Submitted:
31 March 2025
Posted:
31 March 2025
You are already at the latest version
Abstract
Keywords:
Introduction
Materials and Methods
Curcumin
Cell Viability Assay
Poly-Caspase Assay
SA-Beta Galactosidase Assay
RNA Isolation, Library Preparation
RNA-Seq Analysis
Results
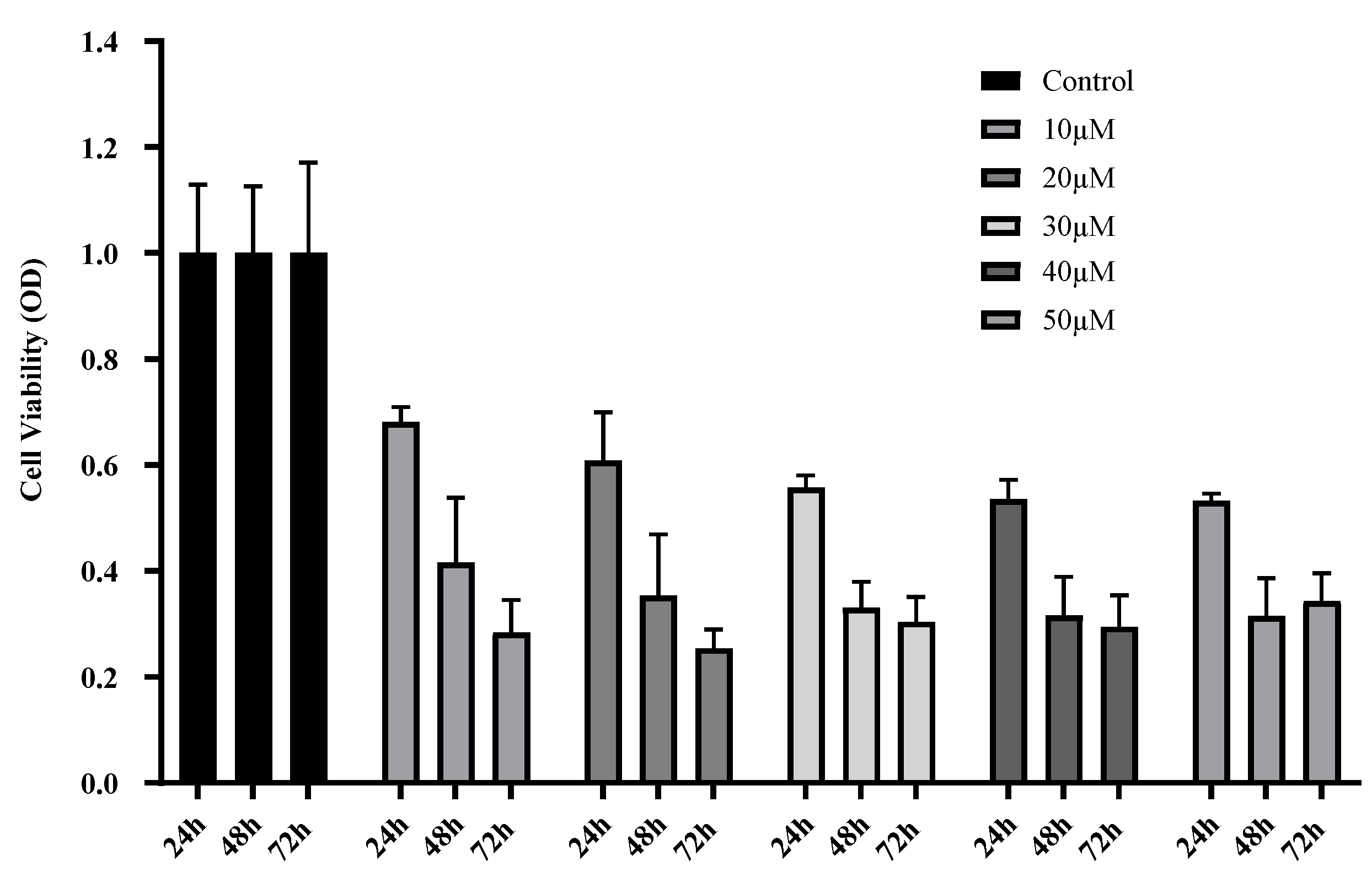
Curcumin Treatment Decreases U87 MG Cell Viability and Proliferation
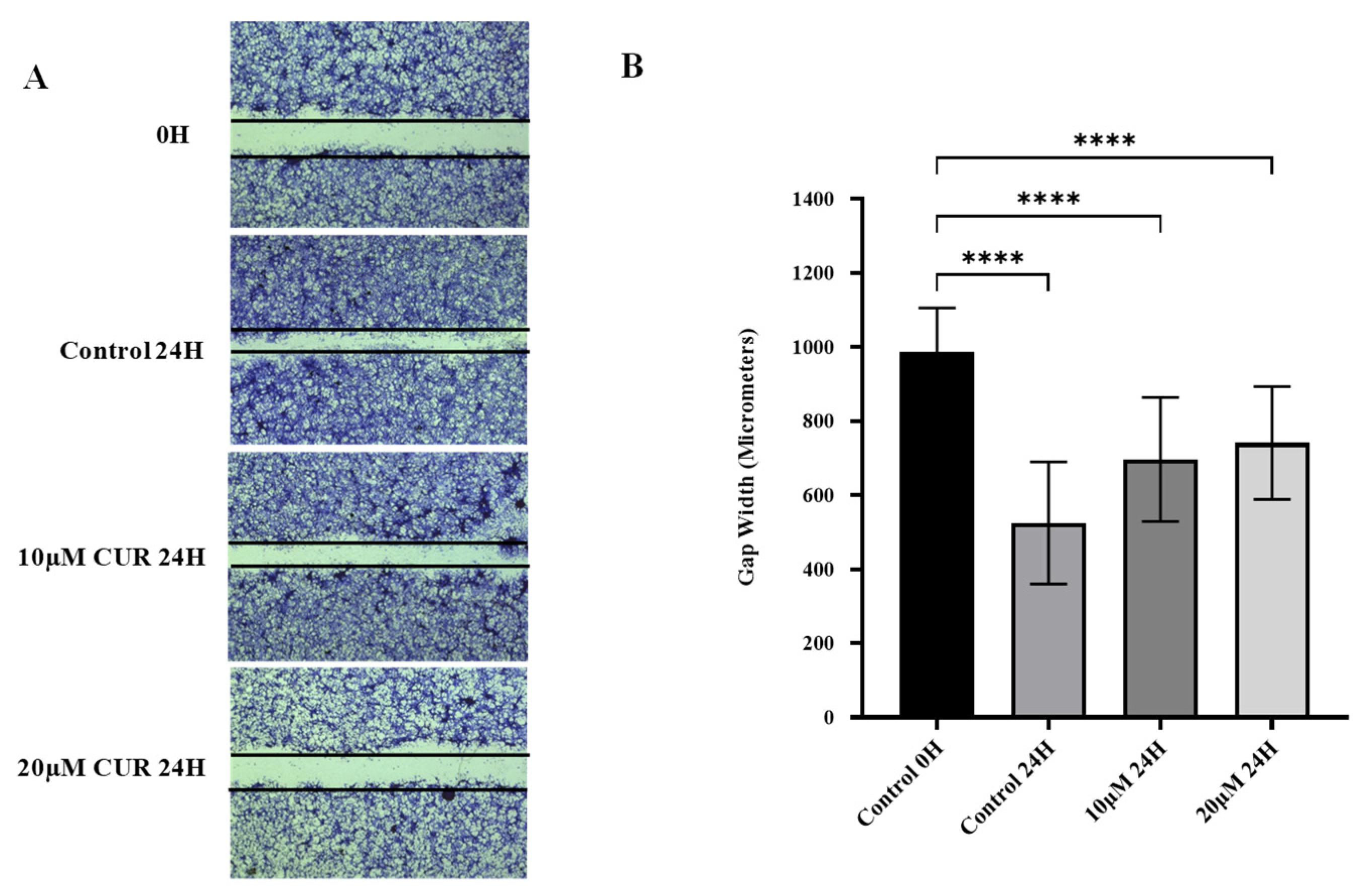
Curcumin Reduced Migration of U87 MG Cells
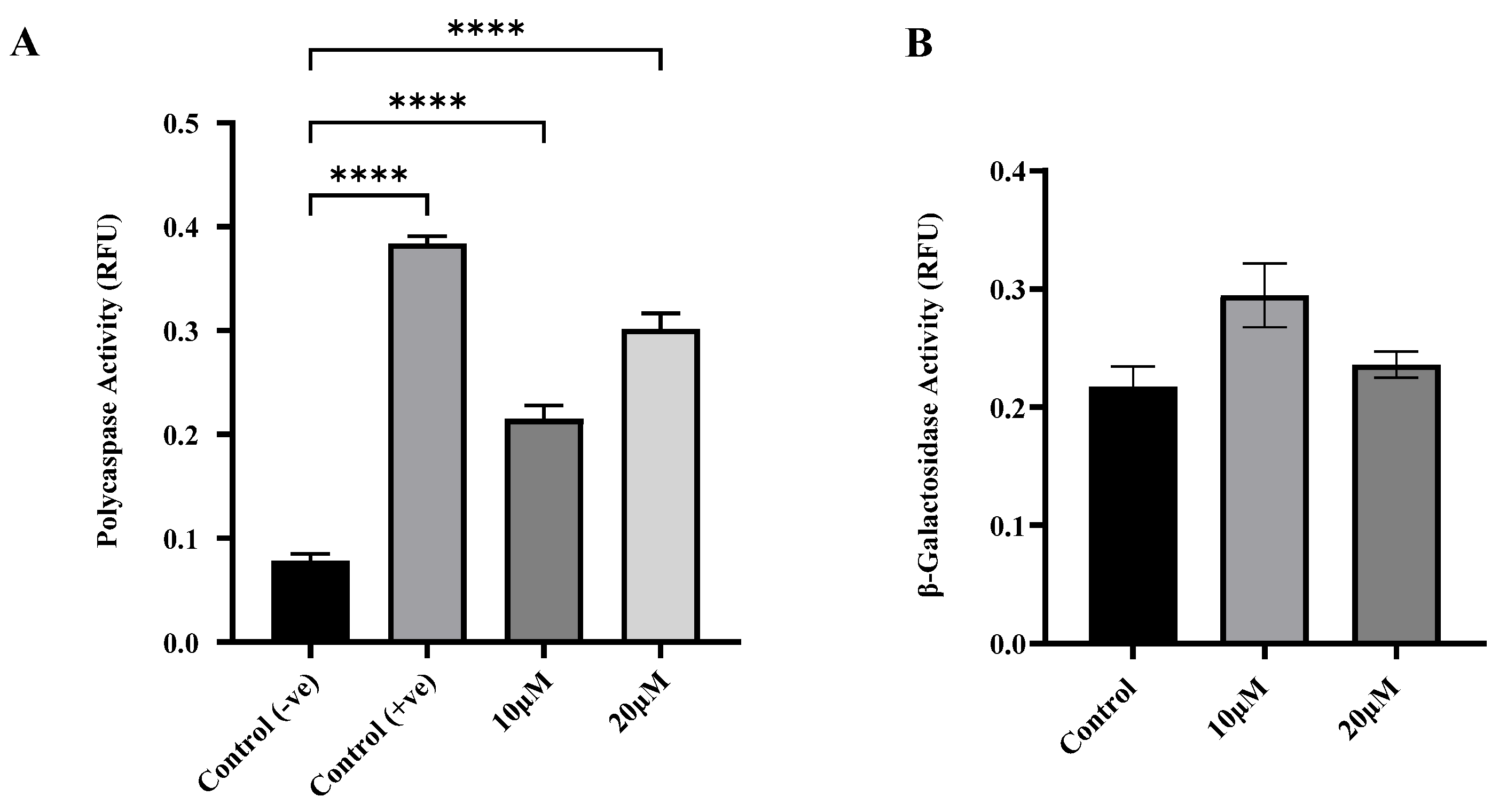
Curcumin induced apoptosis but not senescence in U87 MG cells
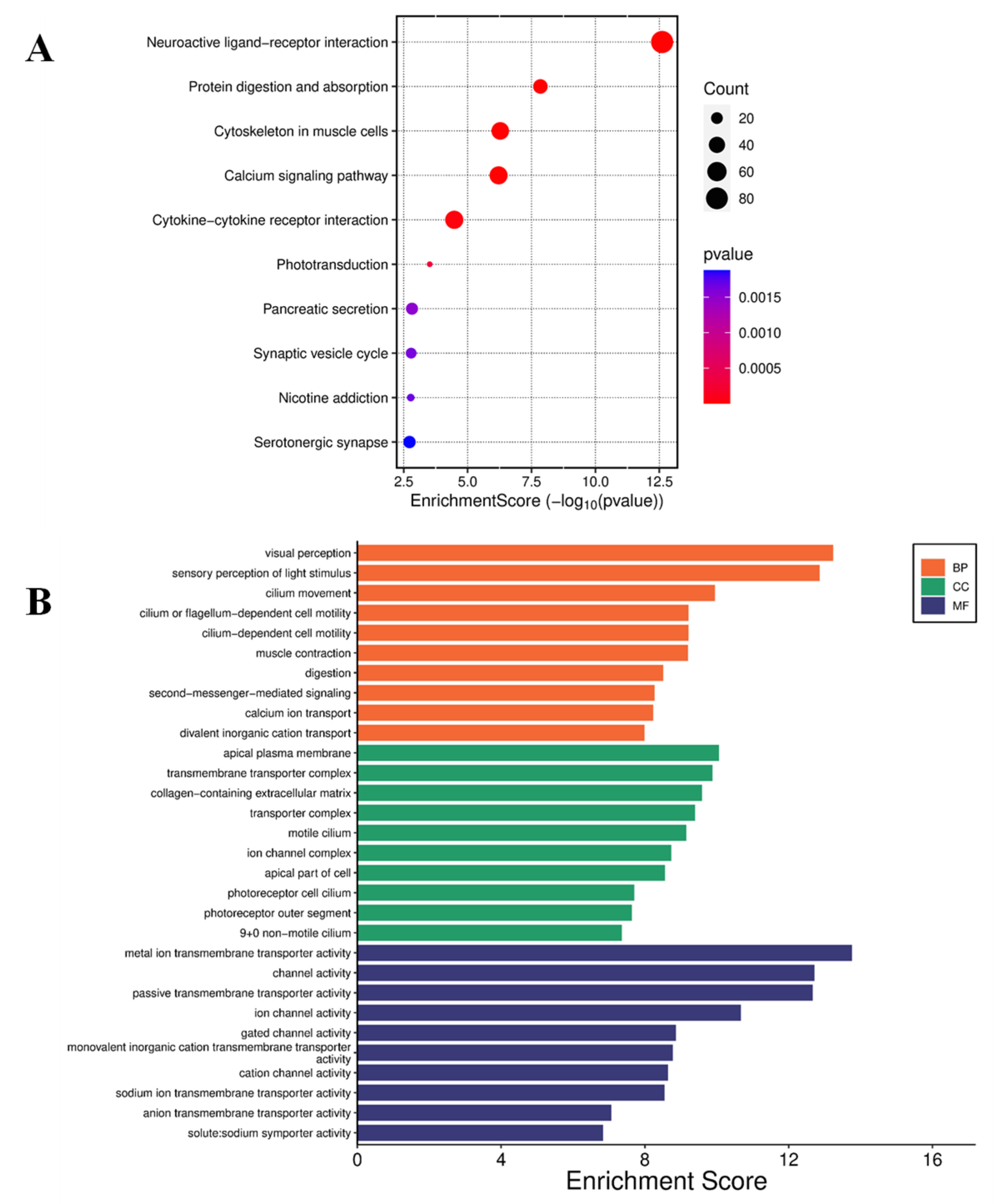
Curcumin Induced Differential Gene Expression in U87 MG cells
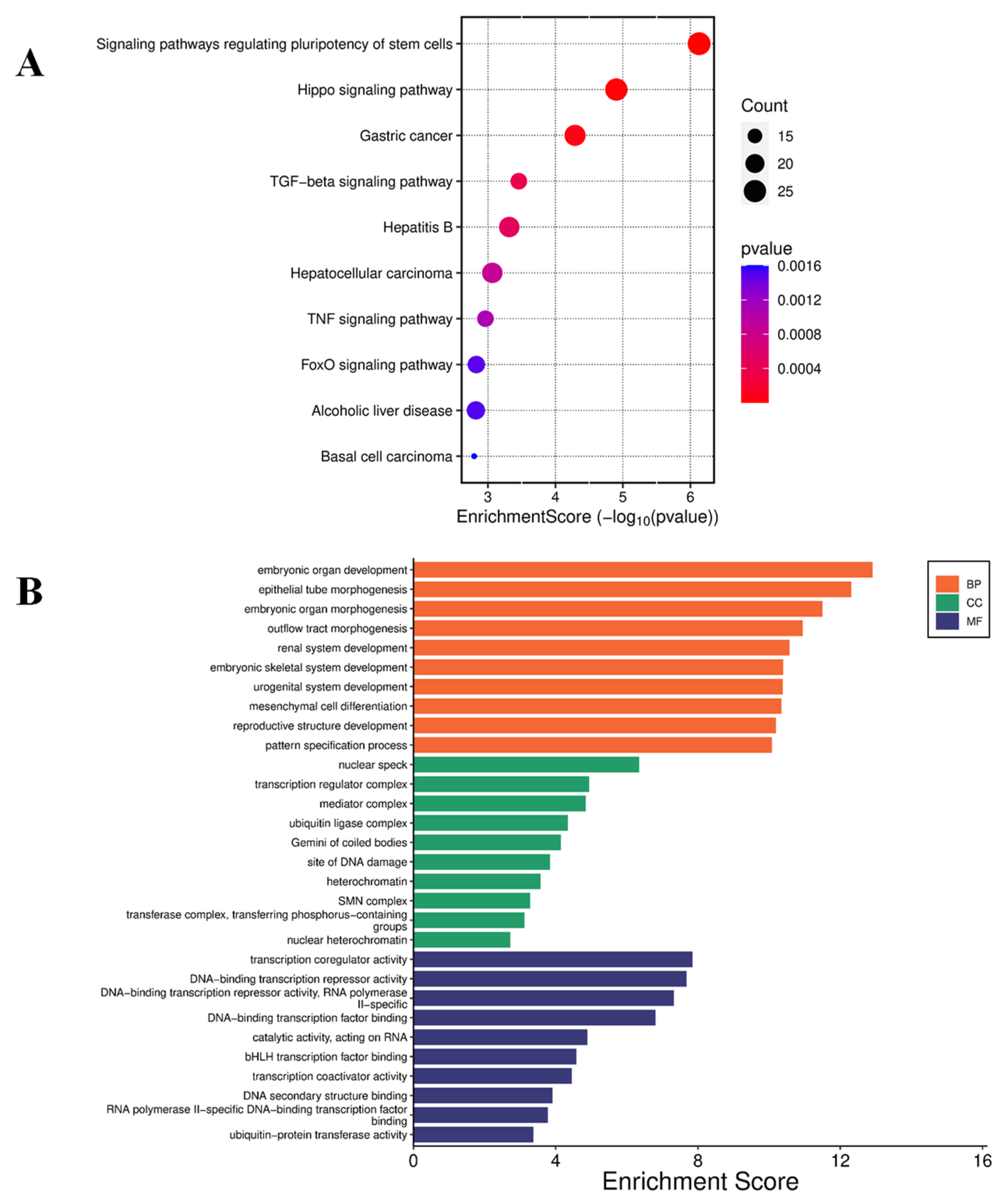
Discussion
Curcumin Suppresses Viability and Migration of U87 MG Cells
Curcumin Triggers Apoptosis in U87 MG Glioblastoma Cells_
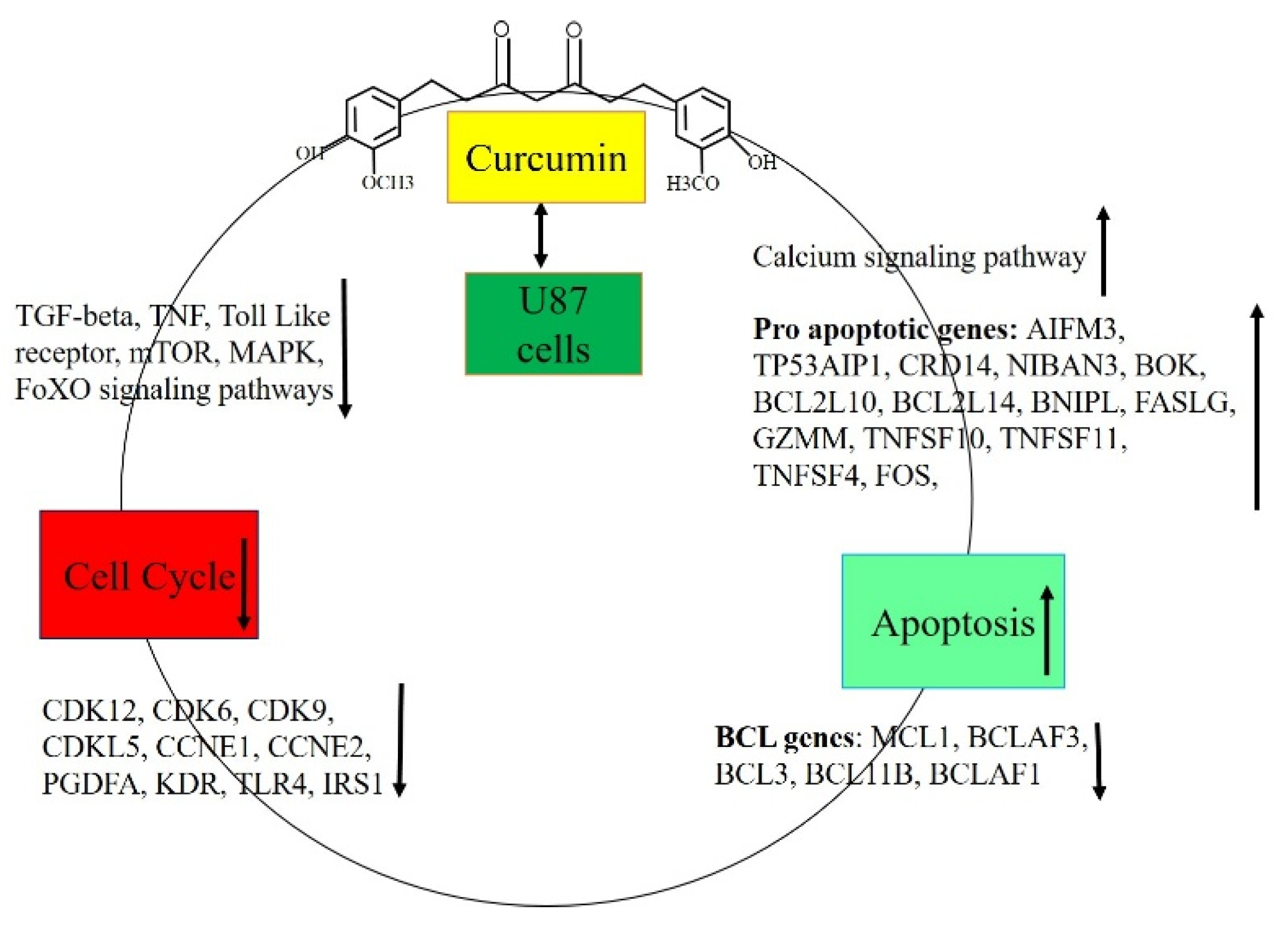
Curcumin treatment altered gene expression in U87 MG cells
Conclusion
Author Contributions
Acknowledgments
Conflicts of Interest
Data availability
References
- Agosti, E.; Zeppieri, M.; De Maria, L.; Tedeschi, C.; Fontanella, M.M.; Panciani, P.P.; Ius, T. Glioblastoma immunotherapy: A systematic review of the present strategies and prospects for advancements. International Journal of Molecular Sciences 2023, 24, 15037. [Google Scholar] [CrossRef]
- Ren, X.; Qin, L. Glioblastoma Management in the Post-COVID-19 Era: Challenges, Strategies, and Adaptations. 2025. [Google Scholar]
- Jackson, G.A.; Adamson, D.C. Similarities in Mechanisms of Ovarian Cancer Metastasis and Brain Glioblastoma Multiforme Invasion Suggest Common Therapeutic Targets. Cells 2025, 14, 171. [Google Scholar] [CrossRef] [PubMed]
- Aleksakhina, S.N.; Kashyap, A.; Imyanitov, E.N. Mechanisms of acquired tumor drug resistance. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer 2019, 1872, 188310. [Google Scholar] [CrossRef] [PubMed]
- Srirangan, P.; Sabina, E.P. Protective effects of herbal compounds against cyclophosphamide-induced organ toxicity: a pathway-centered approach. Drug and Chemical Toxicology 2025, 1–43. [Google Scholar] [CrossRef]
- Reddy, C.S.; Natarajan, P.; Nimmakayala, P.; Hankins, G.R.; Reddy, U.K. From Fruit Waste to Medical Insight: The Comprehensive Role of Watermelon Rind Extract on Renal Adenocarcinoma Cellular and Transcriptomic Dynamics. International Journal of Molecular Sciences 2023, 24, 15615. [Google Scholar] [CrossRef] [PubMed]
- Chinreddy, S.R.; Mashozhera, N.T.; Rashrash, B.; Flores-Iga, G.; Nimmakayala, P.; Hankins, G.R.; Harris, R.T.; Reddy, U.K. Unraveling TRPV1’s Role in Cancer: Expression, Modulation, and Therapeutic Opportunities with Capsaicin. Molecules 2024, 29, 4729. [Google Scholar] [CrossRef]
- Datta, S.; Saha, P.; Dey, S.; Sinha, D. Natural Products as Chemosensitizers for Adjunct Therapy in Cancer Management. Pharmacotherapeutic Botanicals for Cancer Chemoprevention 2020, 67–119. [Google Scholar]
- Islam, M.R.; Rauf, A.; Akash, S.; Trisha, S.I.; Nasim, A.H.; Akter, M.; Dhar, P.S.; Ogaly, H.A.; Hemeg, H.A.; Wilairatana, P. Targeted therapies of curcumin focus on its therapeutic benefits in cancers and human health: Molecular signaling pathway-based approaches and future perspectives. Biomedicine & Pharmacotherapy 2024, 170, 116034. [Google Scholar]
- Banerjee, S.; Singh, S.K.; Chowdhury, I.; Lillard Jr, J.W.; Singh, R. Combinatorial effect of curcumin with docetaxel modulates apoptotic and cell survival molecules in prostate cancer. Frontiers in bioscience (Elite edition) 2017, 9, 235. [Google Scholar]
- Tiryaki, S.; Macit, M.; Zemheri, I.E.; Süt, P.A.; Duman, G.; Telci, D. Anticancer Activity of Soy Lecithin-Based Curcumin in Prostate Cancer. Journal of Applied Polymer Science 2025, e56816. [Google Scholar] [CrossRef]
- Du, B.; Jiang, L.; Xia, Q.; Zhong, L. Synergistic inhibitory effects of curcumin and 5-fluorouracil on the growth of the human colon cancer cell line HT-29. Chemotherapy 2005, 52, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Cespedes-Acuña, C.L.; Yang, Z.; Abu Bakar, M.Z.; Chan, K.W.; Deng, X. Plant foods and their bioactives as dietary enhancers for colon cancer treatment with 5-fluorouracil. Food Reviews International 2025, 1–50. [Google Scholar] [CrossRef]
- Casarcia, N.; Rogers, P.; Guld, E.; Iyer, S.; Li, Y.; Burcher, J.T.; Deliberto, L.K.; Banerjee, S.; Bishayee, A. Phytochemicals for the prevention and treatment of pancreatic cancer: Current progress and future prospects. British Journal of Pharmacology 2023. [Google Scholar] [CrossRef]
- Allegra, A.; Mirabile, G.; Ettari, R.; Pioggia, G.; Gangemi, S. The impact of curcumin on immune response: an immunomodulatory strategy to treat sepsis. International journal of molecular sciences 2022, 23, 14710. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, M.K.; Rane, G.; Kanchi, M.M.; Arfuso, F.; Chinnathambi, A.; Zayed, M.; Alharbi, S.A.; Tan, B.K.; Kumar, A.P.; Sethi, G. The multifaceted role of curcumin in cancer prevention and treatment. Molecules 2015, 20, 2728–2769. [Google Scholar] [CrossRef] [PubMed]
- Dhandapani, K.M.; Mahesh, V.B.; Brann, D.W. Curcumin suppresses growth and chemoresistance of human glioblastoma cells via AP-1 and NFκB transcription factors. Journal of neurochemistry 2007, 102, 522–538. [Google Scholar] [CrossRef]
- Lin, S.-S.; Lai, K.-C.; Hsu, S.-C.; Yang, J.-S.; Kuo, C.-L.; Lin, J.-P.; Ma, Y.-S.; Wu, C.-C.; Chung, J.-G. Curcumin inhibits the migration and invasion of human A549 lung cancer cells through the inhibition of matrix metalloproteinase-2 and-9 and Vascular Endothelial Growth Factor (VEGF). Cancer letters 2009, 285, 127–133. [Google Scholar] [CrossRef]
- Xu, M.X.; Zhao, L.; Deng, C.; Yang, L.; Wang, Y.; Guo, T.; Li, L.; Lin, J.; Zhang, L. Curcumin suppresses proliferation and induces apoptosis of human hepatocellular carcinoma cells via the wnt signaling pathway. International journal of oncology 2013, 43, 1951–1959. [Google Scholar] [CrossRef]
- Wang, N.; Feng, T.; Liu, X.; Liu, Q. Curcumin inhibits migration and invasion of non-small cell lung cancer cells through up-regulation of miR-206 and suppression of PI3K/AKT/mTOR signaling pathway. Acta pharmaceutica 2020, 70, 399–409. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, K.; Liu, J.; Yang, J.; Tian, Y.; Yang, C.; Li, Y.; Shao, M.; Su, W.; Song, N. Curcumin regulates cancer progression: Focus on ncRNAs and molecular signaling pathways. Frontiers in Oncology 2021, 11, 660712. [Google Scholar] [CrossRef] [PubMed]
- Khazei, K.; Jamali, M.; Sarhadi, S.; Dadashpour, M.; Shokrollahzade, S.; Zarghami, N. Transcriptome profiling of curcumin-treated T47D human breast cancer cells by a system-based approach. Gene Reports 2022, 27, 101556. [Google Scholar] [CrossRef]
- Wang, R.; Yu, H.; Chen, P.; Yuan, T.; Zhang, J. Integrated transcriptome and molecular docking to identify the hub superimposed attenuation targets of curcumin in breast cancer cells. International Journal of Molecular Sciences 2023, 24, 12479. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Liang, C.; Yang, H.; Li, X.; Deng, X.; Liang, X.; Li, L.; Huang, Z.; Lu, D.; Ma, Y. Curcumin induces apoptosis and inhibits the growth of adrenocortical carcinoma: Identification of potential candidate genes and pathways by transcriptome analysis. Oncology Letters 2021, 21, 476. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhang, J.; Zhou, Y.; Gao, Q.; Wang, R.; Fu, Y.; Zheng, L.; Yu, H. Transcriptome investigation and in vitro verification of curcumin-induced HO-1 as a feature of ferroptosis in breast cancer cells. Oxidative Medicine and Cellular Longevity 2020, 2020, 3469840. [Google Scholar] [CrossRef]
- Mo, F.; Xiao, Y.; Zeng, H.; Fan, D.; Song, J.; Liu, X.; Luo, M.; Ma, X. Curcumin-induced global profiling of transcriptomes in small cell lung cancer cells. Frontiers in Cell and Developmental Biology 2021, 8, 588299. [Google Scholar] [CrossRef]
- Bolger, A.; Giorgi, F. Trimmomatic: A flexible read trimming tool for Illumina NGS data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.; Anders, S.; Huber, W. Differential analysis of count data–the DESeq2 package. Genome Biol 2014, 15, 10–1186. [Google Scholar]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic acids research 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Salam, R.; Saliou, A.; Bielle, F.; Bertrand, M.; Antoniewski, C.; Carpentier, C.; Alentorn, A.; Capelle, L.; Sanson, M.; Huillard, E. Cellular senescence in malignant cells promotes tumor progression in mouse and patient Glioblastoma. Nature communications 2023, 14, 441. [Google Scholar] [CrossRef]
- Giordano, A.; Tommonaro, G. Curcumin and cancer. Nutrients 2019, 11, 2376. [Google Scholar] [CrossRef]
- Wong, S.C.; Kamarudin, M.N.A.; Naidu, R. Anticancer mechanism of curcumin on human glioblastoma. Nutrients 2021, 13, 950. [Google Scholar] [CrossRef]
- Zoi, V.; Kyritsis, A.P.; Galani, V.; Lazari, D.; Sioka, C.; Voulgaris, S.; Alexiou, G.A. The role of curcumin in cancer: a focus on the PI3K/Akt pathway. Cancers 2024, 16, 1554. [Google Scholar] [CrossRef]
- Hesari, A.; Rezaei, M.; Rezaei, M.; Dashtiahangar, M.; Fathi, M.; Rad, J.G.; Momeni, F.; Avan, A.; Ghasemi, F. Effect of curcumin on glioblastoma cells. Journal of cellular physiology 2019, 234, 10281–10288. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-L.; Liu, Y.-Y.; Ma, Y.-G.; Xue, Y.-X.; Liu, D.-G.; Ren, Y.; Liu, X.-B.; Li, Y.; Li, Z. Curcumin blocks small cell lung cancer cells migration, invasion, angiogenesis, cell cycle and neoplasia through Janus kinase-STAT3 signalling pathway. PloS one 2012, 7, e37960. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.-y.; Zheng, Y.; Jiao, D.-m.; Chen, F.-y.; Hu, H.-z.; Wu, Y.-q.; Song, J.; Yan, J.; Wu, L.-j.; Lv, G.-y. Curcumin inhibits lung cancer cell migration and invasion through Rac1-dependent signaling pathway. The Journal of nutritional biochemistry 2014, 25, 177–185. [Google Scholar] [CrossRef]
- Li, W.; Sun, L.; Lei, J.; Wu, Z.; Ma, Q.; Wang, Z. Curcumin inhibits pancreatic cancer cell invasion and EMT by interfering with tumor-stromal crosstalk under hypoxic conditions via the IL-6/ERK/NF-κB axis. Oncology reports 2020, 44, 382–392. [Google Scholar] [CrossRef]
- Weissenberger, J.; Priester, M.; Bernreuther, C.; Rakel, S.; Glatzel, M.; Seifert, V.; Kögel, D. Dietary curcumin attenuates glioma growth in a syngeneic mouse model by inhibition of the JAK1, 2/STAT3 signaling pathway. Clinical cancer research 2010, 16, 5781–5795. [Google Scholar] [CrossRef]
- Li, J.; Ye, L.; Parr, C.; Douglas-Jones, A.; Kynaston, H.G.; Mansel, R.E.; Jiang, W.G. The aberrant expression of bone morphogenetic protein 12 (BMP-12) in human breast cancer and its potential prognostic value. Gene Therapy and Molecular Biology 2009, 13, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Abdullah Thani, N.A.; Sallis, B.; Nuttall, R.; Schubert, F.R.; Ahsan, M.; Davies, D.; Purewal, S.; Cooper, A.; Rooprai, H.K. Induction of apoptosis and reduction of MMP gene expression in the U373 cell line by polyphenolics in Aronia melanocarpa and by curcumin. Oncology reports 2012, 28, 1435–1442. [Google Scholar] [CrossRef] [PubMed]
- Karmakar, S.; Banik, N.L.; Ray, S.K. Curcumin suppressed anti-apoptotic signals and activated cysteine proteases for apoptosis in human malignant glioblastoma U87MG cells. Neurochemical research 2007, 32, 2103–2113. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Jiao, J.T.; Qian, Y.; Guo, X.Y.; Huang, J.; Dai, M.C.; Zhang, L.; Ding, X.P.; Zong, D.; Shao, J.F. Curcumin induces G2/M arrest and triggers apoptosis via FoxO1 signaling in U87 human glioma cells. Molecular Medicine Reports 2016, 13, 3763–3770. [Google Scholar] [CrossRef]
- Wang, P.; Hao, X.; Li, X.; Yan, Y.; Tian, W.; Xiao, L.; Wang, Z.; Dong, J. Curcumin inhibits adverse psychological stress-induced proliferation and invasion of glioma cells via down-regulating the ERK/MAPK pathway. Journal of Cellular and Molecular Medicine 2021, 25, 7190–7203. [Google Scholar] [CrossRef]
- Wang, L.; Hu, R.; Dai, A. Curcumin Increased the Sensitivity of Non-Small-Cell Lung Cancer to Cisplatin through the Endoplasmic Reticulum Stress Pathway. Evidence-Based Complementary and Alternative Medicine 2022, 2022, 6886366. [Google Scholar] [CrossRef]
- Huang, Y.-F.; Zhu, D.-J.; Chen, X.-W.; Chen, Q.-K.; Luo, Z.-T.; Liu, C.-C.; Wang, G.-X.; Zhang, W.-J.; Liao, N.-Z. Curcumin enhances the effects of irinotecan on colorectal cancer cells through the generation of reactive oxygen species and activation of the endoplasmic reticulum stress pathway. Oncotarget 2017, 8, 40264. [Google Scholar] [CrossRef]
- Su, X.; Chen, S.; Lu, H.; Li, H.; Qin, C. Study on the inhibitory effect of curcumin on GBM and its potential mechanism. Drug Design, Development and Therapy 2781, 2781–2781. [Google Scholar] [CrossRef]
- Huang, T.-Y.; Tsai, T.-H.; Hsu, C.-W.; Hsu, Y.-C. Curcuminoids suppress the growth and induce apoptosis through caspase-3-dependent pathways in glioblastoma multiforme (GBM) 8401 cells. Journal of agricultural and food chemistry 2010, 58, 10639–10645. [Google Scholar] [CrossRef]
- Mosieniak, G.; Adamowicz, M.; Alster, O.; Jaskowiak, H.; Szczepankiewicz, A.A.; Wilczynski, G.M.; Ciechomska, I.A.; Sikora, E. Curcumin induces permanent growth arrest of human colon cancer cells: link between senescence and autophagy. Mechanisms of ageing and development 2012, 133, 444–455. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, R.; Yu, Y.; Liu, J.; Luo, T.; Fan, F. Glioblastoma treatment modalities besides surgery. Journal of Cancer 2019, 10, 4793. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Zeng, M.; Peng, B.; Li, P.; Zhao, S. Transient receptor potential vanilloid-1 (TRPV1) channels act as suppressors of the growth of glioma. Brain Research Bulletin 2024, 211, 110950. [Google Scholar] [CrossRef] [PubMed]
- Pucci, C.; Martinelli, C.; Ciofani, G. Innovative approaches for cancer treatment: Current perspectives and new challenges. ecancermedicalscience 2019, 13, 961. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, B.B.; Sundaram, C.; Malani, N.; Ichikawa, H. Curcumin: the Indian solid gold. The molecular targets and therapeutic uses of curcumin in health and disease 2007, 1–75. [Google Scholar]
- Avci, C.; Dodurga, Y.; Susluer, S.; Sıgva, Z.; Yucebas, M.; Caglar, H.; Akalin, T.; Dalbasti, T.; Oktar, N.; Gunduz, C. Promoter hypermethylation-mediated down-regulation of RUNX3 gene in human brain tumors. Irish journal of medical science 2014, 183, 259–264. [Google Scholar] [CrossRef]
- Chuang, L.; Ito, Y. RUNX3 is multifunctional in carcinogenesis of multiple solid tumors. Oncogene 2010, 29, 2605–2615. [Google Scholar] [CrossRef]
- Li, X.; Zheng, J.; Diao, H.; Liu, Y. RUNX 3 is down-regulated in glioma by Myc-regulated miR-4295. Journal of Cellular and Molecular Medicine 2016, 20, 518–525. [Google Scholar] [CrossRef]
- Steponaitis, G.; Kazlauskas, A.; Vaitkienė, P.; Deltuva, V.P.; Mikuciunas, M.; Skiriutė, D. Oncosuppressive role of RUNX3 in human astrocytomas. Journal of Oncology 2019, 2019, 1232434. [Google Scholar] [CrossRef]
- Mei, P.-J.; Bai, J.; Liu, H.; Li, C.; Wu, Y.-P.; Yu, Z.-Q.; Zheng, J.-N. RUNX3 expression is lost in glioma and its restoration causes drastic suppression of tumor invasion and migration. Journal of cancer research and clinical oncology 2011, 137, 1823–1830. [Google Scholar] [CrossRef]
- Sun, J.; Jia, Z.; Li, B.; Zhang, A.; Wang, G.; Pu, P.; Chen, Z.; Wang, Z.; Yang, W. MiR-19 regulates the proliferation and invasion of glioma by RUNX3 via β-catenin/Tcf-4 signaling. Oncotarget 2017, 8, 110785. [Google Scholar] [CrossRef]
- Sun, J.; Li, B.; Jia, Z.; Zhang, A.; Wang, G.; Chen, Z.; Shang, Z.; Zhang, C.; Cui, J.; Yang, W. RUNX3 inhibits glioma survival and invasion via suppression of the β-catenin/TCF-4 signaling pathway. Journal of neuro-oncology 2018, 140, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Newton, A.C.; Brognard, J. Reversing the paradigm: protein kinase C as a tumor suppressor. Trends in pharmacological sciences 2017, 38, 438–447. [Google Scholar] [CrossRef]
- Pilo, C.A.; Newton, A.C. Two sides of the same coin: protein kinase C γ in cancer and neurodegeneration. Frontiers in cell and developmental biology 2022, 10, 929510. [Google Scholar] [CrossRef] [PubMed]
- Satow, R.; Suzuki, Y.; Asada, S.; Ota, S.; Idogawa, M.; Kubota, S.; Ikeo, N.; Yoneda, A.; Fukami, K. Downregulation of protein kinase C gamma reduces epithelial property and enhances malignant phenotypes in colorectal cancer cells. Iscience 2022, 25. [Google Scholar] [CrossRef]
- Liu, L.; Wang, G.; Wang, L.; Yu, C.; Li, M.; Song, S.; Hao, L.; Ma, L.; Zhang, Z. Computational identification and characterization of glioma candidate biomarkers through multi-omics integrative profiling. Biology Direct 2020, 15, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-Y.; Wang, X.; Wang, X.-J.; Zheng, B.-Z.; Wang, Y.; Liang, B. Curcumin inhibits the growth via Wnt/β-catenin pathway in non-small-cell lung cancer cells. European Review for Medical & Pharmacological Sciences 2018, 22. [Google Scholar]
- Zhang, X.; Dong, S.; Xu, F. Structural and druggability landscape of frizzled G protein-coupled receptors. Trends in Biochemical Sciences 2018, 43, 1033–1046. [Google Scholar] [CrossRef]
- Huang, K.; Xu, H.; Han, L.; Xu, R.; Xu, Z.; Xie, Y. Identification of therapeutic targets and prognostic biomarkers among frizzled family genes in glioma. Frontiers in Molecular Biosciences 2023, 9, 1054614. [Google Scholar] [CrossRef]
- Hirano, H.; Yonezawa, H.; Yunoue, S.; Habu, M.; Uchida, H.; Yoshioka, T.; Kishida, S.; Kishida, M.; Oyoshi, T.; Fujio, S. Immunoreactivity of Wnt5a, Fzd2, Fzd6, and Ryk in glioblastoma: evaluative methodology for DAB chromogenic immunostaining. Brain tumor pathology 2014, 31, 85–93. [Google Scholar] [CrossRef]
- Ran, Y.; Han, S.; Gao, D.; Chen, X.; Liu, C. Interference of FZD2 suppresses proliferation, vasculogenic mimicry and stemness in glioma cells via blocking the Notch/NF-κB signaling pathway. Experimental and Therapeutic Medicine 2024, 28, 373. [Google Scholar] [CrossRef]
- Jia, F.; Zhang, L.; Jiang, Z.; Tan, G.; Wang, Z. FZD1/KLF10-hsa-miR-4762-5p/miR-224-3p-circular RNAs axis as prognostic biomarkers and therapeutic targets for glioblastoma: a comprehensive report. BMC Medical Genomics 2023, 16, 21. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Guo, Z.; Wang, B.; Yang, M.; Yuan, X.; Ji, B.; Wu, Y.; Chen, S. A Computational Framework to Identify Biomarkers for Glioma Recurrence and Potential Drugs Targeting Them. Frontiers in Genetics 2022, 12, 832627. [Google Scholar] [CrossRef]
- Ohka, F.; Shinjo, K.; Deguchi, S.; Matsui, Y.; Okuno, Y.; Katsushima, K.; Suzuki, M.; Kato, A.; Ogiso, N.; Yamamichi, A. Pathogenic epigenetic consequences of genetic alterations in IDH-wild-type diffuse astrocytic gliomas. Cancer research 2019, 79, 4814–4827. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, Z.; Zhang, Y. Expression and prognostic impact of FZDs in pancreatic adenocarcinoma. BMC gastroenterology 2021, 21, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, W.; Zhao, C. Frizzled receptors in tumors, focusing on signaling, roles, modulation mechanisms, and targeted therapies. Oncology Research 2021, 28, 661. [Google Scholar] [CrossRef]
- Yin, S.; Xu, L.; Bonfil, R.D.; Banerjee, S.; Sarkar, F.H.; Sethi, S.; Reddy, K.B. Tumor-initiating cells and FZD8 play a major role in drug resistance in triple-negative breast cancer. Molecular cancer therapeutics 2013, 12, 491–498. [Google Scholar] [CrossRef]
- Caja, L.; Bellomo, C.; Moustakas, A. Transforming growth factor β and bone morphogenetic protein actions in brain tumors. FEBS letters 2015, 589, 1588–1597. [Google Scholar] [CrossRef]
- Tenen, D.G. Disruption of differentiation in human cancer: AML shows the way. Nature reviews cancer 2003, 3, 89–101. [Google Scholar] [CrossRef]
- Hanavadi, S.; Martin, T.; Watkins, G.; Mansel, R.; Jiang, W. The role of growth differentiation factor-9 (GDF-9) and its analog, GDF-9b/BMP-15, in human breast cancer. Annals of surgical oncology 2007, 14, 2159–2166. [Google Scholar] [CrossRef]
- Varadaraj, A.; Patel, P.; Serrao, A.; Bandyopadhay, T.; Lee, N.Y.; Jazaeri, A.A.; Huang, Z.; Murphy, S.K.; Mythreye, K. Epigenetic regulation of GDF2 suppresses anoikis in ovarian and breast epithelia. Neoplasia 2015, 17, 826–838. [Google Scholar] [CrossRef]
- Du, P.; Ye, L.; Li, H.; Ruge, F.; Yang, Y.; Jiang, W.G. Growth differentiation factor-9 expression is inversely correlated with an aggressive behaviour in human bladder cancer cells. International journal of molecular medicine 2012, 29, 428–434. [Google Scholar]
- Codó, P.; Weller, M.; Kaulich, K.; Schraivogel, D.; Silginer, M.; Reifenberger, G.; Meister, G.; Roth, P. Control of glioma cell migration and invasiveness by GDF-15. Oncotarget 2016, 7, 7732. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Li, Z.; Fu, J.; Zhou, R. Growth and differentiation factor 15 regulates PD-L1 expression in glioblastoma. Cancer Management and Research 2019, 2653–2661. [Google Scholar] [CrossRef] [PubMed]
- Kadowaki, M.; Yoshioka, H.; Kamitani, H.; Watanabe, T.; Wade, P.A.; Eling, T.E. DNA methylation-mediated silencing of nonsteroidal anti-inflammatory drug-activated gene (NAG-1/GDF15) in glioma cell lines. International journal of cancer 2012, 130, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Bian, R.; Wang, F.; Wang, Y.; Li, X.; Guo, Y.; Zhang, X.; Luo, G.; Zhan, R. GDF-5 promotes epidermal stem cells proliferation via Foxg1-cyclin D1 signaling. Stem cell research & therapy 2021, 12, 1–11. [Google Scholar]
- Liu, M.; Sui, L.; Fang, Z.; Jiang, W.G.; Ye, L. Aberrant expression of bone morphogenetic proteins in the disease progression and metastasis of breast cancer. Frontiers in Oncology 2023, 13, 1166955. [Google Scholar] [CrossRef]
- Margheri, F.; Schiavone, N.; Papucci, L.; Magnelli, L.; Serratì, S.; Chilla, A.; Laurenzana, A.; Bianchini, F.; Calorini, L.; Torre, E. GDF5 regulates TGFß-dependent angiogenesis in breast carcinoma MCF-7 cells: in vitro and in vivo control by anti-TGFß peptides. PloS one 2012, 7, e50342. [Google Scholar] [CrossRef]
- Zhuo, J.; Li, H.; Zhang, P.; He, C.; Shen, W.; Yang, X.; Lin, Z.; Zhuang, R.; Wei, X.; Zheng, S. Growth differentiation factor 7 alleviates the proliferation and metastasis of hepatocellular carcinoma. Liver Research 2024, 8, 259–268. [Google Scholar] [CrossRef]
- Armandari, I.; Zomerman, W.W.; Plasschaert, S.L.; Smit, M.J.; Martini, T.E.; de Camargo Magalhães, E.S.; Hogeling, S.M.; Rozema-Huizinga, G.C.; Lourens, H.J.; Meeuwsen-de Boer, T.G. CREB signaling activity correlates with differentiation and survival in medulloblastoma. Scientific Reports 2021, 11, 16077. [Google Scholar] [CrossRef]
- Nana, A.W.; Yang, P.-M.; Lin, H.-Y. Overview of transforming growth factor β superfamily involvement in glioblastoma initiation and progression. Asian Pacific Journal of Cancer Prevention 2015, 16, 6813–6823. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




