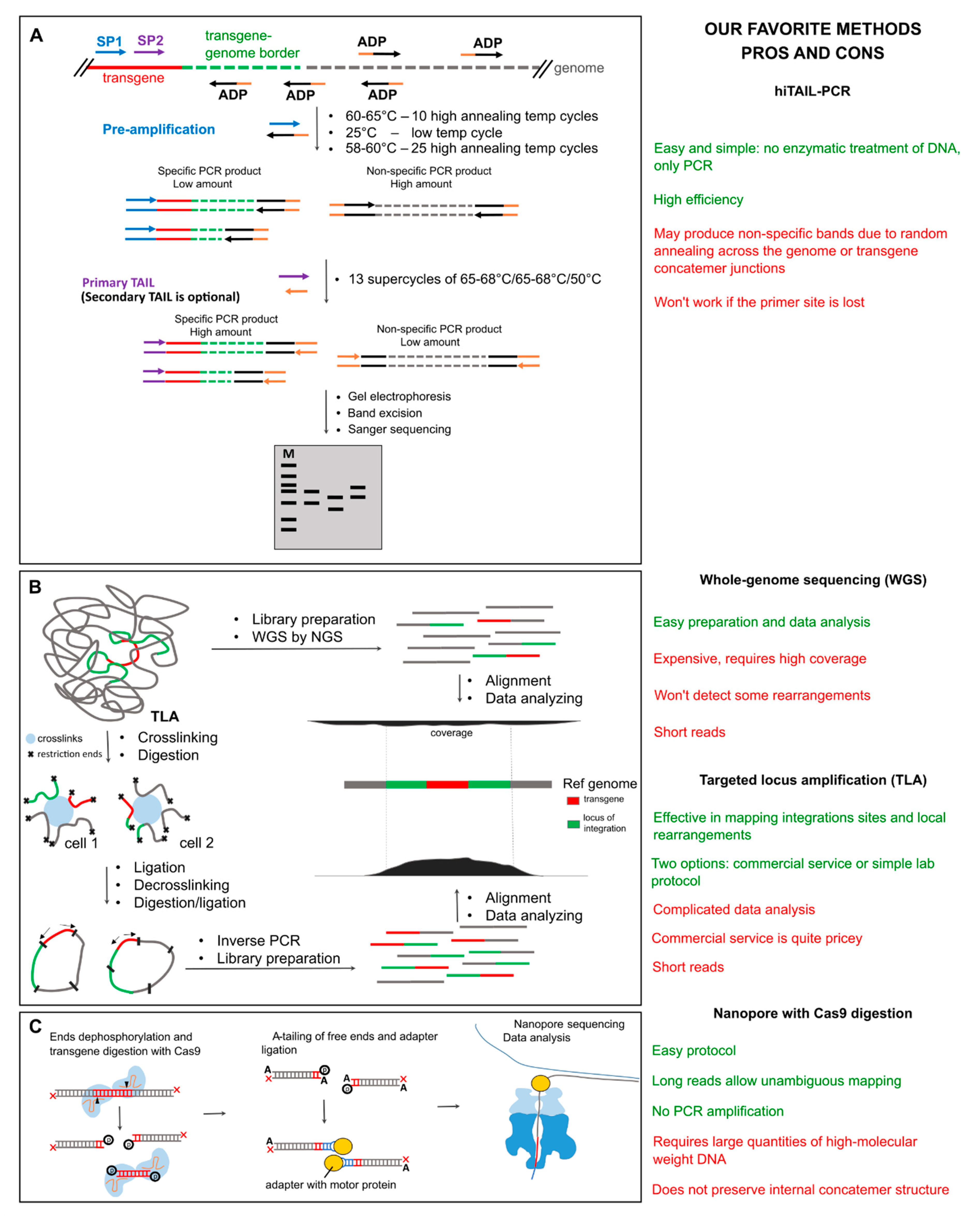
1.2. PCR-Based Methods for Transgene Mapping
1.3. Next-Generation Sequencing and Target Enrichment
1.4. Long-Read Sequencing
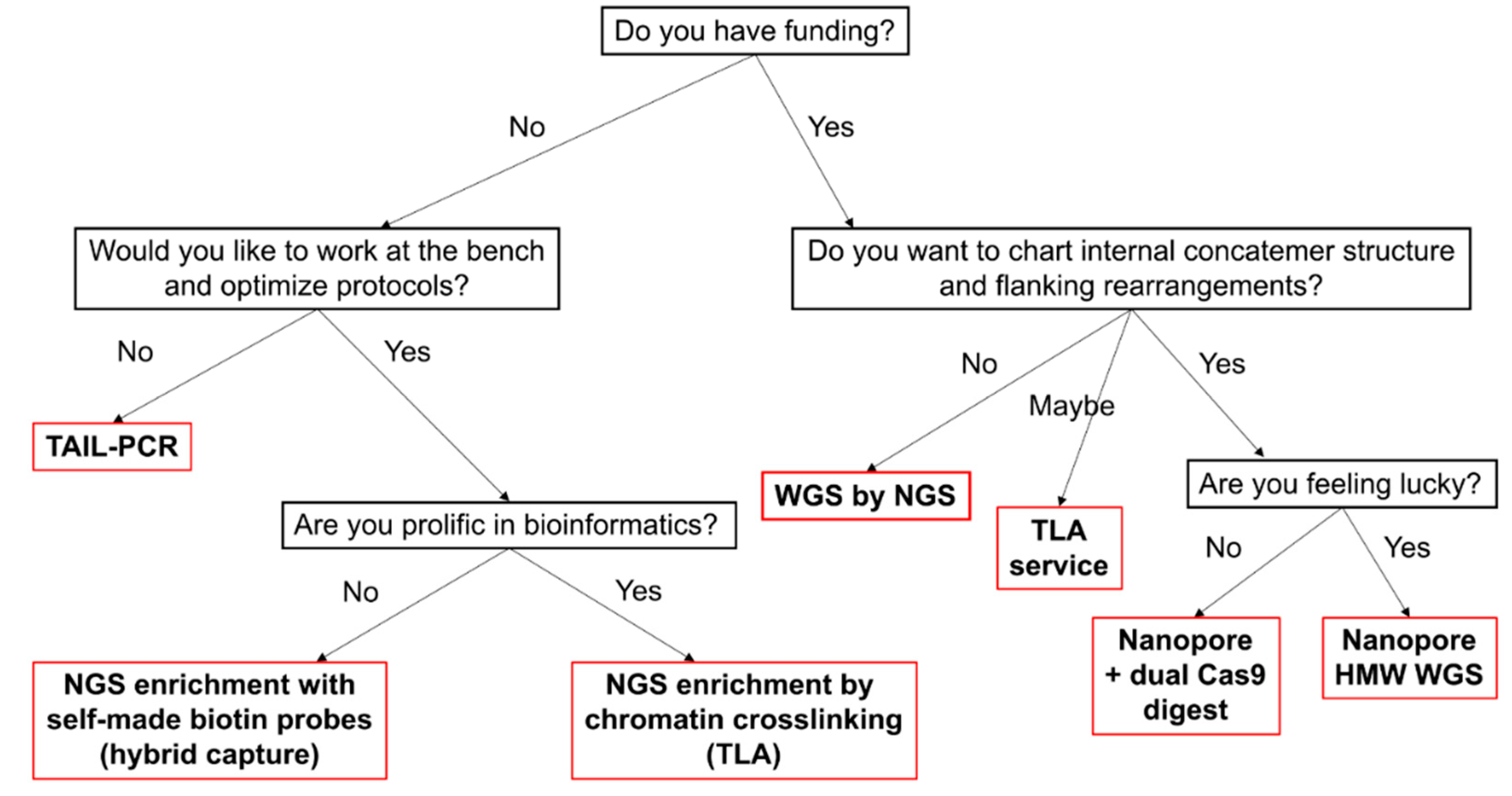
2. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| NGS | Next-generation sequencing |
| LRS | Long-read sequencing |
| TLA | Targeted locus amplification |
| WGS | Whole-genome sequencing |
References
- Tonooka, Y.; Fujishima, M. Comparison and Critical Evaluation of PCR-Mediated Methods to Walk along the Sequence of Genomic DNA. Appl Microbiol Biotechnol 2009, 85, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Leoni, C.; Volpicella, M.; De Leo, F.; Gallerani, R.; Ceci, L.R. Genome Walking in Eukaryotes. The FEBS Journal 2011, 278, 3953–3977. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Lu, S.; Wang, H.; Wang, F.; Xu, W.; Zhu, Y.; Xue, J.; Yang, L. Innovations in Transgene Integration Analysis: A Comprehensive Review of Enrichment and Sequencing Strategies in Biotechnology. ACS Appl. Mater. Interfaces 2025, 17, 2716–2735. [Google Scholar] [CrossRef] [PubMed]
- Vandereyken, K.; Sifrim, A.; Thienpont, B.; Voet, T. Methods and Applications for Single-Cell and Spatial Multi-Omics. Nat Rev Genet 2023, 24, 494–515. [Google Scholar] [CrossRef]
- Nakanishi, T.; Kuroiwa, A.; Yamada, S.; Isotani, A.; Yamashita, A.; Tairaka, A.; Hayashi, T.; Takagi, T.; Ikawa, M.; Matsuda, Y.; et al. FISH Analysis of 142 EGFP Transgene Integration Sites into the Mouse Genome. Genomics 2002, 80, 564–574. [Google Scholar] [CrossRef]
- Bandyopadhyay, A.A.; O’Brien, S.A.; Zhao, L.; Fu, H.; Vishwanathan, N.; Hu, W. Recurring Genomic Structural Variation Leads to Clonal Instability and Loss of Productivity. Biotech & Bioengineering 2019, 116, 41–53. [Google Scholar] [CrossRef]
- Lee, J.S.; Kildegaard, H.F.; Lewis, N.E.; Lee, G.M. Mitigating Clonal Variation in Recombinant Mammalian Cell Lines. Trends in Biotechnology 2019, 37, 931–942. [Google Scholar] [CrossRef]
- Dhiman, H.; Campbell, M.; Melcher, M.; Smith, K.D.; Borth, N. Predicting Favorable Landing Pads for Targeted Integrations in Chinese Hamster Ovary Cell Lines by Learning Stability Characteristics from Random Transgene Integrations. Computational and Structural Biotechnology Journal 2020, 18, 3632–3648. [Google Scholar] [CrossRef]
- Cabrera, A.; Edelstein, H.I.; Glykofrydis, F.; Love, K.S.; Palacios, S.; Tycko, J.; Zhang, M.; Lensch, S.; Shields, C.E.; Livingston, M.; et al. The Sound of Silence: Transgene Silencing in Mammalian Cell Engineering. Cell Systems 2022, 13, 950–973. [Google Scholar] [CrossRef]
- Laboulaye, M.A.; Duan, X.; Qiao, M.; Whitney, I.E.; Sanes, J.R. Mapping Transgene Insertion Sites Reveals Complex Interactions Between Mouse Transgenes and Neighboring Endogenous Genes. Front. Mol. Neurosci. 2018, 11, 385. [Google Scholar] [CrossRef]
- Yan, B.-W.; Zhao, Y.-F.; Cao, W.-G.; Li, N.; Gou, K.-M. Mechanism of Random Integration of Foreign DNA in Transgenic Mice. Transgenic Res 2013, 22, 983–992. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, L.O.; Splinter, E.; Davis, T.L.; Urban, R.; He, H.; Braun, R.E.; Chesler, E.J.; Kumar, V.; Van Min, M.; Ndukum, J.; et al. Large-Scale Discovery of Mouse Transgenic Integration Sites Reveals Frequent Structural Variation and Insertional Mutagenesis. Genome Res. 2019, 29, 494–505. [Google Scholar] [CrossRef] [PubMed]
- Halurkar, M.S.; Inoue, O.; Singh, A.; Mukherjee, R.; Ginugu, M.; Ahn, C.; Bonatto Paese, C.L.; Duszynski, M.; Brugmann, S.A.; Lim, H.-W.; et al. The Widely Used Ucp1-Cre Transgene Elicits Complex Developmental and Metabolic Phenotypes. Nat Commun 2025, 16, 770. [Google Scholar] [CrossRef]
- Cain-Hom, C.; Splinter, E.; van Min, M.; Simonis, M.; van de Heijning, M.; Martinez, M.; Asghari, V.; Cox, J.C.; Warming, S. Efficient Mapping of Transgene Integration Sites and Local Structural Changes in Cre Transgenic Mice Using Targeted Locus Amplification. Nucleic Acids Res 2017, gkw1329. [Google Scholar] [CrossRef]
- Nicholls, P.K.; Bellott, D.W.; Cho, T.-J.; Pyntikova, T.; Page, D.C. Locating and Characterizing a Transgene Integration Site by Nanopore Sequencing. G3 Genes|Genomes|Genetics 2019, 9, 1481–1486. [Google Scholar] [CrossRef]
- Geng, K.; Merino, L.G.; Wedemann, L.; Martens, A.; Sobota, M.; Sanchez, Y.P.; Søndergaard, J.N.; White, R.J.; Kutter, C. Target-Enriched Nanopore Sequencing and de Novo Assembly Reveals Co-Occurrences of Complex on-Target Genomic Rearrangements Induced by CRISPR-Cas9 in Human Cells. Genome Res. 2022, genome;gr.276901.122v2. [CrossRef]
- Guirouilh-Barbat, J.; Lambert, S.; Bertrand, P.; Lopez, B.S. Is Homologous Recombination Really an Error-Free Process? Front. Genet. 2014, 5. [Google Scholar] [CrossRef]
- Smirnov, A.; Battulin, N. Concatenation of Transgenic DNA: Random or Orchestrated? Genes 2021, 12, 1969. [Google Scholar] [CrossRef]
- Chiang, C.; Jacobsen, J.C.; Ernst, C.; Hanscom, C.; Heilbut, A.; Blumenthal, I.; Mills, R.E.; Kirby, A.; Lindgren, A.M.; Rudiger, S.R.; et al. Complex Reorganization and Predominant Non-Homologous Repair Following Chromosomal Breakage in Karyotypically Balanced Germline Rearrangements and Transgenic Integration. Nat Genet 2012, 44, 390–397. [Google Scholar] [CrossRef]
- Hussmann, J.A.; Ling, J.; Ravisankar, P.; Yan, J.; Cirincione, A.; Xu, A.; Simpson, D.; Yang, D.; Bothmer, A.; Cotta-Ramusino, C.; et al. Mapping the Genetic Landscape of DNA Double-Strand Break Repair. Cell 2021, 184, 5653–5669.e25. [Google Scholar] [CrossRef]
- Ohigashi, I.; Yamasaki, Y.; Hirashima, T.; Takahama, Y. Identification of the Transgenic Integration Site in Immunodeficient Tgε26 Human CD3ε Transgenic Mice. PLoS ONE 2010, 5, e14391. [Google Scholar] [CrossRef]
- Sailer, S.; Coassin, S.; Lackner, K.; Fischer, C.; McNeill, E.; Streiter, G.; Kremser, C.; Maglione, M.; Green, C.M.; Moralli, D.; et al. When the Genome Bluffs: A Tandem Duplication Event during Generation of a Novel Agmo Knockout Mouse Model Fools Routine Genotyping. Cell Biosci 2021, 11, 54. [Google Scholar] [CrossRef] [PubMed]
- Low, B.E.; Hosur, V.; Lesbirel, S.; Wiles, M.V. Efficient Targeted Transgenesis of Large Donor DNA into Multiple Mouse Genetic Backgrounds Using Bacteriophage Bxb1 Integrase. Sci Rep 2022, 12, 5424. [Google Scholar] [CrossRef] [PubMed]
- Bryant, W.B.; Yang, A.; Griffin, S.H.; Zhang, W.; Rafiq, A.M.; Han, W.; Deak, F.; Mills, M.K.; Long, X.; Miano, J.M. CRISPR-Cas9 Long-Read Sequencing for Mapping Transgenes in the Mouse Genome. The CRISPR Journal 2023, 6, 163–175. [Google Scholar] [CrossRef]
- Hui, E.K.-W.; Wang, P.-C.; Lo, S.J. PCR-Based Strategies to Clone Unknown DNA Regions from Known Foreign Integrants: An Overview. In PCR Cloning Protocols; Humana Press: New Jersey, 2002; ISBN 978-1-59259-177-0. [Google Scholar]
- Kalendar, R.; Shustov, A.V.; Seppänen, M.M.; Schulman, A.H.; Stoddard, F.L. Palindromic Sequence-Targeted (PST) PCR: A Rapid and Efficient Method for High-Throughput Gene Characterization and Genome Walking. Sci Rep 2019, 9, 17707. [Google Scholar] [CrossRef]
- Hamada, M.; Nishio, N.; Okuno, Y.; Suzuki, S.; Kawashima, N.; Muramatsu, H.; Tsubota, S.; Wilson, M.H.; Morita, D.; Kataoka, S.; et al. Integration Mapping of piggyBac-Mediated CD19 Chimeric Antigen Receptor T Cells Analyzed by Novel Tagmentation-Assisted PCR. EBioMedicine 2018, 34, 18–26. [Google Scholar] [CrossRef]
- Alquezar-Planas, D.E.; Löber, U.; Cui, P.; Quedenau, C.; Chen, W.; Greenwood, A.D. DNA Sonication Inverse PCR for Genome Scale Analysis of Uncharacterized Flanking Sequences. Methods Ecol Evol 2021, 12, 182–195. [Google Scholar] [CrossRef]
- Triglia, T.; Peterson, M.G.; Kemp, D.J. A Procedure for in Vitro Amplification of DNA Segments That Lie Outside the Boundaries of Known Sequences. Nucl Acids Res 1988, 16, 8186–8186. [Google Scholar] [CrossRef]
- Ochman, H.; Gerber, A.S.; Hartl, D.L. Genetic Applications of an Inverse Polymerase Chain Reaction. Genetics 1988, 120, 621–623. [Google Scholar] [CrossRef]
- Schep, R.; Leemans, C.; Brinkman, E.K.; Van Schaik, T.; Van Steensel, B. Protocol: A Multiplexed Reporter Assay to Study Effects of Chromatin Context on DNA Double-Strand Break Repair. Front. Genet. 2022, 12, 785947. [Google Scholar] [CrossRef]
- Smirnov, A.; Fishman, V.; Yunusova, A.; Korablev, A.; Serova, I.; Skryabin, B.V.; Rozhdestvensky, T.S.; Battulin, N. DNA Barcoding Reveals That Injected Transgenes Are Predominantly Processed by Homologous Recombination in Mouse Zygote. Nucleic Acids Research 2019, gkz1085. [Google Scholar] [CrossRef]
- O’Malley, R.C.; Alonso, J.M.; Kim, C.J.; Leisse, T.J.; Ecker, J.R. An Adapter Ligation-Mediated PCR Method for High-Throughput Mapping of T-DNA Inserts in the Arabidopsis Genome. Nat Protoc 2007, 2, 2910–2917. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Zhou, T.; Sun, X.; Sun, Z.; Sheng, X.; Tan, Y.; Liu, L.; Ouyang, N.; Xu, K.; Shi, K.; et al. Cyclic Digestion and Ligation-Mediated PCR Used for Flanking Sequence Walking. Sci Rep 2020, 10, 3434. [Google Scholar] [CrossRef]
- Lung, J.; Hung, M.-S.; Chen, C.-Y.; Yang, T.-M.; Lin, C.-K.; Fang, Y.-H.; Jiang, Y.-Y.; Liao, H.-F.; Lin, Y.-C. An Optimized Ligation-Mediated PCR Method for Chromosome Walking and Fusion Gene Chromosomal Breakpoints Identification. Biology Methods and Protocols 2024, 9, bpae037. [Google Scholar] [CrossRef] [PubMed]
- Uren, A.G.; Mikkers, H.; Kool, J.; Van Der Weyden, L.; Lund, A.H.; Wilson, C.H.; Rance, R.; Jonkers, J.; Van Lohuizen, M.; Berns, A.; et al. A High-Throughput Splinkerette-PCR Method for the Isolation and Sequencing of Retroviral Insertion Sites. Nat Protoc 2009, 4, 789–798. [Google Scholar] [CrossRef]
- Potter, C.J.; Luo, L. Splinkerette PCR for Mapping Transposable Elements in Drosophila. PLoS ONE 2010, 5, e10168. [Google Scholar] [CrossRef]
- Dambrot, C.; Buermans, H.P.J.; Varga, E.; Kosmidis, G.; Langenberg, K.; Casini, S.; Elliott, D.A.; Dinnyes, A.; Atsma, D.E.; Mummery, C.L.; et al. Strategies for Rapidly Mapping Proviral Integration Sites and Assessing Cardiogenic Potential of Nascent Human Induced Pluripotent Stem Cell Clones. Experimental Cell Research 2014, 327, 297–306. [Google Scholar] [CrossRef]
- Jia, W.; Guan, Z.; Shi, S.; Xiang, K.; Chen, P.; Tan, F.; Ullah, N.; Diaby, M.; Guo, M.; Song, C.; et al. The Annotation of Zebrafish Enhancer Trap Lines Generated with PB Transposon. CIMB 2022, 44, 2614–2621. [Google Scholar] [CrossRef]
- Sato, M.; Inada, E.; Saitoh, I.; Nakamura, S.; Watanabe, S. In Vivo Piggybac-Based Gene Delivery towards Murine Pancreatic Parenchyma Confers Sustained Expression of Gene of Interest. IJMS 2019, 20, 3116. [Google Scholar] [CrossRef]
- Han, H.-J.; Kim, D.H.; Baik, J.Y. A Splinkerette PCR-Based Genome Walking Technique for the Identification of Transgene Integration Sites in CHO Cells. Journal of Biotechnology 2023, 371–372, 1–9. [Google Scholar] [CrossRef]
- Schmidt, M.; Schwarzwaelder, K.; Bartholomae, C.; Zaoui, K.; Ball, C.; Pilz, I.; Braun, S.; Glimm, H.; Von Kalle, C. High-Resolution Insertion-Site Analysis by Linear Amplification–Mediated PCR (LAM-PCR). Nat Methods 2007, 4, 1051–1057. [Google Scholar] [CrossRef]
- Gabriel, R.; Eckenberg, R.; Paruzynski, A.; Bartholomae, C.C.; Nowrouzi, A.; Arens, A.; Howe, S.J.; Recchia, A.; Cattoglio, C.; Wang, W.; et al. Comprehensive Genomic Access to Vector Integration in Clinical Gene Therapy. Nat Med 2009, 15, 1431–1436. [Google Scholar] [CrossRef] [PubMed]
- van Haasteren, J.; Munis, A.M.; Gill, D.R.; Hyde, S.C. Genome-Wide Integration Site Detection Using Cas9 Enriched Amplification-Free Long-Range Sequencing. Nucleic Acids Research 2021, 49. [Google Scholar] [CrossRef] [PubMed]
- Singer, T.; Burke, E. High-Throughput TAIL-PCR as a Tool to Identify DNA Flanking Insertions. In Plant Functional Genomics; Humana Press: New Jersey, 2003; ISBN 978-1-59259-413-9. [Google Scholar]
- Liu, Y.-G.; Whittier, R.F. Thermal Asymmetric Interlaced PCR: Automatable Amplification and Sequencing of Insert End Fragments from P1 and YAC Clones for Chromosome Walking. Genomics 1995, 25, 674–681. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-G.; Chen, Y. High-Efficiency Thermal Asymmetric Interlaced PCR for Amplification of Unknown Flanking Sequences. BioTechniques 2007, 43, 649–656. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, W.; Feng, Z.; Hong, Z. A Low Degenerate Primer Pool Improved the Efficiency of High-Efficiency Thermal Asymmetric Interlaced PCR to Amplify T-DNA Flanking Sequences in Arabidopsis Thaliana. 3 Biotech 2018, 8, 14. [Google Scholar] [CrossRef]
- Wu, L.; Di, D.-W.; Zhang, D.; Song, B.; Luo, P.; Guo, G.-Q. Frequent Problems and Their Resolutions by Using Thermal Asymmetric Interlaced PCR (TAIL-PCR) to Clone Genes in Arabidopsis T-DNA Tagged Mutants. Biotechnology & Biotechnological Equipment 2015, 29, 260–267. [Google Scholar] [CrossRef]
- Luo, W.; Li, Z.; Huang, Y.; Han, Y.; Yao, C.; Duan, X.; Ouyang, H.; Li, L. Generation of AQP2-Cre Transgenic Mini-Pigs Specifically Expressing Cre Recombinase in Kidney Collecting Duct Cells. Transgenic Res 2014, 23, 365–375. [Google Scholar] [CrossRef]
- Zhang, R.; Yin, Y.; Zhang, Y.; Li, K.; Zhu, H.; Gong, Q.; Wang, J.; Hu, X.; Li, N. Molecular Characterization of Transgene Integration by Next-Generation Sequencing in Transgenic Cattle. PLoS ONE 2012, 7, e50348. [Google Scholar] [CrossRef] [PubMed]
- Zelensky, A.N.; Schimmel, J.; Kool, H.; Kanaar, R.; Tijsterman, M. Inactivation of Pol θ and C-NHEJ Eliminates off-Target Integration of Exogenous DNA. Nat Commun 2017, 8, 66. [Google Scholar] [CrossRef]
- Kondrychyn, I.; Garcia-Lecea, M.; Emelyanov, A.; Parinov, S.; Korzh, V. Genome-Wide Analysis of Tol2 Transposon Reintegration in Zebrafish. BMC Genomics 2009, 10, 418. [Google Scholar] [CrossRef]
- Johansson, O.N.; Töpel, M.; Pinder, M.I.M.; Kourtchenko, O.; Blomberg, A.; Godhe, A.; Clarke, A.K. Skeletonema Marinoi as a New Genetic Model for Marine Chain-Forming Diatoms. Sci Rep 2019, 9, 5391. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Zhou, Y.; Wang, R.; Wei, X.; Zhang, L.; Dai, Y.; Zhu, Z. Analysis of T-DNA Integration Events in Transgenic Rice. Journal of Plant Physiology 2021, 266, 153527. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Jia, M.; Li, Z.; Liu, X.; Sun, T.; Pei, J.; Wei, C.; Lin, Z.; Li, H. Wristwatch PCR: A Versatile and Efficient Genome Walking Strategy. Front. Bioeng. Biotechnol. 2022, 10, 792848. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Guo, X.; Pan, Z.; Wang, R.; Tian, B.; Li, H. Fork PCR: A Universal and Efficient Genome-Walking Tool. Front. Microbiol. 2023, 14, 1265580. [Google Scholar] [CrossRef]
- Li, H.; Lin, Z.; Guo, X.; Pan, Z.; Pan, H.; Wang, D. Primer Extension Refractory PCR: An Efficient and Reliable Genome Walking Method. Mol Genet Genomics 2024, 299, 27. [Google Scholar] [CrossRef]
- Kalendar, R.; Shustov, A.V.; Schulman, A.H. Palindromic Sequence-Targeted (PST) PCR, Version 2: An Advanced Method for High-Throughput Targeted Gene Characterization and Transposon Display. Front. Plant Sci. 2021, 12, 691940. [Google Scholar] [CrossRef]
- Burkov, I.A.; Serova, I.A.; Battulin, N.R.; Smirnov, A.V.; Babkin, I.V.; Andreeva, L.E.; Dvoryanchikov, G.A.; Serov, O.L. Expression of the Human Granulocyte–Macrophage Colony Stimulating Factor (hGM-CSF) Gene under Control of the 5′-Regulatory Sequence of the Goat Alpha-S1-Casein Gene with and without a MAR Element in Transgenic Mice. Transgenic Res 2013, 22, 949–964. [Google Scholar] [CrossRef]
- Serova, I.A.; Dvoryanchikov, G.A.; Andreeva, L.E.; Burkov, I.A.; Dias, L.P.B.; Battulin, N.R.; Smirnov, A.V.; Serov, O.L. A 3,387 Bp 5′-Flanking Sequence of the Goat Alpha-S1-Casein Gene Provides Correct Tissue-Specific Expression of Human Granulocyte Colony-Stimulating Factor (hG-CSF) in the Mammary Gland of Transgenic Mice. Transgenic Res 2012, 21, 485–498. [Google Scholar] [CrossRef]
- Smirnov, A.V.; Kontsevaya, G.V.; Feofanova, N.A.; Anisimova, M.V.; Serova, I.A.; Gerlinskaya, L.A.; Battulin, N.R.; Moshkin, M.P.; Serov, O.L. Unexpected Phenotypic Effects of a Transgene Integration Causing a Knockout of the Endogenous Contactin-5 Gene in Mice. Transgenic Res 2018, 27, 1–13. [Google Scholar] [CrossRef]
- Le Saux, A.; Houdebine, L.-M.; Jolivet, G. Chromosome Integration of BAC (Bacterial Artificial Chromosome): Evidence of Multiple Rearrangements. Transgenic Res 2010, 19, 923–931. [Google Scholar] [CrossRef]
- Won, M.; Dawid, I.B. PCR Artifact in Testing for Homologous Recombination in Genomic Editing in Zebrafish. PLoS ONE 2017, 12, e0172802. [Google Scholar] [CrossRef] [PubMed]
- Pillai, M.M.; Venkataraman, G.M.; Kosak, S.; Torok-Storb, B. Integration Site Analysis in Transgenic Mice by Thermal Asymmetric Interlaced (TAIL)-PCR: Segregating Multiple-Integrant Founder Lines and Determining Zygosity. Transgenic Res 2008, 17, 749–754. [Google Scholar] [CrossRef] [PubMed]
- Giani, A.M.; Gallo, G.R.; Gianfranceschi, L.; Formenti, G. Long Walk to Genomics: History and Current Approaches to Genome Sequencing and Assembly. Computational and Structural Biotechnology Journal 2020, 18, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Abrams, N.; Zhu, W.; Salinas, E.; Yu, Z.; Palmer, D.C.; Jailwala, P.; Franco, Z.; Roychoudhuri, R.; Stahlberg, E.; et al. Identification of the Genomic Insertion Site of Pmel-1 TCR α and β Transgenes by Next-Generation Sequencing. PLoS ONE 2014, 9, e96650. [Google Scholar] [CrossRef]
- Yong, C.S.M.; Sharkey, J.; Duscio, B.; Venville, B.; Wei, W.-Z.; Jones, R.F.; Slaney, C.Y.; Mir Arnau, G.; Papenfuss, A.T.; Schröder, J.; et al. Embryonic Lethality in Homozygous Human Her-2 Transgenic Mice Due to Disruption of the Pds5b Gene. PLoS ONE 2015, 10, e0136817. [Google Scholar] [CrossRef]
- Srivastava, S.K.; Wolinski, P.; Pereira, A. A Strategy for Genome-Wide Identification of Gene Based Polymorphisms in Rice Reveals Non-Synonymous Variation and Functional Genotypic Markers. PLoS ONE 2014, 9, e105335. [Google Scholar] [CrossRef]
- Owen, J.R.; Hennig, S.L.; McNabb, B.R.; Mansour, T.A.; Smith, J.M.; Lin, J.C.; Young, A.E.; Trott, J.F.; Murray, J.D.; Delany, M.E.; et al. One-Step Generation of a Targeted Knock-in Calf Using the CRISPR-Cas9 System in Bovine Zygotes. BMC Genomics 2021, 22, 118. [Google Scholar] [CrossRef]
- Carlson, D.F.; Lancto, C.A.; Zang, B.; Kim, E.-S.; Walton, M.; Oldeschulte, D.; Seabury, C.; Sonstegard, T.S.; Fahrenkrug, S.C. Production of Hornless Dairy Cattle from Genome-Edited Cell Lines. Nat Biotechnol 2016, 34, 479–481. [Google Scholar] [CrossRef]
- Young, A.E.; Mansour, T.A.; McNabb, B.R.; Owen, J.R.; Trott, J.F.; Brown, C.T.; Van Eenennaam, A.L. Genomic and Phenotypic Analyses of Six Offspring of a Genome-Edited Hornless Bull. Nat Biotechnol 2020, 38, 225–232. [Google Scholar] [CrossRef]
- Niu, L.; He, H.; Zhang, Y.; Yang, J.; Zhao, Q.; Xing, G.; Zhong, X.; Yang, X. Efficient Identification of Genomic Insertions and Flanking Regions through Whole-Genome Sequencing in Three Transgenic Soybean Events. Transgenic Res 2021, 30, 1–9. [Google Scholar] [CrossRef]
- Guo, B.; Guo, Y.; Hong, H.; Qiu, L.-J. Identification of Genomic Insertion and Flanking Sequence of G2-EPSPS and GAT Transgenes in Soybean Using Whole Genome Sequencing Method. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zhang, H.; Zhang, Y.; Shen, P.; Li, X.; Li, R.; Yang, L. A Paired-End Whole-Genome Sequencing Approach Enables Comprehensive Characterization of Transgene Integration in Rice. Commun Biol 2022, 5, 667. [Google Scholar] [CrossRef] [PubMed]
- Kovalic, D.; Garnaat, C.; Guo, L.; Yan, Y.; Groat, J.; Silvanovich, A.; Ralston, L.; Huang, M.; Tian, Q.; Christian, A.; et al. The Use of Next Generation Sequencing and Junction Sequence Analysis Bioinformatics to Achieve Molecular Characterization of Crops Improved Through Modern Biotechnology. The Plant Genome 2012, 5, plantgenome2012.10–0026. [Google Scholar] [CrossRef]
- Volpicella, M.; Leoni, C.; Costanza, A.; Fanizza, I.; Placido, A.; Ceci, L.R. Genome Walking by Next Generation Sequencing Approaches. Biology 2012, 1, 495–507. [Google Scholar] [CrossRef]
- Zhao, S.; Wang, Y.; Zhu, Z.; Chen, P.; Liu, W.; Wang, C.; Lu, H.; Xiang, Y.; Liu, Y.; Qian, Q.; et al. Streamlined Whole-Genome Genotyping through NGS-Enhanced Thermal Asymmetric Interlaced (TAIL)-PCR. Plant Communications 2024, 5, 100983. [Google Scholar] [CrossRef]
- Salnikov, P.A.; Khabarova, A.A.; Koksharova, G.S.; Mungalov, R.V.; Belokopytova, P.S.; Pristyazhnuk, I.E.; Nurislamov, A.R.; Somatich, P.; Gridina, M.M.; Fishman, V.S. Here and There: The Double-Side Transgene Localization. Vestn. VOGiS 2021, 25, 607–612. [Google Scholar] [CrossRef]
- Malekshoar, M.; Azimi, S.A.; Kaki, A.; Mousazadeh, L.; Motaei, J.; Vatankhah, M. CRISPR-Cas9 Targeted Enrichment and Next-Generation Sequencing for Mutation Detection. The Journal of Molecular Diagnostics 2023, 25, 249–262. [Google Scholar] [CrossRef]
- De Vree, P.J.P.; De Wit, E.; Yilmaz, M.; Van De Heijning, M.; Klous, P.; Verstegen, M.J.A.M.; Wan, Y.; Teunissen, H.; Krijger, P.H.L.; Geeven, G.; et al. Targeted Sequencing by Proximity Ligation for Comprehensive Variant Detection and Local Haplotyping. Nat Biotechnol 2014, 32, 1019–1025. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, C.; Kambara, H.; Dambrot, C.; Xie, X.; Zhao, L.; Xu, R.; Oneglia, A.; Liu, F.; Luo, H.R. Identification of the Transgene Integration Site and Host Genome Changes in MRP8-Cre/Ires-EGFP Transgenic Mice by Targeted Locus Amplification. Front. Immunol. 2022, 13, 875991. [Google Scholar] [CrossRef]
- Stadermann, A.; Gamer, M.; Fieder, J.; Lindner, B.; Fehrmann, S.; Schmidt, M.; Schulz, P.; Gorr, I.H. Structural Analysis of Random Transgene Integration in CHO Manufacturing Cell Lines by Targeted Sequencing. Biotech & Bioengineering 2022, 119, 868–880. [Google Scholar] [CrossRef]
- Lefferts, J.W.; Boersma, V.; Hagemeijer, M.C.; Hajo, K.; Beekman, J.M.; Splinter, E. Targeted Locus Amplification and Haplotyping. In Haplotyping; Peters, B.A., Drmanac, R., Eds.; Methods in Molecular Biology; Springer US: New York, NY, 2023; Vol. 2590, pp. 31–48 ISBN 978-1-07-162818-8.
- Tosh, J.L.; Rickman, M.; Rhymes, E.; Norona, F.E.; Clayton, E.; Mucke, L.; Isaacs, A.M.; Fisher, E.M.C.; Wiseman, F.K. The Integration Site of the APP Transgene in the J20 Mouse Model of Alzheimer’s Disease. Wellcome Open Res 2018, 2, 84. [Google Scholar] [CrossRef] [PubMed]
- Hinteregger, B.; Loeffler, T.; Flunkert, S.; Neddens, J.; Birner-Gruenberger, R.; Bayer, T.A.; Madl, T.; Hutter-Paier, B. Transgene Integration Causes RARB Downregulation in Homozygous Tg4–42 Mice. Sci Rep 2020, 10, 6377. [Google Scholar] [CrossRef]
- Wong, A.M.; Patel, T.P.; Altman, E.K.; Tugarinov, N.; Trivellin, G.; Yanovski, J.A. Characterization of the Adiponectin Promoter + Cre Recombinase Insertion in the Tg(Adipoq-Cre)1Evdr Mouse by Targeted Locus Amplification and Droplet Digital PCR. Adipocyte 2021, 10, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Chen, W.; Wei, R.; Qiang, W.; Pearson, J.D.; Yu, T.; Bremner, R.; Chen, D. Mapping Transgene Insertion Sites Reveals the α-Cre Transgene Expression in Both Developing Retina and Olfactory Neurons. Commun Biol 2022, 5, 411. [Google Scholar] [CrossRef]
- Leitner, K.; Motheramgari, K.; Borth, N.; Marx, N. Nanopore Cas9-targeted Sequencing Enables Accurate and Simultaneous Identification of Transgene Integration Sites, Their Structure and Epigenetic Status in Recombinant Chinese Hamster Ovary Cells. Biotech & Bioengineering 2023, 120, 2403–2418. [Google Scholar] [CrossRef]
- Clappier, C.; Böttner, D.; Heinzelmann, D.; Stadermann, A.; Schulz, P.; Schmidt, M.; Lindner, B. Deciphering Integration Loci of CHO Manufacturing Cell Lines Using Long Read Nanopore Sequencing. New Biotechnology 2023, 75, 31–39. [Google Scholar] [CrossRef]
- DuBose, A.J.; Lichtenstein, S.T.; Narisu, N.; Bonnycastle, L.L.; Swift, A.J.; Chines, P.S.; Collins, F.S. Use of Microarray Hybrid Capture and Next-Generation Sequencing to Identify the Anatomy of a Transgene. Nucleic Acids Research 2013, 41, e70–e70. [Google Scholar] [CrossRef]
- Magembe, E.M.; Li, H.; Taheri, A.; Zhou, S.; Ghislain, M. Identification of T-DNA Structure and Insertion Site in Transgenic Crops Using Targeted Capture Sequencing. Front. Plant Sci. 2023, 14, 1156665. [Google Scholar] [CrossRef]
- Ohnuki, N.; Kobayashi, T.; Matsuo, M.; Nishikaku, K.; Kusama, K.; Torii, Y.; Inagaki, Y.; Hori, M.; Imakawa, K.; Satou, Y. A Target Enrichment High Throughput Sequencing System for Characterization of BLV Whole Genome Sequence, Integration Sites, Clonality and Host SNP. Sci Rep 2021, 11, 4521. [Google Scholar] [CrossRef]
- Iwase, S.C.; Miyazato, P.; Katsuya, H.; Islam, S.; Yang, B.T.J.; Ito, J.; Matsuo, M.; Takeuchi, H.; Ishida, T.; Matsuda, K.; et al. HIV-1 DNA-Capture-Seq Is a Useful Tool for the Comprehensive Characterization of HIV-1 Provirus. Sci Rep 2019, 9, 12326. [Google Scholar] [CrossRef]
- Li, X.; Chen, W.; Martin, B.K.; Calderon, D.; Lee, C.; Choi, J.; Chardon, F.M.; McDiarmid, T.A.; Daza, R.M.; Kim, H.; et al. Chromatin Context-Dependent Regulation and Epigenetic Manipulation of Prime Editing. Cell 2024, 187, 2411–2427.e25. [Google Scholar] [CrossRef] [PubMed]
- Siddique, K.; Wei, J.; Li, R.; Zhang, D.; Shi, J. Identification of T-DNA Insertion Site and Flanking Sequence of a Genetically Modified Maize Event IE09S034 Using Next-Generation Sequencing Technology. Mol Biotechnol 2019, 61, 694–702. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Mei, Y.; Ding, L.; Wang, X.; Chen, X.; Wang, J.; Xu, J. Using Combined Methods of Genetic Mapping and Nanopore-Based Sequencing Technology to Analyze the Insertion Positions of G10evo-EPSPS and Cry1Ab/Cry2Aj Transgenes in Maize. Front. Plant Sci. 2021, 12, 690951. [Google Scholar] [CrossRef]
- Van Dijk, E.L.; Jaszczyszyn, Y.; Naquin, D.; Thermes, C. The Third Revolution in Sequencing Technology. Trends in Genetics 2018, 34, 666–681. [Google Scholar] [CrossRef]
- Warburton, P.E.; Sebra, R.P. Long-Read DNA Sequencing: Recent Advances and Remaining Challenges. Annu. Rev. Genom. Hum. Genet. 2023, 24, 109–132. [Google Scholar] [CrossRef]
- Adams, P.E.; Thies, J.L.; Sutton, J.M.; Millwood, J.D.; Caldwell, G.A.; Caldwell, K.A.; Fierst, J.L. Identifying Transgene Insertions in Caenorhabditis Elegans Genomes with Oxford Nanopore Sequencing. PeerJ 2024, 12, e18100. [Google Scholar] [CrossRef]
- Jain, M.; Koren, S.; Miga, K.H.; Quick, J.; Rand, A.C.; Sasani, T.A.; Tyson, J.R.; Beggs, A.D.; Dilthey, A.T.; Fiddes, I.T.; et al. Nanopore Sequencing and Assembly of a Human Genome with Ultra-Long Reads. Nat Biotechnol 2018, 36, 338–345. [Google Scholar] [CrossRef]
- Huang, M.; Kingan, S.; Shoue, D.; Nguyen, O.; Froenicke, L.; Galvin, B.; Lambert, C.; Khan, R.; Maheshwari, C.; Weisz, D.; et al. Improved High Quality Sand Fly Assemblies Enabled by Ultra Low Input Long Read Sequencing. Sci Data 2024, 11, 918. [Google Scholar] [CrossRef]
- Meier, M.J.; Beal, M.A.; Schoenrock, A.; Yauk, C.L.; Marchetti, F. Whole Genome Sequencing of the Mutamouse Model Reveals Strain- and Colony-Level Variation, and Genomic Features of the Transgene Integration Site. Sci Rep 2019, 9, 13775. [Google Scholar] [CrossRef]
- Zhang, H.; Li, R.; Guo, Y.; Zhang, Y.; Zhang, D.; Yang, L. LIFE-Seq: A Universal L Arge I Ntegrated DNA F Ragment E Nrichment Seq Uencing Strategy for Deciphering the Transgene Integration of Genetically Modified Organisms. Plant Biotechnology Journal 2022, 20, 964–976. [Google Scholar] [CrossRef]
- Sheehan, M.; Kumpf, S.W.; Qian, J.; Rubitski, D.M.; Oziolor, E.; Lanz, T.A. Comparison and Cross-Validation of Long-Read and Short-Read Target-Enrichment Sequencing Methods to Assess AAV Vector Integration into Host Genome. Molecular Therapy - Methods & Clinical Development 2024, 32, 101352. [Google Scholar] [CrossRef]
- Suzuki, O.; Koura, M.; Uchio-Yamada, K.; Sasaki, M. Analysis of the Transgene Insertion Pattern in a Transgenic Mouse Strain Using Long-Read Sequencing. Exp. Anim. 2020, 69, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Gilpatrick, T.; Lee, I.; Graham, J.E.; Raimondeau, E.; Bowen, R.; Heron, A.; Downs, B.; Sukumar, S.; Sedlazeck, F.J.; Timp, W. Targeted Nanopore Sequencing with Cas9-Guided Adapter Ligation. Nat Biotechnol 2020, 38, 433–438. [Google Scholar] [CrossRef]
- Smirnov, A.; Nurislamov, A.; Koncevaya, G.; Serova, I.; Kabirova, E.; Chuyko, E.; Maltceva, E.; Savoskin, M.; Zadorozhny, D.; Svyatchenko, V.A.; et al. Characterizing a Lethal CAG-ACE2 Transgenic Mouse Model for SARS-CoV-2 Infection Using Cas9-Enhanced Nanopore Sequencing. Transgenic Res 2024, 33, 453–466. [Google Scholar] [CrossRef]
- Blondal, T.; Gamba, C.; Møller Jagd, L.; Su, L.; Demirov, D.; Guo, S.; Johnston, C.M.; Riising, E.M.; Wu, X.; Mikkelsen, M.J.; et al. Verification of CRISPR Editing and Finding Transgenic Inserts by Xdrop Indirect Sequence Capture Followed by Short- and Long-Read Sequencing. Methods 2021, 191, 68–77. [Google Scholar] [CrossRef]
- Zarka, K.A.; Jagd, L.M.; Douches, D.S. T-DNA Characterization of Genetically Modified 3-R-Gene Late Blight-Resistant Potato Events with a Novel Procedure Utilizing the Samplix Xdrop® Enrichment Technology. Front. Plant Sci. 2024, 15, 1330429. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Y.; Bollas, A.; Wang, Y.; Au, K.F. Nanopore Sequencing Technology, Bioinformatics and Applications. Nat Biotechnol 2021, 39, 1348–1365. [Google Scholar] [CrossRef]
- Li, S.; Jia, S.; Hou, L.; Nguyen, H.; Sato, S.; Holding, D.; Cahoon, E.; Zhang, C.; Clemente, T.; Yu, B. Mapping of Transgenic Alleles in Soybean Using a Nanopore-Based Sequencing Strategy. Journal of Experimental Botany 2019, 70, 3825–3833. [Google Scholar] [CrossRef]
- Giraldo, P.A.; Shinozuka, H.; Spangenberg, G.C.; Smith, K.F.; Cogan, N.O.I. Rapid and Detailed Characterization of Transgene Insertion Sites in Genetically Modified Plants via Nanopore Sequencing. Front. Plant Sci. 2021, 11, 602313. [Google Scholar] [CrossRef]
- Steiert, T.A.; Fuß, J.; Juzenas, S.; Wittig, M.; Hoeppner, M.P.; Vollstedt, M.; Varkalaite, G.; ElAbd, H.; Brockmann, C.; Görg, S.; et al. High-Throughput Method for the Hybridisation-Based Targeted Enrichment of Long Genomic Fragments for PacBio Third-Generation Sequencing. NAR Genomics and Bioinformatics 2022, 4, lqac051. [Google Scholar] [CrossRef]
- Lefoulon, E.; Vaisman, N.; Frydman, H.M.; Sun, L.; Voland, L.; Foster, J.M.; Slatko, B.E. Large Enriched Fragment Targeted Sequencing (LEFT-SEQ) Applied to Capture of Wolbachia Genomes. Sci Rep 2019, 9, 5939. [Google Scholar] [CrossRef]
- Tilleman, L.; Rubben, K.; Van Criekinge, W.; Deforce, D.; Van Nieuwerburgh, F. Haplotyping Pharmacogenes Using TLA Combined with Illumina or Nanopore Sequencing. Sci Rep 2022, 12, 17734. [Google Scholar] [CrossRef]
- Hafford-Tear, N.J.; Tsai, Y.-C.; Sadan, A.N.; Sanchez-Pintado, B.; Zarouchlioti, C.; Maher, G.J.; Liskova, P.; Tuft, S.J.; Hardcastle, A.J.; Clark, T.A.; et al. CRISPR/Cas9-Targeted Enrichment and Long-Read Sequencing of the Fuchs Endothelial Corneal Dystrophy–Associated TCF4 Triplet Repeat. Genetics in Medicine 2019, 21, 2092–2102. [Google Scholar] [CrossRef]
- Tsai, Y.-; Brown, K.; Bernardi, M.; Harting, J.; Clelland, C. Single-Molecule Sequencing of the C9orf72 Repeat Expansion in Patient iPSCs. BIO-PROTOCOL 2024, 14. [Google Scholar] [CrossRef]
- Watson, C.M.; Crinnion, L.A.; Lindsay, H.; Mitchell, R.; Camm, N.; Robinson, R.; Joyce, C.; Tanteles, G.A.; Halloran, D.J.O.; Pena, S.D.J.; et al. Assessing the Utility of Long-Read Nanopore Sequencing for Rapid and Efficient Characterization of Mobile Element Insertions. Laboratory Investigation 2021, 101, 442–449. [Google Scholar] [CrossRef]
- Stangl, C.; De Blank, S.; Renkens, I.; Westera, L.; Verbeek, T.; Valle-Inclan, J.E.; González, R.C.; Henssen, A.G.; Van Roosmalen, M.J.; Stam, R.W.; et al. Partner Independent Fusion Gene Detection by Multiplexed CRISPR-Cas9 Enrichment and Long Read Nanopore Sequencing. Nat Commun 2020, 11, 2861. [Google Scholar] [CrossRef]
- Xu, S.; Shiomi, H.; Yamashita, Y.; Koyama, S.; Horie, T.; Baba, O.; Kimura, M.; Nakashima, Y.; Sowa, N.; Hasegawa, K.; et al. CRISPR-Cas9-Guided Amplification-Free Genomic Diagnosis for Familial Hypercholesterolemia Using Nanopore Sequencing. PLoS ONE 2024, 19, e0297231. [Google Scholar] [CrossRef]
- López-Girona, E.; Davy, M.W.; Albert, N.W.; Hilario, E.; Smart, M.E.M.; Kirk, C.; Thomson, S.J.; Chagné, D. CRISPR-Cas9 Enrichment and Long Read Sequencing for Fine Mapping in Plants. Plant Methods 2020, 16, 121. [Google Scholar] [CrossRef]
- McDonald, T.L.; Zhou, W.; Castro, C.P.; Mumm, C.; Switzenberg, J.A.; Mills, R.E.; Boyle, A.P. Cas9 Targeted Enrichment of Mobile Elements Using Nanopore Sequencing. Nat Commun 2021, 12, 3586. [Google Scholar] [CrossRef]
- Hertel, O.; Neuss, A.; Busche, T.; Brandt, D.; Kalinowski, J.; Bahnemann, J.; Noll, T. Enhancing Stability of Recombinant CHO Cells by CRISPR/Cas9-Mediated Site-Specific Integration into Regions with Distinct Histone Modifications. Front. Bioeng. Biotechnol. 2022, 10, 1010719. [Google Scholar] [CrossRef]
- Reginato, G.; Dello Stritto, M.R.; Wang, Y.; Hao, J.; Pavani, R.; Schmitz, M.; Halder, S.; Morin, V.; Cannavo, E.; Ceppi, I.; et al. HLTF Disrupts Cas9-DNA Post-Cleavage Complexes to Allow DNA Break Processing. Nat Commun 2024, 15, 5789. [Google Scholar] [CrossRef]
- Keraite, I.; Becker, P.; Canevazzi, D.; Frias-López, C.; Dabad, M.; Tonda-Hernandez, R.; Paramonov, I.; Ingham, M.J.; Brun-Heath, I.; Leno, J.; et al. A Method for Multiplexed Full-Length Single-Molecule Sequencing of the Human Mitochondrial Genome. Nat Commun 2022, 13, 5902. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Lan, X.; Zhang, T.; Sun, H.; Ma, S.; Xia, Q. Precise Characterization of Bombyx Mori Fibroin Heavy Chain Gene Using Cpf1-Based Enrichment and Oxford Nanopore Technologies. Insects 2021, 12, 832. [Google Scholar] [CrossRef]
- Madsen, E.B.; Höijer, I.; Kvist, T.; Ameur, A.; Mikkelsen, M.J. Xdrop: Targeted Sequencing of Long DNA Molecules from Low Input Samples Using Droplet Sorting. Human Mutation 2020, 41, 1671–1679. [Google Scholar] [CrossRef] [PubMed]
| Preparation price per sample | Sequencing price per sample |
Total / On-target data (coverage) | Preparation / Run time |
|
|---|---|---|---|---|
| Inverse PCR [30] | 20-30$ | 10-30$ (Sanger) | Several kb | ~ 9-12/3-4 h |
| TAIL-PCR [47] | 40-50$ | 10-30$ (Sanger) | Several kb | ~ 8-12/3-4 h |
|
WGS by NGS (Illumina paired-end 150 bp) [51,67,68,70] |
75-135$ |
NGS Option A: NovaSeq 6000 S4 ~160-250$ NGS Option B: NextSeq 500/550 ~1900-2400$ |
30 GB / >0.01% (coverage >10) |
NGS Option A: ~ 3-5/45 h NGS Option B: ~ 3-5/35 h |
|
NGS + TLA (commercial) [12,13,87] |
1000-2000 $ | NA | Weeks | |
|
NGS + TLA (lab) [82,83,84] |
50-75$ |
NGS Option A: ~35-70$ NGS Option B: ~200-250$ |
3 GB / ~30–70% (coverage >30) |
NGS Option A: ~36-48/35 h NGS Option B: ~36-48/45 h |
|
NGS + hybrid capture (using 120 nt commercial tiling probes) [92,93] |
180-250$ |
NGS Option A: ~10-20$ NGS Option B: ~75-150$ |
1 GB / ~40–80%, up to 95% based on the probes (coverage >30) |
NGS Option A: ~24-36/45 h NGS Option B: ~24-36/35 h |
|
NGS + hybrid capture (probes made in the lab) |
50-60$ |
NGS Option A: ~10-20$ NGS Option B: ~75-150$ |
1 GB / ~80–90%, up to 93% (coverage >50) |
NGS Option A: ~50/45 h NGS Option B: ~50/35 h |
|
NGS + T7 In vitro transcription [95] |
50–70$ |
NGS Option A: ~35-70$ NGS Option B: ~200-250$ |
3GB / ~35–70% (coverage >30) |
NGS Option A: ~6-9/45 h NGS Option B: ~6-9/35 h |
| PacBio WGS [32,105] | ~100–150$ | 900-1600$ | 45-90 GB / >0.01% (coverage >15-25) |
6-10/24-36 h |
| PacBio + hybrid capture (using 120nt commercial probes) [104] | ~350–500$ | 125-200 $ | 5-10 GB / 40–60% (coverage >30) |
30-40/24-36 h |
|
Oxford Nanopore Technologies (ONT) WGS [15,100,106] |
~100–150$ |
ONT Option A: MinION, 2-3 flow cells 1200–2400$ ONT Option B: PromethION (shared) 300-600$ |
60-90 GB / >0.01% (coverage 20-30) |
ONT Option A: ~5-7/24-60 h ONT Option B: ~5-7/48-72 h |
|
ONT + nCATs [23,90,107] |
~160–200$ |
ONT Option A (1 flow cell): 600–800$ ONT Option B: 100-150$ |
30 GB / 10–40%, depends on the gRNA (coverage 20-30) |
ONT Option A: ~7-10/24-60 h ONT Option B: ~7-10/48-72 h |
|
ONT + internal cuts (AFIS-seq, CRISPR-LRS) [24,44,108] |
150-200$ |
ONT Option A (1 flow cell): 600–800$ ONT Option B: 100-150$ |
30 GB / 5-40%, depends on the gRNA (coverage >30) |
ONT Option A: ~7-10/24-60 h ONT Option B: ~7-10/48-72 h |
|
Nanopore + Xdrop (commercial) [16,109,110] |
650-900$ |
ONT Option A (1 flow cell): 600–800$ ONT Option B: 100-150$ |
30 GB / ~60–90% (coverage >100) |
ONT Option A: ~4-5 days/24-60 h ONT Option B: ~4-5 days/48-72 h |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





