1. Introduction
Deep learning has significantly advanced medical image analysis, allowing automated disease classification with high accuracy through architectures such as Convolutional Neural Networks (CNNs) and Vision Transformers (ViTs) [
1,
2]. These techniques excel in human medical imaging, including the detection of tumors and retinal disorders [
3], and are increasingly applied to veterinary imaging, where early diagnosis is critical for animal health [
4]. Canine ocular diseases—such as eyelid tumors, nuclear sclerosis, cataracts, ulcerative keratitis, and epiphora—pose diagnostic challenges due to subtle visual differences and variable presentations. Traditional diagnosis relies heavily on specialized veterinary ophthalmologists, which may not be readily available in many geographic regions, and manual examination is subject to variability between observers [
5].
With a comprehensive data set of 44,637 canine eye images divided into training (75%), validation (10%) and test (15%) sets, there is a growing opportunity to develop robust automated classification methods tailored to this domain. Traditional deep learning approaches in medical imaging are based primarily on spatial features extracted from raw RGB images [
6]. However, these approaches may miss valuable diagnostic features that are more apparent in other domains.
Frequency domain representations, such as those derived from Fourier Transform (FT) and Wavelet Transform (WT), offer complementary insights by capturing global frequency distributions and localized texture variations, respectively [
7,
8]. The Fourier transform decomposes images into constituent frequency components, revealing periodic patterns regardless of spatial location, while wavelets provide localized frequency information, preserving both frequency and spatial details. Although these techniques have shown promise in human imaging applications, such as improving microcalcification detection on mammograms [
9], their integration into the classification of veterinary ocular diseases remains largely unexplored. Existing studies have focused mainly on spatial feature-based models, leaving untapped potential to take advantage of frequency domain information for improved diagnostic precision [
4].
This study proposes a novel multimodal deep learning framework that integrates Vision Transformer models with Fourier and wavelet transforms to classify canine ocular diseases. We hypothesize that combining spatial and frequency domain features can enhance model performance by capturing subtle patterns not fully addressed by conventional methods. Our experimental design includes three distinct approaches: (1) classification using original images alone (baseline), (2) original images combined with Fourier-transformed images, and (3) original images with wavelet-transformed images.
Our key contributions include
Design and implementation of a dual input vision transformer (ViT) architecture that processes both original eye images and their frequency domain representations.
Comprehensive evaluation and demonstration of the efficacy of frequency domain features in veterinary ophthalmology.
Comparative analysis of Fourier and wavelet transformations to improve diagnostic precision in five common canine eye conditions.
The remainder of this paper is organized as follows. Section II presents related work on medical image analysis and multimodal learning; Section III details our methodology, including data preparation, model architecture, and training procedure; Section IV reports experimental results and comparative analysis; Section V discusses implications, limitations, and future directions; and Section VI concludes the article. These advances could improve veterinary diagnostic tools and inspire applications in other medical imaging fields.
2. Related Work
2.1. Deep Learning in Veterinary Medical Imaging
Deep learning applications in veterinary medicine have grown significantly in recent years. Nogueira et al. [
10] applied convolutional neural networks (CNN) to classify radiographic images of canine hip dysplasia with precision comparable to that of board-certified radiologists. Similarly, Nyquist et al. [
11] evaluated a novel artificial intelligence software program for veterinary dental radiography, showing good to excellent agreement with human evaluators in detecting common dental pathologies, such as periodontal conditions, in dogs and cats. Furthermore, Dumortier et al. [
12] employed a CNN based on ResNet50V2 to detect pulmonary abnormalities from lateral thoracic radiographs in cats, achieving an average accuracy of 82% and demonstrating the potential of deep learning in veterinary radiographic analysis. However, most existing approaches rely solely on spatial domain features extracted from original images, potentially missing valuable diagnostic information.
2.2. Multi-Modal Learning in Medical Imaging
Multi-modal learning leverages complementary information from different data sources or representations to improve performance [
13]. In human medical imaging, multimodal approaches have shown superior results compared to single-modal methods. For example, Zhang et al. [
14] combined magnetic resonance and computed tomography data to improve brain tumor segmentation, while Wang et al. [
15] integrated clinical metadata with radiographic images for a more accurate diagnosis of pneumonia. In ophthalmology specifically, Li et al. [
16] combined optical coherence tomography (OCT) and fundus photographs in an AI-based dual-modality analysis to detect diabetic retinopathy, achieving higher sensitivity and specificity compared to single-modal approaches.
2.2.1. Frequency Domain Analysis in Medical Imaging
Frequency domain transformations have long been used in medical image processing to enhance feature extraction. The Fourier transform decomposes an image into its constituent frequency components, uncovering periodic patterns regardless of their spatial location [
7]. In contrast, wavelet transforms offer localized frequency information, preserving both frequency and spatial details [
8]. In medical applications, Rahman et al. [
17] leveraged Fourier features to improve breast cancer detection on mammograms, while Abd El Kader et al. [
18] utilized wavelet transformations in a deep autoencoder model to improve brain tumor classification on magnetic resonance images, achieving high accuracy. Furthermore, Yoon et al. [
19] surveyed domain generalization techniques in medical image analysis, emphasizing how frequency domain methods can improve model robustness across diverse datasets, a critical aspect for extending such approaches to specialized domains like veterinary imaging. Despite these advances in human medicine, the application of frequency domain analysis in veterinary medical imaging remains largely unexplored, presenting an opportunity for novel contributions in this field.
2.3. Vision Transformers in Medical Image Analysis
Since their introduction by Dosovitskiy et al. [
1], Vision Transformers (ViT) have emerged as powerful alternatives to CNN for image analysis tasks. By applying the self-attention mechanism to image patches, ViTs can capture long-range dependencies and global context more effectively than CNNs, which rely on local convolution operations. In medical imaging, ViTs have demonstrated state-of-the-art performance across multiple tasks and modalities. Chen et al. [
20] adapted ViTs for chest X-ray classification, outperforming CNN-based approaches, while Hatamizadeh et al. [
21] proposed a ViT-based architecture for medical image segmentation that achieved superior results on several benchmark datasets. Our work builds on these advances, leveraging the strengths of ViTs within a novel multimodal framework that incorporates information from both the spatial and frequency domains for enhanced classification of canine ophthalmic diseases.
3. Materials and Methods
3.1. Dataset
We used a comprehensive data set of canine eye images collected from veterinary ophthalmology centers [
22]. The data set contains 44,637 images in five common conditions, with the distribution detailed in
Table 1.
The images were captured under standardized clinical conditions using ophthalmoscopes and specialized cameras. All diagnoses were confirmed by board-certified veterinary ophthalmologists. As shown in
Table 1, the data set was split into training sets (75%), validation sets (10%) and testing sets (15%), with stratification to maintain class distribution between groups.
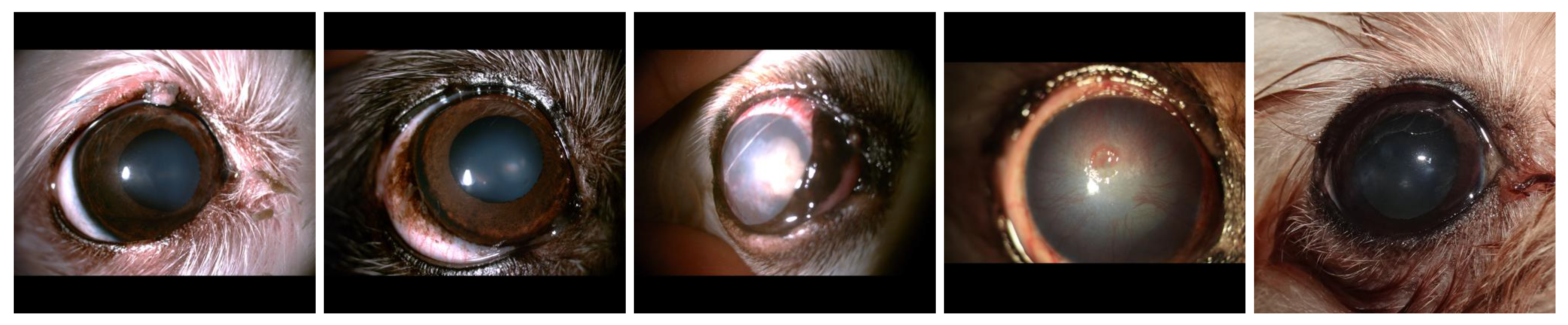
Figure 1.
Representative images from each disease category in our dataset: (a) Eyelid Tumor; (b) Nuclear Sclerosis; (c) Cataract; (d) Ulcerative Keratitis; (e) Epiphora.
Figure 1.
Representative images from each disease category in our dataset: (a) Eyelid Tumor; (b) Nuclear Sclerosis; (c) Cataract; (d) Ulcerative Keratitis; (e) Epiphora.
4. Image Preprocessing and Frequency Domain Transformations
4.1. Spatial Domain Preprocessing
All images were resized to 384×384 pixels using Lanczos resampling to preserve details during downscaling. Single channel (grayscale) images were converted to RGB format. For training augmentation, we applied random horizontal flipping, rotation (±15°), and color jittering (brightness, contrast, and saturation adjustments of up to 20%).
4.1.1. Fourier Transform Preprocessing
For the Fourier domain representation, we applied the 2D Fast Fourier Transform (FFT) to each color channel of the preprocessed images. The resulting frequency spectra were shifted to center the zero-frequency component and converted to a logarithmic scale to enhance visualization of high-frequency patterns:
where is the original image, is its Fourier transform, and is the logarithmically scaled spectrum.
4.1.2. Wavelet Transform Preprocessing
For the wavelet domain representation, we applied a discrete wavelet transform (DWT) using the Daubechies-4 wavelet. The DWT decomposes the image into approximation coefficients (low-frequency content) and detail coefficients (high-frequency content) at multiple scales. We utilized a 3-level decomposition, generating 10 coefficient matrices per color channel. These coefficients were normalized and combined into RGB-like images for input to the network.
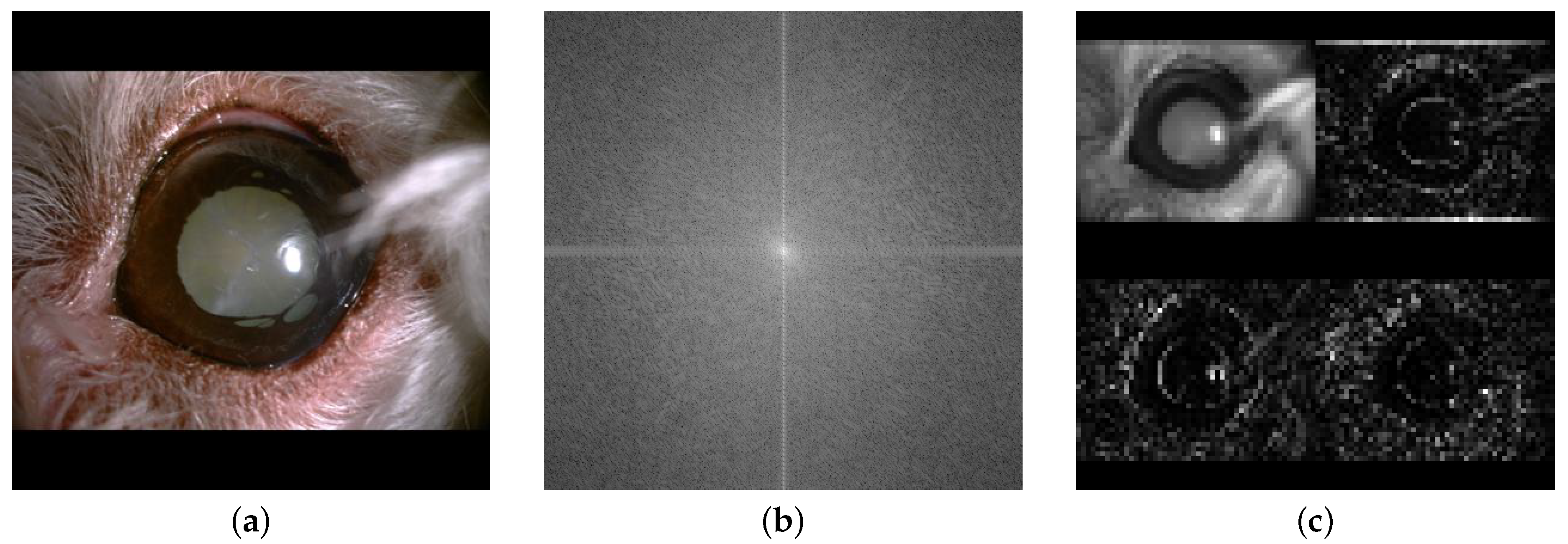
Figure 2.
Visual comparison of image representations of a canine eye with cataract: (a) Original image; (b) Fourier transform; (c) Wavelet transform.
Figure 2.
Visual comparison of image representations of a canine eye with cataract: (a) Original image; (b) Fourier transform; (c) Wavelet transform.
5. Model Architecture
We developed three distinct model architectures for comparative evaluation:
Baseline Model: A single-input Vision Transformer (ViT) that processes only the original images
Fourier Multi-modal Model: A dual-input ViT that processes both original images and their Fourier transformations
Wavelet Multi-modal Model: A dual-input ViT that processes both original images and their wavelet transformations
5.1. Baseline Model
The baseline model used a pre-trained ViT-Base architecture (patch size 16×16, 12 transformer layers, 12 attention heads, embedding dimension 768) fine-tuned for our specific classification task. The model was implemented using the HuggingFace Transformers library, with the pre-trained weights from the ’google/vit-base-patch16-384’ checkpoint.
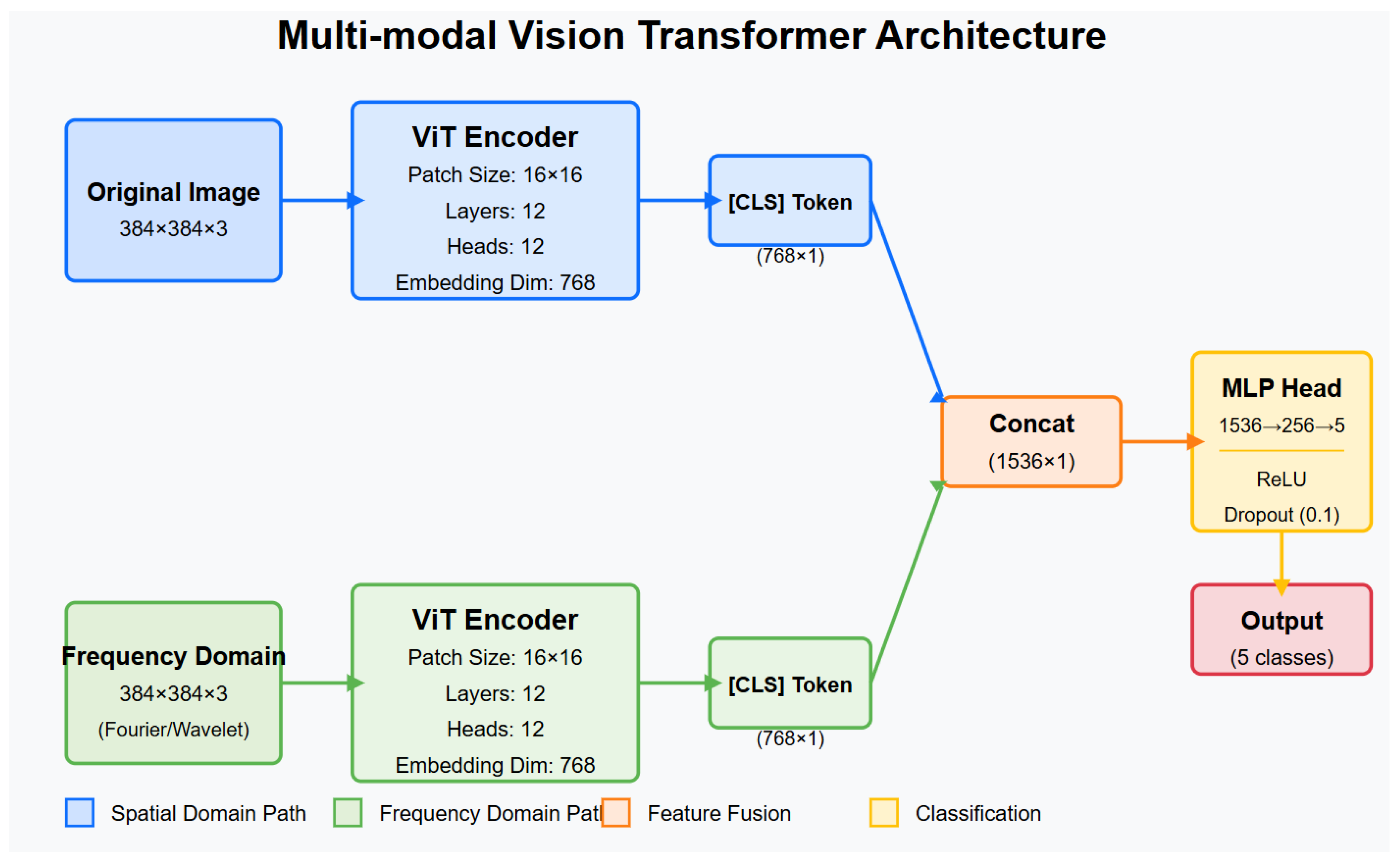
5.2. Multi-Modal Models
The multi-modal architectures (both Fourier and wavelet variants) consisted of:
-
Two parallel ViT-Base encoders:
A fusion module to combine the embeddings from both encoders
A classification head to produce the final predictions
The fusion module concatenated the [CLS] token embeddings (768 dimensional vectors) from both encoders, resulting in a representation of 1536 dimensional. This combined representation was then passed through a multilayer perceptron (MLP) with architecture:
Linear layer: 1536 → 256 units with ReLU activation
Dropout layer (rate = 0.1)
Linear layer: 256 → 5 units (one per class)
Figure 3.
Architecture diagram of the proposed multi-modal ViT model, showing parallel processing streams for spatial and frequency domain inputs with subsequent feature fusion.
Figure 3.
Architecture diagram of the proposed multi-modal ViT model, showing parallel processing streams for spatial and frequency domain inputs with subsequent feature fusion.
6. Training Procedure
All models were trained using similar hyperparameters to ensure a fair comparison:
- Optimizer: Adam with learning rate - Learning rate schedule: Step decay (gamma = 0.1) every 5 epochs - Loss function: Cross-entropy with class weights inversely proportional to class frequencies - Batch size: 32 - Training duration: Up to 25 epochs with early stopping (patience = 5 epochs) based on validation accuracy - Hardware: NVIDIA H100 80GB GPUs (multi-GPU training with DataParallel)
To address class imbalance, we applied inverse frequency weighting in the loss function:
where is the weight of the class c and is the frequency of class c in the training set.
The code was implemented in PyTorch, and all experiments were performed with the same random seed to ensure reproducibility. For model evaluation, we used accuracy, F1 score, ROC-AUC, and confusion matrices as the primary performance metrics.
7. Results
7.1. Classification Performance
Table 2 summarizes the performance metrics of the three models on the test set.
The Fourier-based multimodal model achieved the highest overall accuracy (86. 52%) and the F1 score (0.854), outperforming both the baseline single-modality model and the wavelet-based model. All models demonstrated excellent ROC-AUC scores (>0.98), indicating high discriminative power across all classes of disease.
7.2. Class-Wise Performance
Table 3 presents the class-wise accuracy and F1 scores for each model.
The Fourier-based multimodal model showed the best performance for four of five disease classes, with particularly notable improvements in eyelid tumor classification (+4. 43% accracy) and cataract detection (+2. 23% accuracy) compared to baseline. The wavelet-based model performed best for Nuclear Sclerosis, though by a small margin. In all models, Ulcerative Keratitis was the most accurately classified condition, while Nuclear Sclerosis proved to be the most challenging.
7.3. Confusion Matrix Analysis
The confusion matrices in
Figure 4 reveal the error patterns of each model. All three models showed similar trends of confusion, with nuclear sclerosis frequently misclassified as other conditions, particularly Epiphora and Cataract. The Fourier-based model (
Figure 4b) demonstrated reduced confusion, particularly for the classes of eyelid tumors and cataracts compared to baseline (
Figure 4a).
A notable pattern across all models was the relatively high accuracy for Ulcerative Keratitis (>92%), likely due to its distinctive visual presentation with clear corneal defects. The integration of frequency domain information in both multimodal approaches was particularly beneficial in differentiating between conditions with subtle visual differences, such as Eyelid Tumor and Cataract, where the Fourier-based model showed the most substantial improvements over the baseline.
8. Discussion
8.1. Interpretation of Results
Our experimental results demonstrate that incorporating frequency domain information consistently improves classification performance for canine ophthalmic diseases. The multimodal approaches outperformed the baseline model in all key metrics, confirming our hypothesis that frequency transformations can reveal complementary diagnostic features that are not readily apparent in the spatial domain.
The Fourier-based model’s superior performance, particularly for Eyelid Tumor and Cataract classes, suggests that global frequency patterns may be especially relevant for these conditions. Eyelid tumors often present with distinctive textural changes, whereas cataracts typically manifest as opacity patterns that may be more discernible in the frequency domain. For Nuclear Sclerosis, the slight advantage of the wavelet-based model indicates that localized frequency information might better capture the subtle, gradual changes characteristic of this condition.
Ulcerative Keratitis showed consistently high performance in all models, likely due to its distinctive presentation with clear corneal defects that are readily identifiable in both spatial and frequency domains. The similar performance across Epiphora models suggests that its key diagnostic features are equally well represented in all domains.
8.2. Technical Implications and Potential Applications
The improved performance metrics of our multimodal approach demonstrate technical advantages that could be applied in various contexts:
Model Architecture Efficiency: Our dual-input ViT architecture shows that integrating complementary data representations can yield performance improvements without requiring more complex backbone networks.
Frequency Domain Effectiveness: The consistent performance gains, particularly with Fourier transforms, suggest that frequency domain information could benefit other medical image classification tasks beyond veterinary applications.
Automated Analysis Framework: The proposed framework provides a pipeline for automated image analysis that could be adapted to other diagnostic imaging tasks where consistent evaluation is valuable.
Interpretable Results: The class-wise performance variations offer insights into which conditions benefit most from frequency domain analysis, potentially guiding future research directions.
The significant improvements in specific categories (eyelid tumors +4.43% and cataracts +2.23%) highlight the value of targeted multimodal approaches for challenging classification cases, which could be extended to other computer vision applications requiring fine-grained discrimination.
8.3. Limitations
Despite the promising results, several limitations should be acknowledged:
Dataset Composition: Although comprehensive, our data set may not represent the full diversity of presentations in different dog breeds, ages, and disease stages.
Computational Requirements: The dual encoder architecture significantly increases computational demands during both training and inference, potentially limiting deployment on resource-constrained devices.
Binary Approach to Disease: Our classification framework treats each condition as present or absent, whereas real-world cases often involve gradations of severity or co-occurring conditions.
Model Interpretability: The attention mechanisms of ViTs offer some interpretability, but more work is needed to provide clinically meaningful explanations for the predictions.
8.4. Future Directions
Several promising avenues for future research emerge from this work.
Model Optimization: Investigating more efficient fusion strategies and lightweight architectures to reduce computational requirements.
Additional Modalities: Incorporating clinical metadata, breed information, or other imaging modalities (e.g. ultrasound) could further enhance performance.
Explainable AI: Developing visualization techniques to highlight the specific features that contribute to diagnoses, enhancing clinician trust and adoption.
Severity Grading: Extending the framework to assess the severity of the disease, not just the presence, would provide more nuanced clinical information.
Comprehensive Benchmark Framework: Developing standardized evaluation protocols and benchmark datasets to systematically assess model performance across diverse image qualities, acquisition devices, and environmental conditions. This would enable objective comparison between different computational approaches and establish reliable metrics for real-world deployment readiness.
9. Conclusion
This study presented a novel multimodal deep learning approach for the classification of canine ophthalmic diseases, utilizing information from the spatial and frequency domains through a dual input vision transformer architecture. Our results demonstrate that the integration of Fourier transformations with original images yields superior performance compared to both single-modality approaches and wavelet-based alternatives.
The proposed Fourier multimodal model achieved 86.52% precision, 0.854 F1 score and 0.983 ROC-AUC in five common canine eye conditions, with particular improvements in the detection of eyelid tumors (+4.43%) and cataracts (+2.23%). These findings highlight the value of frequency domain analysis in medical imaging applications and demonstrate the potential of multimodal approaches for complex classification tasks.
Future work will focus on model optimization, computational efficiency improvements, and developing robust benchmark frameworks to evaluate performance across diverse conditions. By advancing the technical capabilities of automated classification systems, this research contributes to the growing field of AI-assisted image analysis and provides a foundation for further computational innovations in medical imaging domains.
Acknowledgments
This research was supported by the MSIT (Ministry of Science ICT), Korea, under the National Program for Excellence in SW, supervised by the IITP (Institute of Information & Communications Technology Planning & Evaluation) in 2022 (2022-0-01091, 1711175863).
Abbreviations
The following abbreviations are used in this manuscript:
| ViT |
Vision Transformer |
| CNN |
Convolutional Neural Network |
| FT |
Fourier Transform |
| WT |
Wavelet Transform |
| FFT |
Fast Fourier Transform |
| DWT |
Discrete Wavelet Transform |
| ROC-AUC |
Receiver Operating Characteristic - Area Under Curve |
| MLP |
Multilayer Perceptron |
References
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint 2020. arXiv:2010.11929.
- Shamshad, F.; Khan, S.; Zamir, S.W.; Khan, M.H.; Hayat, M.; Khan, F.S.; Fu, H. Transformers in medical imaging: A survey. Medical image analysis 2023, 88, 102802.
- Takahashi, S.; Sakaguchi, Y.; Kouno, N.; Takasawa, K.; Ishizu, K.; Akagi, Y.; Aoyama, R.; Teraya, N.; Bolatkan, A.; Shinkai, N.; et al. Comparison of vision transformers and convolutional neural networks in medical image analysis: a systematic review. Journal of Medical Systems 2024, 48, 84. [CrossRef]
- Xiao, S.; Dhand, N.K.; Wang, Z.; Hu, K.; Thomson, P.C.; House, J.K.; Khatkar, M.S. Review of applications of deep learning in veterinary diagnostics and animal health. Frontiers in Veterinary Science 2025, 12, 1511522. [CrossRef]
- Boroffka, S.A.; Voorhout, G.; Verbruggen, A.M.; Teske, E. Intraobserver and interobserver repeatability of ocular biometric measurements obtained by means of B-mode ultrasonography in dogs. American journal of veterinary research 2006, 67, 1743–1749. [CrossRef]
- Zhou, S.K.; Greenspan, H.; Davatzikos, C.; Duncan, J.S.; Van Ginneken, B.; Madabhushi, A.; Prince, J.L.; Rueckert, D.; Summers, R.M. A review of deep learning in medical imaging: Imaging traits, technology trends, case studies with progress highlights, and future promises. Proceedings of the IEEE 2021, 109, 820–838. [CrossRef]
- Bracewell, R.N. The fourier transform. Scientific American 1989, 260, 86–95.
- Mallat, S.G. A theory for multiresolution signal decomposition: the wavelet representation. IEEE transactions on pattern analysis and machine intelligence 1989, 11, 674–693. [CrossRef]
- Mini, M.; Devassia, V.; Thomas, T. Multiplexed wavelet transform technique for detection of microcalcification in digitized mammograms. Journal of digital imaging 2004, 17, 285–291. [CrossRef]
- Gomes, D.A.; Alves-Pimenta, M.S.; Ginja, M.; Filipe, V. Predicting canine hip dysplasia in x-ray images using deep learning. In Proceedings of the International Conference on Optimization, Learning Algorithms and Applications. Springer, 2021, pp. 393–400.
- Nyquist, M.L.; Fink, L.A.; Mauldin, G.E.; Coffman, C.R. Evaluation of a novel veterinary dental radiography artificial intelligence software program. Journal of Veterinary Dentistry 2025, 42, 118–127. [CrossRef]
- Dumortier, L.; Guépin, F.; Delignette-Muller, M.L.; Boulocher, C.; Grenier, T. Deep learning in veterinary medicine, an approach based on CNN to detect pulmonary abnormalities from lateral thoracic radiographs in cats. Scientific reports 2022, 12, 11418. [CrossRef]
- Baltrušaitis, T.; Ahuja, C.; Morency, L.P. Multimodal machine learning: A survey and taxonomy. IEEE transactions on pattern analysis and machine intelligence 2018, 41, 423–443.
- Zhou, T.; Ruan, S.; Guo, Y.; Canu, S. A multi-modality fusion network based on attention mechanism for brain tumor segmentation. In Proceedings of the 2020 IEEE 17th international symposium on biomedical imaging (ISBI). IEEE, 2020, pp. 377–380.
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In Proceedings of the Proceedings of the IEEE conference on computer vision and pattern recognition, 2017, pp. 2097–2106.
- Liu, R.; Li, Q.; Xu, F.; Wang, S.; He, J.; Cao, Y.; Shi, F.; Chen, X.; Chen, J. Application of artificial intelligence-based dual-modality analysis combining fundus photography and optical coherence tomography in diabetic retinopathy screening in a community hospital. BioMedical Engineering OnLine 2022, 21, 47. [CrossRef]
- Rahman, M.M.; Bhattacharya, P.; Desai, B.C. A framework for medical image retrieval using machine learning and statistical similarity matching techniques with relevance feedback. IEEE transactions on Information Technology in Biomedicine 2007, 11, 58–69. [CrossRef]
- Abd El Kader, I.; Xu, G.; Shuai, Z.; Saminu, S.; Javaid, I.; Ahmad, I.S.; Kamhi, S. Brain tumor detection and classification on MR images by a deep wavelet auto-encoder model. diagnostics 2021, 11, 1589. [CrossRef]
- Yoon, J.S.; Oh, K.; Shin, Y.; Mazurowski, M.A.; Suk, H.I. Domain generalization for medical image analysis: A survey. arXiv preprint arXiv:2310.08598 2023.
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306 2021.
- Hatamizadeh, A.; Tang, Y.; Nath, V.; Yang, D.; Myronenko, A.; Landman, B.; Roth, H.R.; Xu, D. Unetr: Transformers for 3d medical image segmentation. In Proceedings of the Proceedings of the IEEE/CVF winter conference on applications of computer vision, 2022, pp. 574–584.
- Hub, A. Canine Ophthalmic Disease Dataset. Available online: https://aihub.or.kr/aihubdata/data/view.do?currMenu=&topMenu=&aihubDataSe=data&dataSetSn=562 (accessed on 3 Feb 2025), 2021. AI Hub Open Dataset.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).