Submitted:
31 March 2025
Posted:
01 April 2025
You are already at the latest version
Abstract
Keywords:
I. Introduction
2. Methods
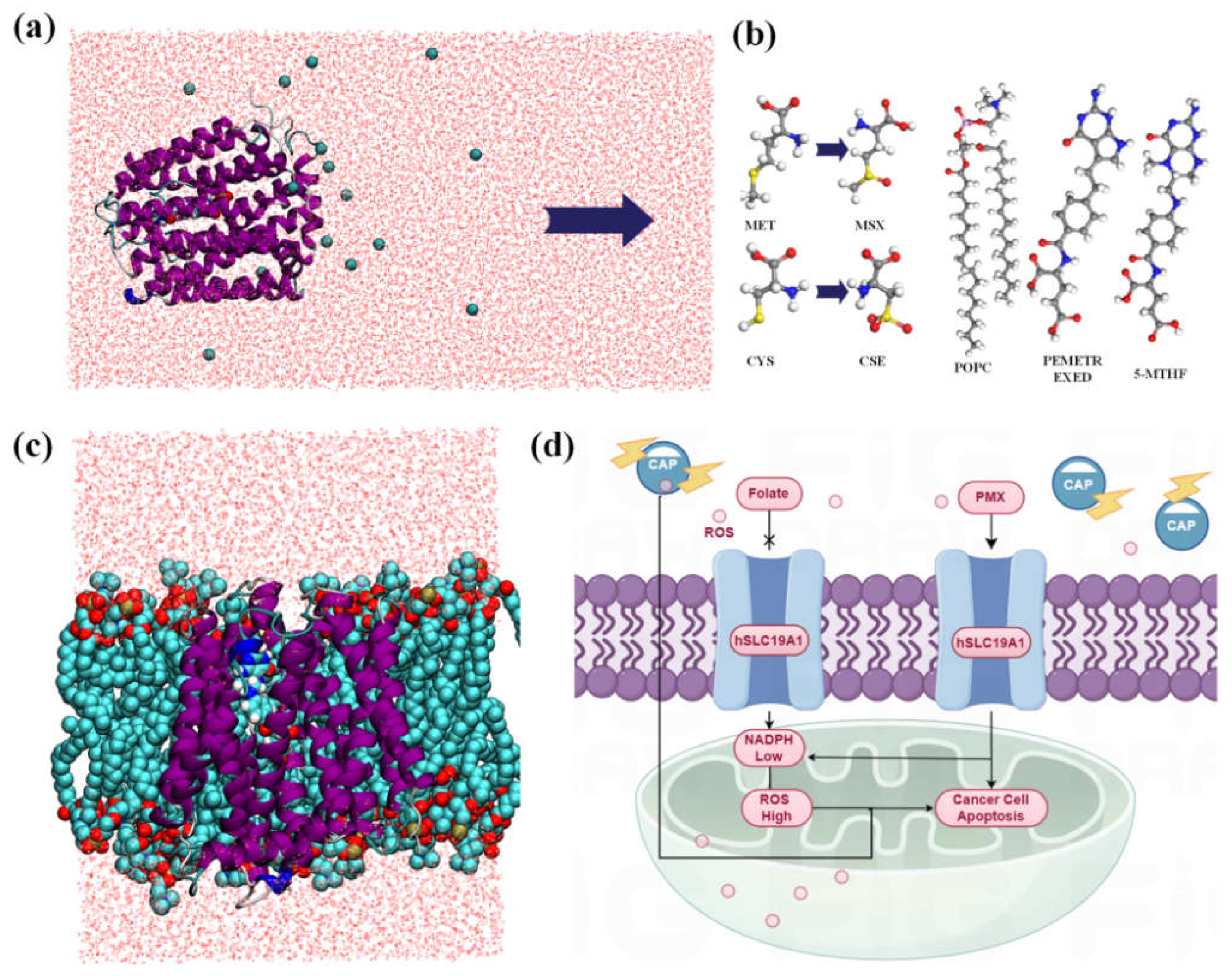
2.1. Protein Modeling
2.2. Simulation Details
2.3. Free Energy Calculations
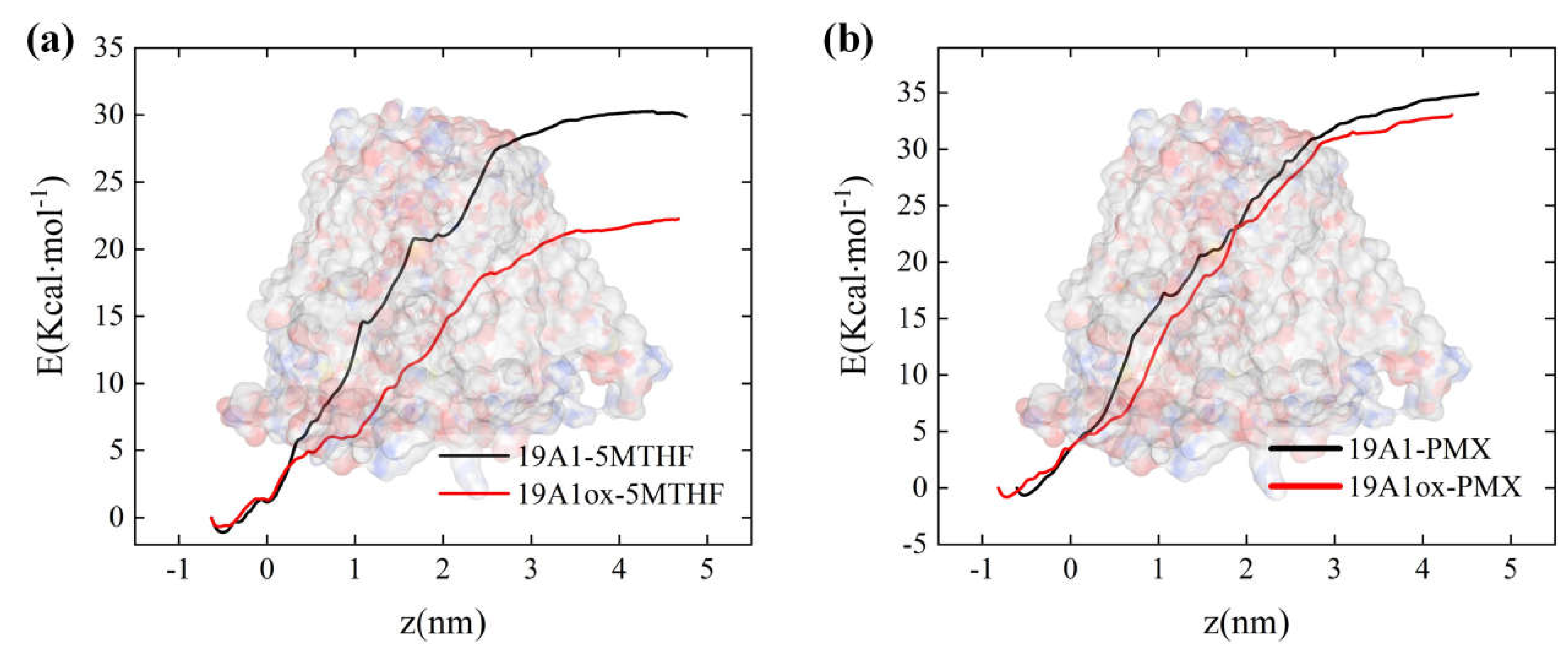
3. Results and Discussion
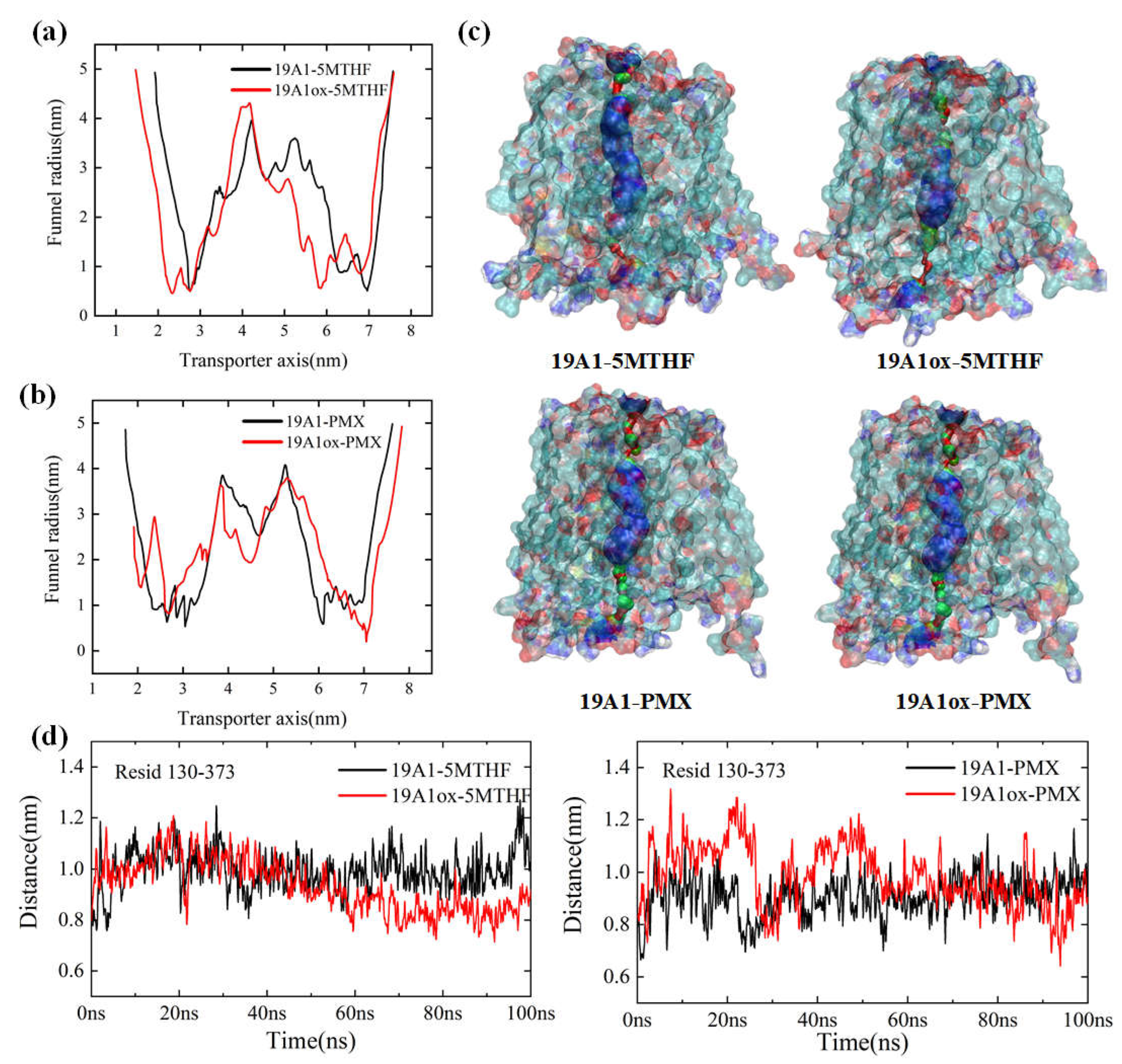
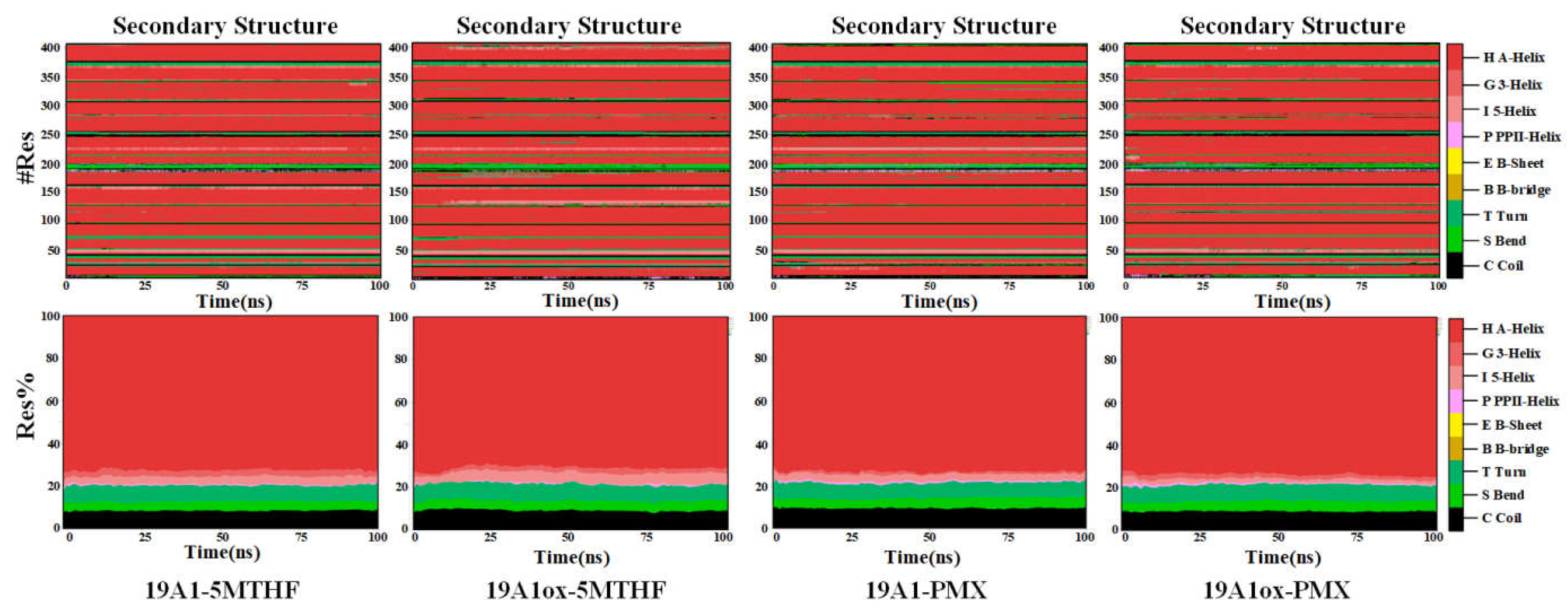
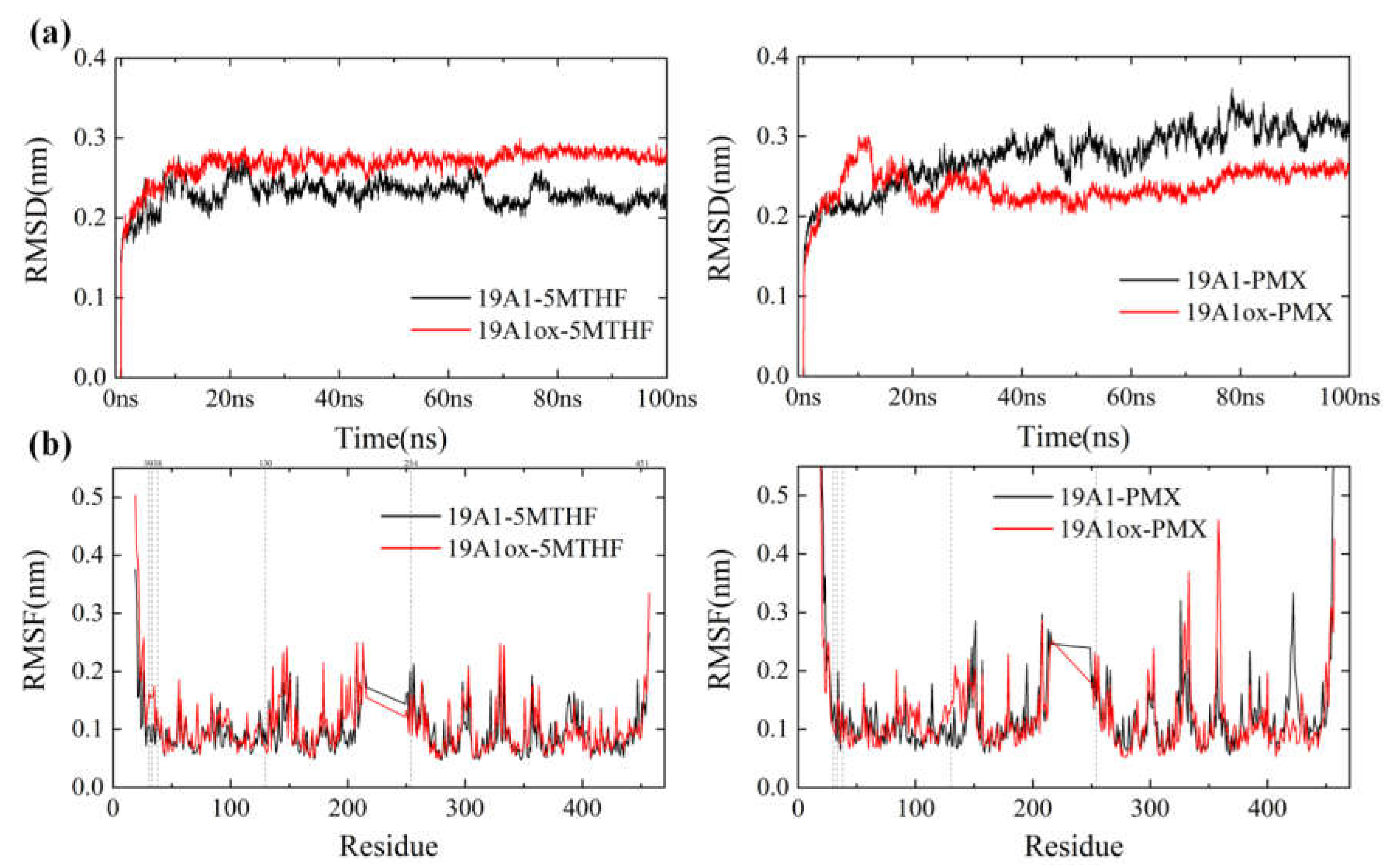
3.1. Changes in Protein Structure
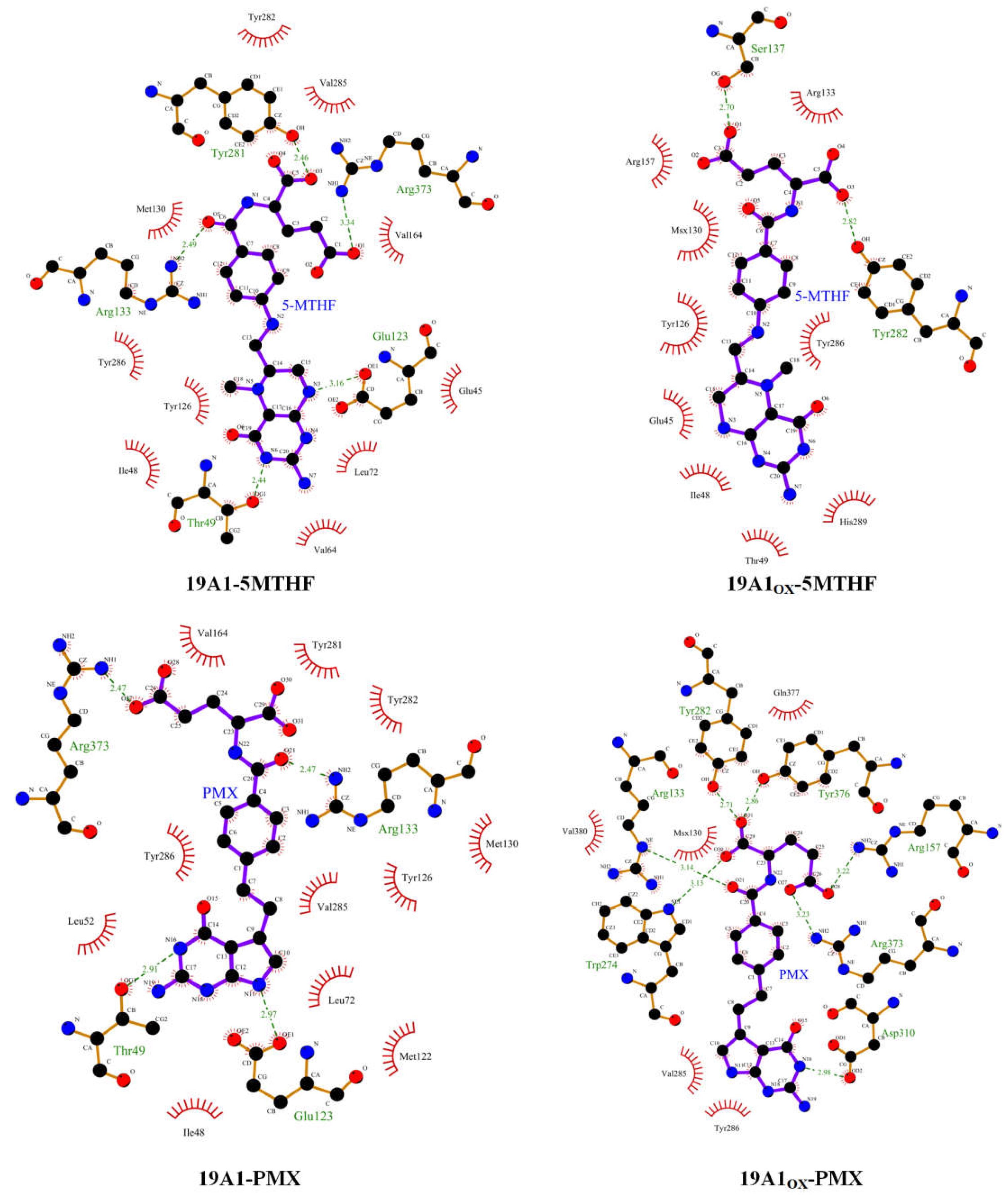
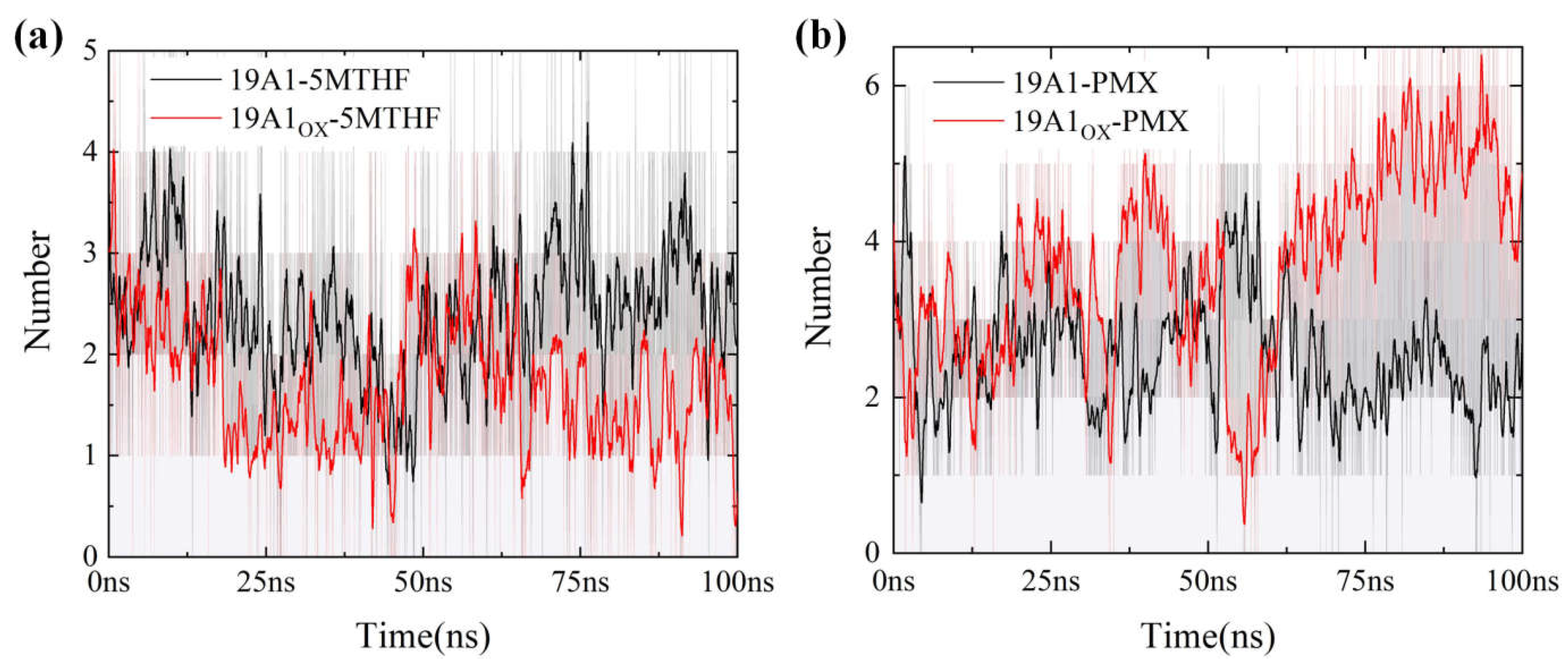
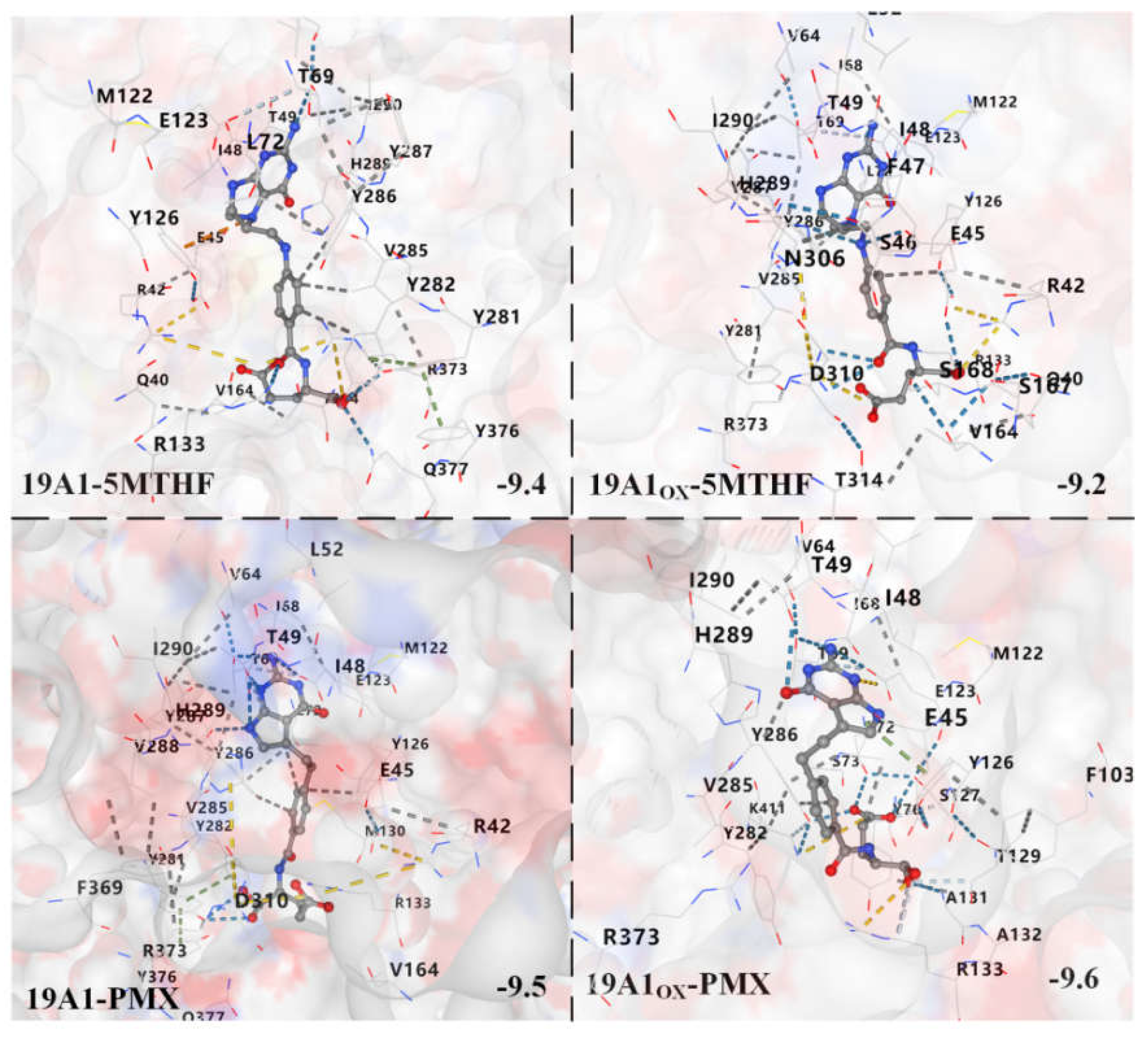
3.2. Analysis of Changes in Protein-Ligand Interactions
4. Conclusions
Declaration of competing interest
Data availability
Acknowledgments
References
- Ferlay J, Colombet M, Soerjomataram I, et al. Cancer statistics for the year 2020: An overview. International journal of cancer 2021, 149, 778–789. [Google Scholar]
- Epstein J B, Thariat J, Bensadoun R J, et al. Oral complications of cancer and cancer therapy: from cancer treatment to survivorship. CA: a cancer journal for clinicians 2012, 62, 400–422. [Google Scholar]
- Keidar M, Walk R, Shashurin A, et al. Cold plasma selectivity and the possibility of a paradigm shift in cancer therapy. British journal of cancer 2011, 105, 1295–1301. [Google Scholar]
- HARBOR C S, LI N Y. Cold spring harbor symposia on quantitative biology[C]//Cold Spring Harb Symp Quant Biol. sn 1979, 43, 1197–1208. [Google Scholar]
- Lee H J, Shon C H, Kim Y S, et al. Degradation of adhesion molecules of G361 melanoma cells by a non-thermal atmospheric pressure microplasma. New Journal of Physics 2009, 11, 115026. [Google Scholar]
- Utsumi F, Kajiyama H, Nakamura K, et al. Effect of indirect nonequilibrium atmospheric pressure plasma on anti-proliferative activity against chronic chemo-resistant ovarian cancer cells in vitro and in vivo. PloS one 2013, 8, e81576. [Google Scholar]
- Mohades S, Barekzi N, Laroussi M. Efficacy of low temperature plasma against SCaBER cancer cells. Plasma Processes and Polymers 2014, 11, 1150–1155. [Google Scholar]
- Perrotti V, Caponio V C A, Muzio L L, et al. Open questions in cold atmospheric plasma treatment in head and neck cancer: a systematic review. International Journal of Molecular Sciences 2022, 23, 10238. [Google Scholar]
- Lin A, Chernets N, Han J, et al. Non-equilibrium dielectric barrier discharge treatment of mesenchymal stem cells: charges and reactive oxygen species play the major role in cell death. Plasma Processes and Polymers 2015, 12, 1117–1127. [Google Scholar]
- Remon J, Besse B, Aix S P, et al. Osimertinib treatment based on plasma T790M monitoring in patients with EGFR-mutant non-small-cell lung cancer (NSCLC): EORTC Lung Cancer Group 1613 APPLE phase II randomized clinical trial. Annals of Oncology 2023, 34, 468–476. [Google Scholar] [CrossRef]
- Yun J H, Yang Y H, Han C H, et al. Non-thermal atmospheric pressure plasma induces selective cancer cell apoptosis by modulating redox homeostasis. Cell Communication and Signaling 2024, 22, 452. [Google Scholar] [CrossRef] [PubMed]
- Hayes J D, Dinkova-Kostova A T, Tew K D. Oxidative stress in cancer. Cancer cell 2020, 38, 167–197. [Google Scholar] [CrossRef] [PubMed]
- Chen L, Zhang Z, Hoshino A, et al. NADPH production by the oxidative pentose-phosphate pathway supports folate metabolism. Nature metabolism 2019, 1, 404–415. [Google Scholar]
- Ebisch I M W, Thomas C M G, Peters W H M, et al. The importance of folate, zinc and antioxidants in the pathogenesis and prevention of subfertility. Human reproduction update 2007, 13, 163–174. [Google Scholar]
- Ducker G S, Rabinowitz J D. One-carbon metabolism in health and disease. Cell metabolism 2017, 25, 27–42. [Google Scholar] [CrossRef]
- Hsu H C, Chang W M, Wu J Y, et al. Folate deficiency triggered apoptosis of synoviocytes: Role of overproduction of reactive oxygen species generated via NADPH oxidase/mitochondrial complex II and calcium perturbation. PLoS One 2016, 11, e0146440. [Google Scholar]
- Lee D, Xu I M J, Chiu D K C, et al. Folate cycle enzyme MTHFD1L confers metabolic advantages in hepatocellular carcinoma. The Journal of clinical investigation 2017, 127, 1856–1872. [Google Scholar]
- Ducker G S, Chen L, Morscher R J, et al. Reversal of cytosolic one-carbon flux compensates for loss of the mitochondrial folate pathway. Cell metabolism 2016, 23, 1140–1153. [Google Scholar] [CrossRef]
- Ye Y L, Chan Y T, Liu H C, et al. Depleted folate pool and dysfunctional mitochondria associated with defective mitochondrial folate proteins sensitize Chinese ovary cell mutants to tert-butylhydroperoxide-induced oxidative stress and apoptosis. The Journal of Nutritional Biochemistry 2010, 21, 793–800. [Google Scholar]
- Chern C L, Huang R F S, Chen Y H, et al. Folate deficiency-induced oxidative stress and apoptosis are mediated via homocysteine-dependent overproduction of hydrogen peroxide and enhanced activation of NF-κB in human Hep G2 cells. Biomedicine & pharmacotherapy 2001, 55, 434–442. [Google Scholar]
- Zhao S, Xiong Z, Mao X, et al. Atmospheric pressure room temperature plasma jets facilitate oxidative and nitrative stress and lead to endoplasmic reticulum stress dependent apoptosis in HepG2 cells. PloS one 2013, 8, e73665. [Google Scholar]
- Bauer G, Sersenová D, Graves D B, et al. Cold atmospheric plasma and plasma-activated medium trigger RONS-based tumor cell apoptosis. Scientific reports 2019, 9, 14210. [Google Scholar]
- Bekeschus S, Eisenmann S, Sagwal S K, et al. xCT (SLC7A11) expression confers intrinsic resistance to physical plasma treatment in tumor cells. Redox Biology 2020, 30, 101423. [Google Scholar]
- Moniruzzaman R, Rehman M U, Zhao Q L, et al. Roles of intracellular and extracellular ROS formation in apoptosis induced by cold atmospheric helium plasma and X-irradiation in the presence of sulfasalazine. Free Radical Biology and Medicine 2018, 129, 537–547. [Google Scholar]
- Matherly L H, Hou Z, Deng Y. Human reduced folate carrier: translation of basic biology to cancer etiology and therapy. Cancer and metastasis reviews 2007, 26, 111–128. [Google Scholar]
- Rezaei M, Ghasemitarei M, Razzokov J, et al. In silico study of the impact of oxidation on pyruvate transmission across the hVDAC1 protein channel. Archives of Biochemistry and Biophysics 2024, 751, 109835. [Google Scholar]
- Ghasemitarei M, Yusupov M, Razzokov J, et al. Effect of oxidative stress on cystine transportation by xC‾ antiporter. Archives of biochemistry and biophysics 2019, 674, 108114. [Google Scholar]
- Margreitter C, Petrov D, Zagrovic B. Vienna-PTM web server: a toolkit for MD simulations of protein post-translational modifications. Nucleic acids research 2013, 41, W422–W426. [Google Scholar]
- Margreitter C, Reif M M, Oostenbrink C. Update on phosphate and charged post-translationally modified amino acid parameters in the GROMOS force field. Journal of computational chemistry 2017, 38, 714–720. [Google Scholar]
- Petrov D, Margreitter C, Grandits M, et al. A systematic framework for molecular dynamics simulations of protein post-translational modifications. PLoS computational biology 2013, 9, e1003154. [Google Scholar]
- Xu G, Chance M R. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chemical reviews 2007, 107, 3514–3543. [Google Scholar] [CrossRef] [PubMed]
- Takai E, Kitamura T, Kuwabara J, et al. Chemical modification of amino acids by atmospheric-pressure cold plasma in aqueous solution. Journal of Physics D: Applied Physics 2014, 47, 285403. [Google Scholar] [CrossRef]
- Zhou R, Zhou R, Zhuang J, et al. Interaction of atmospheric-pressure air microplasmas with amino acids as fundamental processes in aqueous solution. PloS one 2016, 11, e0155584. [Google Scholar]
- Malde A K, Zuo L, Breeze M; et al. An automated force field topology builder (ATB) and repository: version 1.0. Journal of chemical theory and computation 2011, 7, 4026–4037. [Google Scholar]
- Stroet M, Caron B, Engler M S, et al. OFraMP: a fragment-based tool to facilitate the parametrization of large molecules. Journal of computer-aided molecular design 2023, 37, 357–371. [Google Scholar] [CrossRef]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. Journal of molecular graphics 1996, 14, 33–38. [Google Scholar] [CrossRef]
- https://jerkwin.github.
- Smart O S, Goodfellow J M, Wallace B A. The pore dimensions of gramicidin A. Biophysical journal 1993, 65, 2455–2460. [Google Scholar] [CrossRef]
- Wallace A C, Laskowski R A, Thornton J M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein engineering, design and selection 1995, 8, 127–134. [Google Scholar] [CrossRef]
- Laskowski R A, Swindells M B. LigPlot+: multiple ligand–protein interaction diagrams for drug discovery.
- Liu Y, Yang X, Gan J, et al. CB-Dock2: improved protein–ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic acids research 2022, 50, W159–W164. [Google Scholar] [CrossRef]
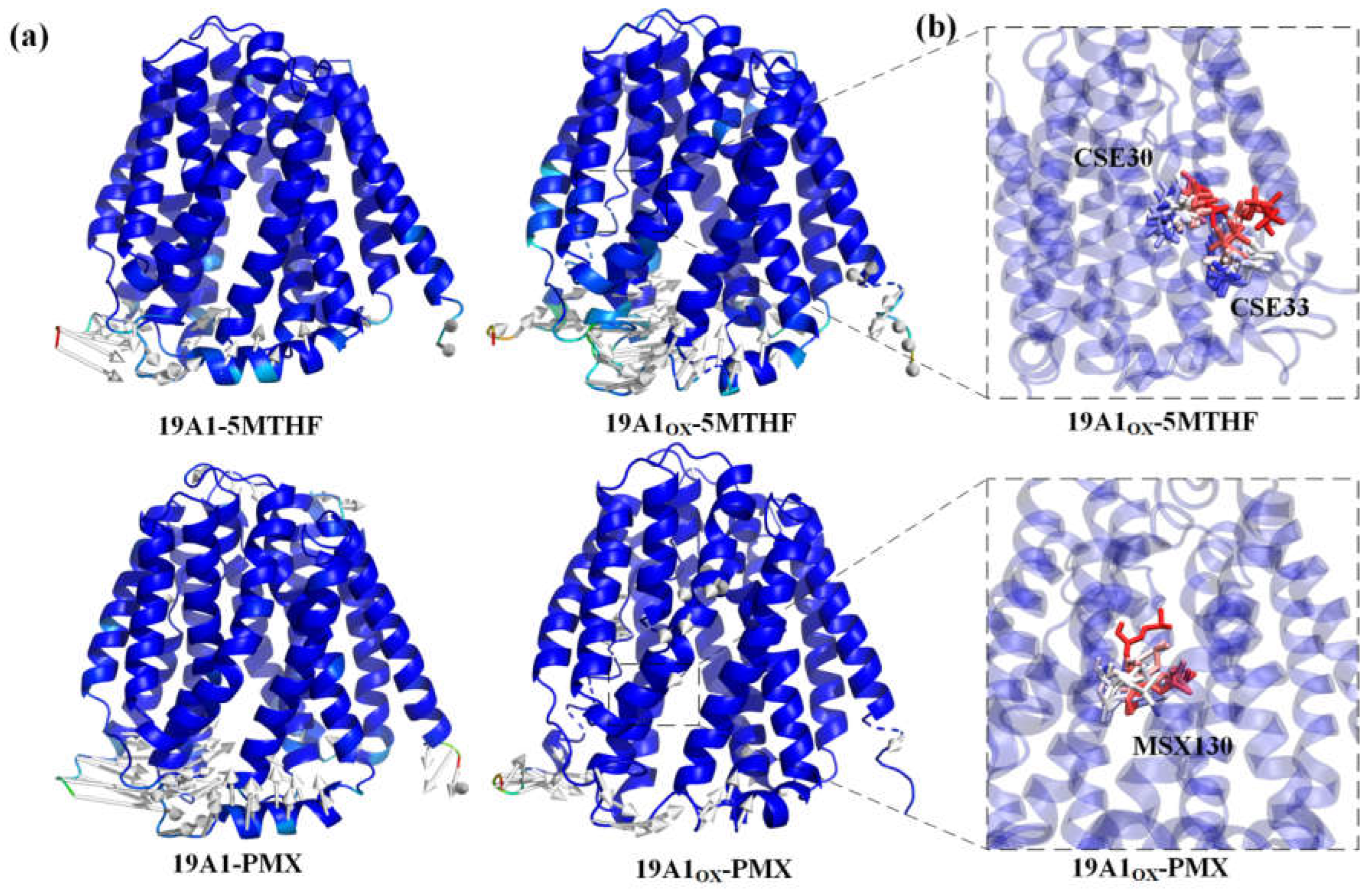
| Residue type |
Met | Met | Met | Met | Met | Cys | Cys | Cys | Cys |
|---|---|---|---|---|---|---|---|---|---|
| Resid Number |
38 | 119 | 122 | 130 | 254 | 30 | 33 | 365 | 396 |
| SASA | 0.715 | 0.015 | 0.073 | 0.350 | 0.696 | 0.813 | 0.239 | 0.009 | 0.050 |
| 19A1-5MTHF | 19A1ox-5MTHF | 19A1-PMX | 19A1ox-PMX | |
|---|---|---|---|---|
| Protein | hSLC19A1 | hSLC19A1OX | hSLC19A1 | hSLC19A1OX |
| Ligand | 5-MTHF | 5-MTHF | Pemetrexed | Pemetrexed |
| Hydrogen bonding rate | Average number of hydrogen bonds |
||||||
|---|---|---|---|---|---|---|---|
| OX1 | Tyr281 | Gln40 | Tyr282 | Arg133 | Tyr286 | Arg373 | 2.4197 |
| 98.95% | 53.47% | 32.93% | 27.24% | 11.94% | 10.29% | ||
| OX2 | Tyr282 | Tyr281 | Ser137 | Gln377 | Arg133 | Arg157 | 1.7067 |
| 70.36% | 36.28% | 23.54% | 15.74% | 7.05% | 5.95%- | ||
| OX3 | Tyr281 | Lys411 | Thr404 | Glu123 | Tyr282 | Glu45 | 2.5364 |
| 82.21% | 68.67% | 32.08% | 27.49% | 11.84% | 8.30% | ||
| OX4 | Tyr282 | Tyr376 | Tyr281 | Gln377 | Arg373 | Glu123 | 3.6851 |
| 79.71% | 78.11% | 71.46% | 52.62% | 30.13% | 21.59% | ||
| 19A1-5MTHF | 19A1ox-5MTHF | 19A1-PMX | 19A1ox-PMX | |
|---|---|---|---|---|
| MM | -532.891 | -570.111 | -591.315 | -573.037 |
| PB | 416.934 | 512.568 | 485.418 | 460.364 |
| SA | -32.761 | -34.905 | -28.211 | -27.893 |
| COU | -331.411 | -365.373 | -449.946 | -423.8 |
| VDW | -201.480 | -204.738 | -141.369 | -149.237 |
| Tds | 25.101 | 21.878 | 16.025 | 19.384 |
| dG(kcal/mol) | -28.023 | -16.866 | -28.223 | -28.963 |
| 19A1-5MTHF | 19A1ox-5MTHF | 19A1-PMX | 19A1ox-PMX | |
|---|---|---|---|---|
| Cys30 | 0.095 | 0.857 | 0.157 | 2.228 |
| Cys33 | 0.361 | 0.995 | 0.47 | 9.619 |
| Met38 | 0.024 | 0.041 | -0.184 | -0.051 |
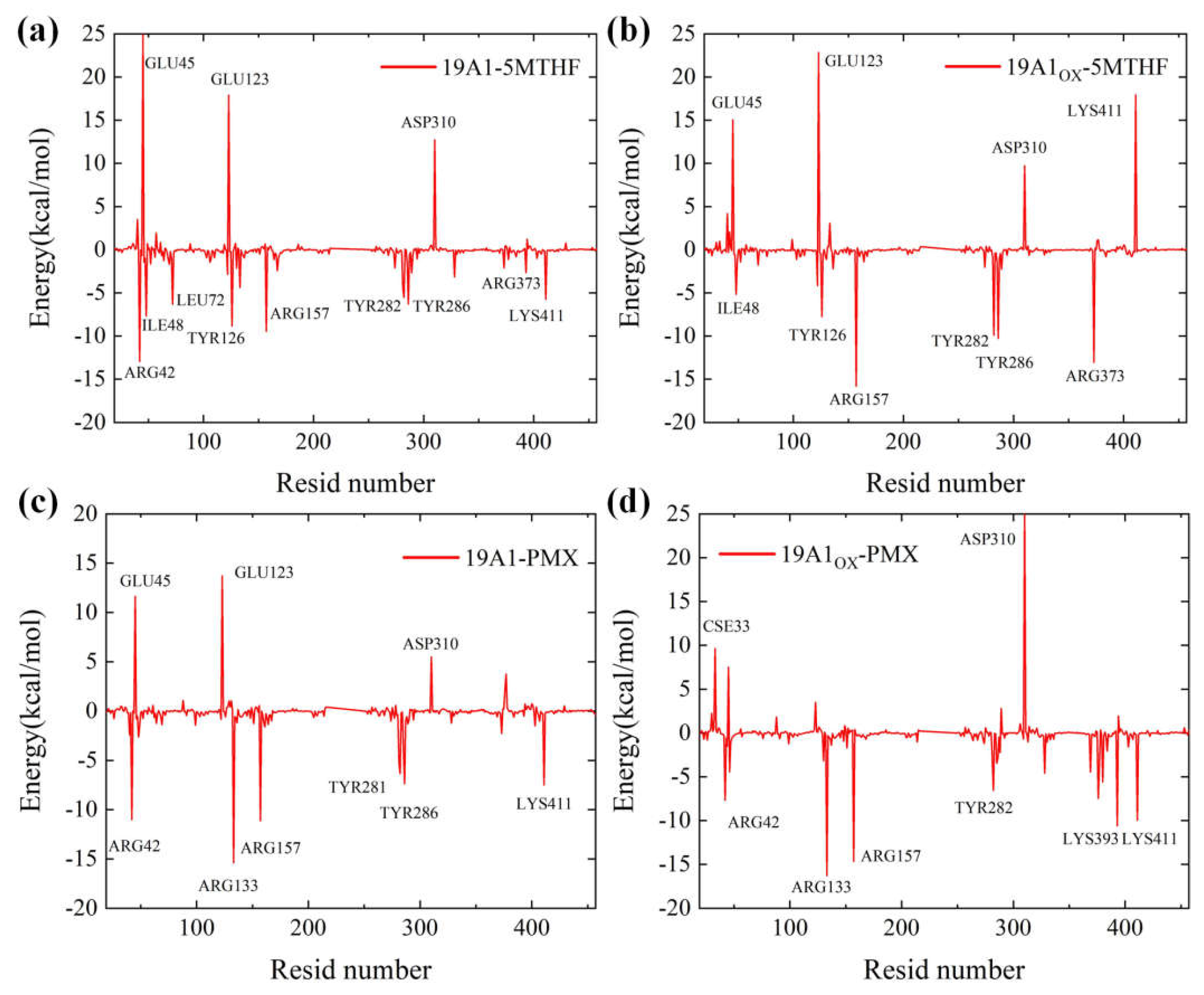
| Arg42 | -12.945 | 2.088 | -11.012 | -7.685 |
| Glu45 | 25.177 | 15.048 | 11.624 | 7.523 |
| Tyr126 | -8.849 | -7.746 | -0.602 | 0.688 |
| Met130 | -2.145 | -1.131 | 0.397 | -3.177 |
| Arg133 | -4.388 | 3.084 | -15.397 | -16.281 |
| Arg157 | -9.451 | -15.801 | -11.123 | -14.658 |
| Met254 | 0.014 | 0.023 | 0.070 | -0.078 |
| Tyr281 | -4.430 | -0.925 | -5.203 | -2.791 |
| Tyr282 | -5.548 | -9.930 | -6.362 | -6.548 |
| Asp310 | 12.721 | 9.721 | 5.493 | 25.672 |
| Arg373 | -2.116 | -13.064 | -2.269 | -0.414 |
| Lys411 | -5.756 | 17.921 | -7.507 | -9.973 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





