Submitted:
04 April 2025
Posted:
08 April 2025
You are already at the latest version
Abstract
Keywords:
Introduction
2. Materials and Methods
2.1. Plant Materials and Pathogen Isolates
2.2. Plant Inoculation, Infection Assessment and Leaf-Tissue Sampling
2.3. Sample Preparation for Metabolomic Analysis Using CIL LC-MS
2.4. Metabolome Quantification
2.5. LC−MS Analysis
2.6. LC-MS Raw Data Extraction and Processing
2.7. Validating the Potential Involvement of Selected Metabolites in Resistance
2.8. Data Analysis
3. Results
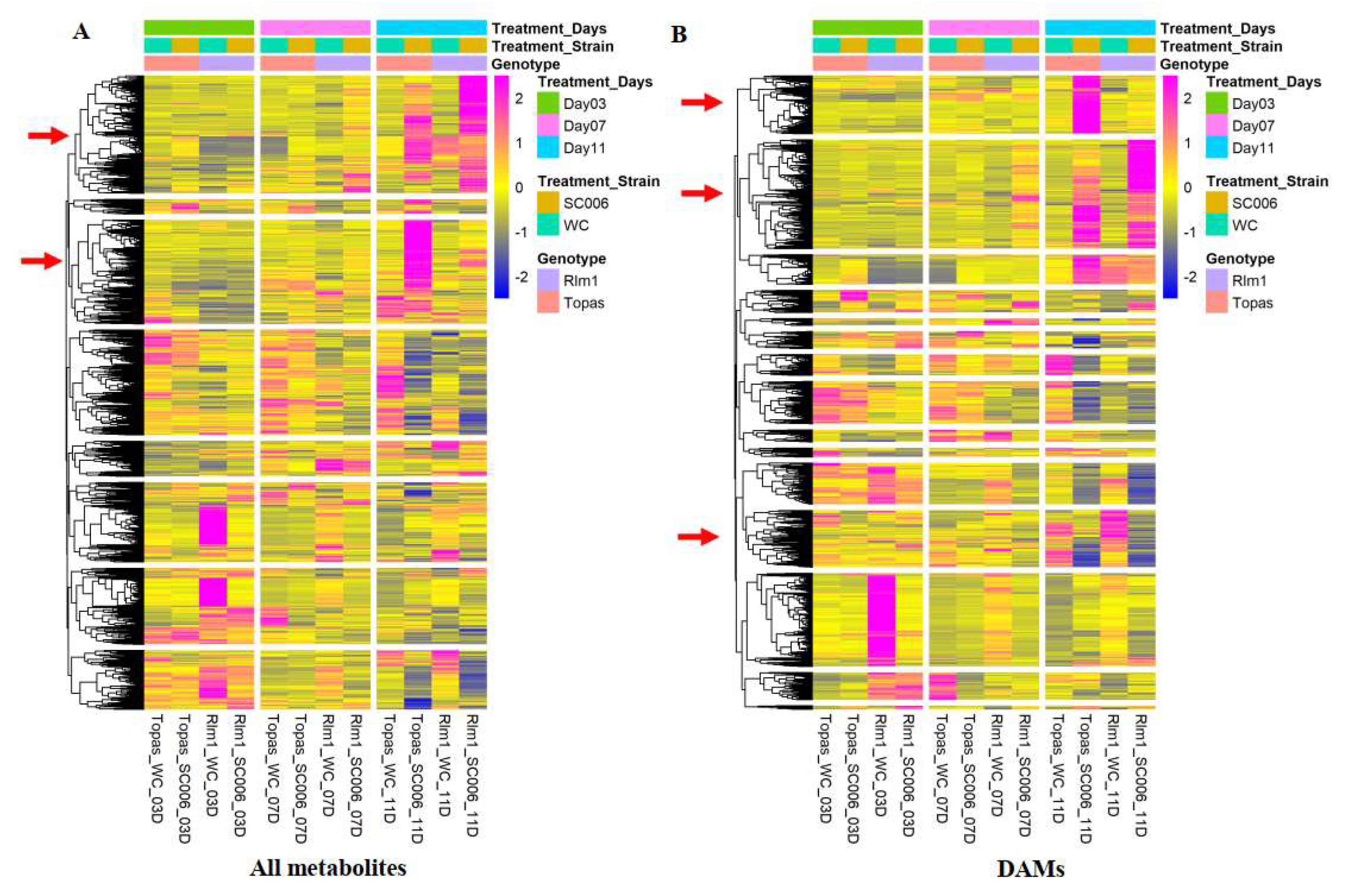
3.1. Multivariate Analysis of Metabolomic Data
3.2. Univariate Analysis of Metabolomic Data
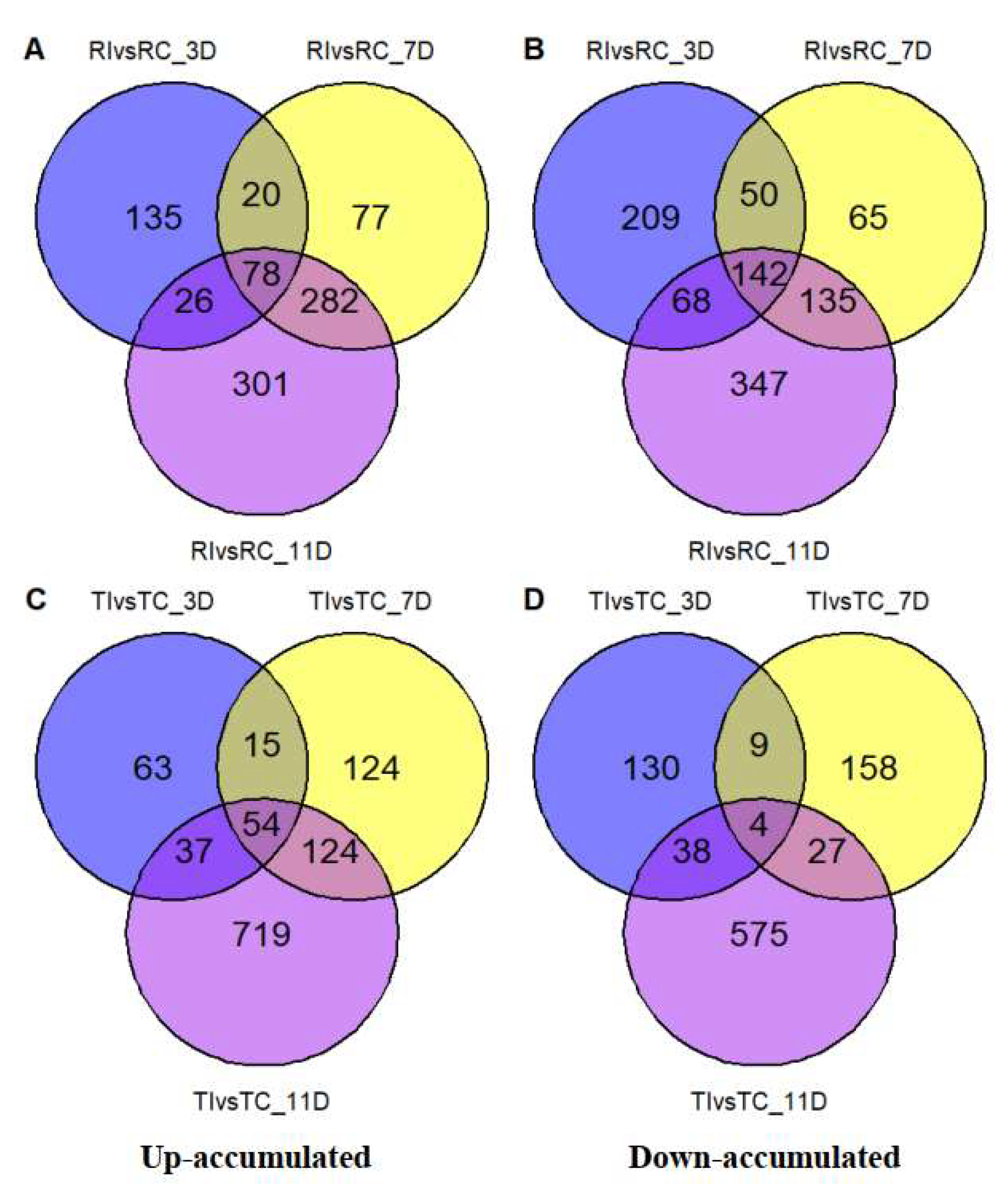
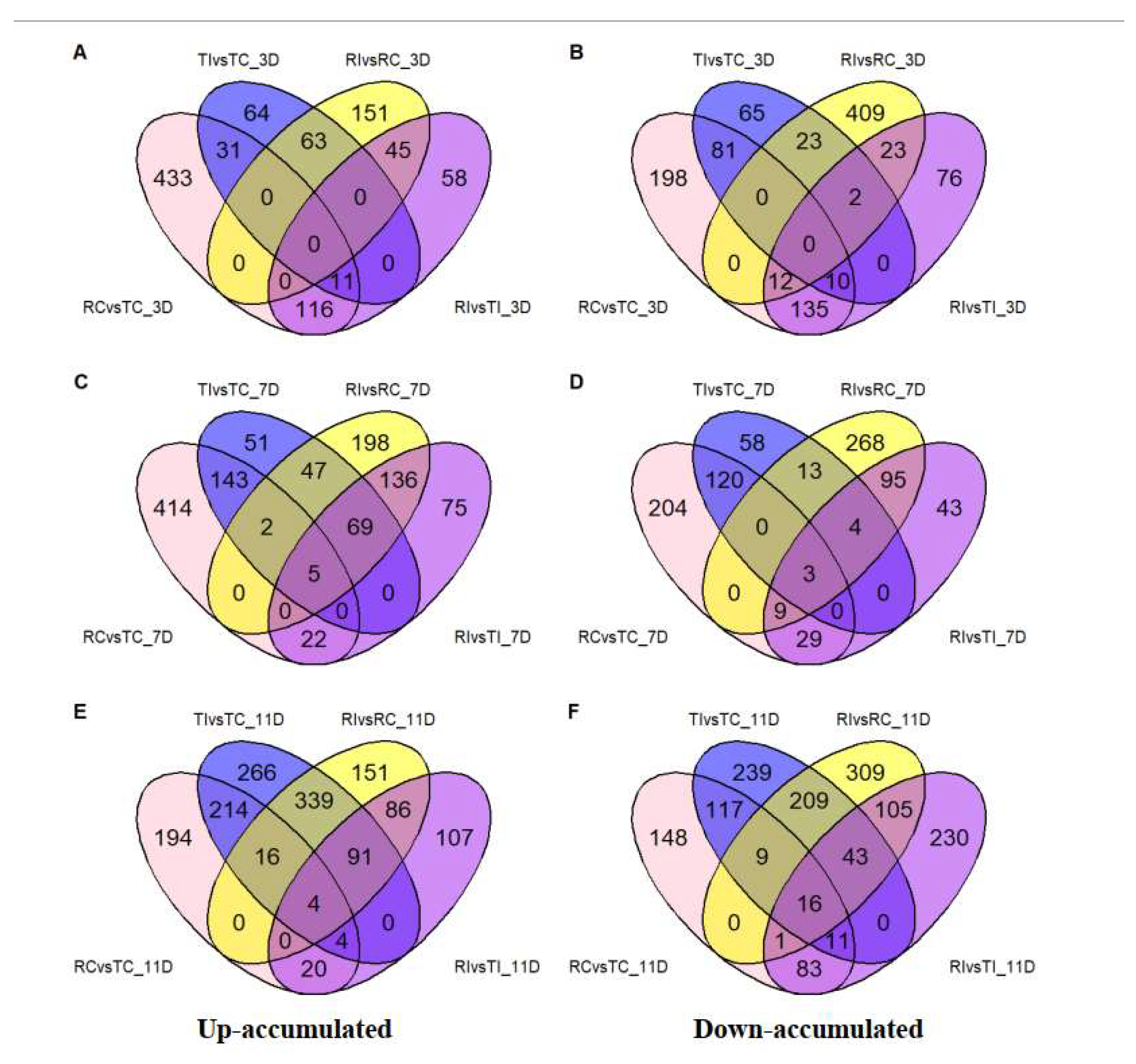
3.3. DAMs in Relation to Inoculation and Resistance
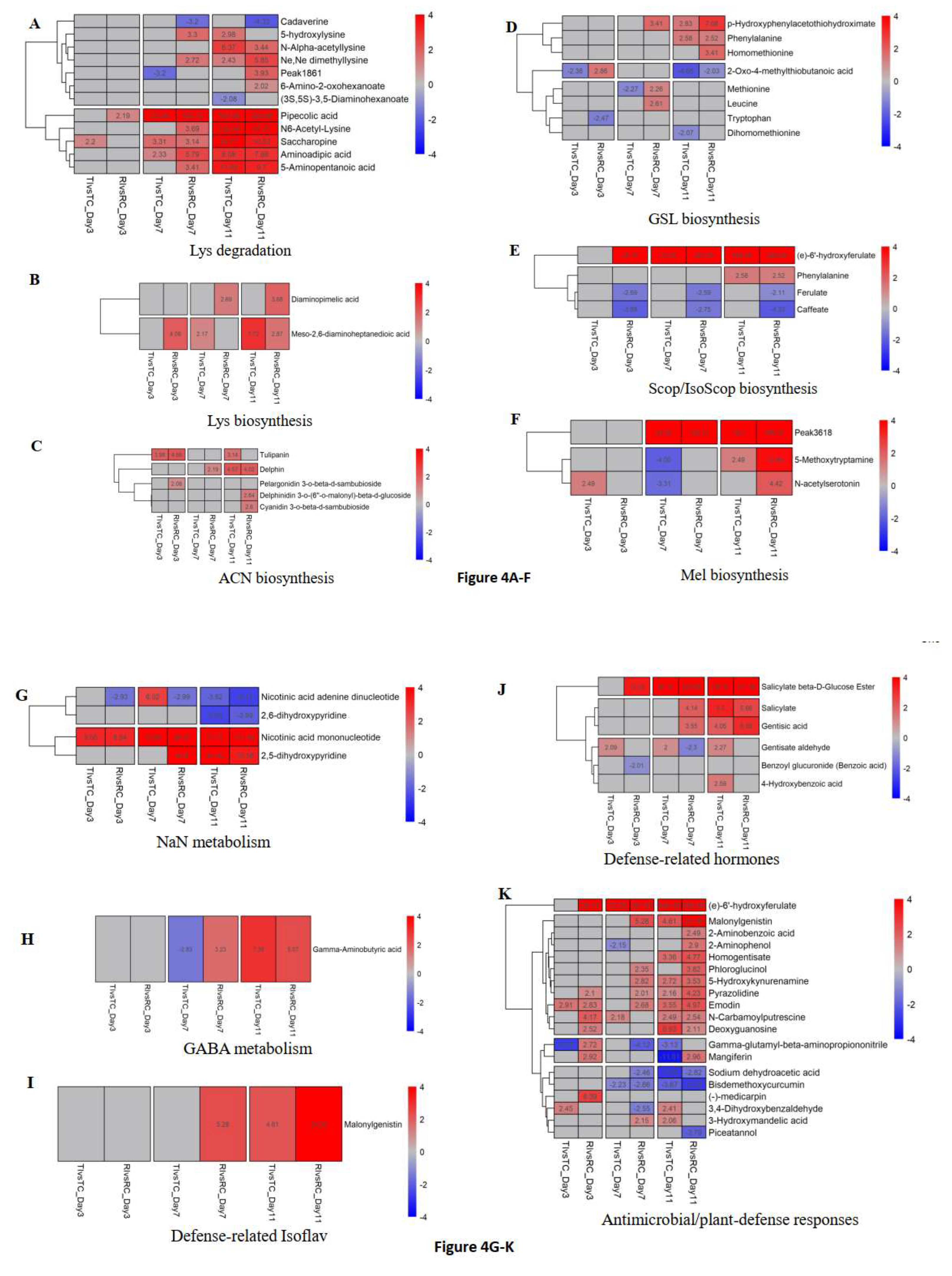
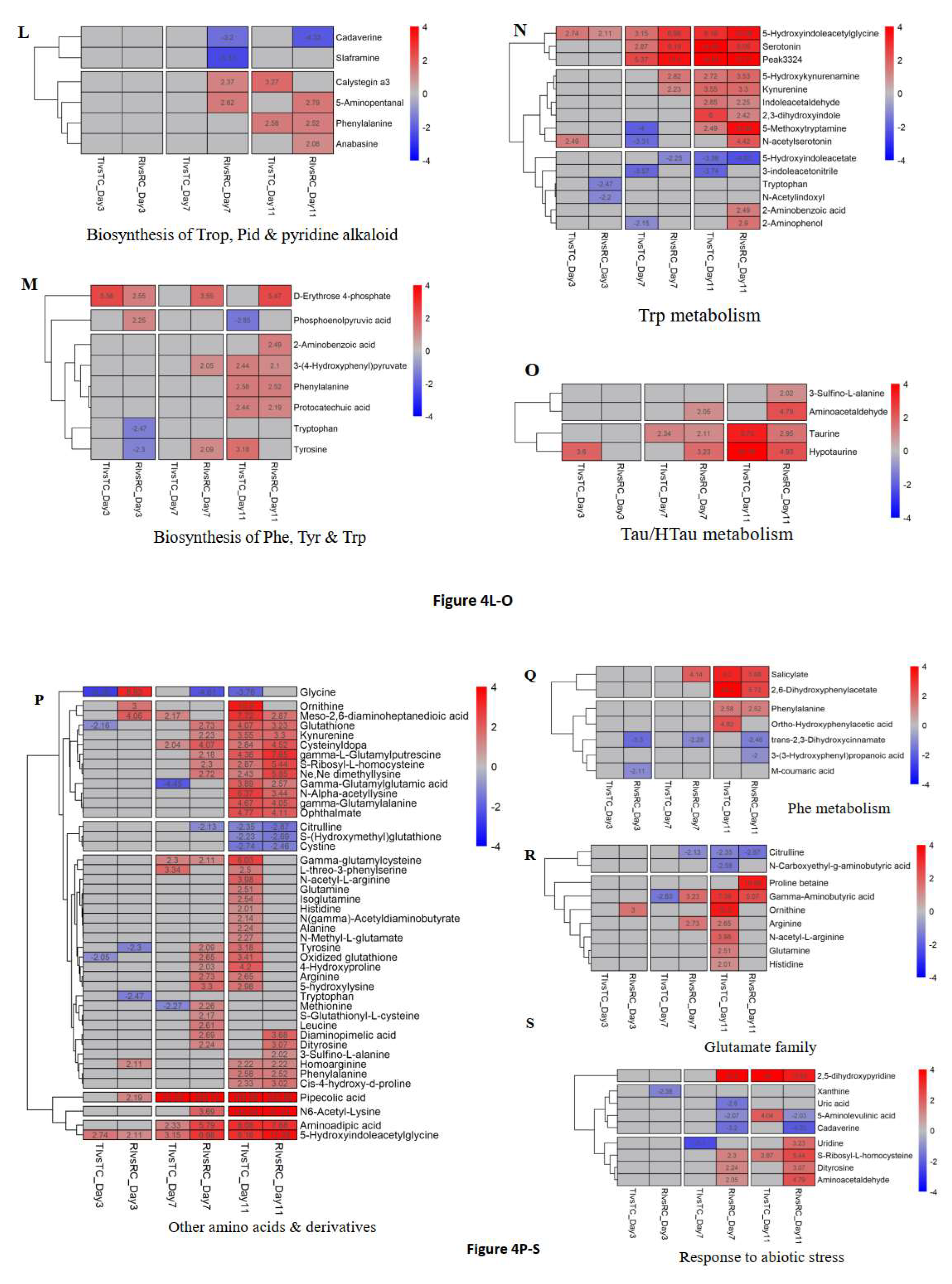
3.4. Prominent DAMs and Their Related Pathways
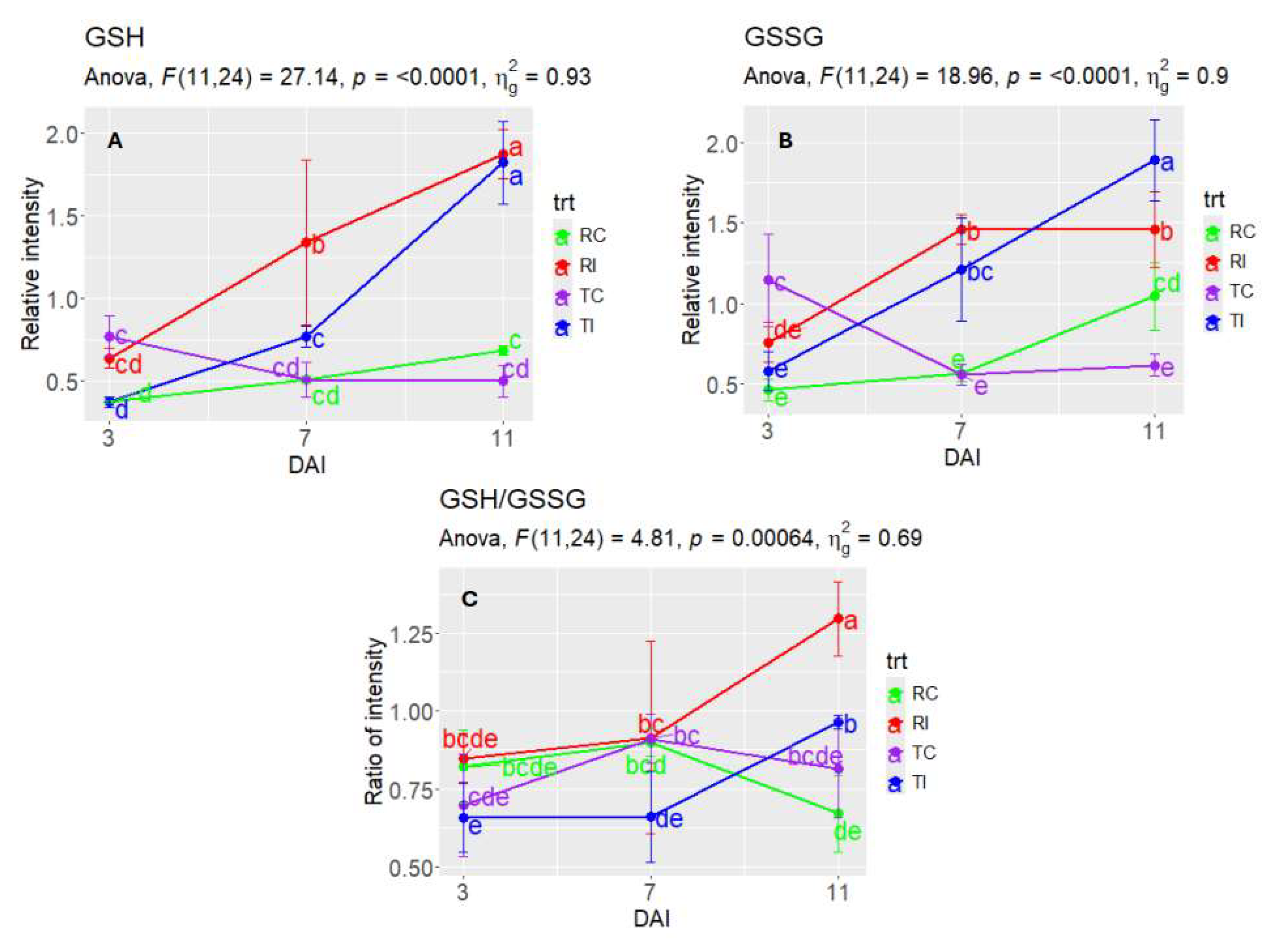
3.5. Metabolites/Pathways Potentially Related to Rlm1-Mediated Resistance
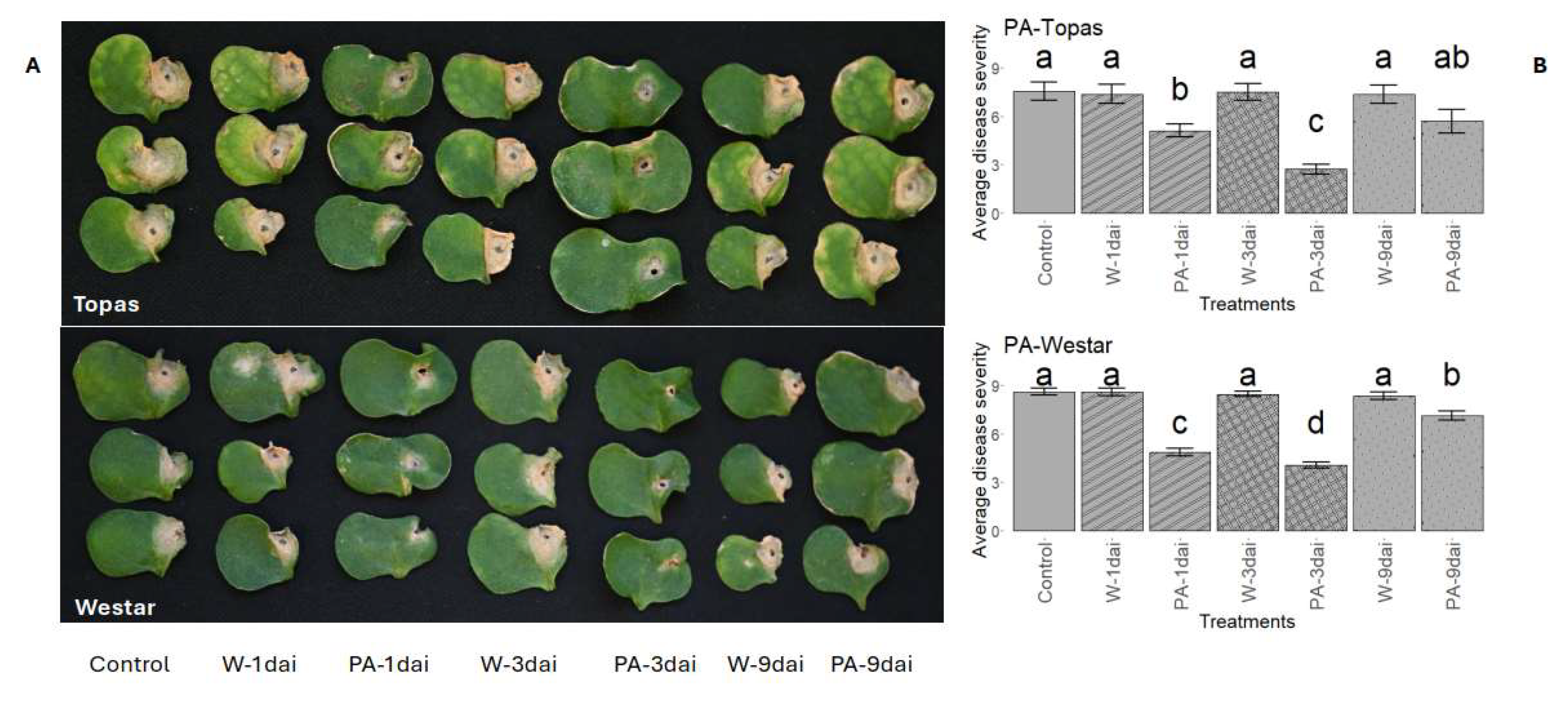
3.6. Validating DAM Candidates for Their Potential Roles in Rlm1-Mediated Resistance
4. Discussion
5. Conclusion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, G., C. Wu, B. Li, H. Su, S. Zhen, and Y. An, Detection of Leptosphaeria maculans from imported Canola seeds/Nachweis von Leptosphaeria maculons in importiertem Rapssaatgut. Journal of Plant Diseases and Protection, 2010. 117(4): p. 173-176.
- Zhang, X., G. Peng, H.R. Kutcher, M.-H. Balesdent, R. Delourme, and W.D. Fernando, Breakdown of Rlm3 resistance in the Brassica napus–Leptosphaeria maculans pathosystem in western Canada. European Journal of Plant Pathology, 2016. 145: p. 659-674. [CrossRef]
- Hubbard, M., C. Zhai, and G. Peng, Exploring mechanisms of quantitative resistance to Leptosphaeria maculans (Blackleg) in the cotyledons of canola (Brassica napus) based on transcriptomic and microscopic analyses. Plants, 2020. 9(7): p. 864. [CrossRef]
- Hubbard, M. and G. Peng, Quantitative resistance against an isolate of Leptosphaeria maculans (blackleg) in selected Canadian canola cultivars remains effective under increased temperatures. Plant pathology, 2018. 67(6): p. 1329-1338. [CrossRef]
- Liban, S., D. Cross, H. Kutcher, G. Peng, and W. Fernando, Race structure and frequency of avirulence genes in the western Canadian Leptosphaeria maculans pathogen population, the causal agent of blackleg in brassica species. Plant Pathology, 2016. 65(7): p. 1161-1169.
- Soomro, W., R. Kutcher, F. Yu, S.-F. Hwang, D. Fernando, S.E. Strelkov, and G. Peng, The race structure of Leptosphaeria maculans in western Canada between 2012 and 2014 and its influence on blackleg of canola. Canadian Journal of Plant Pathology, 2021. 43(3): p. 480-493. [CrossRef]
- Liu, F., Z. Zou, G. Peng, and W. Dilantha Fernando, Leptosphaeria maculans isolates reveal their allele frequency in Western Canada. Plant Disease, 2021. 105(05): p. 1440-1447. [CrossRef]
- Rouxel, T., E. Willner, L. Coudard, and M.-H. Balesdent, Screening and identification of resistance to Leptosphaeria maculans (stem canker) in Brassica napus accessions. Euphytica, 2003. 133(2): p. 219-231. [CrossRef]
- Rouxel, T. and M. Balesdent, The stem canker (blackleg) fungus, Leptosphaeria maculans, enters the genomic era. Molecular plant pathology, 2005. 6(3): p. 225-241. [CrossRef]
- Borhan, M.H., A.P. Van de Wouw, and N.J. Larkan, Molecular interactions between Leptosphaeria maculans and Brassica species. Annual Review of Phytopathology, 2022. 60: p. 237-257.
- Chu, M., T. Song, K.C. Falk, X. Zhang, X. Liu, A. Chang, R. Lahlali, L. McGregor, B.D. Gossen, and F. Yu, Fine mapping of Rcr1 and analyses of its effect on transcriptome patterns during infection by Plasmodiophora brassicae. BMC genomics, 2014. 15: p. 1-20. [CrossRef]
- Song, T., M. Chu, R. Lahlali, F. Yu, and G. Peng, Shotgun label-free proteomic analysis of clubroot (Plasmodiophora brassicae) resistance conferred by the gene Rcr1 in Brassica rapa. Frontiers in plant science, 2016. 7: p. 1013. [CrossRef]
- Fudal, I., S. Ross, L. Gout, F. Blaise, M. Kuhn, M. Eckert, L. Cattolico, S. Bernard-Samain, M. Balesdent, and T. Rouxel, Heterochromatin-like regions as ecological niches for avirulence genes in the Leptosphaeria maculans genome: map-based cloning of AvrLm6. Molecular Plant-Microbe Interactions, 2007. 20(4): p. 459-470.
- Zhai, C., X. Liu, T. Song, F. Yu, and G. Peng, Genome-wide transcriptome reveals mechanisms underlying Rlm1-mediated blackleg resistance on canola. Scientific Reports, 2021. 11(1): p. 4407. [CrossRef]
- Jones, J.D. and J.L. Dangl, The plant immune system. nature, 2006. 444(7117): p. 323-329. [CrossRef]
- AbuQamar, S., K. Moustafa, and L.S. Tran, Mechanisms and strategies of plant defense against Botrytis cinerea. Critical reviews in biotechnology, 2017. 37(2): p. 262-274. [CrossRef]
- Bezerra-Neto, J.P., F.C. Araújo, J.R. Ferreira-Neto, R.L. Silva, A.N. Borges, M.K. Matos, J.B. Silva, M.D. Silva, E.A. Kido, and A.M. Benko-Iseppon, NBS-LRR genes—Plant health sentinels: Structure, roles, evolution and biotechnological applications, Applied Plant Biotechnology for Improving Resistance to Biotic Stress. 2020, Elsevier. p. 63-120.
- Wang, J.W. and J.Y. Wu, Effective elicitors and process strategies for enhancement of secondary metabolite production in hairy root cultures, Biotechnology of Hairy Root Systems. 2013, Springer. p. 55-89.
- Kumar, A., R. Irchhaiya, A. Yadav, N. Gupta, S. Kumar, N. Gupta, S. Kumar, V. Yadav, A. Prakash, and H. Gurjar, Metabolites in plants and its classification. World J Pharm Pharm Sci, 2015. 4(1): p. 287-305.
- Lobo, M., N. Hounsome, and B. Hounsome, Biochemistry of vegetables: secondary metabolites in vegetables—terpenoids, phenolics, alkaloids, and sulfur-containing compounds. Handbook of vegetables and vegetable processing, 2018: p. 47-82.
- Singh, R., Medicinal plants: A review. Journal of Plant Sciences, 2015. 3(1): p. 50.
- Pusztahelyi, T., I.J. Holb, and I. Pócsi, Secondary metabolites in fungus-plant interactions. Frontiers in plant science, 2015. 6: p. 573. [CrossRef]
- Wink, M., Plant breeding: importance of plant secondary metabolites for protection against pathogens and herbivores. Theoretical and applied genetics, 1988. 75(2): p. 225-233. [CrossRef]
- Cevallos-Cevallos, J.M. and J.I. Reyes-De-Corcuera, Metabolomics in food science, Advances in food and nutrition research. 2012, Elsevier. p. 1-24.
- Luo, X., X. Gu, and L. Li, Development of a simple and efficient method of harvesting and lysing adherent mammalian cells for chemical isotope labeling LC-MS-based cellular metabolomics. Analytica chimica acta, 2018. 1037: p. 97-106. [CrossRef]
- Guo, K. and L. Li, Differential 12C-/13C-isotope dansylation labeling and fast liquid chromatography/mass spectrometry for absolute and relative quantification of the metabolome. Analytical chemistry, 2009. 81(10): p. 3919-3932. [CrossRef]
- Luo, X., S. Zhao, T. Huan, D. Sun, R.M.N. Friis, M.C. Schultz, and L. Li, High-performance chemical isotope labeling liquid chromatography–mass spectrometry for profiling the metabolomic reprogramming elicited by ammonium limitation in yeast. Journal of proteome research, 2016. 15(5): p. 1602-1612. [CrossRef]
- Shen, W., W. Han, Y. Li, Z. Meng, L. Cai, and L. Li, Development of chemical isotope labeling liquid chromatography mass spectrometry for silkworm hemolymph metabolomics. Analytica chimica acta, 2016. 942: p. 1-11. [CrossRef]
- Tunsagool, P., X. Wang, W. Leelasuphakul, W. Jutidamrongphan, N. Phaonakrop, J. Jaresitthikunchai, S. Roytrakul, G. Chen, and L. Li, Metabolomic study of stress responses leading to plant resistance in mandarin fruit mediated by preventive applications of Bacillus subtilis cyclic lipopeptides. Postharvest Biology and Technology, 2019. 156: p. 110946. [CrossRef]
- Larkan, N.J., F. Yu, D.J. Lydiate, S.R. Rimmer, and M.H. Borhan, Single R gene introgression lines for accurate dissection of the Brassica-Leptosphaeria pathosystem. Frontiers in Plant Science, 2016. 7: p. 1771. [CrossRef]
- Fu, F., X. Liu, R. Wang, C. Zhai, G. Peng, F. Yu, and W.D. Fernando, Fine mapping of Brassica napus blackleg resistance gene Rlm1 through bulked segregant RNA sequencing. Scientific Reports, 2019. 9(1): p. 14600. [CrossRef]
- Sharpe, A., I. Parkin, D. Keith, and D. Lydiate, Frequent nonreciprocal translocations in the amphidiploid genome of oilseed rape (Brassica napus). Genome, 1995. 38(6): p. 1112-1121. [CrossRef]
- Yu, F., D.J. Lydiate, R. Gugel, A. Sharpe, and S. Rimmer, Introgression of Brassica rapa subsp. sylvestris blackleg resistance into B. napus. Molecular breeding, 2012. 30: p. 1495-1506. [CrossRef]
- Chen, Y. and W. Fernando, Prevalence of pathogenicity groups of Leptosphaeria maculans in western Canada and North Dakota, USA. Canadian Journal of Plant Pathology, 2006. 28(4): p. 533-539. [CrossRef]
- Koch, E., H. Badawy, and H. Hoppe, Differences between aggressive and non-aggressive single spore lines of Leptosphaeria maculans in cultural characteristics and phytotoxin production. Journal of phytopathology, 1989. 124(1): p. 52-62. [CrossRef]
- Zhao, S., X. Luo, and L. Li, Chemical isotope labeling LC-MS for high coverage and quantitative profiling of the hydroxyl submetabolome in metabolomics. Analytical chemistry, 2016. 88(21): p. 10617-10623. [CrossRef]
- Chambers, M.C., B. Maclean, R. Burke, D. Amodei, D.L. Ruderman, S. Neumann, L. Gatto, B. Fischer, B. Pratt, and J. Egertson, A cross-platform toolkit for mass spectrometry and proteomics. Nature biotechnology, 2012. 30(10): p. 918-920. [CrossRef]
- Zhou, R., C.-L. Tseng, T. Huan, and L. Li, IsoMS: automated processing of LC-MS data generated by a chemical isotope labeling metabolomics platform. Analytical chemistry, 2014. 86(10): p. 4675-4679. [CrossRef]
- Warrack, B.M., S. Hnatyshyn, K.-H. Ott, M.D. Reily, M. Sanders, H. Zhang, and D.M. Drexler, Normalization strategies for metabonomic analysis of urine samples. Journal of Chromatography B, 2009. 877(5-6): p. 547-552. [CrossRef]
- Cheng, Z. and L. Li, Development of Chemical Isotope Labeling Liquid Chromatography Orbitrap Mass Spectrometry for Comprehensive Analysis of Dipeptides. Analytical Chemistry, 2023. 95(16): p. 6629-6636. [CrossRef]
- Dahabiyeh, L.A., A.K. Malkawi, X. Wang, D. Colak, A.H. Mujamammi, E.M. Sabi, L. Li, M. Dasouki, and A.M. Abdel Rahman, Dexamethasone-induced perturbations in tissue metabolomics revealed by chemical isotope labeling LC-MS analysis. Metabolites, 2020. 10(2): p. 42. [CrossRef]
- Zhao, S., H. Li, W. Han, W. Chan, and L. Li, Metabolomic coverage of chemical-group-submetabolome analysis: group classification and four-channel chemical isotope labeling LC-MS. Analytical Chemistry, 2019. 91(18): p. 12108-12115. [CrossRef]
- Li, L., R. Li, J. Zhou, A. Zuniga, A.E. Stanislaus, Y. Wu, T. Huan, J. Zheng, Y. Shi, and D.S. Wishart, MyCompoundID: using an evidence-based metabolome library for metabolite identification. Analytical chemistry, 2013. 85(6): p. 3401-3408. [CrossRef]
- Bayoumi, S.A., M.G. Rowan, J.R. Beeching, and I.S. Blagbrough, Investigation of biosynthetic pathways to hydroxycoumarins during post-harvest physiological deterioration in cassava roots by using stable isotope labelling. ChemBioChem, 2008. 9(18): p. 3013-3022. [CrossRef]
- Zhao, Y., N. Wang, Z. Sui, C. Huang, Z. Zeng, and L. Kong, The molecular and structural basis of O-methylation reaction in coumarin biosynthesis in Peucedanum praeruptorum Dunn. International Journal of Molecular Sciences, 2019. 20(7): p. 1533. [CrossRef]
- R Core Team, R: A language and environment for statistical computing. Foundation for Statistical Computing, Vienna, Austria, 2024.
- Love, M.I., W. Huber, and S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology, 2014. 15: p. 1-21.
- Wickham, H., ggplot2. Wiley interdisciplinary reviews: computational statistics, 2011. 3(2): p. 180-185.
- Wickham, H., ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York, 2016.
- Wickham, H., W. Chang, L. Henry, T.L. Pedersen, K. Takahashi, C. Wilke, K. Woo, H. Yutani, D. Dunnington, and T. van den Brand, Create elegant data visualisations using the grammar of graphics. Version 3.5.1. https://ggplot2.tidyverse.org. 2024, CRAN.
- Yan, L., ggvenn: Draw Venn Diagram by 'ggplot2'. R package version 0.1.10. https://CRAN.R-project.org/package=ggvenn. 2023, CRAN.
- Kolde, R., Pretty Heatmaps. R package version 1.0.12. https://CRAN.R-project.org/package=pheatmap. 2019, CRAN.
- Kassambara, A., rstatix: Pipe-friendly framework for basic statistical tests. R package version 0.7.2. https://CRAN.R-project.org/package=rstatix. 2023, CRAN.
- Youssef, S.A. and K.A. Tartoura, Compost enhances plant resistance against the bacterial wilt pathogen Ralstonia solanacearum via up-regulation of ascorbate-glutathione redox cycle. European Journal of Plant Pathology, 2013. 137: p. 821-834. [CrossRef]
- de Mendiburu, F., Agricolae: Statistical Procedures for Agricultural Research. R package version 1.3-7. https://CRAN.R-project.org/package=agricolae. 2023, CRAN.
- Pang, Z., Y. Lu, G. Zhou, F. Hui, L. Xu, C. Viau, A.F. Spigelman, P.E. MacDonald, D.S. Wishart, and S. Li, MetaboAnalyst 6.0: towards a unified platform for metabolomics data processing, analysis and interpretation. Nucleic Acids Research, 2024: p. gkae253.
- Wobbrock, J.O., L. Findlater, D. Gergle, and J.J. Higgins. The aligned rank transform for nonparametric factorial analyses using only anova procedures. Proceedings of the ACM Conference on Human Factors in Computing Systems (CHI 2011). 2011. Vancouver, British Columbia (May 7-12, 2011) New York: ACM Press, pp. 143-146. [CrossRef]
- Mangiafico, S.S., Aligned Ranks Transformation ANOVA. Summary and Analysis of Extension Program Evaluation in R. New Brunswick (NJ): Rutgers Cooperative Extension, 2016: p. 315-327.
- Kay, M., L.A. Elkin, J.J. Higgins, and J.O. Wobbrock, An aligned rank transform procedure for multifactor contrast tests. R package version 0.11.1. https://github.com/mjskay/ARTool. 2021, CRAN.
- Kay, M. and J.O. Wobbrock, Aligned Rank Transform. Version 0.10.7. https://github.com/mjskay/ARTool. 2020, CRAN.
- Lenth, R.V., emmeans: Estimated marginal means, aka least-squares means. R package version 1.10.4. https://CRAN.R-project.org/package=emmeans. 2024, CRAN.
- Mangiafico, S., rcompanion: Functions to Support Extension Education Program Evaluation. version 2.4. 36. Rutgers Cooperative Extension. New Brunswick, New Jersey. https://CRAN.R-project.org/package=rcompanion. 2024, CRAN.
- Chamoun, R., K.A. Aliferis, and S. Jabaji, Identification of signatory secondary metabolites during mycoparasitism of Rhizoctonia solani by Stachybotrys elegans. Frontiers in microbiology, 2015. 6: p. 138210.
- Moulin, M., C. Deleu, F. Larher, and A. Bouchereau, The lysine-ketoglutarate reductase–saccharopine dehydrogenase is involved in the osmo-induced synthesis of pipecolic acid in rapeseed leaf tissues. Plant Physiology and Biochemistry, 2006. 44(7-9): p. 474-482. [CrossRef]
- Mano, Y. and K. Nemoto, The pathway of auxin biosynthesis in plants. Journal of experimental Botany, 2012. 63(8): p. 2853-2872. [CrossRef]
- Jiang, Z., H. Zhang, P. Jiao, X. Wei, S. Liu, S. Guan, and Y. Ma, The Integration of Metabolomics and Transcriptomics Provides New Insights for the Identification of Genes Key to Auxin Synthesis at Different Growth Stages of Maize. International Journal of Molecular Sciences, 2022. 23(21): p. 13195. [CrossRef]
- Schalk, M., F. Cabello-Hurtado, M.-A.s. Pierrel, R. Atanassova, P. Saindrenan, and D.l. Werck-Reichhart, Piperonylic acid, a selective, mechanism-based inactivator of the trans-cinnamate 4-hydroxylase: a new tool to control the flux of metabolites in the phenylpropanoid pathway. Plant Physiology, 1998. 118(1): p. 209-218.
- da Costa, T.P.S., C.J. Hall, S. Panjikar, J.A. Wyllie, R.M. Christoff, S. Bayat, M.D. Hulett, B.M. Abbott, A.R. Gendall, and M.A. Perugini, Towards novel herbicide modes of action by inhibiting lysine biosynthesis in plants. Elife, 2021. 10: p. e69444.
- Dzierzbicka, K., Synthesis of 2, 6-diaminopimelic acid (DAP) and its analogues. Polish Journal of Chemistry, 2007. 81(4): p. 455-473.
- Dempsey, D.M.A. and D.F. Klessig, SOS–too many signals for systemic acquired resistance? Trends in plant science, 2012. 17(9): p. 538-545.
- Návarová, H., F. Bernsdorff, A.-C. Döring, and J. Zeier, Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. The Plant Cell, 2012. 24(12): p. 5123-5141. [CrossRef]
- Vranova, V., L. Lojkova, K. Rejsek, and P. Formanek, Significance of the natural occurrence of L-versus D-pipecolic acid: a review. Chirality, 2013. 25(12): p. 823-831.
- Liu, Y., Y. Li, Y. Bi, Q. Jiang, R. Mao, Z. Liu, Y. Huang, M. Zhang, and D.B. Prusky, Induction of defense response against Alternaria rot in Zaosu pear fruit by exogenous L-lysine through regulating ROS metabolism and activating defense-related proteins. Postharvest Biology and Technology, 2021. 179: p. 111567. [CrossRef]
- Prabhu, B.R. and N.B. Mulchandani, Biosynthesis of piperlongumine. Phytochemistry, 1985. 24(11): p. 2589-2591. [CrossRef]
- Yang, H. and U. Ludewig, Lysine catabolism, amino acid transport, and systemic acquired resistance: what is the link? Plant signaling & behavior, 2014. 9(7): p. e28933.
- Shan, L. and P. He, Pipped at the post: pipecolic acid derivative identified as SAR regulator. Cell, 2018. 173(2): p. 286-287. [CrossRef]
- Nawrath, C. and J.-P. Métraux, Salicylic acid induction–deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. The Plant Cell, 1999. 11(8): p. 1393-1404.
- El-Shetehy, M., C. Wang, M. Shine, K. Yu, A. Kachroo, and P. Kachroo, Nitric oxide and reactive oxygen species are required for systemic acquired resistance in plants. Plant Signaling & Behavior, 2015. 10(9): p. e998544. [CrossRef]
- Vogel-Adghough, D., E. Stahl, H. Návarová, and J. Zeier, Pipecolic acid enhances resistance to bacterial infection and primes salicylic acid and nicotine accumulation in tobacco. Plant signaling & behavior, 2013. 8(11): p. e26366. [CrossRef]
- Liu, S., L. Xie, J. Su, B. Tian, A. Fang, Y. Yu, C. Bi, and Y. Yang, Integrated metabolo-transcriptomics reveals the defense response of homogentisic acid in wheat against Puccinia striiformis f. sp. tritici. Journal of Agricultural and Food Chemistry, 2022. 70(12): p. 3719-3729. [CrossRef]
- Arruda, P. and P. Barreto, Lysine catabolism through the saccharopine pathway: enzymes and intermediates involved in plant responses to abiotic and biotic stress. Frontiers in plant science, 2020. 11: p. 535796.
- Yang, H., S. Postel, B. Kemmerling, and U. Ludewig, Altered growth and improved resistance of Arabidopsis against Pseudomonas syringae by overexpression of the basic amino acid transporter AtCAT1. Plant, cell & environment, 2014. 37(6): p. 1404-1414.
- Delaney, T.P., S. Uknes, B. Vernooij, L. Friedrich, K. Weymann, D. Negrotto, T. Gaffney, M. Gut-Rella, H. Kessmann, and E. Ward, A central role of salicylic acid in plant disease resistance. Science, 1994. 266(5188): p. 1247-1250. [CrossRef]
- Alazem, M. and N.S. Lin, Roles of plant hormones in the regulation of host–virus interactions. Molecular plant pathology, 2015. 16(5): p. 529-540. [CrossRef]
- Grant, J.J., A. Chini, D. Basu, and G.J. Loake, Targeted activation tagging of the Arabidopsis NBS-LRR gene, ADR1, conveys resistance to virulent pathogens. Molecular Plant-Microbe Interactions, 2003. 16(8): p. 669-680.
- Zhu, X., A. Soliman, M.R. Islam, L.R. Adam, and F. Daayf, Verticillium dahliae’s isochorismatase hydrolase is a virulence factor that contributes to interference with potato’s salicylate and jasmonate defense signaling. Frontiers in Plant Science, 2017. 8: p. 399. [CrossRef]
- Ryals, J.A., U.H. Neuenschwander, M.G. Willits, A. Molina, H.-Y. Steiner, and M.D. Hunt, Systemic acquired resistance. The plant cell, 1996. 8(10): p. 1809.
- Pontier, D., Z.H. Miao, and E. Lam, Trans-dominant suppression of plant TGA factors reveals their negative and positive roles in plant defense responses. The Plant Journal, 2001. 27(6): p. 529-538. [CrossRef]
- Rochon, A., P. Boyle, T. Wignes, P.R. Fobert, and C. Després, The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C-terminal cysteines. The Plant Cell, 2006. 18(12): p. 3670-3685. [CrossRef]
- Cameron, R.K., N.L. Paiva, C.J. Lamb, and R.A. Dixon, Accumulation of salicylic acid and PR-1 gene transcripts in relation to the systemic acquired resistance (SAR) response induced by Pseudomonas syringae pv. tomato in Arabidopsis. Physiological and Molecular Plant Pathology, 1999. 55(2): p. 121-130. [CrossRef]
- Lincoln, J.E., J.P. Sanchez, K. Zumstein, and D.G. Gilchrist, Plant and animal PR1 family members inhibit programmed cell death and suppress bacterial pathogens in plant tissues. Molecular Plant Pathology, 2018. 19(9): p. 2111-2123. [CrossRef]
- Jain, D. and J.P. Khurana, Role of pathogenesis-related (PR) proteins in plant defense mechanism. Molecular aspects of plant-pathogen interaction, 2018: p. 265-281.
- Yi, S.Y., K. Shirasu, J.S. Moon, S.-G. Lee, and S.-Y. Kwon, The activated SA and JA signaling pathways have an influence on flg22-triggered oxidative burst and callose deposition. PloS one, 2014. 9(2): p. e88951. [CrossRef]
- War, A.R., M.G. Paulraj, M.Y. War, and S. Ignacimuthu, Role of salicylic acid in induction of plant defense system in chickpea (Cicer arietinum L.). Plant signaling & behavior, 2011. 6(11): p. 1787-1792. [CrossRef]
- Neuenschwander, U., B. Vernooij, L. Friedrich, S. Uknes, H. Kessmann, and J. Ryals, Is hydrogen peroxide a second messenger of salicylic acid in systemic acquired resistance? The Plant Journal, 1995. 8(2): p. 227-233. [CrossRef]
- Herrera-Vásquez, A., P. Salinas, and L. Holuigue, Salicylic acid and reactive oxygen species interplay in the transcriptional control of defense genes expression. Frontiers in plant science, 2015. 6: p. 171. [CrossRef]
- Kovács, J., P. Poór, Á. Szepesi, and I. Tari, Salicylic acid induced cysteine protease activity during programmed cell death in tomato plants. Acta Biologica Hungarica, 2016. 67(2): p. 148-158. [CrossRef]
- Greenberg, J.T., A. Guo, D.F. Klessig, and F.M. Ausubel, Programmed cell death in plants: a pathogen-triggered response activated coordinately with multiple defense functions. Cell, 1994. 77(4): p. 551-563. [CrossRef]
- Mittler, R., O. Del Pozo, L. Meisel, and E. Lam, Pathogen-induced programmed cell death in plants, a possible defense mechanism. Developmental genetics, 1997. 21(4): p. 279-289.
- Garattini, E., R. Mendel, M.J. Romão, R. Wright, and M. Terao, Mammalian molybdo-flavoenzymes, an expanding family of proteins: structure, genetics, regulation, function and pathophysiology. Biochemical Journal, 2003. 372(1): p. 15-32. [CrossRef]
- Bellés, J.M., R. Garro, J. Fayos, P. Navarro, J. Primo, and V. Conejero, Gentisic acid as a pathogen-inducible signal, additional to salicylic acid for activation of plant defenses in tomato. Molecular Plant-Microbe Interactions, 1999. 12(3): p. 227-235. [CrossRef]
- Campos, L., P. Granell, S. Tárraga, P. López-Gresa, V. Conejero, J.M. Bellés, I. Rodrigo, and P. Lisón, Salicylic acid and gentisic acid induce RNA silencing-related genes and plant resistance to RNA pathogens. Plant physiology and biochemistry, 2014. 77: p. 35-43. [CrossRef]
- Bellés, J.M., R. Garro, V. Pallás, J. Fayos, I. Rodrigo, and V. Conejero, Accumulation of gentisic acid as associated with systemic infections but not with the hypersensitive response in plant-pathogen interactions. Planta, 2006. 223: p. 500-511. [CrossRef]
- Yalpani, N., J. León, M.A. Lawton, and I. Raskin, Pathway of salicylic acid biosynthesis in healthy and virus-inoculated tobacco. Plant physiology, 1993. 103(2): p. 315-321. [CrossRef]
- Agerbirk, N. and C.E. Olsen, Glucosinolate hydrolysis products in the crucifer Barbarea vulgaris include a thiazolidine-2-one from a specific phenolic isomer as well as oxazolidine-2-thiones. Phytochemistry, 2015. 115: p. 143-151. [CrossRef]
- Brader, G., M.D. Mikkelsen, B.A. Halkier, and E. Tapio Palva, Altering glucosinolate profiles modulates disease resistance in plants. The Plant Journal, 2006. 46(5): p. 758-767. [CrossRef]
- Rodrigues, A.S. and E.A.S. Rosa, Effect of post-harvest treatments on the level of glucosinolates in broccoli. Journal of the Science of Food and Agriculture, 1999. 79(7): p. 1028-1032. [CrossRef]
- Sanchez-Vallet, A., B. Ramos, P. Bednarek, G. López, M. Piślewska-Bednarek, P. Schulze-Lefert, and A. Molina, Tryptophan-derived secondary metabolites in Arabidopsis thaliana confer non-host resistance to necrotrophic Plectosphaerella cucumerina fungi. The Plant Journal, 2010. 63(1): p. 115-127. [CrossRef]
- Smolinska, U., G. Knudsen, M. Morra, and V. Borek, Inhibition of Aphanomyces euteiches f. sp. pisi by volatiles produced by hydrolysis of Brassica napus seed meal. Plant disease, 1997. 81(3): p. 288-292.
- Van Eylen, D., N. Bellostas, B.W. Strobel, I. Oey, M. Hendrickx, A. Van Loey, H. Sørensen, and J.C. Sørensen, Influence of pressure/temperature treatments on glucosinolate conversion in broccoli (Brassica oleraceae L. cv Italica) heads. Food Chemistry, 2009. 112(3): p. 646-653. [CrossRef]
- Fahey, J.W., A.T. Zalcmann, and P. Talalay, The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry, 2001. 56(1): p. 5-51. [CrossRef]
- Singh, A., D. Guest, and L. Copeland, Associations Between Glucosinolates, White Rust, and Plant Defense Activators inBrassicaPlants: A Review. International Journal of Vegetable Science, 2014. 21(3): p. 297-313.
- Zhai, K., D. Liang, H. Li, F. Jiao, B. Yan, J. Liu, Z. Lei, L. Huang, X. Gong, and X. Wang, NLRs guard metabolism to coordinate pattern-and effector-triggered immunity. Nature, 2022. 601(7892): p. 245-251. [CrossRef]
- Escaray, F., A. Felipo-Benavent, and P. Vera, Linking plant metabolism and immunity through methionine biosynthesis. Molecular Plant, 2022. 15(1): p. 6-8. [CrossRef]
- Yan, X., L. Ma, H. Pang, P. Wang, L. Liu, Y. Cheng, J. Cheng, Y. Guo, and Q. Li, METHIONINE SYNTHASE1 is involved in chromatin silencing by maintaining DNA and histone methylation. Plant physiology, 2019. 181(1): p. 249-261. [CrossRef]
- Mäkinen, K. and S. De, The significance of methionine cycle enzymes in plant virus infections. Current opinion in plant biology, 2019. 50: p. 67-75. [CrossRef]
- Byeon, Y., H.J. Lee, H.Y. Lee, and K. Back, Cloning and functional characterization of the Arabidopsis N-acetylserotonin O-methyltransferase responsible for melatonin synthesis. J Pineal Res, 2016. 60(1): p. 65-73.
- Byeon, Y., G.H. Choi, H.Y. Lee, and K. Back, Melatonin biosynthesis requires N-acetylserotonin methyltransferase activity of caffeic acid O-methyltransferase in rice. J Exp Bot, 2015. 66(21): p. 6917-25. [CrossRef]
- Wei, Y., G. Liu, Y. Bai, F. Xia, C. He, H. Shi, and C. Foyer, Two transcriptional activators of N-acetylserotonin O-methyltransferase 2 and melatonin biosynthesis in cassava. J Exp Bot, 2017. 68(17): p. 4997-5006. [CrossRef]
- Kang, K., K. Lee, S. Park, Y.S. Kim, and K. Back, Enhanced production of melatonin by ectopic overexpression of human serotonin N-acetyltransferase plays a role in cold resistance in transgenic rice seedlings. J Pineal Res, 2010. 49(2): p. 176-82. [CrossRef]
- Zhou, W., Y. Wang, B. Li, L. Petijova, S. Hu, Q. Zhang, J. Niu, D. Wang, S. Wang, Y. Dong, E. Cellarova, and Z. Wang, Whole-genome sequence data of Hypericum perforatum and functional characterization of melatonin biosynthesis by N-acetylserotonin O-methyltransferase. J Pineal Res, 2021. 70(2): p. e12709. [CrossRef]
- Wang, L., C. Feng, X. Zheng, Y. Guo, F. Zhou, D. Shan, X. Liu, and J. Kong, Plant mitochondria synthesize melatonin and enhance the tolerance of plants to drought stress. J Pineal Res, 2017. 63(3). [CrossRef]
- Zheng, X., D.X. Tan, A.C. Allan, B. Zuo, Y. Zhao, R.J. Reiter, L. Wang, Z. Wang, Y. Guo, J. Zhou, D. Shan, Q. Li, Z. Han, and J. Kong, Chloroplastic biosynthesis of melatonin and its involvement in protection of plants from salt stress. Sci Rep, 2017. 7: p. 41236. [CrossRef]
- Zhu, Y., M.J. Guo, J.B. Song, S.Y. Zhang, R. Guo, D.R. Hou, C.Y. Hao, H.L. An, and X. Huang, Roles of Endogenous Melatonin in Resistance to Botrytis cinerea Infection in an Arabidopsis Model. Front Plant Sci, 2021. 12: p. 683228. [CrossRef]
- Zhou, K., Y. Li, L. Hu, J. Zhang, H. Yue, S. Yang, Y. Liu, X. Gong, and F. Ma, Overexpression of MdASMT9, an N-acetylserotonin methyltransferase gene, increases melatonin biosynthesis and improves water-use efficiency in transgenic apple. Tree Physiol, 2022. 42(5): p. 1114-1126. [CrossRef]
- Lee, H.Y., Y. Byeon, and K. Back, Melatonin as a signal molecule triggering defense responses against pathogen attack in Arabidopsis and tobacco. J Pineal Res, 2014. 57(3): p. 262-8. [CrossRef]
- Kong, M., T. Sheng, J. Liang, Q. Ali, Q. Gu, H. Wu, J. Chen, J. Liu, and X. Gao, Melatonin and Its Homologs Induce Immune Responses via Receptors trP47363-trP13076 in Nicotiana benthamiana. Front Plant Sci, 2021. 12: p. 691835. [CrossRef]
- Li, M., X. Zhang, J. Li, M. Ali, Y. Wang, X. Liu, F. Li, and X. Li, GABA primes defense responses against Botrytis cinerea in tomato fruit by modulating ethylene and JA signaling pathways. Postharvest Biology and Technology, 2024. 208: p. 112665. [CrossRef]
- Xuan Phong, H., Q. Le Viet, L. Minh Chau, D. Long, H. Bui, N.N. Thanh, D. Tan Phat, and L.D. Truong, Isolation and selection of lactic acid bacteria with the capacity of producing γ-aminobutyric acid (GABA) and antimicrobial activity: Its application in fermented meat product. Current Nutrition & Food Science, 2023. 19(8): p. 831-837. [CrossRef]
- Guo, Z., J. Lv, X. Dong, N. Du, and F. Piao, Gamma-aminobutyric acid improves phenanthrene phytotoxicity tolerance in cucumber through the glutathione-dependent system of antioxidant defense. Ecotoxicology and environmental safety, 2021. 217: p. 112254. [CrossRef]
- Hijaz, F., Y. Nehela, and N. Killiny, Application of gamma-aminobutyric acid increased the level of phytohormones in Citrus sinensis. Planta, 2018. 248: p. 909-918. [CrossRef]
- Ghosh, S., P. Kanwar, and G. Jha, Alterations in rice chloroplast integrity, photosynthesis and metabolome associated with pathogenesis of Rhizoctonia solani. Scientific reports, 2017. 7(1): p. 1-12. [CrossRef]
- Deng, X., X. Xu, Y. Liu, Y. Zhang, L. Yang, S. Zhang, and J. Xu, Induction of γ-aminobutyric acid plays a positive role to Arabidopsis resistance against Pseudomonas syringae. Journal of integrative plant biology, 2020. 62(11): p. 1797-1812.
- Rani, M. and G. Jha, Host gamma-aminobutyric acid metabolic pathway is involved in resistance against Rhizoctonia solani. Phytopathology®, 2021. 111(7): p. 1207-1218. [CrossRef]
- Meher, H.C., V.T. Gajbhiye, and G. Singh, Salicylic acid-induced glutathione status in tomato crop and resistance to root-knot nematode, Meloidogyne incognita (Kofoid & White) Chitwood. Journal of xenobiotics, 2011. 1(1): p. e5. [CrossRef]
- Meher, H.C., V.T. Gajbhiye, G. Singh, and G. Chawla, Altered metabolomic profile of selected metabolites and improved resistance of Cicer arietinum (L.) against Meloidogyne incognita (Kofoid & White) Chitwood following seed soaking with salicylic acid, benzothiadiazole or nicotinic acid. Acta Physiologiae Plantarum, 2015. 37: p. 1-12. [CrossRef]
- Cui, W., P. Yao, J. Pan, C. Dai, H. Cao, Z. Chen, S. Zhang, S. Xu, and W. Shen, Transcriptome analysis reveals insight into molecular hydrogen-induced cadmium tolerance in alfalfa: The prominent role of sulfur and (homo) glutathione metabolism. BMC plant Biology, 2020. 20(1): p. 1-19. [CrossRef]
- Gullner, G., T. Komives, L. Király, and P. Schröder, Glutathione S-transferase enzymes in plant-pathogen interactions. Frontiers in plant science, 2018. 9: p. 1836. [CrossRef]
- Hiruma, K., S. Fukunaga, P. Bednarek, M. Piślewska-Bednarek, S. Watanabe, Y. Narusaka, K. Shirasu, and Y. Takano, Glutathione and tryptophan metabolism are required for Arabidopsis immunity during the hypersensitive response to hemibiotrophs. Proceedings of the National Academy of Sciences, 2013. 110(23): p. 9589-9594. [CrossRef]
- Liu, X., S. Zhang, R.J. Whitworth, J.J. Stuart, and M.-S. Chen, Unbalanced activation of glutathione metabolic pathways suggests potential involvement in plant defense against the gall midge Mayetiola destructor in wheat. Scientific Reports, 2015. 5(1): p. 8092. [CrossRef]
- Chen, Y.-P., L.-P. Xing, G.-J. Wu, H.-Z. Wang, X.-E. Wang, A.-Z. Cao, and P.-D. Chen, Plastidial glutathione reductase from Haynaldia villosa is an enhancer of powdery mildew resistance in wheat (Triticum aestivum). Plant and cell physiology, 2007. 48(12): p. 1702-1712. [CrossRef]
- Sova, M., Antioxidant and antimicrobial activities of cinnamic acid derivatives. Mini reviews in medicinal chemistry, 2012. 12(8): p. 749-767. [CrossRef]
- Muroi, A., A. Ishihara, C. Tanaka, A. Ishizuka, J. Takabayashi, H. Miyoshi, and T. Nishioka, Accumulation of hydroxycinnamic acid amides induced by pathogen infection and identification of agmatine coumaroyltransferase in Arabidopsis thaliana. Planta, 2009. 230: p. 517-527. [CrossRef]
- Guo, M., C. Li, R. Huang, L. Qu, J. Liu, C. Zhang, and Y. Ge, Ferulic acid enhanced resistance against blue mold of Malus domestica by regulating reactive oxygen species and phenylpropanoid metabolism. Postharvest Biology and Technology, 2023. 202: p. 112378. [CrossRef]
- Gozzo, F., Systemic acquired resistance in crop protection: from nature to a chemical approach. Journal of Agricultural and Food Chemistry, 2003. 51(16): p. 4487-4503. [CrossRef]
- Huang, G.-Y., C. Cui, Z.-P. Wang, Y.-Q. Li, L.-X. Xiong, L.-Z. Wang, S.-J. Yu, Z.-M. Li, and W.-G. Zhao, Synthesis and characteristics of (Hydrogenated) ferulic acid derivatives as potential antiviral agents with insecticidal activity. Chemistry Central Journal, 2013. 7(1): p. 1-12. [CrossRef]
- Wu, Z., J. Zhang, J. Chen, J. Pan, L. Zhao, D. Liu, A. Zhang, J. Chen, D. Hu, and B. Song, Design, synthesis, antiviral bioactivity and three-dimensional quantitative structure–activity relationship study of novel ferulic acid ester derivatives containing quinazoline moiety. Pest management science, 2017. 73(10): p. 2079-2089.
- Kwon, Y.-S., A. Kobayashi, S.-I. Kajiyama, K. Kawazu, H. Kanzaki, and C.-M. Kim, Antimicrobial constituents of Angelica dahurica roots. Phytochemistry, 1997. 44(5): p. 887-889. [CrossRef]
- Fang, L., B. Kraus, J. Lehmann, J. Heilmann, Y. Zhang, and M. Decker, Design and synthesis of tacrine–ferulic acid hybrids as multi-potent anti-Alzheimer drug candidates. Bioorganic & medicinal chemistry letters, 2008. 18(9): p. 2905-2909. [CrossRef]
- He, F., P. Wei, G. Yu, S. Guo, Z. Zheng, S. Chen, A. Dai, R. Zhang, Z. Wu, and J. Wu, Synthesis of trans-methyl ferulate bearing an oxadiazole ether as potential activators for controlling plant virus. Bioorganic Chemistry, 2021. 115: p. 105248. [CrossRef]
- Gan, X., D. Hu, Y. Wang, L. Yu, and B. Song, Novel trans-ferulic acid derivatives containing a chalcone moiety as potential activator for plant resistance induction. Journal of Agricultural and Food Chemistry, 2017. 65(22): p. 4367-4377. [CrossRef]
- Zhao, X., P. Li, X. Liu, T. Xu, Y. Zhang, H. Meng, and T. Xia, High temperature increased lignin contents of poplar (Populus spp) stem via inducing the synthesis caffeate and coniferaldehyde. 2021.
- Tang, Y., Z. Zhang, Y. Lei, G. Hu, J. Liu, M. Hao, A. Chen, Q. Peng, and J. Wu, Cotton WATs modulate SA biosynthesis and local lignin deposition participating in plant resistance against Verticillium dahliae. Frontiers in Plant Science, 2019. 10: p. 526. [CrossRef]
- Wang, J.-Z., C.-H. Yan, X.-R. Zhang, Q.-B. Tu, Y. Xu, S. Sheng, F.-A. Wu, and J. Wang, A novel nanoparticle loaded with methyl caffeate and caffeic acid phenethyl ester against Ralstonia solanacearum—a plant pathogenic bacteria. RSC advances, 2020. 10(7): p. 3978-3990. [CrossRef]
- Vogt, T., Phenylpropanoid biosynthesis. Molecular plant, 2010. 3(1): p. 2-20.
- McCalla, D. and A. Neish, Metabolism of phenylpropanoid compounds in Salvia: II. Biosynthesis of phenolic cinnamic acids. Canadian Journal of Biochemistry and Physiology, 1959. 37(4): p. 537-547.
- Dixon, R.A., L. Achnine, P. Kota, C.J. Liu, M.S. Reddy, and L. Wang, The phenylpropanoid pathway and plant defence—a genomics perspective. Molecular plant pathology, 2002. 3(5): p. 371-390. [CrossRef]
- Denancé, N., P. Ranocha, N. Oria, X. Barlet, M.P. Rivière, K.A. Yadeta, L. Hoffmann, F. Perreau, G. Clément, and A. Maia-Grondard, Arabidopsis wat1 (walls are thin1)-mediated resistance to the bacterial vascular pathogen, Ralstonia solanacearum, is accompanied by cross-regulation of salicylic acid and tryptophan metabolism. The Plant Journal, 2013. 73(2): p. 225-239.
- Wiklund, P. and J. Bergman, The chemistry of anthranilic acid. Current Organic Synthesis, 2006. 3(3): p. 379-402.
- Winter, A., A hypothetical route for the biogenisis of IAA. Planta, 1966. 71: p. 229-239. [CrossRef]
- Doyle, S.M., A. Rigal, P. Grones, M. Karady, D.K. Barange, M. Majda, B. Parizkova, M. Karampelias, M. Zwiewka, A. Pencik, F. Almqvist, K. Ljung, O. Novak, and S. Robert, A role for the auxin precursor anthranilic acid in root gravitropism via regulation of PIN-FORMED protein polarity and relocalisation in Arabidopsis. New Phytol, 2019. 223(3): p. 1420-1432. [CrossRef]
- Yang, S.Y., M.R. Park, I.S. Kim, Y.C. Kim, J.W. Yang, and C.-M. Ryu, 2-Aminobenzoic acid of Bacillus sp. BS107 as an ISR determinant against Pectobacterium carotovorum subsp. carotovotrum SCC1 in tobacco. European Journal of Plant Pathology, 2011. 129: p. 371-378. [CrossRef]
- Hossain, M., M. Hossain, R. Islam, A. Alam, K. Zahan, S. Sarkar, and M. Farooque, Antimicrobial and cytotoxic activities of 2-aminobenzoic acid and 2-aminophenol and their coordination complexes with Magnesium (Mg-II). Pak J Biol Sci, 2004. 7(1): p. 25-27. [CrossRef]
- Zhang, Z., X. Bi, X. Du, H. Liu, T. An, Y. Zhao, H. Yu, Y. Chen, and J. Wen, Comparative metabolomics reveal the participation of soybean unique rhizosphere metabolites in susceptibility and resistance of host soybean to Phytophthora sojae. Plant and Soil, 2022: p. 1-15.
- Iriti, M. and F. Faoro, Benzothiadiazole (BTH) induces cell-death independent resistance in Phaseolus vulgaris against Uromyces appendiculatus. Journal of Phytopathology, 2003. 151(3): p. 171-180. [CrossRef]
- Iriti, M., M. Rossoni, M. Borgo, and F. Faoro, Benzothiadiazole enhances resveratrol and anthocyanin biosynthesis in grapevine, meanwhile improving resistance to Botrytis cinerea. Journal of agricultural and food chemistry, 2004. 52(14): p. 4406-4413. [CrossRef]
- Takahashi, Y., The role of polyamines in plant disease resistance. Environmental Control in Biology, 2016. 54(1): p. 17-21. [CrossRef]
- Kusano, T., K. Yamaguchi, T. Berberich, and Y. Takahashi, Advances in polyamine research in 2007. Journal of plant research, 2007. 120: p. 345-350.
- Takahashi, Y., T. Berberich, K. Yamashita, Y. Uehara, A. Miyazaki, and T. Kusano, Identification of tobacco HIN1 and two closely related genes as spermine-responsive genes and their differential expression during the Tobacco mosaic virus-induced hypersensitive response and during leaf-and flower-senescence. Plant molecular biology, 2004. 54: p. 613-622. [CrossRef]
- Yamakawa, H., H. Kamada, M. Satoh, and Y. Ohashi, Spermine is a salicylate-independent endogenous inducer for both tobacco acidic pathogenesis-related proteins and resistance against tobacco mosaic virus infection. Plant Physiology, 1998. 118(4): p. 1213-1222. [CrossRef]
- da Graça, J.P., T.E. Ueda, T. Janegitz, S.S. Vieira, M.C. Salvador, M.C. de Oliveira, S.M. Zingaretti, S.J. Powers, J.A. Pickett, and M.A. Birkett, The natural plant stress elicitor cis-jasmone causes cultivar-dependent reduction in growth of the stink bug, Euschistus heros and associated changes in flavonoid concentrations in soybean, Glycine max. Phytochemistry, 2016. 131: p. 84-91. [CrossRef]
- Stevenson, P., H. Turner, and M. Haware, Phytoalexin accumulation in the roots of chickpea (Cicer arietinumL.) seedlings associated with resistance to fusarium wilt (Fusarium oxysporumf. sp. ciceri). Physiological and Molecular Plant Pathology, 1997. 50(3): p. 167-178. [CrossRef]
- Gupta, A., P. Awasthi, N. Sharma, S. Parveen, R.P. Vats, N. Singh, Y. Kumar, A. Goel, and D. Chandran, Medicarpin confers powdery mildew resistance in Medicago truncatula and activates the salicylic acid signalling pathway. Molecular Plant Pathology, 2022. 23(7): p. 966-983. [CrossRef]
- Kumar, R.S., M. Moydeen, S.S. Al-Deyab, A. Manilal, and A. Idhayadhulla, Synthesis of new morpholine-connected pyrazolidine derivatives and their antimicrobial, antioxidant, and cytotoxic activities. Bioorganic & Medicinal Chemistry Letters, 2017. 27(1): p. 66-71.
- Jyotshna, P. Khare, and K. Shanker, Mangiferin: A review of sources and interventions for biological activities. Biofactors, 2016. 42(5): p. 504-514. [CrossRef]
- Ghosal, S., K. Biswas, D.K. Chakrabarti, and K. Basu Chaudhary, Control of Fusarium wilt of safflower by mangiferin. Phytopathology, 1977. 67(4): p. 548-550.
- Gao, X., K. Li, Z. Ma, H. Zou, H. Jin, and J. Wang, Cucumber Fusarium wilt resistance induced by intercropping with celery differs from that induced by the cucumber genotype and is related to sulfur-containing allelochemicals. Scientia Horticulturae, 2020. 271: p. 109475. [CrossRef]
- Godard, S., I. Slacanin, O. Viret, and K. Gindro, Induction of defence mechanisms in grapevine leaves by emodin-and anthraquinone-rich plant extracts and their conferred resistance to downy mildew. Plant Physiology and Biochemistry, 2009. 47(9): p. 827-837. [CrossRef]
- Zhao, Y., G. Han, Y. Li, and H. Lv, Changes in quality characteristics and metabolites composition of wheat under different storage temperatures. Journal of Stored Products Research, 2024. 105: p. 102229. [CrossRef]
- Li, X., J. Zhang, S. Lin, Y. Xing, X. Zhang, M. Ye, Y. Chang, H. Guo, and X. Sun, (+)-Catechin, epicatechin and epigallocatechin gallate are important inducible defensive compounds against Ectropis grisescens in tea plants. Plant, Cell & Environment, 2022. 45(2): p. 496-511. [CrossRef]
- Piispanen, J., U. Bergmann, J. Karhu, T. Kauppila, and J. Kaitera, Variation of compounds in leaves of susceptible and resistant alternate hosts of Cronartium pini and C. ribicola. European Journal of Plant Pathology, 2023: p. 1-16.
- Jo, J., J. Lee, Y. Ahn, Y.S. Hwang, J. Park, J. Lee, and J. Choi, Metabolome and transcriptome analyses of plants grown in naturally attenuated soil after hydrogen fluoride exposure. Journal of Hazardous Materials, 2022. 437: p. 129323. [CrossRef]
- Wu, H., L. Wu, J. Wang, Q. Zhu, S. Lin, J. Xu, C. Zheng, J. Chen, X. Qin, and C. Fang, Mixed phenolic acids mediated proliferation of pathogens Talaromyces helicus and Kosakonia sacchari in continuously monocultured Radix pseudostellariae rhizosphere soil. Frontiers in Microbiology, 2016. 7: p. 335. [CrossRef]
- Schoch, G.A., G.N. Nikov, W.L. Alworth, and D. Werck-Reichhart, Chemical inactivation of the cinnamate 4-hydroxylase allows for the accumulation of salicylic acid in elicited cells. Plant physiology, 2002. 130(2): p. 1022-1031. [CrossRef]
- Desmedt, W., W. Jonckheere, V.H. Nguyen, M. Ameye, N. De Zutter, K. De Kock, J. Debode, T. Van Leeuwen, K. Audenaert, and B. Vanholme, The phenylpropanoid pathway inhibitor piperonylic acid induces broad-spectrum pest and disease resistance in plants. Plant, Cell & Environment, 2021. 44(9): p. 3122-3139. [CrossRef]

| Common name | Abbreviation | Chemical name | Mol. Formula | Concentration1 | Supplier |
| Pipecolic acid | PA | Piperidine-2-carboxylic acid | C6H11NO2 | 40 mM | Tokyo Chemical Industry (TCI) |
| Salicylic acid (sodium salt) | SA | Sodium 2-hydroxybenzoate | C7H5NaO3 | 1 mM | Thermo Fisher |
| Gentisic acid (sodium salt hydrate) | GA | 2,5-Dihydroxybenzoic acid sodium salt | C7H5O4Na | 10 mM | Sigma Aldrich |
| Glutathione | GSH | γ-L-glutamyl-L-cysteinylglycine | C6H11NO2 | 20 mM | Thermo Fisher |
| Lysine | Lys | (S)-2,6-Diaminocaproic acid | C6H14N2O2 | 10 mM | Sigma Aldrich |
| Diaminopimelic acid | DAP | 2,6-Diaminopimelic acid | C7H14N2O4 | 30 mM | Sigma Aldrich |
| Ferulic acid | FA | Trans-ferulic acid | C10H10O4 | 1 mM2 | Sigma Aldrich |
| Caffeic acid | CFA | (E)-3-(3,4-dihydroxyphenyl) prop-2-enoic acid | C9H8O4 | 10 mM3 | Sigma Aldrich |
| Benzoic acid | BA | Benzoic acid | C7H6O2 | 10 mM4 | Thermo Fisher |
| Piperonylic acid | PipA | 1,3-benzodioxole-5-carboxylic acid | C8H6O4 | 3 mM5 | Sigma Aldrich |
| Pathways | Total Compounds | Hits | Raw p | Impact |
| Flavone and flavonol biosynthesis | 10 | 4 | 5.2649e-05 | 0.5 |
| Isoquinoline alkaloid biosynthesis | 6 | 1 | 0.013614 | 0.5 |
| Arginine and proline metabolism | 32 | 3 | 0.11938 | 0.32738 |
| Biosynthesis of various plant secondary metabolites | 29 | 1 | 0.24485 | 0.24 |
| Glycine, serine and threonine metabolism | 33 | 2 | 0.018356 | 0.22375 |
| Lysine biosynthesis | 9 | 1 | 0.000103 | 0.16216 |
| Arginine biosynthesis | 18 | 1 | 0.17605 | 0.13981 |
| Tryptophan metabolism | 29 | 1 | 0.010177 | 0.10687 |
| Glyoxylate and dicarboxylate metabolism | 29 | 1 | 0.11936 | 0.10147 |
| Tyrosine metabolism | 17 | 1 | 0.013614 | 0.10056 |
| Phenylpropanoid biosynthesis | 43 | 2 | 0.00128 | 0.09634 |
| Glutathione metabolism | 26 | 1 | 0.11936 | 0.07114 |
| Pyrimidine metabolism | 41 | 1 | 0.021007 | 0.02929 |
| Purine metabolism | 73 | 3 | 0.000316 | 0.02344 |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 22 | 2 | 0.011081 | 0.02002 |
| Flavonoid biosynthesis | 47 | 2 | 4.2424e-05 | 0.00338 |
| Cysteine and methionine metabolism | 47 | 1 | 0.000562 | 0.00265 |
| Lipoic acid metabolism | 24 | 1 | 0.11936 | 0.0016 |
| D-Amino acid metabolism | 7 | 1 | 0.000103 | 0 |
| Indole alkaloid biosynthesis | 4 | 1 | 0.010177 | 0 |
| Glucosinolate biosynthesis | 65 | 1 | 0.010177 | 0 |
| Ubiquinone and other terpenoid-quinone biosynthesis | 47 | 1 | 0.013614 | 0 |
| Anthocyanin biosynthesis | 11 | 1 | 0.014562 | 0 |
| Lysine degradation | 20 | 1 | 0.038832 | 0 |
| Thiamine metabolism | 22 | 1 | 0.11936 | 0 |
| Cyanoamino acid metabolism | 29 | 2 | 0.16748 | 0 |
| Pathways | Total Compounds | Hits | Raw p | Impact |
| Taurine and hypotaurine metabolism | 5 | 2 | 0.004092 | 1 |
| Glutathione metabolism | 26 | 5 | 0.000843 | 0.51637 |
| Isoquinoline alkaloid biosynthesis | 6 | 3 | 0.000207 | 0.5 |
| Tyrosine metabolism | 17 | 4 | 0.001364 | 0.32961 |
| Glycine, serine and threonine metabolism | 33 | 1 | 7.41E-05 | 0.22375 |
| Arginine biosynthesis | 18 | 2 | 0.020249 | 0.17088 |
| Lysine degradation | 20 | 3 | 5.38E-05 | 0.16667 |
| Lysine biosynthesis | 9 | 1 | 0.000818 | 0.16216 |
| Flavone and flavonol biosynthesis | 10 | 2 | 0.00029 | 0.15 |
| Arginine and proline metabolism | 32 | 3 | 0.040159 | 0.14584 |
| Butanoate metabolism | 17 | 1 | 0.13621 | 0.13636 |
| Cysteine and methionine metabolism | 47 | 2 | 0.000311 | 0.13181 |
| Alanine, aspartate and glutamate metabolism | 22 | 1 | 0.13621 | 0.1295 |
| Purine metabolism | 73 | 3 | 0.001022 | 0.10374 |
| Glyoxylate and dicarboxylate metabolism | 29 | 1 | 7.41E-05 | 0.10147 |
| Phenylpropanoid biosynthesis | 43 | 1 | 0.017894 | 0.05935 |
| Pyrimidine metabolism | 41 | 1 | 0.000913 | 0.02929 |
| Folate biosynthesis | 31 | 1 | 0.39026 | 0.02624 |
| Porphyrin metabolism | 48 | 1 | 0.001198 | 0.02261 |
| Ubiquinone and other terpenoid-quinone biosynthesis | 47 | 2 | 4.84E-05 | 0.02209 |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 22 | 2 | 4.84E-05 | 0.02002 |
| Tryptophan metabolism | 29 | 3 | 0.000722 | 0.01527 |
| Lipoic acid metabolism | 24 | 1 | 7.41E-05 | 0.0016 |
| Thiamine metabolism | 22 | 1 | 7.41E-05 | 0 |
| Cyanoamino acid metabolism | 29 | 2 | 8.89E-05 | 0 |
| Glucosinolate biosynthesis | 65 | 3 | 0.000128 | 0 |
| Flavonoid biosynthesis | 47 | 1 | 0.000193 | 0 |
| Biosynthesis of various plant secondary metabolites | 29 | 1 | 0.000248 | 0 |
| Valine, leucine and isoleucine degradation | 37 | 1 | 0.000662 | 0 |
| Valine, leucine and isoleucine biosynthesis | 22 | 1 | 0.000662 | 0 |
| D-Amino acid metabolism | 7 | 1 | 0.000818 | 0 |
| Tropane, piperidine and pyridine alkaloid biosynthesis | 9 | 2 | 0.003453 | 0 |
| Anthocyanin biosynthesis | 11 | 1 | 0.012109 | 0 |
| Zeatin biosynthesis | 21 | 1 | 0.044967 | 0 |
| Pathways | Total Compounds | Hits | Raw p | Impact |
| Taurine and hypotaurine metabolism | 5 | 3 | 0.003645 | 1 |
| Phenylalanine metabolism | 12 | 1 | 0.000113 | 0.42308 |
| Glutathione metabolism | 26 | 3 | 9.13E-05 | 0.40276 |
| Tyrosine metabolism | 17 | 5 | 0.000141 | 0.39665 |
| Phenylpropanoid biosynthesis | 43 | 8 | 9.23E-05 | 0.28583 |
| Ubiquinone and other terpenoid-quinone biosynthesis | 47 | 2 | 0.000443 | 0.1998 |
| beta-Alanine metabolism | 18 | 2 | 0.022568 | 0.19444 |
| Lysine degradation | 20 | 3 | 0.000743 | 0.16667 |
| Lysine biosynthesis | 9 | 1 | 0.000192 | 0.16216 |
| Arginine and proline metabolism | 32 | 3 | 3.65E-06 | 0.15774 |
| Butanoate metabolism | 17 | 1 | 0.000299 | 0.13636 |
| Alanine, aspartate and glutamate metabolism | 22 | 1 | 0.000299 | 0.1295 |
| Purine metabolism | 73 | 3 | 0.003473 | 0.09255 |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 22 | 3 | 1.11E-05 | 0.09159 |
| Arginine biosynthesis | 18 | 1 | 0.004743 | 0.08641 |
| Pyrimidine metabolism | 41 | 2 | 0.002791 | 0.07198 |
| Flavonoid biosynthesis | 47 | 5 | 0.00363 | 0.06956 |
| Cysteine and methionine metabolism | 47 | 5 | 0.000575 | 0.05644 |
| Glucosinolate biosynthesis | 65 | 3 | 1.36E-06 | 0.04236 |
| Tryptophan metabolism | 29 | 4 | 4.93E-05 | 0.03054 |
| Pantothenate and CoA biosynthesis | 25 | 1 | 0.16968 | 0.02796 |
| Porphyrin metabolism | 48 | 1 | 0.013372 | 0.02261 |
| Flavone and flavonol biosynthesis | 10 | 2 | 6.24E-05 | 0 |
| Cyanoamino acid metabolism | 29 | 1 | 0.000113 | 0 |
| Tropane, piperidine and pyridine alkaloid biosynthesis | 9 | 3 | 0.000161 | 0 |
| D-Amino acid metabolism | 7 | 1 | 0.000192 | 0 |
| Anthocyanin biosynthesis | 11 | 2 | 0.00027 | 0 |
| Glycine, serine and threonine metabolism | 33 | 1 | 0.002717 | 0 |
| Zeatin biosynthesis | 21 | 1 | 0.005646 | 0 |
| Isoquinoline alkaloid biosynthesis | 6 | 2 | 0.006486 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





