Submitted:
11 April 2025
Posted:
14 April 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
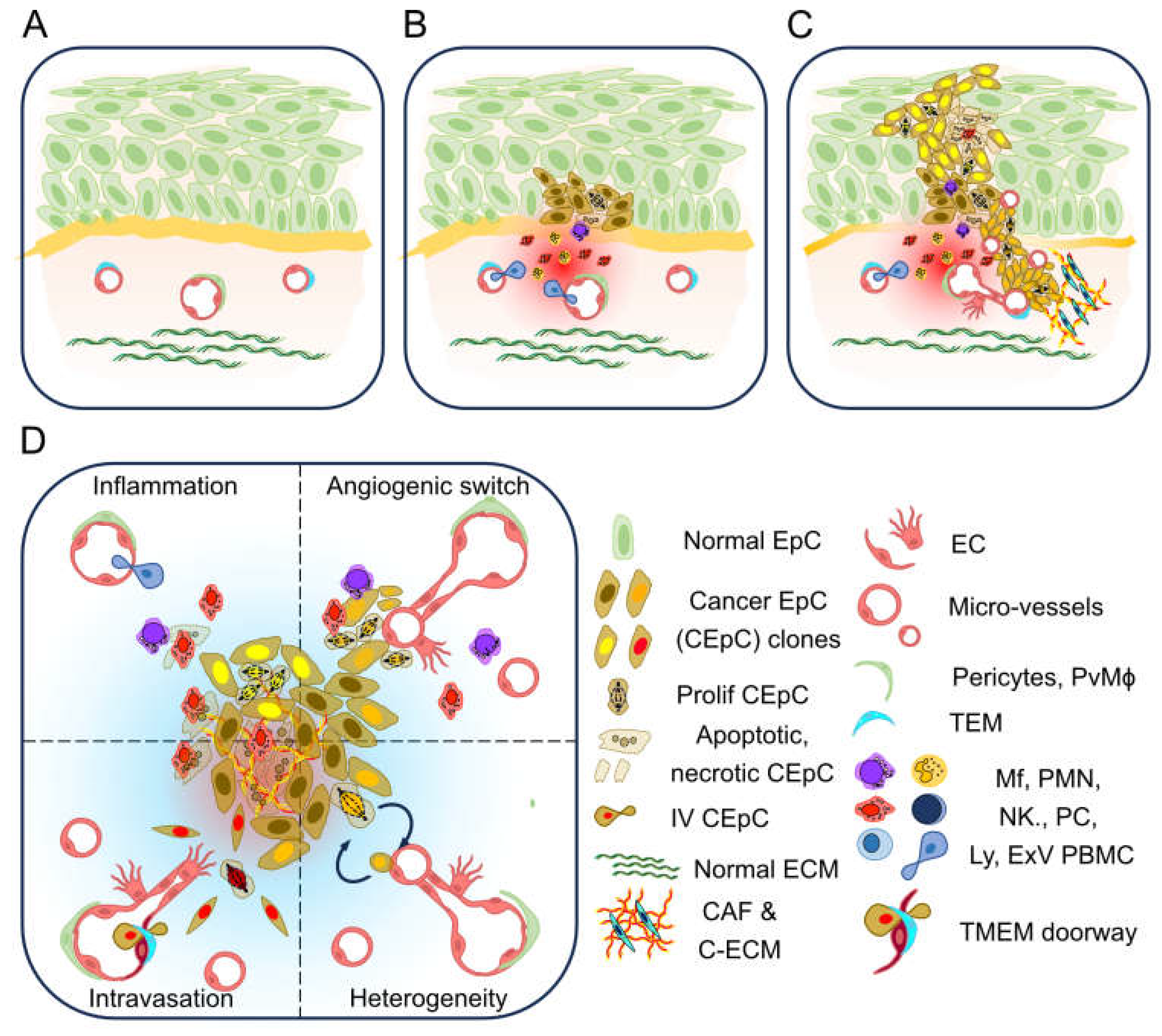
1.1. Tumour Associated Micro Vasculature
1.2. Therapeutic Implications
2. Non-Animal Technologies
2.1. Static Cultures
2.2. Ex Vivo Culture of Tumour Explants
2.3. Dynamic Cultures
3. Modelling Tumour Vasculature and Angiogenesis
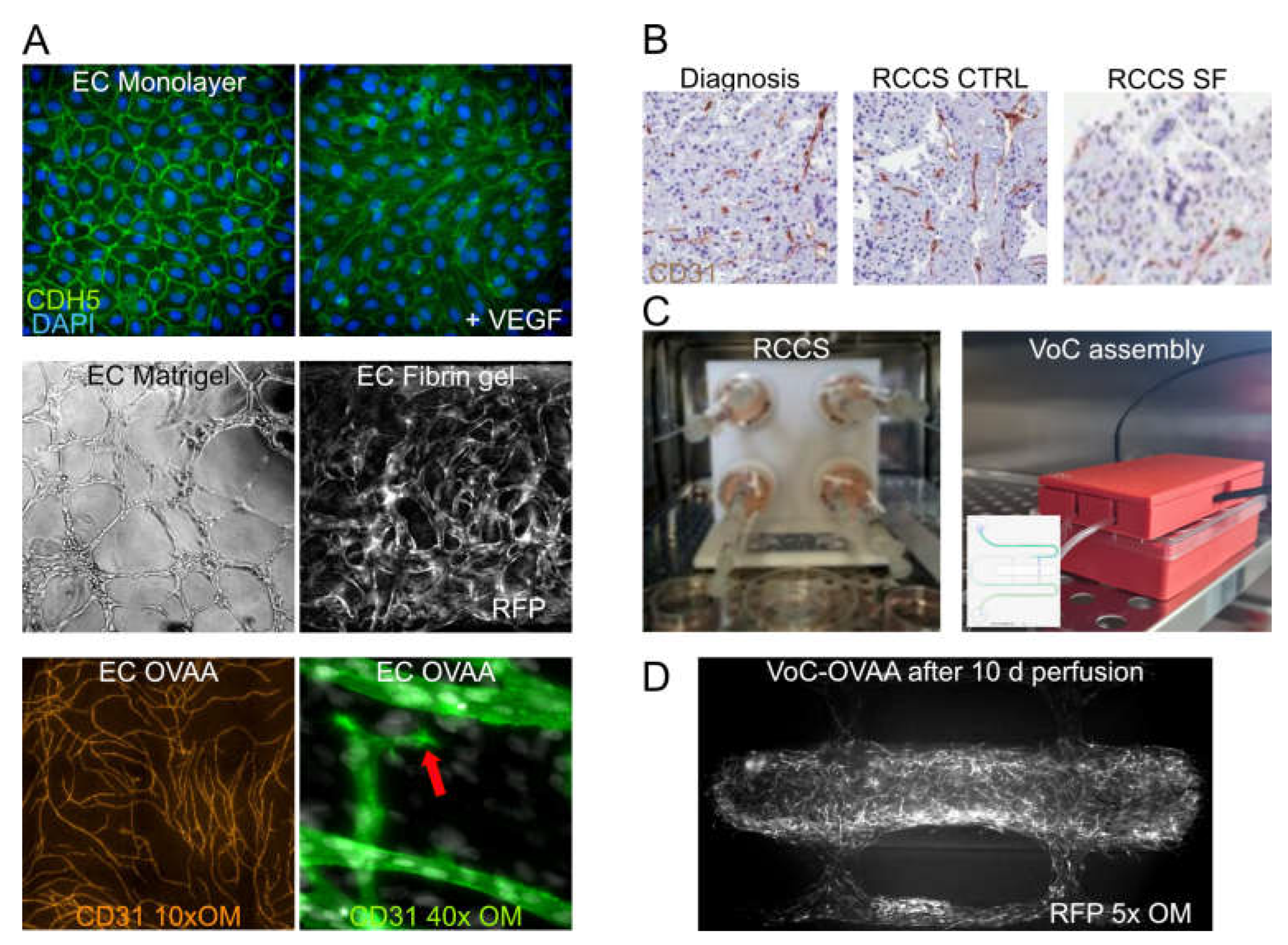
3.1. EC Cultures and PCS
3.2. In Vitro Tools to Study Vascular Permeability and Trans-Endothelial Cells Migration
3.3. In Vitro Tools to Study Angiogenesis
3.4. Vascularised Organoids and 3D Dynamic Cultures
3.5. Microphysiologic Systems to Perfuse Engineered Microvasculature
3.6. Spontaneous Tumour Models in Companion Animals
3.7. Computational Pathology and Artificial Intelligence
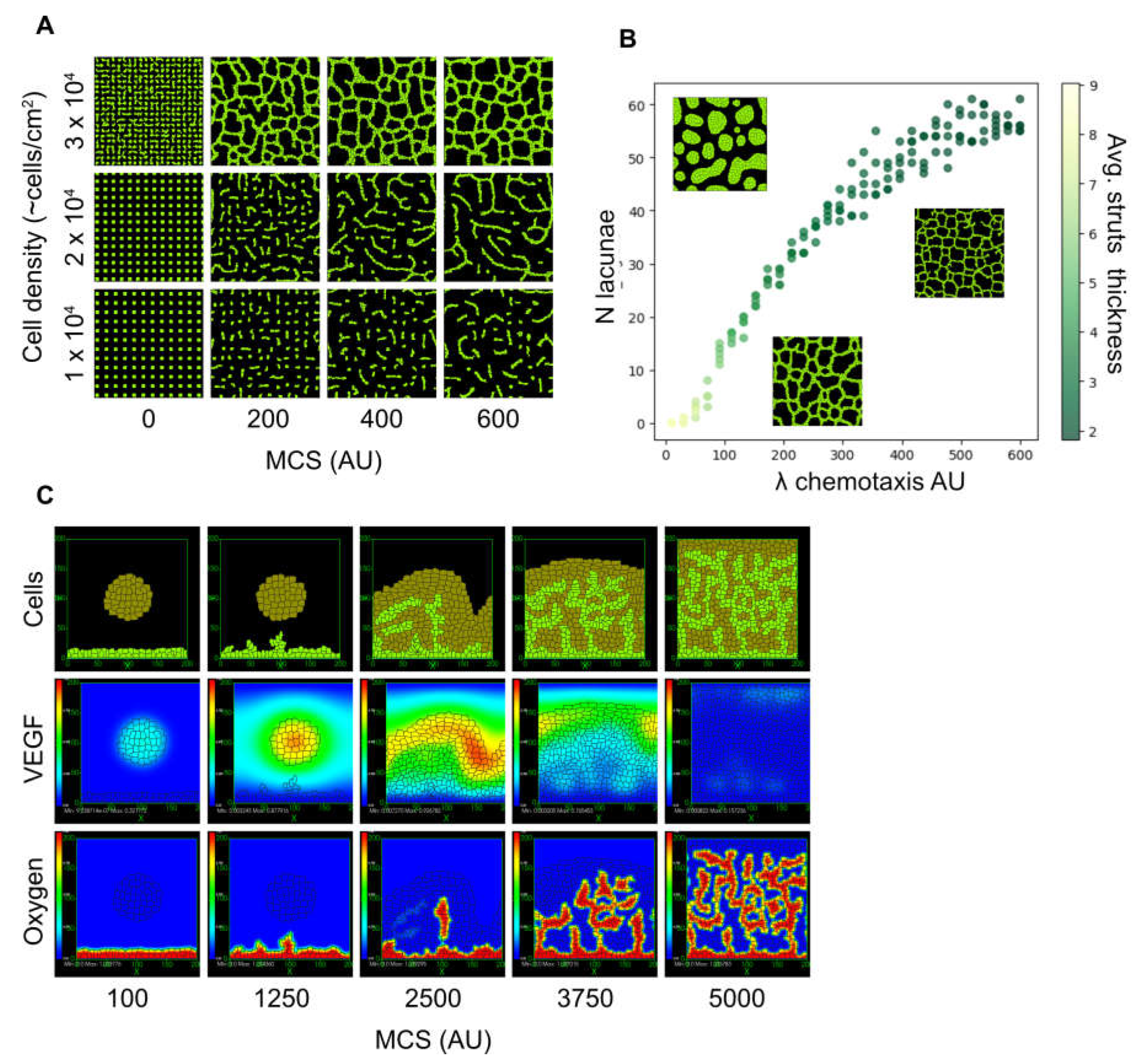
3.8. Mechanistic Modelling
4. Conclusions/Perspectives
Acknowledgments
References
- Hanahan, D. & Weinberg, R. A. The hallmarks of cancer. Cell 100, 57–70 (2000). [CrossRef]
- de Visser, K. E. & Joyce, J. A. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell 41, 374–403 (2023). [CrossRef]
- Mantovani, A., Allavena, P., Marchesi, F. & Garlanda, C. Macrophages as tools and targets in cancer therapy. Nat Rev Drug Discov 21, 799–820 (2022). [CrossRef]
- Chen, Y., McAndrews, K. M. & Kalluri, R. Clinical and therapeutic relevance of cancer-associated fibroblasts. Nat Rev Clin Oncol 18, 792–804 (2021). [CrossRef]
- Arner, E. N. & Rathmell, J. C. Metabolic programming and immune suppression in the tumor microenvironment. Cancer Cell 41, 421–433 (2023). [CrossRef]
- Mantovani, A., Allavena, P., Sica, A. & Balkwill, F. Cancer-related inflammation. Nature 454, 436–444 (2008). [CrossRef]
- Colotta, F., Allavena, P., Sica, A., Garlanda, C. & Mantovani, A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis 30, 1073–1081 (2009). [CrossRef]
- Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). [CrossRef]
- van Seijen, M. et al. Ductal carcinoma in situ: to treat or not to treat, that is the question. Br J Cancer 121, 285–292 (2019). [CrossRef]
- Knoblauch, M., Kühn, F., von Ehrlich-Treuenstätt, V., Werner, J. & Renz, B. W. Diagnostic and Therapeutic Management of Early Colorectal Cancer. Visc Med 39, 10–16 (2023). [CrossRef]
- Folkman, J., Merler, E., Abernathy, C. & Williams, G. Isolation of a tumor factor responsible for angiogenesis. J Exp Med 133, 275–288 (1971). [CrossRef]
- Folkman, J. Tumor angiogenesis: therapeutic implications. N Engl J Med 285, 1182–1186 (1971). [CrossRef]
- Leroi, N., Lallemand, F., Coucke, P., Noel, A. & Martinive, P. Impacts of Ionizing Radiation on the Different Compartments of the Tumor Microenvironment. Front Pharmacol 7, 78 (2016). [CrossRef]
- Minchinton, A. I. & Tannock, I. F. Drug penetration in solid tumours. Nat Rev Cancer 6, 583–592 (2006). [CrossRef]
- Deyell, M., Garris, C. S. & Laughney, A. M. Cancer metastasis as a non-healing wound. Br J Cancer 124, 1491–1502 (2021). [CrossRef]
- Whiteside, T. L. Tumor-Derived Exosomes and Their Role in Cancer Progression. Adv Clin Chem 74, 103–141 (2016). [CrossRef]
- Patras, L., Shaashua, L., Matei, I. & Lyden, D. Immune determinants of the pre-metastatic niche. Cancer Cell 41, 546–572 (2023). [CrossRef]
- Fidler, I. J. & Nicolson, G. L. Organ selectivity for implantation survival and growth of B16 melanoma variant tumor lines. J Natl Cancer Inst 57, 1199–1202 (1976). [CrossRef]
- Wicks, E. E. & Semenza, G. L. Hypoxia-inducible factors: cancer progression and clinical translation. J Clin Invest 132, e159839 (2022). [CrossRef]
- Mole, D. R. & Ratcliffe, P. J. Cellular oxygen sensing in health and disease. Pediatr Nephrol 23, 681–694 (2008). [CrossRef]
- Kalucka, J. et al. Single-Cell Transcriptome Atlas of Murine Endothelial Cells. Cell 180, 764-779.e20 (2020). [CrossRef]
- Eelen, G., Treps, L., Li, X. & Carmeliet, P. Basic and Therapeutic Aspects of Angiogenesis Updated. Circ Res 127, 310–329 (2020). [CrossRef]
- Li, X., Sun, X. & Carmeliet, P. Hallmarks of Endothelial Cell Metabolism in Health and Disease. Cell Metab. 30, 414–433 (2019). [CrossRef]
- Butler, J. M., Kobayashi, H. & Rafii, S. Instructive role of the vascular niche in promoting tumour growth and tissue repair by angiocrine factors. Nat Rev Cancer 10, 138–146 (2010). [CrossRef]
- Gajewski, T. F., Schreiber, H. & Fu, Y.-X. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol 14, 1014–1022 (2013). [CrossRef]
- Whiteside, T. L., Demaria, S., Rodriguez-Ruiz, M. E., Zarour, H. M. & Melero, I. Emerging Opportunities and Challenges in Cancer Immunotherapy. Clin Cancer Res 22, 1845–1855 (2016). [CrossRef]
- Mantovani, A., Marchesi, F., Jaillon, S., Garlanda, C. & Allavena, P. Tumor-associated myeloid cells: diversity and therapeutic targeting. Cell Mol Immunol 18, 566–578 (2021). [CrossRef]
- Kang, Y. & Pantel, K. Tumor cell dissemination: emerging biological insights from animal models and cancer patients. Cancer Cell 23, 573–581 (2013). [CrossRef]
- Coste, A. et al. Hematogenous Dissemination of Breast Cancer Cells From Lymph Nodes Is Mediated by Tumor MicroEnvironment of Metastasis Doorways. Front Oncol 10, 571100 (2020). [CrossRef]
- Lambert, A. W., Pattabiraman, D. R. & Weinberg, R. A. Emerging Biological Principles of Metastasis. Cell 168, 670–691 (2017). [CrossRef]
- Massagué, J. & Ganesh, K. Metastasis-Initiating Cells and Ecosystems. Cancer Discov 11, 971–994 (2021). [CrossRef]
- Potente, M., Gerhardt, H. & Carmeliet, P. Basic and therapeutic aspects of angiogenesis. Cell 146, 873–887 (2011). [CrossRef]
- Ganesh, K. & Massagué, J. Targeting metastatic cancer. Nat Med 27, 34–44 (2021). [CrossRef]
- Fan, P. et al. Alleviating hypoxia to improve cancer immunotherapy. Oncogene (2023) doi:10.1038/s41388-023-02869-2. [CrossRef]
- Jain, R. K. Antiangiogenesis strategies revisited: from starving tumors to alleviating hypoxia. Cancer Cell 26, 605–622 (2014). [CrossRef]
- Huang, Y. et al. Improving immune-vascular crosstalk for cancer immunotherapy. Nat Rev Immunol 18, 195–203 (2018). [CrossRef]
- Zitvogel, L., Pitt, J. M., Daillère, R., Smyth, M. J. & Kroemer, G. Mouse models in oncoimmunology. Nat Rev Cancer 16, 759–773 (2016). [CrossRef]
- Bédard, P. et al. Innovative Human Three-Dimensional Tissue-Engineered Models as an Alternative to Animal Testing. Bioengineering (Basel) 7, 115 (2020). [CrossRef]
- Moutinho, S. Researchers and regulators plan for a future without lab animals. Nat Med 29, 2151–2154 (2023). [CrossRef]
- Pampaloni, F., Reynaud, E. G. & Stelzer, E. H. K. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol 8, 839–845 (2007). [CrossRef]
- Kapałczyńska, M. et al. 2D and 3D cell cultures – a comparison of different types of cancer cell cultures. Arch Med Sci 14, 910–919 (2018). [CrossRef]
- Zimmer, J., Castriconi, R. & Scaglione, S. Editorial: Recent 3D Tumor Models for Testing Immune-Mediated Therapies. Front Immunol 12, 798493 (2021). [CrossRef]
- Allemang, A. et al. Assessing the genotoxicity and carcinogenicity of 2-chloroethanol through structure activity relationships and in vitro testing approaches. Food Chem Toxicol 168, 113290 (2022). [CrossRef]
- Lee, G. Y., Kenny, P. A., Lee, E. H. & Bissell, M. J. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods 4, 359–365 (2007). [CrossRef]
- Roskelley, C. D., Desprez, P. Y. & Bissell, M. J. Extracellular matrix-dependent tissue-specific gene expression in mammary epithelial cells requires both physical and biochemical signal transduction. Proc Natl Acad Sci U S A 91, 12378–12382 (1994). [CrossRef]
- Chang, T. T. & Hughes-Fulford, M. Monolayer and spheroid culture of human liver hepatocellular carcinoma cell line cells demonstrate distinct global gene expression patterns and functional phenotypes. Tissue Eng Part A 15, 559–567 (2009). [CrossRef]
- Riedl, A. et al. Comparison of cancer cells in 2D vs 3D culture reveals differences in AKT-mTOR-S6K signaling and drug responses. J Cell Sci 130, 203–218 (2017). [CrossRef]
- van Renterghem, A. W. J., van de Haar, J. & Voest, E. E. Functional precision oncology using patient-derived assays: bridging genotype and phenotype. Nat Rev Clin Oncol 20, 305–317 (2023). [CrossRef]
- Weiswald, L.-B., Bellet, D. & Dangles-Marie, V. Spherical cancer models in tumor biology. Neoplasia 17, 1–15 (2015). [CrossRef]
- Bray, L. J., Hutmacher, D. W. & Bock, N. Addressing Patient Specificity in the Engineering of Tumor Models. Front Bioeng Biotechnol 7, 217 (2019). [CrossRef]
- Gunti, S., Hoke, A. T. K., Vu, K. P. & London, N. R. Organoid and Spheroid Tumor Models: Techniques and Applications. Cancers (Basel) 13, 874 (2021). [CrossRef]
- Lee, K.-H. & Kim, T.-H. Recent Advances in Multicellular Tumor Spheroid Generation for Drug Screening. Biosensors (Basel) 11, 445 (2021). [CrossRef]
- Nunes, A. S., Barros, A. S., Costa, E. C., Moreira, A. F. & Correia, I. J. 3D tumor spheroids as in vitro models to mimic in vivo human solid tumors resistance to therapeutic drugs. Biotechnol Bioeng 116, 206–226 (2019). [CrossRef]
- Carletti, E., Motta, A. & Migliaresi, C. Scaffolds for tissue engineering and 3D cell culture. Methods Mol Biol 695, 17–39 (2011). [CrossRef]
- Drost, J. & Clevers, H. Organoids in cancer research. Nat Rev Cancer 18, 407–418 (2018). [CrossRef]
- Datta, P., Dey, M., Ataie, Z., Unutmaz, D. & Ozbolat, I. T. 3D bioprinting for reconstituting the cancer microenvironment. NPJ Precis Oncol 4, 18 (2020). [CrossRef]
- Augustine, R. et al. 3D Bioprinted cancer models: Revolutionizing personalized cancer therapy. Transl Oncol 14, 101015 (2021). [CrossRef]
- Powley, I. R. et al. Patient-derived explants (PDEs) as a powerful preclinical platform for anti-cancer drug and biomarker discovery. Br J Cancer 122, 735–744 (2020). [CrossRef]
- Kirby, A. J. et al. Multicellular ‘hotspots’ harbor high-grade potential in lower-grade gliomas. Neurooncol Adv 3, vdab026 (2021). [CrossRef]
- Navran, S. The application of low shear modeled microgravity to 3-D cell biology and tissue engineering. Biotechnol Annu Rev 14, 275–296 (2008). [CrossRef]
- Selden, C. & Fuller, B. Role of Bioreactor Technology in Tissue Engineering for Clinical Use and Therapeutic Target Design. Bioengineering (Basel) 5, 32 (2018). [CrossRef]
- Muraro, M. G. et al. Ex-vivo assessment of drug response on breast cancer primary tissue with preserved microenvironments. Oncoimmunology 6, e1331798 (2017). [CrossRef]
- Manfredonia, C. et al. Maintenance of Primary Human Colorectal Cancer Microenvironment Using a Perfusion Bioreactor-Based 3D Culture System. Adv Biosyst 3, e1800300 (2019). [CrossRef]
- Ferreira, L. P., Gaspar, V. M. & Mano, J. F. Design of spherically structured 3D in vitro tumor models -Advances and prospects. Acta Biomater 75, 11–34 (2018). [CrossRef]
- Belloni, D. et al. Modeling multiple myeloma-bone marrow interactions and response to drugs in a 3D surrogate microenvironment. Haematologica 103, 707–716 (2018).
- Grimm, D. et al. Growing tissues in real and simulated microgravity: new methods for tissue engineering. Tissue Eng Part B Rev 20, 555–566 (2014). [CrossRef]
- Guzzeloni, V., Veschini, L., Pedica, F., Ferrero, E. & Ferrarini, M. 3D Models as a Tool to Assess the Anti-Tumor Efficacy of Therapeutic Antibodies: Advantages and Limitations. Antibodies (Basel) 11, 46 (2022). [CrossRef]
- Ferrarini, M. et al. 3D-Dynamic Culture Models of Multiple Myeloma. Methods Mol Biol 1612, 177–190 (2017). [CrossRef]
- Ferrero, E. et al. Immunometabolic activation of macrophages leads to cytokine production in the pathogenesis of KRAS-mutated histiocytosis. Rheumatology (Oxford) 61, e93–e96 (2022). [CrossRef]
- Holton, A. B. et al. Microfluidic Biopsy Trapping Device for the Real-Time Monitoring of Tumor Microenvironment. PLoS One 12, e0169797 (2017). [CrossRef]
- Marei, I. et al. 3D Tissue-Engineered Vascular Drug Screening Platforms: Promise and Considerations. Front Cardiovasc Med 9, 847554 (2022). [CrossRef]
- Simons, M. et al. State-of-the-Art Methods for Evaluation of Angiogenesis and Tissue Vascularization: A Scientific Statement From the American Heart Association. Circ. Res. 116, 99–132 (2015). [CrossRef]
- Oh, J. E., Jung, C. & Yoon, Y.-S. Human Induced Pluripotent Stem Cell-Derived Vascular Cells: Recent Progress and Future Directions. J Cardiovasc Dev Dis 8, 148 (2021). [CrossRef]
- Palpant, N. J. et al. Generating high-purity cardiac and endothelial derivatives from patterned mesoderm using human pluripotent stem cells. Nature Protocols 12, 15–31 (2016). [CrossRef]
- Kumar, M. et al. A fully defined matrix to support a pluripotent stem cell derived multi-cell-liver steatohepatitis and fibrosis model. Biomaterials 276, 121006 (2021). [CrossRef]
- Ferrero, E. et al. CD14+ CD34+ peripheral blood mononuclear cells migrate across endothelium and give rise to immunostimulatory dendritic cells. J Immunol 160, 2675–2683 (1998).
- Langheim, S. et al. Increased expression and secretion of resistin in epicardial adipose tissue of patients with acute coronary syndrome. Am. J. Physiol. Heart Circ. Physiol. 298, H746-753 (2010). [CrossRef]
- Chesnais, F. et al. High-content image analysis to study phenotypic heterogeneity in endothelial cell monolayers. J Cell Sci 135, jcs259104 (2022). [CrossRef]
- Veschini, L. et al. High-Content Imaging to Phenotype Human Primary and iPSC-Derived Cells. Methods Mol Biol 2185, 423–445 (2021). [CrossRef]
- Kerns, S. J. et al. Human immunocompetent Organ-on-Chip platforms allow safety profiling of tumor-targeted T-cell bispecific antibodies. eLife 10, e67106 (2021). [CrossRef]
- Ewart, L. et al. Performance assessment and economic analysis of a human Liver-Chip for predictive toxicology. Commun Med 2, 1–16 (2022). [CrossRef]
- Pediaditakis, I. et al. A microengineered Brain-Chip to model neuroinflammation in humans. iScience 25, (2022). [CrossRef]
- Ronaldson-Bouchard, K. et al. Engineering complexity in human tissue models of cancer. Advanced Drug Delivery Reviews 184, 114181 (2022). [CrossRef]
- Ronaldson-Bouchard, K. et al. A multi-organ chip with matured tissue niches linked by vascular flow. Nat Biomed Eng 6, 351–371 (2022). [CrossRef]
- Nakatsu, M. N., Davis, J. & Hughes, C. C. W. Optimized fibrin gel bead assay for the study of angiogenesis. J Vis Exp 186 (2007). [CrossRef]
- Jauhiainen, S. et al. ErbB signaling is a potential therapeutic target for vascular lesions with fibrous component. Elife 12, e82543 (2023). [CrossRef]
- Francis, C. R., Kincross, H. & Kushner, E. J. Rab35 governs apicobasal polarity through regulation of actin dynamics during sprouting angiogenesis. Nat Commun 13, 5276 (2022). [CrossRef]
- Hetheridge, C., Mavria, G. & Mellor, H. Uses of the in vitro endothelial-fibroblast organotypic co-culture assay in angiogenesis research. Biochem. Soc. Trans. 39, 1597–1600 (2011). [CrossRef]
- Strobel, H. A., Moss, S. M. & Hoying, J. B. Vascularized Tissue Organoids. Bioengineering (Basel) 10, 124 (2023). [CrossRef]
- Yu, J. Vascularized Organoids: A More Complete Model. Int J Stem Cells 14, 127–137 (2021). [CrossRef]
- Orlova, V. V. et al. Generation, expansion and functional analysis of endothelial cells and pericytes derived from human pluripotent stem cells. Nat Protoc 9, 1514–1531 (2014). [CrossRef]
- Kumar, A. et al. Specification and Diversification of Pericytes and Smooth Muscle Cells from Mesenchymoangioblasts. Cell Rep 19, 1902–1916 (2017). [CrossRef]
- Ferrarini, M. et al. Ex-vivo dynamic 3-D culture of human tissues in the RCCSTM bioreactor allows the study of Multiple Myeloma biology and response to therapy. PLoS One 8, e71613 (2013). [CrossRef]
- Osaki, T., Sivathanu, V. & Kamm, R. D. Vascularized microfluidic organ-chips for drug screening, disease models and tissue engineering. Curr. Opin. Biotechnol. 52, 116–123 (2018). [CrossRef]
- Chen, M. B. et al. On-chip human microvasculature assay for visualization and quantification of tumor cell extravasation dynamics. Nat Protoc 12, 865–880 (2017).
- Yamamoto, K. et al. Construction of Continuous Capillary Networks Stabilized by Pericyte-like Perivascular Cells. Tissue Eng Part A 25, 499–510 (2019). [CrossRef]
- Boussommier-Calleja, A. et al. The effects of monocytes on tumor cell extravasation in a 3D vascularized microfluidic model. Biomaterials 198, 180–193 (2019). [CrossRef]
- Sobrino, A. et al. 3D microtumors in vitro supported by perfused vascular networks. Sci Rep 6, 31589 (2016). [CrossRef]
- Hachey, S. J. et al. A human vascularized microtumor model of patient-derived colorectal cancer recapitulates clinical disease. Transl Res 255, 97–108 (2023). [CrossRef]
- Chesnais, F. et al. Continuously perfusable, customisable, and matrix-free vasculature on a chip platform. Lab Chip 23, 761–772 (2023). [CrossRef]
- Razavirad, A., Rismanchi, S., Mortazavi, P. & Muhammadnejad, A. Canine Mammary Tumors as a Potential Model for Human Breast Cancer in Comparative Oncology. Veterinary Medicine International 2024, 9319651 (2024). [CrossRef]
- Palma, S. D., McConnell, A., Verganti, S. & Starkey, M. Review on Canine Oral Melanoma: An Undervalued Authentic Genetic Model of Human Oral Melanoma? Vet Pathol 58, 881–889 (2021). [CrossRef]
- Wong, K. et al. Cross-species genomic landscape comparison of human mucosal melanoma with canine oral and equine melanoma. Nat Commun 10, 353 (2019). [CrossRef]
- Oh, J. H. & Cho, J.-Y. Comparative oncology: overcoming human cancer through companion animal studies. Exp Mol Med 55, 725–734 (2023). [CrossRef]
- Onaciu, A. et al. Spontaneous and Induced Animal Models for Cancer Research. Diagnostics (Basel) 10, 660 (2020). [CrossRef]
- Mariano, L. C., Warnakulasuriya, S., Straif, K. & Monteiro, L. Secondhand smoke exposure and oral cancer risk: a systematic review and meta-analysis. Tob Control 31, 597–607 (2022). [CrossRef]
- Zaccone, R. et al. Environmental risk factors for the development of oral squamous cell carcinoma in cats. J Vet Intern Med 36, 1398–1408 (2022). [CrossRef]
- Sarver, A. L., Makielski, K. M., DePauw, T. A., Schulte, A. J. & Modiano, J. F. Increased risk of cancer in dogs and humans: a consequence of recent extension of lifespan beyond evolutionarily-determined limitations? Aging Cancer 3, 3–19 (2022). [CrossRef]
- Carvalho, M. I. et al. EGFR and microvessel density in canine malignant mammary tumours. Res Vet Sci 95, 1094–1099 (2013). [CrossRef]
- Queiroga, F. L., Pires, I., Parente, M., Gregório, H. & Lopes, C. S. COX-2 over-expression correlates with VEGF and tumour angiogenesis in canine mammary cancer. Vet J 189, 77–82 (2011). [CrossRef]
- Laufer-Amorim, R. et al. Comprehensive Genomic Profiling of Androgen-Receptor-Negative Canine Prostate Cancer. Int J Mol Sci 20, 1555 (2019). [CrossRef]
- Nordby, Y. et al. Stromal expression of VEGF-A and VEGFR-2 in prostate tissue is associated with biochemical and clinical recurrence after radical prostatectomy. Prostate 75, 1682–1693 (2015). [CrossRef]
- Warszawik-Hendzel, O., Słowińska, M., Olszewska, M. & Rudnicka, L. Melanoma of the oral cavity: pathogenesis, dermoscopy, clinical features, staging and management. J Dermatol Case Rep 8, 60–66 (2014). [CrossRef]
- Bergman, P. J. Canine oral melanoma. Clin Tech Small Anim Pract 22, 55–60 (2007). [CrossRef]
- Patel, V. et al. The impact of intensity-modulated radiation treatment on dento-alveolar microvasculature in pharyngeal cancer implant patients. J Oral Rehabil 47, 1411–1421 (2020). [CrossRef]
- Nafe, R. & Schlote, W. Histomorphometry of brain tumours. Neuropathol Appl Neurobiol 30, 315–328 (2004). [CrossRef]
- Mahmood, H. et al. Use of artificial intelligence in diagnosis of head and neck precancerous and cancerous lesions: A systematic review. Oral Oncol 110, 104885 (2020). [CrossRef]
- Veschini, L. et al. The vasostatin-1 fragment of chromogranin A preserves a quiescent phenotype in hypoxia-driven endothelial cells and regulates tumor neovascularization. FASEB J. 25, 3906–3914 (2011). [CrossRef]
- Madusanka, N., Jayalath, P., Fernando, D., Yasakethu, L. & Lee, B.-I. Impact of H&E Stain Normalization on Deep Learning Models in Cancer Image Classification: Performance, Complexity, and Trade-Offs. Cancers (Basel) 15, 4144 (2023). [CrossRef]
- Hue, J., Valinciute, Z., Thavaraj, S. & Veschini, L. Multifactorial estimation of clinical outcome in HPV-associated oropharyngeal squamous cell carcinoma via automated image analysis of routine diagnostic H&E slides and neural network modelling. Oral Oncol 141, 106399 (2023). [CrossRef]
- Graham, S. et al. CoNIC Challenge: Pushing the frontiers of nuclear detection, segmentation, classification and counting. Med Image Anal 92, 103047 (2024). [CrossRef]
- Graham, S. et al. Hover-Net: Simultaneous segmentation and classification of nuclei in multi-tissue histology images. Medical Image Analysis 58, 101563 (2019). [CrossRef]
- Cabral, B. P., Braga, L. A. M., Syed-Abdul, S. & Mota, F. B. Future of Artificial Intelligence Applications in Cancer Care: A Global Cross-Sectional Survey of Researchers. Curr Oncol 30, 3432–3446 (2023). [CrossRef]
- Timakova, A. et al. Artificial Intelligence Assists in the Detection of Blood Vessels in Whole Slide Images: Practical Benefits for Oncological Pathology. Biomolecules 13, 1327 (2023). [CrossRef]
- Gniadek, T., Kang, J., Theparee, T. & Krive, J. Framework for Classifying Explainable Artificial Intelligence (XAI) Algorithms in Clinical Medicine. Online J Public Health Inform 15, e50934 (2023). [CrossRef]
- Ali, S. et al. The enlightening role of explainable artificial intelligence in medical & healthcare domains: A systematic literature review. Comput Biol Med 166, 107555 (2023). [CrossRef]
- Wang, X. et al. Application of physiologically based pharmacokinetics modeling in the research of small-molecule targeted anti-cancer drugs. Cancer Chemother Pharmacol 92, 253–270 (2023). [CrossRef]
- Calzone, L., Noël, V., Barillot, E., Kroemer, G. & Stoll, G. Modeling signaling pathways in biology with MaBoSS: From one single cell to a dynamic population of heterogeneous interacting cells. Comput Struct Biotechnol J 20, 5661–5671 (2022). [CrossRef]
- Choi, K. et al. Tellurium: An extensible python-based modeling environment for systems and synthetic biology. Biosystems 171, 74–79 (2018). [CrossRef]
- Graner, F. & Glazier, J. A. Simulation of biological cell sorting using a two-dimensional extended Potts model. Phys Rev Lett 69, 2013–2016 (1992). [CrossRef]
- Starruß, J., de Back, W., Brusch, L. & Deutsch, A. Morpheus: a user-friendly modeling environment for multiscale and multicellular systems biology. Bioinformatics 30, 1331–1332 (2014). [CrossRef]
- Swat, M. H. et al. Multi-scale modeling of tissues using CompuCell3D. Methods Cell Biol 110, 325–366 (2012). [CrossRef]
- Shirinifard, A. et al. 3D multi-cell simulation of tumor growth and angiogenesis. PLoS One 4, e7190 (2009). [CrossRef]
- Merks, R. M. H. & Glazier, J. A. Dynamic mechanisms of blood vessel growth. Nonlinearity 19, C1–C10 (2006). [CrossRef]
- Merks, R. M. H., Perryn, E. D., Shirinifard, A. & Glazier, J. A. Contact-Inhibited Chemotaxis in De Novo and Sprouting Blood-Vessel Growth. PLOS Computational Biology 4, e1000163 (2008). [CrossRef]
- Niarakis, A. et al. Immune digital twins for complex human pathologies: applications, limitations, and challenges. NPJ Syst Biol Appl 10, 141 (2024). [CrossRef]
- Yang, H. M. Mathematical modeling of solid cancer growth with angiogenesis. Theor Biol Med Model 9, 2 (2012). [CrossRef]
- Moffett, A. S., Deng, Y. & Levine, H. Modeling the Role of Immune Cell Conversion in the Tumor-Immune Microenvironment. Bull Math Biol 85, 93 (2023). [CrossRef]
- Uatay, A. et al. Physiological Indirect Response Model to Omics-Powered Quantitative Systems Pharmacology Model. J Pharm Sci S0022-3549(23)00438–0 (2023). [CrossRef]
- Pereira, M. et al. A Comprehensive Look at In Vitro Angiogenesis Image Analysis Software. Int J Mol Sci 24, 17625 (2023). [CrossRef]
- Yan, H. H. N. et al. A Comprehensive Human Gastric Cancer Organoid Biobank Captures Tumor Subtype Heterogeneity and Enables Therapeutic Screening. Cell Stem Cell 23, 882-897.e11 (2018). [CrossRef]
- Oyoshi, H. et al. Comprehensive single-cell analysis demonstrates radiotherapy-induced infiltration of macrophages expressing immunosuppressive genes into tumor in esophageal squamous cell carcinoma. Sci Adv 9, eadh9069 (2023). [CrossRef]
- Shen, Y., Ni, S., Li, S. & Lv, B. Role of stemness-related genes TIMP1, PGF, and SNAI1 in the prognosis of colorectal cancer through single-cell RNA-seq. Cancer Med 12, 11611–11623 (2023). [CrossRef]
- Laubenbacher, R. et al. Building digital twins of the human immune system: toward a roadmap. npj Digit. Med. 5, 1–5 (2022). [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




