Submitted:
03 April 2025
Posted:
04 April 2025
You are already at the latest version
Abstract
Keywords:
Introduction
Materials and Methods
Animal Care
Experimental Design and Animal Handling
Sample Collection
Measurement of Hematological Parameters
Measurement of Plasma Stress-Related Hormones, Haptoglobin, and TNF-α
Measurement of Intestinal Morphology
Measurement of Apoptosis in Ileal Cells
Measurement of Antioxidant and Immune Indices in Ileum
mRNA Library Construction and Sequencing
Statistical Analysis
Results
Hematological Responses
Plasma Stress-Related Hormones, Haptoglobin, and TNF-α
Intestinal Morphology
Apoptosis of Ileal Cells
Antioxidant and Immune Indices in Ileum
RNA Sequencing (RNA-seq) Data Mapping and Annotation
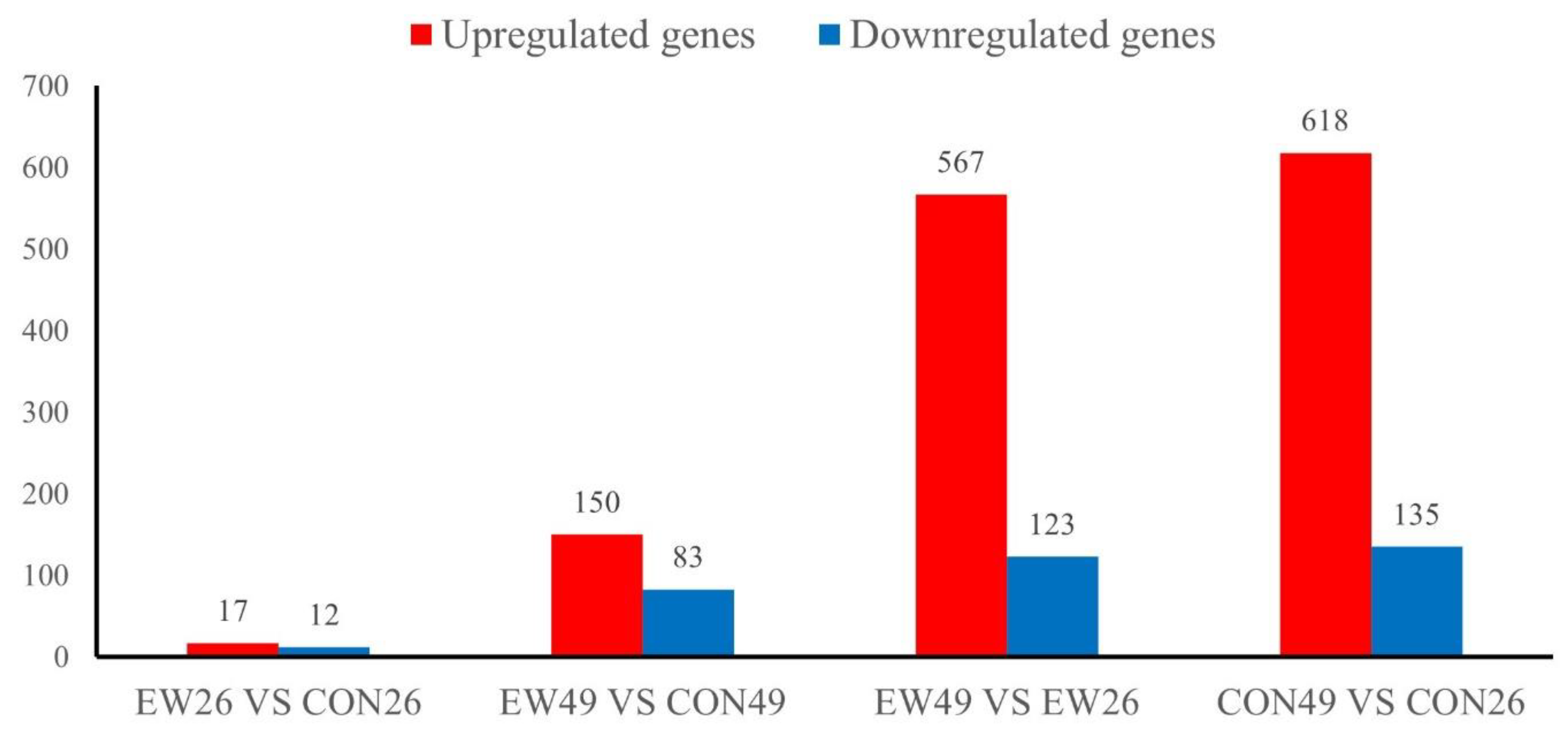
Differentially Expressed Genes (DEGs)
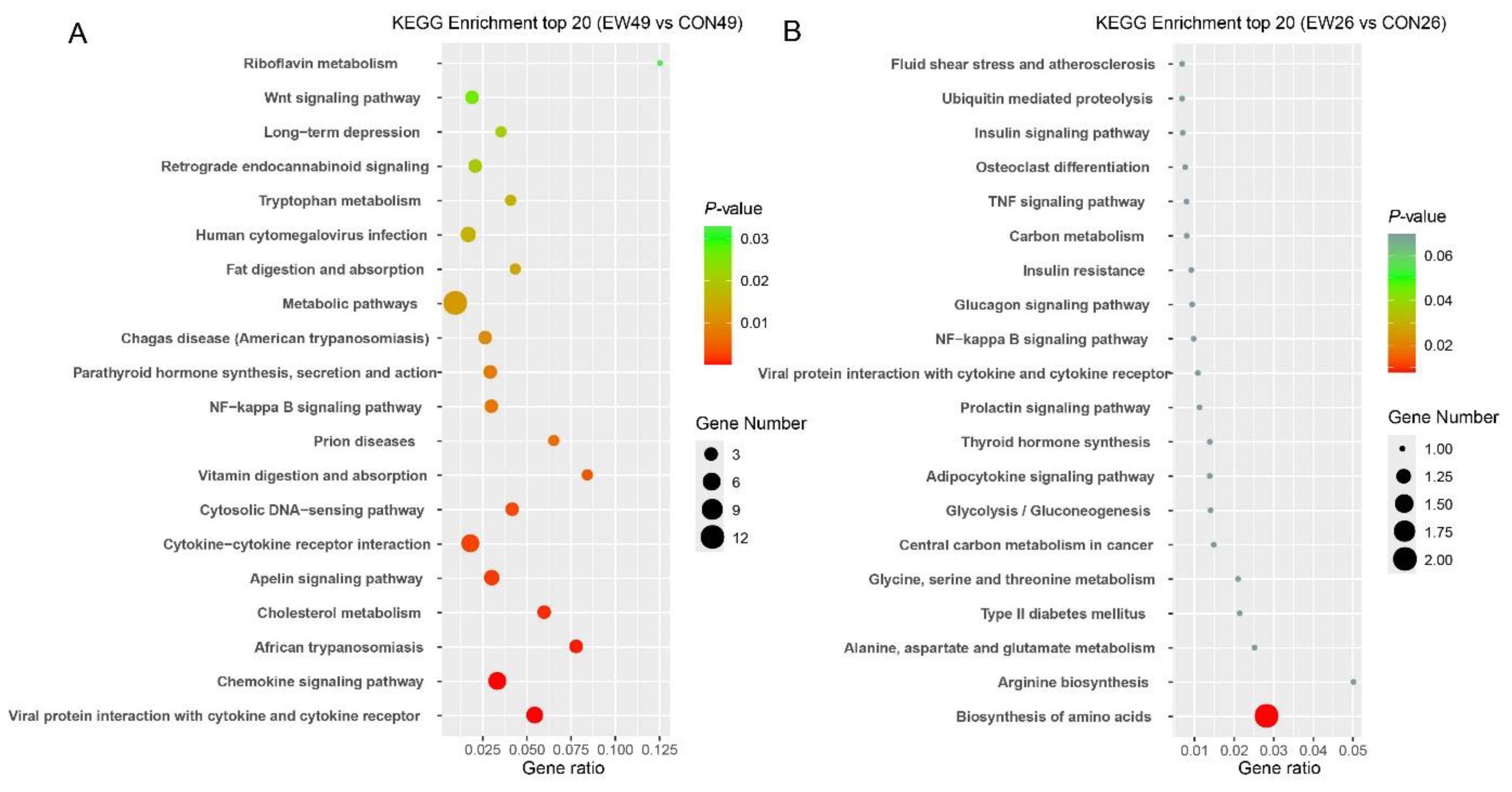
KEGG Pathway Analysis of DEGs
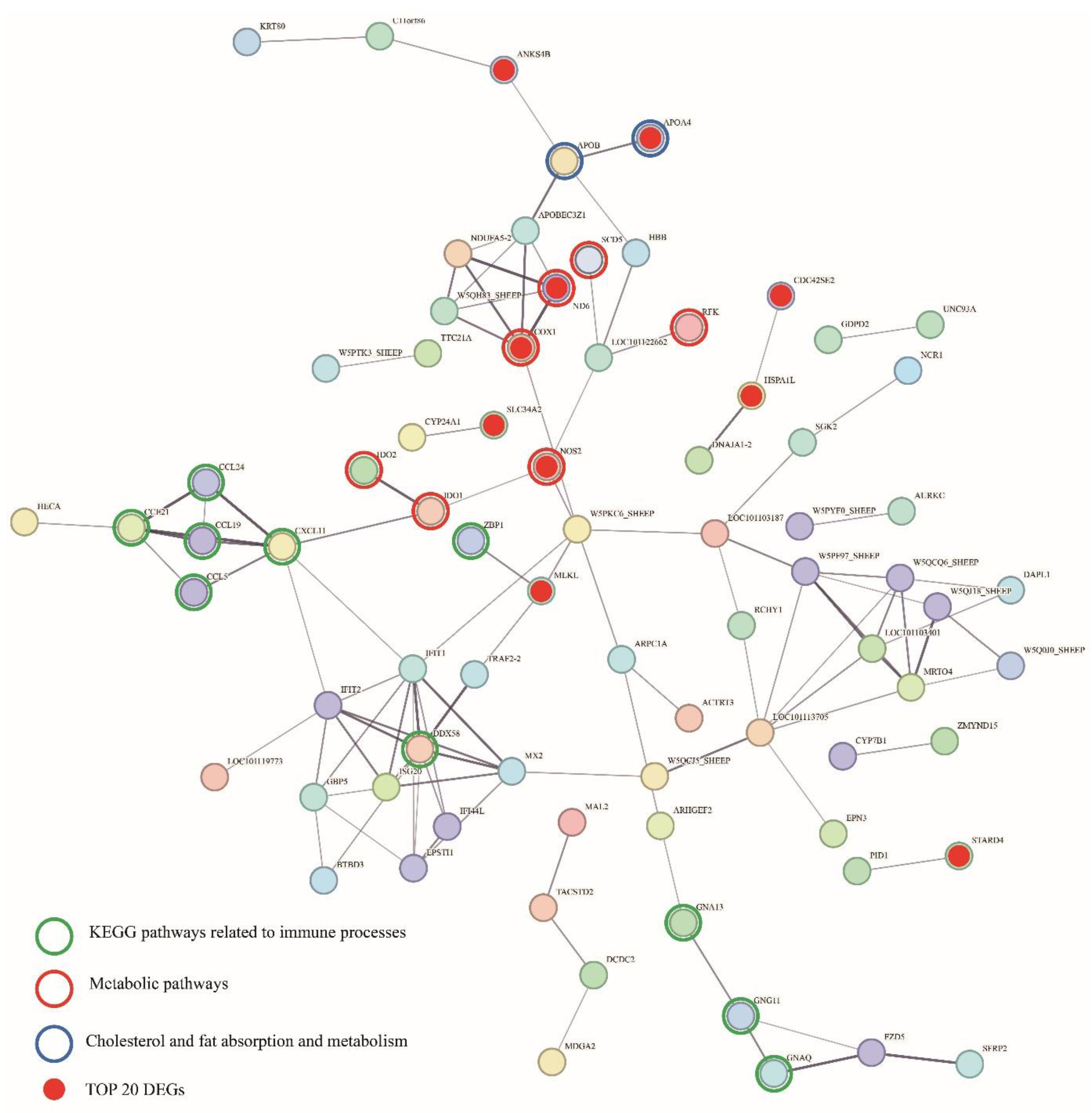
Protein-Protein Interaction (PPI) Network of DEGs
Discussion
Conclusion
Funding
Acknowledgments
Conflicts of Interest
References
- Schichowski C, Moors E, Gauly M. Influence of weaning age and an experimental Haemonchus contortus infection on behaviour and growth rates of lambs. Appl Anim Behav Sci 2010;125:103-8. [CrossRef]
- Fazio E, Medica P, Cravana C, Ferlazzo A. Short- and Long-term Effects of Weaning on Adrenocortical and Functional Response of Lambs. Acta Sci Vet 2014;42.
- Mialon MM, Boivin X, Durand D, et al. Short- and mid-term effects on performance, health and qualitative behavioural assessment of Romane lambs in different milk feeding conditions. Animal 2021;15:100157. [CrossRef]
- Beard SC, Schmied JD, Hodgins DC, Mallard BA. The effects of timing of high immune response phenotyping in relation to weaning on immune responses of crossbred beef calves. J Anim Sci 2023;101. [CrossRef]
- Lambertz C, Farke-Rover A, Gauly M. Effects of sex and age on behavior and weight gain in beef calves after abrupt weaning. Anim Sci J 2015;86:345-50.
- Napolitano F, Annicchiarico G, Caroprese M, et al. Lambs prevented from suckling their mothers display behavioral, immune and endocrine disturbances. Physiol Behav 2003;78:81-9. [CrossRef]
- Li C, Wang G, Zhang Q, et al. Developmental changes of nutrient digestion in young lambs are influenced by weaning and associated with intestinal microbiota. Anim Biotechnol 2023;34:1362-76. [CrossRef]
- Han C, Li M, Li F, et al. Temporary sensory separation of lamb groups from ewes affects behaviors and serum levels of stress-related indicators of small-tailed Han lambs. Physiol Behav 2024;277:114504. [CrossRef]
- Hickey MC, Drennan M, Earley B. The effect of abrupt weaning of suckler calves on the plasma concentrations of cortisol, catecholamines, leukocytes, acute-phase proteins and in vitro interferon-gamma production. J Anim Sci 2003;81:2847-55. [CrossRef]
- Nicolaides NC, Kyratzi E, Lamprokostopoulou A, Chrousos GP, Charmandari E. Stress, the stress system and the role of glucocorticoids. Neuroimmunomodulat 2015;22:6-19. [CrossRef]
- Gao X, Cao Q, Cheng Y, et al. Chronic stress promotes colitis by disturbing the gut microbiota and triggering immune system response. Proc Natl Acad Sci U S A 2018;115:E2960-e9. [CrossRef]
- He Y, Liu N, Ji Y, Tso P, Wu Z. Weaning Stress in Piglets Alters the Expression of Intestinal Proteins Involved in Fat Absorption. J Nutr 2022;152:2387-95. [CrossRef]
- Wang X, Niu L, Wang Y, et al. Combining 16S rRNA Sequencing and Metabolomics Data to Decipher the Interactions between Gut Microbiota, Host Immunity, and Metabolites in Diarrheic Young Small Ruminants. Int J Mol Sci 2023;24. [CrossRef]
- Zhong T, Wang Y, Wang X, et al. Diarrhea in suckling lambs is associated with changes in gut microbiota, serum immunological and biochemical parameters in an intensive production system. Front Microbiol 2022;13:1020657. [CrossRef]
- Han X, Hu X, Jin W, Liu G. Dietary nutrition, intestinal microbiota dysbiosis and post-weaning diarrhea in piglets. Anim Nutr 2024;17:188-207. [CrossRef]
- Upadhaya SD, Kim IH. The Impact of Weaning Stress on Gut Health and the Mechanistic Aspects of Several Feed Additives Contributing to Improved Gut Health Function in Weanling Piglets-A Review. Animals (Basel) 2021;11. [CrossRef]
- Zhuang Y, Wu H, Wang X, et al. Resveratrol Attenuates Oxidative Stress-Induced Intestinal Barrier Injury through PI3K/Akt-Mediated Nrf2 Signaling Pathway. Oxid Med Cell Longev 2019;2019:7591840. [CrossRef]
- Frazee AC, Pertea G, Jaffe AE, et al. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat Biotechnol 2015;33:243-6. [CrossRef]
- Hulbert LE, Moisa SJ. Stress, immunity, and the management of calves. J Dairy Sci 2016. [CrossRef]
- O'Connor DB, Thayer JF, Vedhara K. Stress and Health: A Review of Psychobiological Processes. Annu Rev Psychol 2021;72:663-88. [CrossRef]
- Ledezma-Torres RA, Sanchez-Davila F, Rodriguez-Miranda DA, et al. Sexual performance and semen quality of pubertal lambs treated with different weaning methods. Arch Anim Breed 2022;65:259-65. [CrossRef]
- Millington, GW. The role of proopiomelanocortin (POMC) neurones in feeding behaviour. Nutr Metab (Lond) 2007;4:18. [CrossRef]
- Lynch EM, Earley B, McGee M, Doyle S. Effect of abrupt weaning at housing on leukocyte distribution, functional activity of neutrophils, and acute phase protein response of beef calves. Bmc Vet Res 2010;6:39. [CrossRef]
- Carroll JA, Arthington JD, Chase CC, Jr. Early weaning alters the acute-phase reaction to an endotoxin challenge in beef calves. J Anim Sci 2009;87:4167-72. [CrossRef]
- O'Loughlin A, McGee M, Doyle S, Earley B. Biomarker responses to weaning stress in beef calves. Res Vet Sci 2014;97:458-63.
- Ceja G, Boerman JP, Neves RC, Jorgensen MW, Johnson JS. l-Glutamine supplementation reduces gastrointestinal permeability and biomarkers of physiological stress in preweaning Holstein heifer calves. J Dairy Sci 2023;106:9663-76. [CrossRef]
- Chi H, Pepper M, Thomas PG. Principles and therapeutic applications of adaptive immunity. Cell 2024;187:2052-78. [CrossRef]
- Boivin X, Nowak R, Garcia AT. The presence of the dam affects the efficiency of gentling and feeding on the early establishment of the stockperson-lamb relationship. Appl Anim Behav Sci 2001;72:89-103.
- McCoard SA, Cristobal-Carballo O, Knol FW, et al. Impact of early weaning on small intestine, metabolic, immune and endocrine system development, growth and body composition in artificially reared lambs. J Anim Sci 2020;98. [CrossRef]
- Hu CH, Xiao K, Luan ZS, Song J. Early weaning increases intestinal permeability, alters expression of cytokine and tight junction proteins, and activates mitogen-activated protein kinases in pigs. J Anim Sci 2013;91:1094-101.
- Han L, Tao H, Kang L, et al. Transcriptome and iTRAQ-Based Proteome Reveal the Molecular Mechanism of Intestinal Injury Induced by Weaning Ewe's Milk in Lambs. Front Vet Sci 2022;9:809188. [CrossRef]
- Barker N, van de Wetering M, Clevers H. The intestinal stem cell. Genes Dev 2008;22:1856-64. [CrossRef]
- Zhu MH, Sung TS, Kurahashi M, et al. Na+-K+-Cl- cotransporter (NKCC) maintains the chloride gradient to sustain pacemaker activity in interstitial cells of Cajal. Am J Physiol Gastrointest Liver Physiol 2016;311:G1037-G46. [CrossRef]
- Shyer AE, Huycke TR, Lee C, Mahadevan L, Tabin CJ. Bending gradients: how the intestinal stem cell gets its home. Cell 2015;161:569-80. [CrossRef]
- Wood KM, Palmer SI, Steele MA, Metcalf JA, Penner GB. The influence of age and weaning on permeability of the gastrointestinal tract in Holstein bull calves. J Dairy Sci 2015;98:7226-37. [CrossRef]
- Dunière L, Ruiz P, Lebbaoui Y, et al. Effects of rearing mode on gastro-intestinal microbiota and development, immunocompetence, sanitary status and growth performance of lambs from birth to two months of age. Anim Microbiome 2023;5:34. [CrossRef]
- Fu ZL, Yang Y, Ma L, et al. Dynamics of oxidative stress and immune responses in neonatal calves during diarrhea. J Dairy Sci 2024;107:1286-98. [CrossRef]
- Pi J, Zhang Q, Fu J, et al. ROS signaling, oxidative stress and Nrf2 in pancreatic beta-cell function. Toxicol Appl Pharmacol 2010;244:77-83. [CrossRef]
- Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative stress, inflammation, and cancer: how are they linked? Free Radic Biol Med 2010;49:1603-16. [CrossRef]
- Dantzer R, O'Connor JC, Freund GG, Johnson RW, Kelley KW. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat Rev Neurosci 2008;9:46-56. [CrossRef]
- Liu M, Ma J, Xu J, et al. Fecal microbiota transplantation alleviates intestinal inflammatory diarrhea caused by oxidative stress and pyroptosis via reducing gut microbiota-derived lipopolysaccharides. Int J Biol Macromol 2024;261:129696. [CrossRef]
- Zhu LH, Zhao KL, Chen XL, Xu JX. Impact of weaning and an antioxidant blend on intestinal barrier function and antioxidant status in pigs. J Anim Sci 2012;90:2581-9.
- Yin J, Wu MM, Xiao H, et al. Development of an antioxidant system after early weaning in piglets. J Anim Sci 2014;92:612-9. [CrossRef]
- Chen Z, Wang G, Wang W, et al. Relationship between jejunum ATPase activity and antioxidant function on the growth performance, feed conversion efficiency, and jejunum microbiota in Hu sheep (Ovis aries). Bmc Vet Res 2024;20:242. [CrossRef]
- Wang F, Kohan AB, Lo C-M, et al. Apolipoprotein A-IV: a protein intimately involved in metabolism. J Lipid Res 2015;56:1403-18. [CrossRef]
- Nishihara K, van Niekerk J, He Z, et al. Reduction in mucosa thickness is associated with changes in immune function in the colon mucosa during the weaning transition in Holstein bull dairy calves. Genomics 2023;115:110680. [CrossRef]
- Sorokin, A. Nitric Oxide Synthase and Cyclooxygenase Pathways: A Complex Interplay in Cellular Signaling. Curr Med Chem 2016;23:2559-78. [CrossRef]
- Romani L, Fallarino F, De Luca A, et al. Defective tryptophan catabolism underlies inflammation in mouse chronic granulomatous disease. Nature 2008;451:211-5. [CrossRef]
- Tokunaga R, Zhang W, Naseem M, et al. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation - A target for novel cancer therapy. Cancer Treat Rev 2018;63:40-7. [CrossRef]
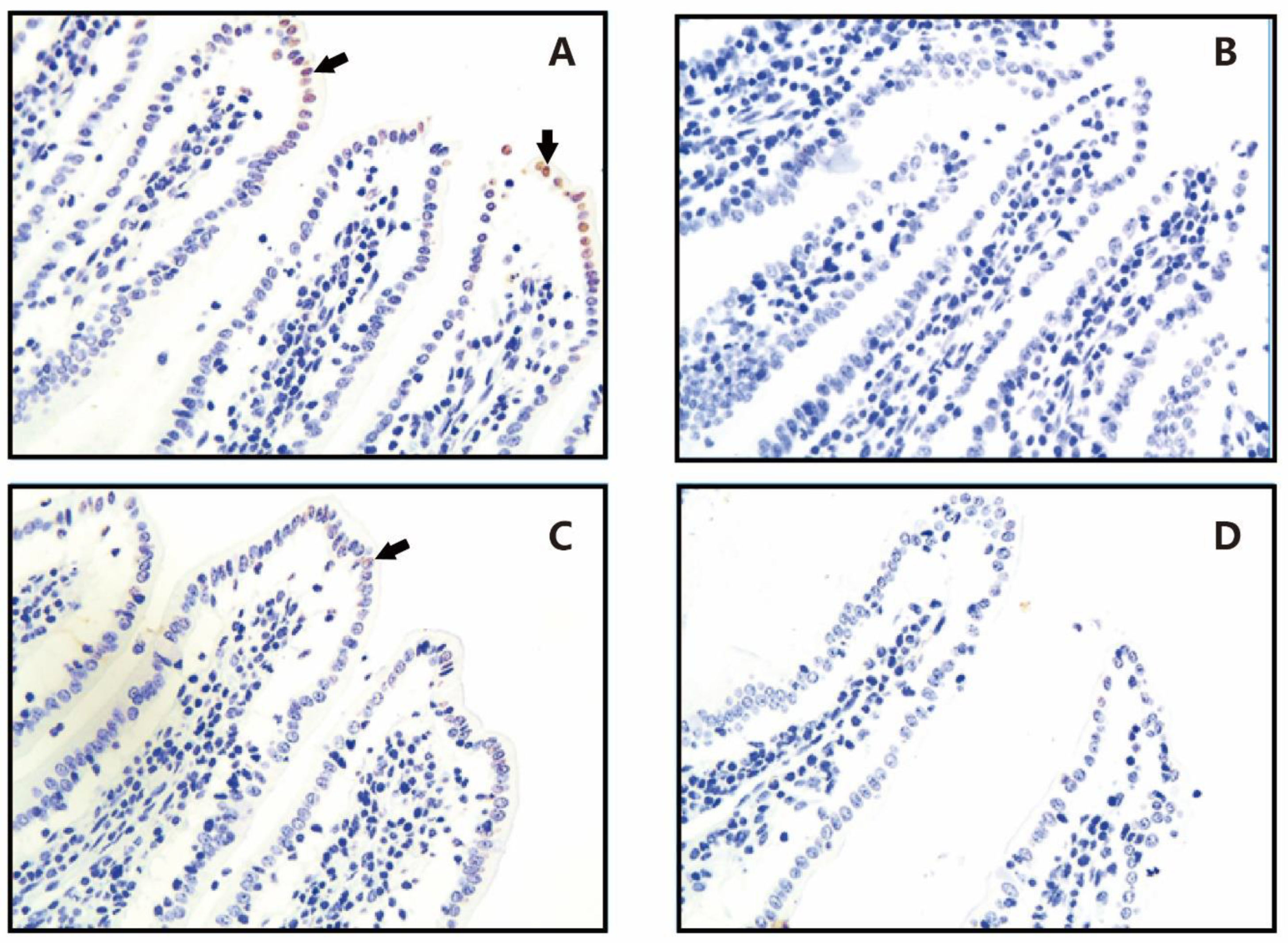
| Items | Treatment | Days post-weaning (d) | SEM | P value | ||||||||
| 0 | 1 | 2 | 3 | 7 | 14 | 28 | Weaning | Age | Weaning × Age | |||
| WBC (×109cells/L) | CON | 8.29 | 8.59 | 9.03 | 8.31 | 9.20 | 8.28 | 9.86 | 0.223 | 0.024 | 0.328 | 0.464 |
| EW | 7.95b | 10.92a* | 10.15ab | 10.66a* | 9.79ab | 9.53ab | 9.68ab | |||||
| LYM(×109cells/L) | CON | 3.52b | 3.64b | 3.62b | 3.32b | 4.01b | 4.32b | 5.47a | 0.108 | 0.041 | 0.004 | 0.528 |
| EW | 3.47b | 4.45ab | 4.37ab | 4.46ab* | 4.53ab | 4.69ab | 5.06a | |||||
| NEU(×109cells/L) | CON | 3.49 | 3.71 | 4.06 | 3.69 | 3.68 | 2.75 | 3.30 | 0.150 | 0.145 | 0.097 | 0.642 |
| EW | 3.24b | 5.18a* | 4.41ab | 4.69ab | 3.75ab | 3.35b | 3.14b | |||||
| NEU/LYM | CON | 1.02ab | 1.04ab | 1.21a | 1.12ab | 1.03ab | 0.66ab | 0.61b | 0.043 | 0.945 | 0.006 | 0.858 |
| EW | 1.00ab | 1.31a | 1.10ab | 1.11ab | 0.88ab | 0.74ab | 0.61 b | |||||
| RBC(×109cells/L) | CON | 8.47b | 8.00b | 8.02b | 7.91b | 7.94b | 8.37b | 9.22a | 0.068 | 0.432 | 0.004 | 0.449 |
| EW | 8.04b | 8.49ab | 8.34ab | 8.23ab | 8.25ab | 8.25ab | 9.08a | |||||
| Hb (g/L) | CON | 122.64ab | 113.86ab | 111.83ab | 109.79b | 111.94ab | 116.67ab | 127.43a | 1.066 | 0.586 | 0.024 | 0.293 |
| EW | 113.08 | 120.45 | 115.50 | 113.29 | 109.93 | 112.25 | 121.50 | |||||
| Items | Treatment | Days post-weaning (d) | SEM | P value | ||||||
| 0 | 1 | 2 | 3 | 7 | Weaning | Age | Weaning×Age | |||
| CORT (µg/mL) | CON | 116.43 | 114.62 | 115.92 | 125.68 | 115.82 | 1.297 | 0.225 | 0.383 | 0.232 |
| EW | 118.22 | 129.29* | 124.04 | 122.82 | 123.79 | |||||
| HPT (µg/mL) | CON | 51.02 | 50.75 | 52.02 | 49.15 | 44.94 | 0.826 | 0.391 | 0.109 | 0.960 |
| EW | 51.55 | 54.16* | 53.39 | 50.54 | 48.37 | |||||
| NE (µg/mL) | CON | 1468.23 | 1488.63 | 1508.55 | 1544.86 | 1456.08 | 16.074 | 0.378 | 0.049 | 0.320 |
| EW | 1417.75b | 1542.85ab | 1670.66a* | 1555.05ab | 1458.10b | |||||
| TNF-α (µg/mL) | CON | 97.46 | 94.74 | 94.24 | 95.93 | 90.75 | 0.999 | 0.454 | 0.202 | 0.382 |
| EW | 97.37ab | 103.94a* | 93.15b | 95.45ab | 94.93ab | |||||
| Items | Day 26 | Day 49 | P value | ||||||
| CON | EW | CON | EW | SEM | Weaning | Age | Weaning × Age | ||
| Duodenum | Villi height (μm) | 210.68 | 238.47 | 452.96 | 437.60 | 8.376 | 0.714 | <0.001 | 0.211 |
| Villi width (μm) | 75.88 | 81.21 | 173.27 | 155.40 | 3.717 | 0.818 | <0.001 | 0.115 | |
| Crypt depth (μm) | 120.00 | 149.87* | 220.67 | 212.29 | 5.020 | 0.296 | <0.001 | 0.070 | |
| Muscle layer thickness (μm) | 97.52 | 116.00* | 112.89 | 136.13* | 4.752 | 4.752 | 0.075 | 0.804 | |
| Jejunum | Villi height (μm) | 430.20 | 412.50 | 447.90 | 413.95 | 11.172 | 0.269 | 0.675 | 0.722 |
| Villi width (μm) | 93.08 | 101.92 | 114.62 | 127.15 | 3.437 | 0.144 | 0.005 | 0.793 | |
| Crypt depth (μm) | 126.95 | 195.33* | 159.18 | 181.18 | 7.699 | 0.012 | 0.567 | 0.156 | |
| Muscle layer thickness (μm) | 99.58 | 87.90 | 93.93 | 86.20 | 5.041 | 0.248 | 0.909 | 0.664 | |
| Ileum | Villi height (μm) | 458.10* | 397.97 | 427.54* | 372.47 | 11.047 | 0.028 | 0.151 | 0.737 |
| Villi width (μm) | 103.74 | 108.57 | 114.86 | 114.07 | 4.240 | 0.814 | 0.339 | 0.744 | |
| Crypt depth (μm) | 123.16 | 189.65* | 173.68 | 192.76 | 7.160 | 0.008 | 0.077 | 0.114 | |
| Muscle layer thickness (μm) | 112.20 | 103.55 | 93.86 | 88.27 | 8.142 | 0.306 | 0.116 | 0.926 | |
| Colon | Villi height (μm) | 429.80 | 464.20 | 480.06 | 478.86 | 11.77 | 0.491 | 0.188 | 0.461 |
| Villi width (μm) | 43.80 | 48.94 | 50.86 | 48.38 | 1.654 | 0.694 | 0.342 | 0.267 | |
| Crypt depth (μm) | 63.46 | 71.48 | 74.94 | 77.44 | 7.067 | 0.715 | 0.546 | 0.848 | |
| Muscle layer thickness (μm) | 174.82 | 187.52 | 202.28 | 189.24 | 10.187 | 0.993 | 0.484 | 0.537 | |
| Items | Day 26 | Day 49 | P value | |||||
| CON | EW | CON | EW | SEM | Weaning | Age | Weaning×Age | |
| SOD (U/mg) | 0.12 | 0.10 | 0.11 | 0.11 | 0.006 | 0.382 | 0.961 | 0.732 |
| GSH-Px (U/mg) | 1.23 | 1.23 | 1.12 | 1.18 | 0.043 | 0.763 | 0364 | 0.716 |
| MDA (nmol/g) | 8.53 | 8.29 | 10.13 | 22.30* | 1.735 | 0.098 | 0.036 | 0.089 |
| IgA (mg/g) | 1.31* | 0.85 | 1.18* | 0.99 | 0.060 | 0.014 | 0.904 | 0.268 |
| Gene ID | Gene Symbol | Description | Group | log2FC | P value | Q value | |
| CON | EW | ||||||
| MSTRG.7028 | APOA4 | Apolipoprotein A4 | 6.67 | 37.14 | 2.45 | <0.001 | <0.001 |
| MSTRG.2698 | SLC10A2 | Solute carrier family 10 member 2 | 5.90 | 18.23 | 1.60 | <0.001 | <0.001 |
| MSTRG.12128 | HSPA1L | Heat shock 70 kda protein 1-like | 0.69 | <0.01 | -9.98 | <0.001 | <0.001 |
| MSTRG.5151 | PRNP | Major prion protein | 0.17 | <0.01 | -8.91 | <0.001 | 0.010 |
| MSTRG.9443 | OXSR1 | Serine/threonine-protein kinase OSR1 | <0.01 | 4.77 | 11.29 | <0.001 | 0.015 |
| MSTRG.2150 | PDE9A | Phosphodiesterase 9A | 11.77 | 27.12 | 1.18 | <0.001 | 0.015 |
| MSTRG.2965 | NOS2 | Nitric oxide synthase 2 | 0.74 | 4.74 | 2.64 | <0.001 | 0.019 |
| MSTRG.19179 | CDC42SE2 | CDC42 small effector 2 | 8.52 | 20.11 | 1.21 | <0.001 | 0.019 |
| MSTRG.22642 | COX1 | Cytochrome c oxidase subunit 1 | 1195.86 | 6.55 | -7.53 | <0.001 | 0.020 |
| MSTRG.12142 | C6orf47 | Uncharacterized protein c6orf47 | 1.04 | <0.01 | -10.56 | <0.001 | 0.020 |
| MSTRG.2916 | ZNF830 | Zinc finger protein 830 | <0.01 | 0.64 | 8.68 | <0.001 | 0.032 |
| MSTRG.19825 | STARD4 | StAR related lipid transfer domain containing 4 | 0.81 | 2.71 | 1.71 | <0.001 | 0.032 |
| MSTRG.20048 | SLC34A2 | Solute carrier family 34 member 2 | 14.21 | 33.44 | 1.21 | <0.001 | 0.032 |
| MSTRG.12237 | TRIM15 | Tripartite motif containing 15 | 0.79 | 2.33 | 1.52 | <0.001 | 0.047 |
| MSTRG.22661 | ND6 | NADH-ubiquinone oxidoreductase chain 6 | <0.01 | 4.74 | 8.59 | <0.001 | 0.049 |
| MSTRG.16608 | SOAT2 | Sterol O-acyltransferase 2 | 0.38 | 1.59 | 2.03 | <0.001 | 0.052 |
| MSTRG.12735 | MS4A18 | Membrane spanning 4-domains A18 | 1.69 | 5.27 | 1.61 | <0.001 | 0.069 |
| MSTRG.9443 | AREG | Amphiregulin | 1.21 | 0.85 | -2.09 | 0.000 | 0.070 |
| MSTRG.14252 | ANKS4B | Ankyrin repeat and sterile alpha motif domain containing 4B | 1.04 | 3.51 | 1.72 | 0.001 | 0.154 |
| MSTRG.5689 | MLKL | Mixed lineage kinase domain like pseudokinase | 2.80 | 5.71 | 1.00 | 0.001 | 0.154 |
| Gene ID | Gene Symbol | Description | Group | log2FC | P value | Q value | |
| CON | EW | ||||||
| MSTRG.24683 | ZBTB33 | Zinc finger and BTB domain containing 33 | 3.31 | 1.7 | -2.08 | <0.001 | 0.922 |
| MSTRG.16036 | RAB11FIP1 | RAB11 family interacting protein 1 | 2.99 | 1.17 | -1.66 | <0.001 | 0.922 |
| MSTRG.21722 | AGGF1 | Angiogenic factor with G-patch and FHA domains 1 | 1.57 | 5.03 | 1.58 | 0.001 | 0.922 |
| MSTRG.4224 | GNA13 | G protein subunit alpha 13 | 50.13 | 31.05 | -4.00 | 0.009 | 0.922 |
| MSTRG.6324 | CMTR2 | CAP methyltransferase 2 | 3.73 | 2.67 | -1.53 | 0.010 | 0.922 |
| MSTRG.12043 | NYAP2 | Neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 | 1.1 | 3.04 | 1.31 | 0.014 | 0.922 |
| MSTRG.10771 | CCL19 | C-C motif chemokine ligand 19 | 5.33 | 10.22 | 1.05 | 0.017 | 0.922 |
| MSTRG.19058 | PRR15 | Proline rich 15 | 3.57 | 2.02 | -1.05 | 0.018 | 0.922 |
| MSTRG.16341 | ASS1 | Argininosuccinate synthase 1 | 4.96 | 13.22 | 1.32 | 0.026 | 0.922 |
| MSTRG.6232 | CES2 | Carboxylesterase 2 | 21.13 | 13.63 | -1.02 | 0.027 | 0.922 |
| MSTRG.18314 | CCND2 | Cyclin D2 | 6.25 | <0.001 | -1.99 | 0.029 | 0.922 |
| MSTRG.22347 | DUOXA2 | Dual oxidase maturation factor 2 | 1.19 | 9.26 | 1.77 | 0.033 | 0.922 |
| MSTRG.22348 | DUOX2 | Dual oxidase 2 | 1.7 | 12.07 | 1.63 | 0.038 | 0.922 |
| MSTRG.5089 | PGAM1 | Phosphoglycerate mutase 1 | 17.72 | 34.79 | 2.36 | 0.041 | 0.922 |
| MSTRG.3081 | CCL8 | Phosphoglycerate mutase 1 | 1.79 | 6.12 | 1.09 | 0.043 | 0.922 |
| MSTRG.21722 | CRAMP1 | CAMP-regulated antimicrobial peptide 1 | 2.77 | 2.05 | -1.02 | 0.044 | 0.922 |
| MSTRG.24683 | SOCS3 | Suppressor of cytokine signaling 3 | 2.32 | 7.51 | 1.06 | 0.046 | 0.922 |
| MSTRG.16036 | RBM15 | Suppressor of cytokine signaling 3 | 4.78 | 2.46 | -1.39 | 0.046 | 0.922 |
| MSTRG.4224 | ISG20 | Interferon-stimulated exonuclease gene 20 | 2.47 | 7.96 | 1.14 | 0.049 | 0.922 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





