Submitted:
14 April 2025
Posted:
15 April 2025
You are already at the latest version
Abstract
Keywords:
Introduction
Methods
Results
Discussion
Conclusion
Author Contributions
Fund Sources
Data Availability Statement
Conflict of Interest
Statement of Ethics
References
- Tianzhu Song, Dingming Huang, Dongzhe Song, The potential regulatory role of BMP9 in inflammatory responses, Genes & Diseases, Volume 9, Issue 6, 2022, Pages 1566-1578, ISSN 2352-3042. [CrossRef]
- Zhenquan Wei, Richard M. Salmon, Paul D. Upton, Nicholas W. Morrell, Wei Li, Regulation of Bone Morphogenetic Protein 9 (BMP9) by Redox-dependent Proteolysis*, Journal of Biological Chemistry, Volume 289, Issue 45,2014,Pages 31150-31159,ISSN 0021-9258, . [CrossRef]
- Wei Z, Salmon RM, Upton PD, Morrell NW, Li W. Regulation of bone morphogenetic protein 9 (BMP9) by redox-dependent proteolysis. J Biol Chem. 2014 Nov 7;289(45):31150-9. Epub 2014 Sep 18. [CrossRef] [PubMed] [PubMed Central]
- Bakr A, Hey J, Sigismondo G, Liu CS, Sadik A, Goyal A, Cross A, Iyer RL, Müller P, Trauernicht M, Breuer K, Lutsik P, Opitz CA, Krijgsveld J, Weichenhan D, Plass C, Popanda O, Schmezer P. ID3 promotes homologous recombination via non-transcriptional and transcriptional mechanisms and its loss confers sensitivity to PARP inhibition. Nucleic Acids Res. 2021 Nov 18;49(20):11666-11689; Erratum in: Nucleic Acids Res. 2023 Nov 10;51(20):11407. https://doi.org/10.1093/nar/gkad877. [CrossRef] [PubMed] [PubMed Central]
- Avecilla, V. Effect of Transcriptional Regulator ID3 on Pulmonary Arterial Hypertension and Hereditary Hemorrhagic Telangiectasia. Int J Vasc Med. 2019 Jul 11; 2019:2123906.
- Avecilla V, Doke M, Appunni S, Rubens M, Ramamoorthy V, Das JK. Pathophysiological Features of Remodeling in Vascular Diseases: Impact of Inhibitor of DNA-Binding/Differentiation-3 and Estrogenic Endocrine Disruptors. Medical Sciences. 2025; 13(1):2. [CrossRef]
- Avecilla, V. Gene Interactions in Cerebral Cavernous Malformations: A Brief Report. Preprints 2024, 2024050746. [CrossRef]
- Mostafa S, Pakvasa M, Coalson E, Zhu A, Alverdy A, Castillo H, Fan J, Li A, Feng Y, Wu D, Bishop E, Du S, Spezia M, Li A, Hagag O, Deng A, Liu W, Li M, Ho SS, Athiviraham A, Lee MJ, Wolf JM, Ameer GA, Luu HH, Haydon RC, Strelzow J, Hynes K, He TC, Reid RR. The wonders of BMP9: From mesenchymal stem cell differentiation, angiogenesis, neurogenesis, tumorigenesis, and metabolism to regenerative medicine. Genes Dis. 2019 Jul 24;6(3):201-223. [CrossRef] [PubMed Central]
- Ma J, van der Zon G, Gonçalves MAFV, van Dinther M, Thorikay M, Sanchez-Duffhues G, Ten Dijke P. TGF-β-Induced Endothelial to Mesenchymal Transition Is Determined by a Balance Between SNAIL and ID Factors. Front Cell Dev Biol. 2021 Feb 12;9:616610. [CrossRef] [PubMed] [PubMed Central]
- Yang J, Li X, Li Y, Southwood M, Ye L, Long L, Al-Lamki RS, Morrell NW. Id proteins are critical downstream effectors of BMP signaling in human pulmonary arterial smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2013 Aug 15;305(4):L312-21. Epub 2013 Jun 14. [CrossRef] [PubMed] [PubMed Central]
- Young K, Conley B, Romero D, Tweedie E, O'Neill C, Pinz I, Brogan L, Lindner V, Liaw L, Vary CP. BMP9 regulates endoglin-dependent chemokine responses in endothelial cells. Blood. 2012 Nov 15;120(20):4263-73. Epub 2012 Sep 26. Erratum in: Blood. 2018 Dec 6;132(23):2526. https://doi.org/10.1182/blood-2018-10-882589. [CrossRef] [PubMed] [PubMed Central]
- Avecilla, Vincent E., "ID3, Estrogenic Chemicals, and the Pathogenesis of Tumor-Like Proliferative Vascular Lesions" (2017). FIU Electronic Theses and Dissertations. 3519. https://digitalcommons.fiu.edu/etd/3519.
- Tanya Barrett, Stephen E. Wilhite, Pierre Ledoux, Carlos Evangelista, Irene F. Kim, Maxim Tomashevsky, Kimberly A. Marshall, Katherine H. Phillippy, Patti M. Sherman, Michelle Holko, Andrey Yefanov, Hyeseung Lee, Naigong Zhang, Cynthia L. Robertson, Nadezhda Serova, Sean Davis, Alexandra Soboleva, NCBI GEO: archive for functional genomics data sets—update, Nucleic Acids Research, Volume 41, Issue D1, 1 January 2013, Pages D991–D995. [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47.
- 15. David Warde-Farley, Sylva L. Donaldson, Ovi Comes, Khalid Zuberi, Rashad Badrawi, Pauline Chao, Max Franz, Chris Grouios, Farzana Kazi, Christian Tannus Lopes, Anson Maitland, Sara Mostafavi, Jason Montojo, Quentin Shao, George Wright, Gary D. Bader, Quaid Morris, The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function, Nucleic Acids Research, Volume 38, Issue suppl_2, 1 July 2010, Pages W214–W220, . [CrossRef]
- Kim JH, Peacock MR, George SC, Hughes CC. BMP9 induces EphrinB2 expression in endothelial cells through an Alk1-BMPRII/ActRII-ID1/ID3-dependent pathway: implications for hereditary hemorrhagic telangiectasia type II. Angiogenesis. 2012 Sep;15(3):497-509. Epub 2012 May 24. [CrossRef] [PubMed] [PubMed Central]
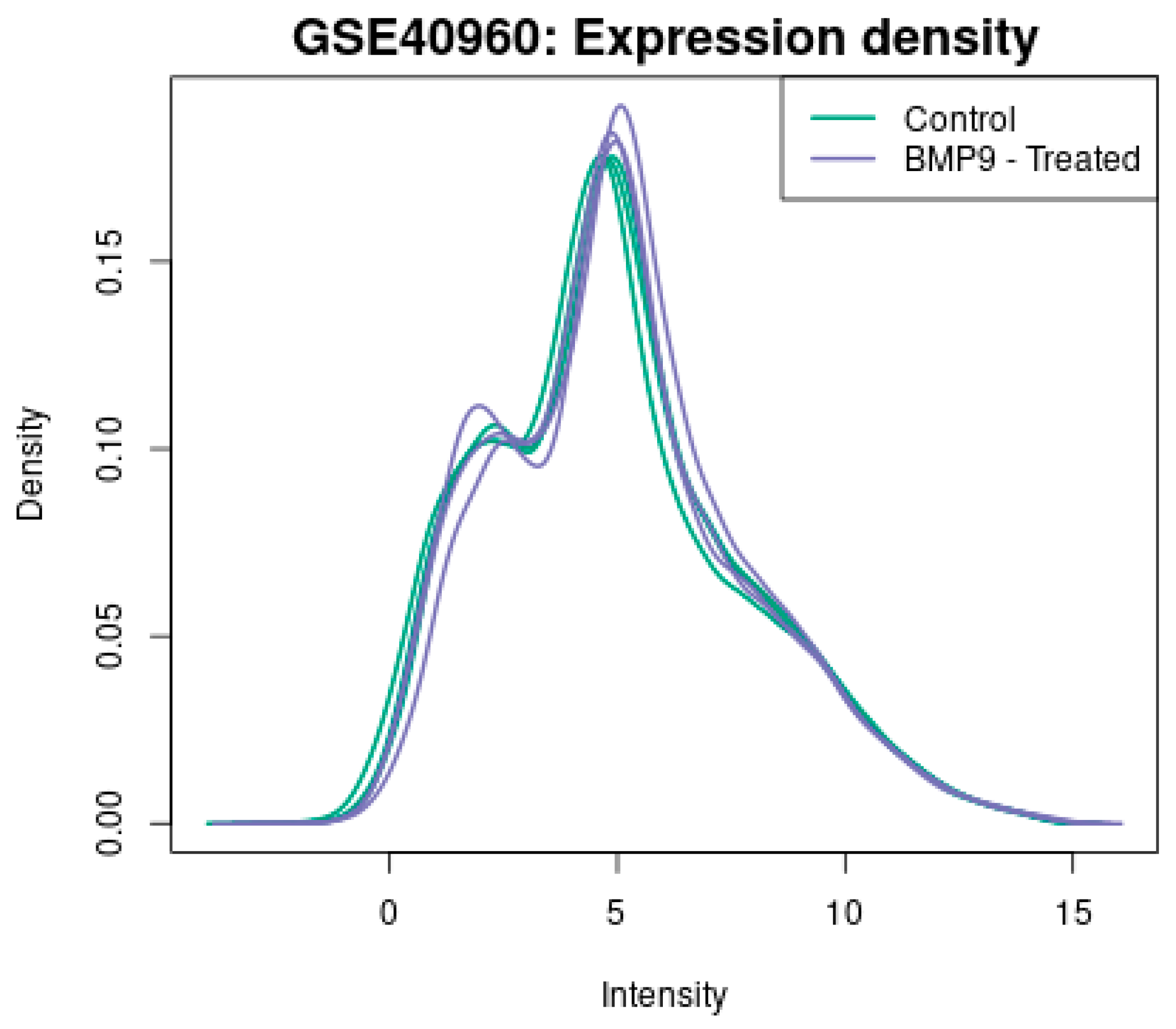
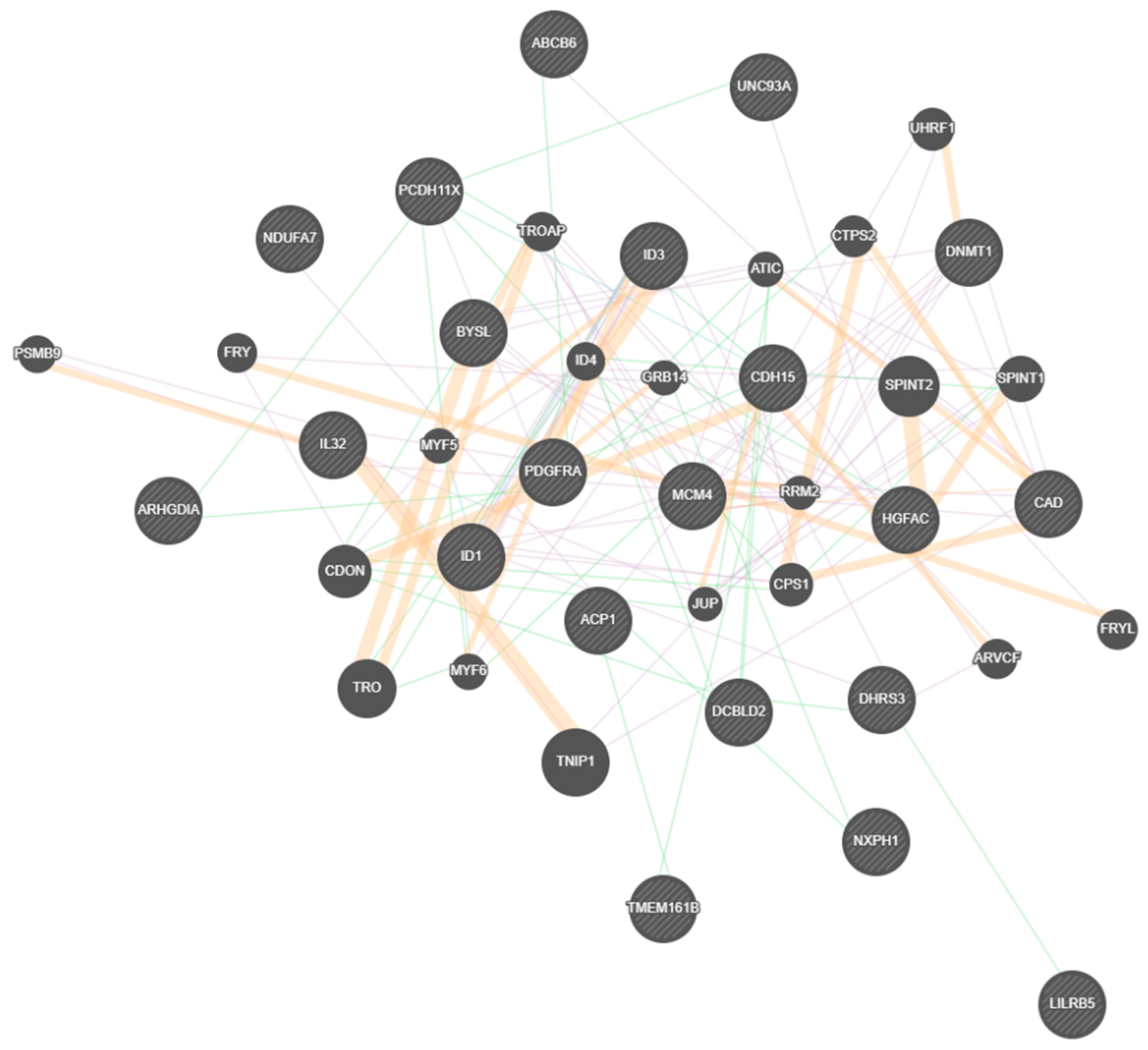
| Gene | Fold Change |
|---|---|
| CCDC144B | 23.89865699 |
| NXPH1 | 16.22331458 |
| PCDH11X | 16.08455194 |
| LOC285835 | 15.70455983 |
| TAAR1 | 14.77448089 |
| ZPBP2 | 14.44781726 |
| ID1 | 13.76492843 |
| LILRB5 | 12.90671705 |
| HGFAC | 12.26808441 |
| TMEM161B | 12.14174311 |
| LIAS | 0.081963901 |
| IL32 | 0.091010293 |
| C10orf44 | 0.092433913 |
| UNC93A | 0.099472054 |
| PDGFRA | 0.100139557 |
| ARHGDIA | 0.103217557 |
| PRR16 | 0.105769262 |
| LOC100132891 | 0.109581022 |
| FLJ30838 | 0.11039034 |
| IZUMO1 | 0.110476869 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





