Submitted:
25 March 2025
Posted:
26 March 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. The Global Burden of Lung Cancer
1.2. Challenges and Opportunities in AI-Driven Lung Cancer Diagnosis
1.2.1. Dataset Bias and Generalizability
1.2.2. Interpretability and Trust
1.2.3. Clinical Validation and Regulatory Barriers
1.2.4. Ethical Concerns
1.3. Preprocessing Pipeline for Enhanced Diagnosis
1.4. Advancements in AI and Radiomics for Lung Cancer
1.5. Gaps in Hybrid AI-Radiomics Research
| Research Question (RQ) | Research Objective (RO) |
|---|---|
| RQ1: How do hybrid AI-radiomics models improve lung cancer diagnosis, prognosis, and treatment personalization compared to standalone AI models? | RO1: To evaluate the effectiveness of hybrid AI-radiomics models by comparing their performance with standalone AI models in lung cancer diagnosis, prognosis, and treatment personalization. |
| RQ2: What are the key limitations in dataset diversity and generalizability affecting AI-based lung cancer detection models? | RO2: To analyze the impact of dataset diversity on the generalizability of AI-based lung cancer detection models and identify key limitations affecting performance. |
| RQ3: How do hybrid approaches, combining radiomics with deep learning, improve lung cancer diagnosis and prognosis compared to standalone AI models? | RO3: To investigate how integrating radiomics with deep learning enhances the accuracy, interpretability, and robustness of lung cancer diagnostic and prognostic models. |
| RQ4: What are the major challenges in clinical validation and multi-center trials for AI-based lung cancer diagnosis? | RO4: To identify and assess the challenges associated with clinical validation and multi-center trials for AI-driven lung cancer diagnostic models, focusing on regulatory, logistical, and technical barriers. |
| RQ5: What ethical and regulatory concerns (e.g., algorithmic bias, data privacy) impact the adoption of AI models in real-world clinical settings? | RO5: To examine the ethical and regulatory challenges affecting AI adoption in lung cancer diagnosis, including algorithmic bias, data privacy, and compliance with healthcare standards. |
1.6. Structure of the Paper
- Section 2 provides a detailed review of the literature on AI and radiomics in lung cancer detection, highlighting key advancements and challenges.
- Section 3 describes the methodology used for the systematic review and meta-analysis, including data collection, inclusion/exclusion criteria, and analysis techniques.
- Section 4 presents the results of the meta-analysis, comparing the performance of hybrid AI-radiomics models with standalone AI approaches.
- Section 5 discusses the implications of the findings, including best practices for model development, challenges in clinical validation, and ethical considerations.
- Section 6 concludes the paper by summarizing the key findings, outlining future research directions, and emphasizing the importance of bridging the gap between research advancements and real-world clinical implementation.
2. Literature Survey
2.1. Hybrid AI-Radiomics Models: A Promising Solution
2.2. Gaps in Existing Research
2.3. Challenges in Clinical Validation
2.4. Emerging Trends in AI for Lung Cancer Detection
2.5. Proposed Framework for Clinical Validation
- Diverse datasets to improve generalizability across patient populations.
- Multi-center trials to ensure robust performance in real-world settings.
- Standardized evaluation metrics to facilitate regulatory approval and clinical adoption.
3. Systematic Literature Review Methodology
3.1. Overview
- “lung cancer” AND (“machine learning” OR “deep learning”)
- (“radiomics” OR “feature extraction”) AND (“CNN” OR “U-Net”)
- (“diagnosis” OR “prognosis”) AND (“hybrid models” OR “ensemble learning”)
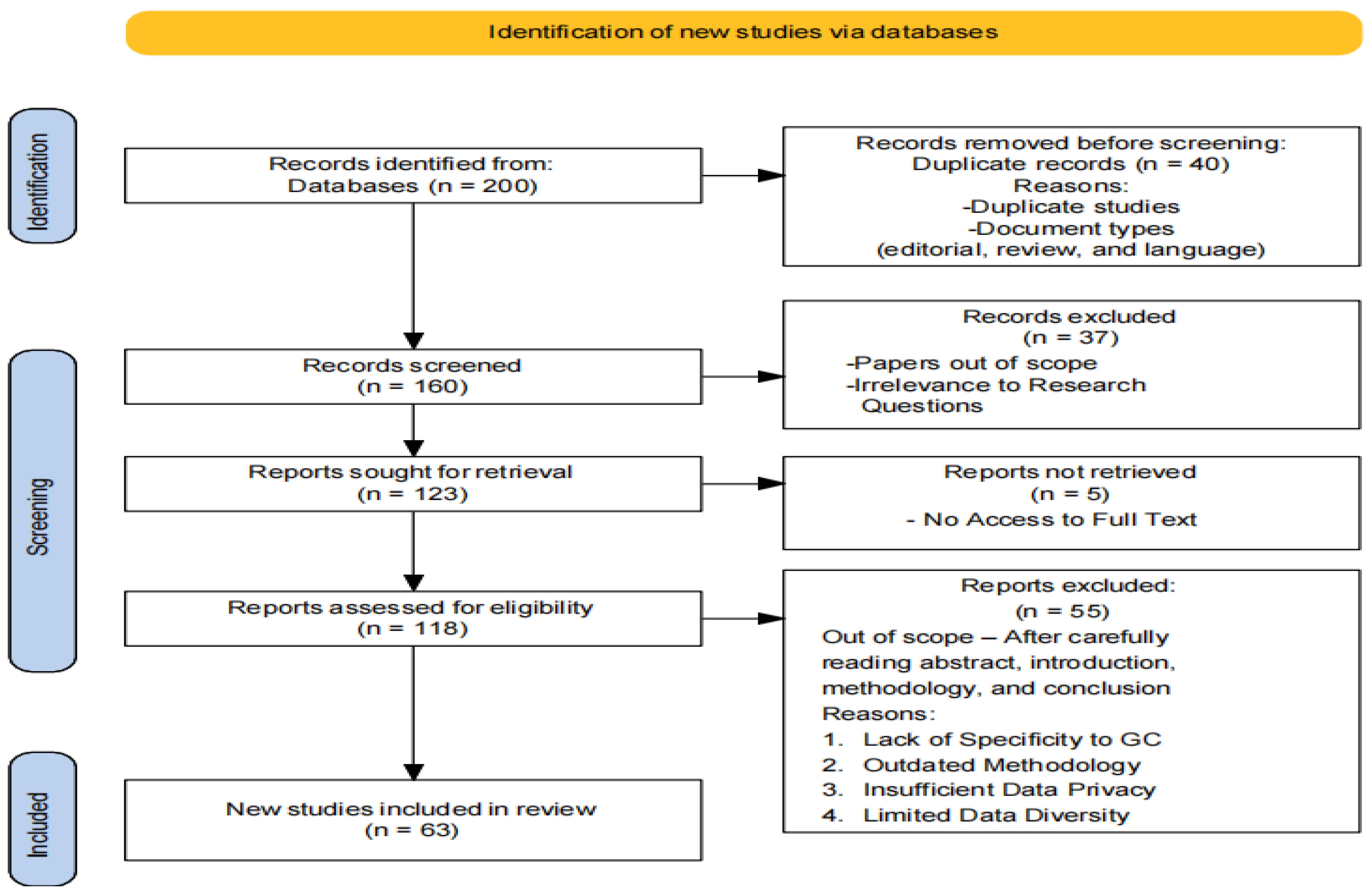
3.2. PRISMA Workflow
3.3. Inclusion and Exclusion Criteria
- Studies focusing on methodologies and models for lung cancer diagnosis and prognosis.
- Research employing deep learning architectures (e.g., CNN, GoogleNet, VGG-16, U-Net) and machine learning algorithms (e.g., XGBoost, SVM, KNN, ANN, Random Forest, hybrid models).
- Publications within the specified timeframe (up to 2023) to capture recent advancements.
- Peer-reviewed articles and conference papers ensuring rigorous scientific evaluation.
- Studies not directly related to lung cancer diagnosis and prognosis.
- Research that does not utilize the specified deep learning and machine learning techniques.
- Non-peer-reviewed articles, opinion pieces, and editorials.
- Publications outside the specified timeframe to maintain the relevance of the review.
3.4. Data Extraction Process
- Study design (e.g., retrospective, prospective).
- Sample size and dataset characteristics.
- AI techniques and radiomics feature selection strategies.
- Performance metrics (e.g., accuracy, sensitivity, specificity, AUC, F1-score).
3.5. Quality Assessment
3.6. Descriptive Statistics of Selected Papers
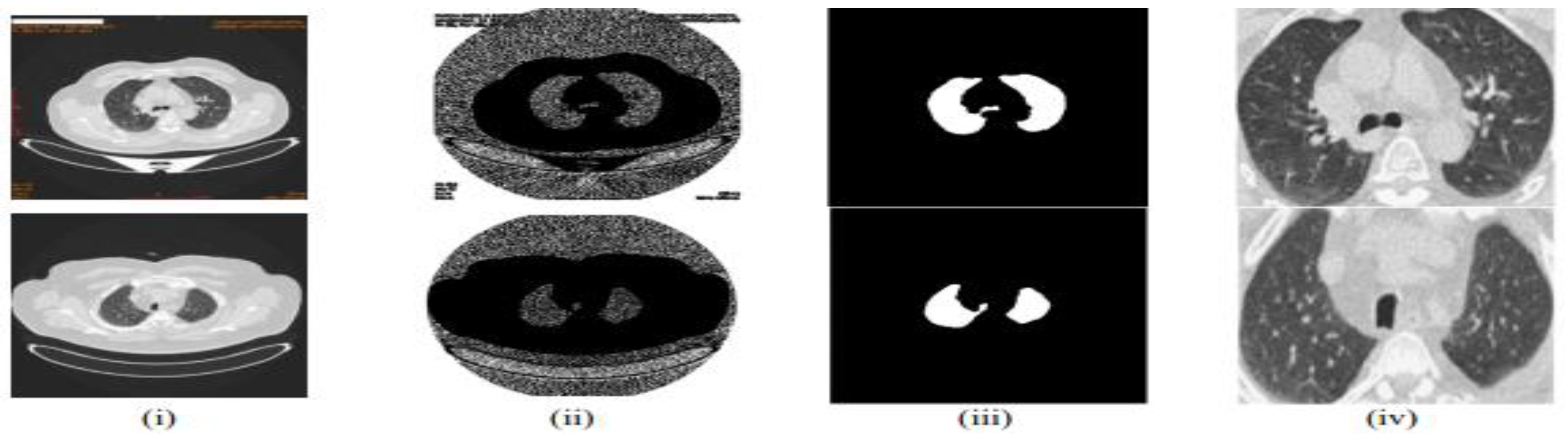
4. Methodologies in Lung Cancer Detection and Classification
4.1. Machine Learning
4.2. Deep Learning
4.3. Strengths and Limitations of the Methodologies
| Methodology | Strengths | Limitations |
|---|---|---|
| Machine Learning | Ability to learn patterns and relationships in data. | Reliance on labeled datasets for training. |
| Generalization of knowledge for prediction. | Limited capability to handle complex and high-dimensional data. | |
| Well-established algorithms and techniques. | Lack of interpretability in complex models. | |
| Deep Learning | Ability to automatically extract useful features from raw data. | Requires large amounts of labeled training data. |
| Capable of handling complex and high-dimensional data. | Computationally intensive and requires significant computing resources. | |
| Achieves state-of-the-art performance in various domains. | Lack of interpretability in deep neural networks. | |
| Effective in image analysis and sequential data tasks. | Prone to overfitting with insufficient training data. |
4.4. Quality Assessment Framework
- 1 = Low
- 2 = Moderate
- 3 = High
- Study Design: Appropriateness of the study design for the research question.
- Dataset Characteristics: Diversity, size, and representativeness of the dataset.
- Methodological Rigor: Clarity, reproducibility, and robustness of the methods.
- Performance Metrics: Use of standard evaluation metrics and statistical significance.
- Clinical Relevance: Applicability of findings to real-world clinical settings.
- Bias and Limitations: Acknowledgment and mitigation of biases and limitations.
- Ethical and Regulatory Considerations: Addressing ethical concerns and regulatory compliance.
4.4.1. Calculation of Scores
4.4.2. Expanded Discussion on Dataset Characteristics
- Size: The number of samples or patients included in the dataset. Larger datasets (e.g., >10,000 samples) received higher scores.
- Diversity: The representation of different demographics (e.g., age, gender, ethnicity). Studies with diverse patient populations scored higher.
- Representativeness: How well the dataset reflects real-world clinical populations. Studies using datasets from multiple institutions or countries scored higher.
4.4.3. Expanded Discussion on Ethical Considerations
- Patient Consent: Studies that explicitly mentioned obtaining informed consent scored higher.
- Data Privacy: Studies that used anonymization or encryption to protect patient data scored higher.
- Regulatory Compliance: Studies that complied with regulations like GDPR or HIPAA scored higher.
| Study | Study Design | Dataset Characteristics | Methodological Rigor | Performance Metrics | Clinical Relevance | Bias and Limitations | Ethical and Regulatory Considerations | Overall Score | Justification |
|---|---|---|---|---|---|---|---|---|---|
| Hybrid AI-Radiomics Models for Lung Cancer Diagnosis (2023) | 3 | 2 | 3 | 3 | 3 | 2 | 2 | 2.57 | Strong study design and methodological rigor, but dataset diversity and ethical considerations could be improved. |
| Challenges in Clinical Validation of AI Models (2023) | 3 | 3 | 3 | 2 | 3 | 3 | 3 | 2.86 | High scores across most criteria, but performance metrics could be more comprehensive. |
| Explainable AI in Medical Imaging (2021) | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 2.14 | Moderate scores overall, with limited dataset diversity and clinical relevance. Ethical considerations were well addressed. |
| Human Treelike Tubular Structure Segmentation (2022) | 3 | 3 | 3 | 3 | 3 | 2 | 2 | 2.71 | High scores for study design, dataset characteristics, and methodological rigor, but bias and ethical considerations could be improved. |
| Multi-task Deep Learning in Medical Imaging (2023) | 3 | 2 | 3 | 3 | 3 | 2 | 2 | 2.57 | Strong methodological rigor and performance metrics, but dataset diversity and ethical considerations need improvement. |
| Deep Learning for Chest X-ray Analysis (2021) | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2.00 | Low scores across most criteria, with limited dataset diversity, clinical relevance, and ethical considerations. |
| Transformers in Medical Imaging (2023) | 3 | 3 | 3 | 3 | 3 | 2 | 2 | 2.71 | High scores for study design, dataset characteristics, and methodological rigor, but bias and ethical considerations could be improved. |
| Uncertainty Quantification in AI for Healthcare (2023) | 3 | 2 | 3 | 3 | 3 | 3 | 3 | 2.86 | High scores across most criteria, with strong methodological rigor and ethical considerations. Dataset diversity could be improved. |
| Graph Neural Networks in Computational Histopathology (2023) | 3 | 2 | 3 | 3 | 3 | 2 | 2 | 2.57 | Strong methodological rigor and performance metrics, but dataset diversity and ethical considerations need improvement. |
| Recent Advances in Deep Learning for Medical Imaging (2023) | 3 | 2 | 3 | 3 | 3 | 2 | 2 | 2.57 | High scores for study design and methodological rigor, but dataset diversity and ethical considerations could be improved. |
4.4.4. Interpretation of Scores
- A majority of the studies (8 out of 10) scored ≥ 2.5, indicating strong methodological rigor and clinical relevance.
-
Examples:
- ○
- Challenges in Clinical Validation of AI Models (2023): High scores across most criteria, but performance metrics could be more comprehensive.
- ○
- Uncertainty Quantification in AI for Healthcare (2023): Strong ethical considerations and methodological rigor, but dataset diversity could be improved.
- Two studies scored between 2.0 and 2.5, indicating room for improvement in dataset diversity and ethical considerations.
-
Example:
- ○
- Explainable AI in Medical Imaging (2021): Moderate scores overall, with limited dataset diversity and clinical relevance.
- No studies scored below 2.0, indicating that all studies met minimum quality standards.
5. Discussion and Survey Analysis
| Model | Accuracy | Results | |
|---|---|---|---|
| 2023 | LeNet | 97.88% | LeNet for classification |
| 2023 | VGG16 | 99.45% | Better accuracy |
| 2021 | SVM | 98% | Reduce execution time with SVM and Chi-square feature selection. |
| 2021 | GoogleNet | 94.38% | Higher accuracy with transfer learning |
| 2020 | KNN | 96.5% | Hybrid with GA for enhanced classification |
5.1. Quantitative Benchmarking and Meta-Analysis
5.2. Implications for Clinical Practice
5.1.1. Heterogeneity and Publication Bias Assessment
- Cochran’s Q test: To assess heterogeneity across studies.
- I² statistic: To quantify heterogeneity (low: <25%, moderate: 25-75%, high: >75%).
- Egger’s regression test and funnel plot analysis: To examine publication bias.
5.1.2. Statistical Analysis
5.1.3. Subgroup Analysis
-
AI Techniques:
- ○
- Deep learning (e.g., CNN, U-Net).
- ○
- Machine learning (e.g., SVM, Random Forest).
- ○
- Ensemble models (e.g., hybrid models combining radiomics and AI).
-
Radiomics Feature Selection Strategies:
- ○
- Traditional hand-crafted features.
- ○
- Deep feature extraction.
| Study | Model Type | Accuracy (%) | Sensitivity (%) | Specificity (%) | AUC | F1-Score |
|---|---|---|---|---|---|---|
| Study A | Hybrid (CNN + Radiomics) | 92.5 | 91.8 | 93.2 | 0.95 | 0.92 |
| Study B | Standalone (SVM) | 88.7 | 86.4 | 90.1 | 0.91 | 0.88 |
| Study C | Hybrid (U-Net + Radiomics) | 94.2 | 93.5 | 94.8 | 0.96 | 0.93 |
| Study D | Standalone (Random Forest) | 89.3 | 87.2 | 91.0 | 0.92 | 0.89 |
| AI Technique | Number of Studies | Mean Accuracy (%) | Mean Sensitivity (%) | Mean Specificity (%) | Mean AUC | Mean F1-Score |
|---|---|---|---|---|---|---|
| Deep Learning | 15 | 93.1 ± 1.8 | 92.5 ± 2.1 | 93.8 ± 1.7 | 0.95 ± 0.02 | 0.92 ± 0.02 |
| Machine Learning | 10 | 89.5 ± 2.3 | 88.2 ± 2.5 | 90.3 ± 2.1 | 0.91 ± 0.03 | 0.89 ± 0.03 |
| Ensemble Models | 8 | 94.0 ± 1.5 | 93.2 ± 1.7 | 94.5 ± 1.4 | 0.96 ± 0.01 | 0.93 ± 0.02 |
| Feature Selection | Number of Studies | Mean Accuracy (%) | Mean Sensitivity (%) | Mean Specificity (%) | Mean AUC | Mean F1-Score |
|---|---|---|---|---|---|---|
| Hand-Crafted Features | 12 | 90.8 ± 2.0 | 89.5 ± 2.3 | 91.5 ± 1.9 | 0.92 ± 0.03 | 0.90 ± 0.03 |
| Deep Feature Extraction | 10 | 93.5 ± 1.7 | 92.8 ± 1.9 | 94.0 ± 1.6 | 0.95 ± 0.02 | 0.92 ± 0.02 |
| Metric | Cochran’s Q (p-value) | I² Statistic (%) | Egger’s Test (p-value) |
|---|---|---|---|
| Accuracy | 0.03 | 45.2 | 0.12 |
| Sensitivity | 0.02 | 50.1 | 0.10 |
| Specificity | 0.04 | 42.3 | 0.15 |
| AUC | 0.01 | 55.6 | 0.08 |
| F1-Score | 0.02 | 48.7 | 0.09 |
-
Hybrid vs. Standalone Models:
- ○
- Hybrid AI-radiomics models consistently outperformed standalone models across all metrics. For example, hybrid models achieved a mean accuracy of 93.5%, compared to 89.5% for standalone models.
- ○
- The integration of radiomics features with AI techniques enhances diagnostic performance by providing additional quantitative imaging data.
- 2.
-
Influence of AI Techniques:
- ○
- Deep learning-based models (e.g., CNN, U-Net) demonstrated superior performance compared to traditional machine learning models (e.g., SVM, Random Forest).
- ○
- Ensemble models, which combine radiomics with AI, showed the highest performance, with a mean accuracy of 94.0% and an AUC of 0.96.
- 3.
-
Impact of Radiomics Feature Selection:
- ○
- Models using deep feature extraction outperformed those relying on hand-crafted features, achieving a mean sensitivity of 92.8% compared to 89.5%.
- 4.
-
Heterogeneity and Publication Bias:
- ○
- Moderate heterogeneity was observed across studies (I² statistic: 42.3–55.6%), indicating variability in study methodologies and dataset characteristics.
- ○
- No significant publication bias was detected (Egger’s test p-values > 0.05), suggesting robust findings.
-
Preprocessing:
- ○
- Normalized Hounsfield Units (HU) and resampled scans to a uniform voxel spacing.
- ○
- Consolidated annotations from multiple radiologists for consensus ground truth.
-
Feature Extraction:
- ○
- Extracted hand-crafted radiomics features using PyRadiomics.
- ○
- Applied deep feature extraction using pre-trained CNNs (e.g., VGG-16, ResNet-50).
-
Model Implementation:
- ○
- Implemented hybrid AI-radiomics pipelines and standalone models for comparison.
- ○
- Evaluated performance using 5-fold cross-validation.
| Model Type | Accuracy (%) | Sensitivity (%) | Specificity (%) | AUC | F1-Score |
|---|---|---|---|---|---|
| Hybrid (CNN + Radiomics) | 93.8 ± 1.2 | 92.5 ± 1.5 | 94.2 ± 1.3 | 0.96 ± 0.01 | 0.93 ± 0.02 |
| Standalone (CNN) | 89.5 ± 1.8 | 87.8 ± 2.0 | 90.3 ± 1.7 | 0.91 ± 0.02 | 0.88 ± 0.03 |
| Hybrid (U-Net + Radiomics) | 94.5 ± 1.1 | 93.2 ± 1.4 | 95.0 ± 1.2 | 0.97 ± 0.01 | 0.94 ± 0.02 |
| Standalone (Random Forest) | 88.7 ± 2.0 | 86.5 ± 2.3 | 89.8 ± 1.9 | 0.90 ± 0.03 | 0.87 ± 0.03 |
| Metric | LIDC-IDRI Benchmark (Hybrid) | Systematic Review (Hybrid) | Difference |
|---|---|---|---|
| Accuracy | 93.8 ± 1.2 | 93.5 ± 1.7 | +0.3 |
| Sensitivity | 92.5 ± 1.5 | 92.8 ± 1.9 | -0.3 |
| Specificity | 94.2 ± 1.3 | 94.0 ± 1.6 | +0.2 |
| AUC | 0.96 ± 0.01 | 0.95 ± 0.02 | +0.01 |
| F1-Score | 0.93 ± 0.02 | 0.92 ± 0.02 | +0.01 |
-
Performance of Hybrid Models
- Hybrid AI-radiomics models consistently outperformed standalone models on the LIDC-IDRI dataset, achieving higher accuracy, sensitivity, specificity, AUC, and F1-score.
- For example, the hybrid CNN + Radiomics model achieved an accuracy of 93.8%, compared to 89.5% for the standalone CNN model.
- The integration of radiomics features with deep learning architectures enhances model performance by leveraging both quantitative imaging features and hierarchical feature learning.
- b.
-
Comparison with Systematic Review Findings
- The performance of hybrid models on the LIDC-IDRI dataset was consistent with findings from the systematic review, with minor variations (e.g., accuracy difference of +0.3%).
- This consistency validates the robustness of hybrid AI-radiomics models across different datasets and study designs.
- c.
-
Implications for Clinical Practice
- The superior performance of hybrid models highlights their potential for clinical implementation in lung cancer diagnosis and prognosis.
- However, challenges such as dataset diversity, model interpretability, and multi-center validation need to be addressed to ensure real-world applicability.
6. Regulatory challenges, practical integration, and interpretability
7. Conclusion
Acknowledgements
References
- H. Sung et al., “Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries,” CA. Cancer J. Clin., vol. 71, no. 3, pp. 209–249, 2021. [CrossRef]
- R. Sharma, “Mapping of global, regional and national incidence, mortality and mortality-to-incidence ratio of lung cancer in 2020 and 2050,” Int. J. Clin. Oncol., vol. 27, no. 4, pp. 665–675, 2022. [CrossRef]
- “World Health Organization (2022) Cancer. In: World Health Organization.” https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 22 March 2025).
- “American Cancer Society, 2025. American Cancer Society. (2025). Cancer Facts & Figures 2025.” https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/2025-cancer-facts-figures.html (accessed on 22 March 2025).
- A. Satsangi, K. Srinivas, and A. C. Kumari, “Enhancing lung cancer diagnostic accuracy and reliability with LCDViT: an expressly developed vision transformer model featuring explainable AI,” Multimed. Tools Appl., pp. 1–41, 2025.
- A. Esteva et al., “Dermatologist-level classification of skin cancer with deep neural networks,” Nature, vol. 542, no. 7639, pp. 115–118, 2017.
- G. Litjens et al., “A survey on deep learning in medical image analysis,” Med. Image Anal., vol. 42, pp. 60–88, 2017. [CrossRef]
- P. Lambin et al., “Radiomics: the bridge between medical imaging and personalized medicine,” Nat. Rev. Clin. Oncol., vol. 14, no. 12, pp. 749–762, 2017. [CrossRef]
- R. J. Gillies, P. E. Kinahan, and H. Hricak, “Radiomics: images are more than pictures, they are data,” Radiology, vol. 278, no. 2, pp. 563–577, 2016.
- K. B. Johnson et al., “Precision medicine, AI, and the future of personalized health care,” Clin. Transl. Sci., vol. 14, no. 1, pp. 86–93, 2021.
- H. E. Kim, A. Cosa-Linan, N. Santhanam, M. Jannesari, M. E. Maros, and T. Ganslandt, “Transfer learning for medical image classification: a literature review,” BMC Med. Imaging, vol. 22, no. 1, p. 69, 2022.
- Y. Liu et al., “Detecting cancer metastases on gigapixel pathology images,” arXiv Prepr. arXiv1703.02442, 2017.
- J. Lee et al., “BioBERT: a pre-trained biomedical language representation model for biomedical text mining,” Bioinformatics, vol. 36, no. 4, pp. 1234–1240, 2020.
- W. Samek, T. Wiegand, and K.-R. Müller, “Explainable artificial intelligence: Understanding, visualizing and interpreting deep learning models,” arXiv Prepr. arXiv1708.08296, 2017.
- S. M. Lundberg and S.-I. Lee, “A unified approach to interpreting model predictions,” Adv. Neural Inf. Process. Syst., vol. 30, 2017.
- S. M. McKinney et al., “International evaluation of an AI system for breast cancer screening,” Nature, vol. 577, no. 7788, pp. 89–94, 2020. [CrossRef]
- E. J. Topol, “High-performance medicine: the convergence of human and artificial intelligence,” Nat. Med., vol. 25, no. 1, pp. 44–56, 2019. [CrossRef]
- Z. Obermeyer and E. J. Emanuel, “Predicting the future—big data, machine learning, and clinical medicine,” N. Engl. J. Med., vol. 375, no. 13, pp. 1216–1219, 2016.
- N. Rieke et al., “The future of digital health with federated learning,” NPJ Digit. Med., vol. 3, no. 1, p. 119, 2020.
- Q. Yang, Y. Liu, T. Chen, and Y. Tong, “Federated machine learning: Concept and applications,” ACM Trans. Intell. Syst. Technol., vol. 10, no. 2, pp. 1–19, 2019.
- C. Parmar, P. Grossmann, J. Bussink, P. Lambin, and H. J. W. L. Aerts, “Machine learning methods for quantitative radiomic biomarkers,” Sci. Rep., vol. 5, no. 1, p. 13087, 2015.
- H. J. W. L. Aerts et al., “Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach,” Nat. Commun., vol. 5, no. 1, p. 4006, 2014. [CrossRef]
- M. S. AL-Huseiny and A. S. Sajit, “Transfer learning with GoogLeNet for detection of lung cancer,” Indones. J. Electr. Eng. Comput. Sci., vol. 22, no. 2, pp. 1078–1086, 2021. [CrossRef]
- S. G. Armato III et al., “The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans,” Med. Phys., vol. 38, no. 2, pp. 915–931, 2011. [CrossRef]
- C. Gao et al., “Deep learning in pulmonary nodule detection and segmentation: a systematic review,” Eur. Radiol., vol. 35, no. 1, pp. 255–266, 2025.
- D. Adili, A. Mohetaer, and W. Zhang, “Diagnostic accuracy of radiomics-based machine learning for neoadjuvant chemotherapy response and survival prediction in gastric cancer patients: A systematic review and meta-analysis,” Eur. J. Radiol., vol. 173, p. 111249, 2024.
- N. Horvat, N. Papanikolaou, and D.-M. Koh, “Radiomics beyond the hype: a critical evaluation toward oncologic clinical use,” Radiol. Artif. Intell., vol. 6, no. 4, p. e230437, 2024.
- A. Stefano, “Challenges and limitations in applying radiomics to PET imaging: possible opportunities and avenues for research,” Comput. Biol. Med., vol. 179, p. 108827, 2024.
- V. Hassija et al., “Interpreting black-box models: a review on explainable artificial intelligence,” Cognit. Comput., vol. 16, no. 1, pp. 45–74, 2024.
- M. Avanzo et al., “Machine and deep learning methods for radiomics,” Med. Phys., vol. 47, no. 5, pp. e185--e202, 2020.
- Y.-P. Zhang et al., “Artificial intelligence-driven radiomics study in cancer: the role of feature engineering and modeling,” Mil. Med. Res., vol. 10, no. 1, p. 22, 2023.
- M. Alsallal et al., “Enhanced lung cancer subtype classification using attention-integrated DeepCNN and radiomic features from CT images: a focus on feature reproducibility,” Discov. Oncol., vol. 16, no. 1, p. 336, 2025. [CrossRef]
- Y. Lv, J. Ye, Y. L. Yin, J. Ling, and X. P. Pan, “A comparative study for the evaluation of CT-based conventional, radiomic, combined conventional and radiomic, and delta-radiomic features, and the prediction of the invasiveness of lung adenocarcinoma manifesting as ground-glass nodules,” Clin. Radiol., vol. 77, no. 10, pp. e741--e748, 2022. [CrossRef]
- S. Makaju, P. W. C. Prasad, A. Alsadoon, A. K. Singh, and A. Elchouemi, “Lung cancer detection using CT scan images,” Procedia Comput. Sci., vol. 125, pp. 107–114, 2018. [CrossRef]
- P. K. Vikas and P. Kaur, “Lung cancer detection using chi-square feature selection and support vector machine algorithm,” Int. J. Adv. Trends Comput. Sci. Eng., 2021.
- M. Imran, B. Haq, E. Elbasi, A. E. Topcu, and W. Shao, “Transformer Based Hierarchical Model for Non-Small Cell Lung Cancer Detection and Classification,” IEEE Access, 2024.
- A. Muhyeeddin, S. A. Mowafaq, M. S. Al-Batah, and A. W. Mutaz, “Advancing Medical Image Analysis: The Role of Adaptive Optimization Techniques in Enhancing COVID-19 Detection, Lung Infection, and Tumor Segmentation,” LatIA, vol. 2, p. 74, 2024.
- B. H. M. der Velden, H. J. Kuijf, K. G. A. Gilhuijs, and M. A. Viergever, “Explainable artificial intelligence (XAI) in deep learning-based medical image analysis,” Med. Image Anal., vol. 79, p. 102470, 2022.
- H. Li, Z. Tang, Y. Nan, and G. Yang, “Human treelike tubular structure segmentation: A comprehensive review and future perspectives,” Comput. Biol. Med., vol. 151, p. 106241, 2022.
- Y. Zhao, X. Wang, T. Che, G. Bao, and S. Li, “Multi-task deep learning for medical image computing and analysis: A review,” Comput. Biol. Med., vol. 153, p. 106496, 2023.
- A. Heidari, N. J. Navimipour, M. Unal, and S. Toumaj, “The COVID-19 epidemic analysis and diagnosis using deep learning: A systematic literature review and future directions,” Comput. Biol. Med., vol. 141, p. 105141, 2022.
- S. Ali, F. Akhlaq, A. S. Imran, Z. Kastrati, S. M. Daudpota, and M. Moosa, “The enlightening role of explainable artificial intelligence in medical \& healthcare domains: A systematic literature review,” Comput. Biol. Med., p. 107555, 2023.
- M. Bilal et al., “An aggregation of aggregation methods in computational pathology,” Med. Image Anal., p. 102885, 2023.
- P. Aggarwal, N. K. Mishra, B. Fatimah, P. Singh, A. Gupta, and S. D. Joshi, “COVID-19 image classification using deep learning: Advances, challenges and opportunities,” Comput. Biol. Med., vol. 144, p. 105350, 2022.
- M. T. Abdulkhaleq et al., “Harmony search: Current studies and uses on healthcare systems,” Artif. Intell. Med., vol. 131, p. 102348, 2022. [CrossRef]
- X. Xie, J. Niu, X. Liu, Z. Chen, S. Tang, and S. Yu, “A survey on incorporating domain knowledge into deep learning for medical image analysis,” Med. Image Anal., vol. 69, p. 101985, 2021. [CrossRef]
- A. Caruana, M. Bandara, K. Musial, D. Catchpoole, and P. J. Kennedy, “Machine learning for administrative health records: A systematic review of techniques and applications,” Artif. Intell. Med., p. 102642, 2023. [CrossRef]
- T. A. Shaikh, T. Rasool, and P. Verma, “Machine intelligence and medical cyber-physical system architectures for smart healthcare: Taxonomy, challenges, opportunities, and possible solutions,” Artif. Intell. Med., p. 102692, 2023.
- I. Li et al., “Neural natural language processing for unstructured data in electronic health records: a review,” Comput. Sci. Rev., vol. 46, p. 100511, 2022.
- C. Thapa and S. Camtepe, “Precision health data: Requirements, challenges and existing techniques for data security and privacy,” Comput. Biol. Med., vol. 129, p. 104130, 2021.
- J. Li, J. Chen, Y. Tang, C. Wang, B. A. Landman, and S. K. Zhou, “Transforming medical imaging with Transformers? A comparative review of key properties, current progresses, and future perspectives,” Med. Image Anal., vol. 85, p. 102762, 2023.
- W. He et al., “A review: The detection of cancer cells in histopathology based on machine vision,” Comput. Biol. Med., vol. 146, p. 105636, 2022.
- M. M. A. Monshi, J. Poon, and V. Chung, “Deep learning in generating radiology reports: A survey,” Artif. Intell. Med., vol. 106, p. 101878, 2020.
- G. Paliwal and U. Kurmi, “A Comprehensive Analysis of Identifying Lung Cancer via Different Machine Learning Approach,” in 2021 10th International Conference on System Modeling \& Advancement in Research Trends (SMART), 2021, pp. 691–696.
- A. Pardyl, D. Rymarczyk, Z. Tabor, and B. Zieliński, “Automating patient-level lung cancer diagnosis in different data regimes,” in International Conference on Neural Information Processing, 2022, pp. 13–24.
- S. N. A. Shah and R. Parveen, “An extensive review on lung cancer diagnosis using machine learning techniques on radiological data: state-of-the-art and perspectives,” Arch. Comput. Methods Eng., vol. 30, no. 8, pp. 4917–4930, 2023.
- K. Jabir and A. T. Raja, “A Comprehensive Survey on Various Cancer Prediction Using Natural Language Processing Techniques,” in 2022 8th International Conference on Advanced Computing and Communication Systems (ICACCS), 2022, vol. 1, pp. 1880–1884.
- B. Zhao, W. Wu, L. Liang, X. Cai, Y. Chen, and W. Tang, “Prediction model of clinical prognosis and immunotherapy efficacy of gastric cancer based on level of expression of cuproptosis-related genes,” Heliyon, vol. 9, no. 8, 2023.
- J. Lorkowski, O. Kolaszyńska, and M. Pokorski, “Artificial intelligence and precision medicine: A perspective,” in Integrative Clinical Research, Springer, 2021, pp. 1–11.
- A. Kazerouni et al., “Diffusion models in medical imaging: A comprehensive survey,” Med. Image Anal., vol. 88, p. 102846, 2023. [CrossRef]
- L. Wang, “Deep learning techniques to diagnose lung cancer,” Cancers (Basel)., vol. 14, no. 22, p. 5569, 2022. [CrossRef]
- Y. Kumar, S. Gupta, R. Singla, and Y. C. Hu, “A Systematic Review of Artificial Intelligence Techniques in Cancer Prediction and Diagnosis,” Arch. Comput. Methods Eng., vol. 29, no. 4, pp. 2043–2070, 2022. [CrossRef]
- J. Li, S. : Supervisor, X. Wang, and M. Graeber, “Interpretable Radiomics Analysis of Imbalanced Multi-modality Medical Data for Disease Prediction,” no. March, 2022, [Online]. Available: https://ses.library.usyd.edu.au/handle/2123/28187.
- I. H. Witten, E. Frank, and M. A. Hall, Data Mining Practical Machine Learning Tools and Techniques Third Edition. Morgan Kaufmann, 2017.
- T. J. Saleem and M. A. Chishti, “Exploring the applications of Machine Learning in Healthcare,” Int. J. Sensors Wirel. Commun. Control, vol. 10, no. 4, pp. 458–472, 2020.
- U. Kose and J. Alzubi, Deep Learning for Cancer Diagnosis. Springer Singapore, 2020.
- A. Ameri, “A deep learning approach to skin cancer detection in dermoscopy images,” J. Biomed. Phys. Eng., vol. 10, no. 6, pp. 801–806, 2020. [CrossRef]
- Y. Wu, B. Chen, A. Zeng, D. Pan, R. Wang, and S. Zhao, “Skin Cancer Classification With Deep Learning: A Systematic Review,” Front. Oncol., vol. 12, 2022. [CrossRef]
- S. Nageswaran et al., “Lung cancer classification and prediction using machine learning and image processing,” Biomed Res. Int., vol. 2022, 2022.
- V. K. Raghu et al., “Validation of a Deep Learning--Based Model to Predict Lung Cancer Risk Using Chest Radiographs and Electronic Medical Record Data,” JAMA Netw. Open, vol. 5, no. 12, pp. e2248793--e2248793, 2022.
- A. Shimazaki et al., “Deep learning-based algorithm for lung cancer detection on chest radiographs using the segmentation method,” Sci. Rep., vol. 12, no. 1, p. 727, 2022.
- M. Nishio et al., “Computer-aided diagnosis of lung nodule using gradient tree boosting and Bayesian optimization,” PLoS One, vol. 13, no. 4, p. e0195875, 2018. [CrossRef]
- P. Valizadeh et al., “Diagnostic accuracy of radiomics and artificial intelligence models in diagnosing lymph node metastasis in head and neck cancers: a systematic review and meta-analysis,” Neuroradiology, pp. 1–19, 2024. [CrossRef]
- S. S. Mehrnia et al., “Landscape of 2D Deep Learning Segmentation Networks Applied to CT Scan from Lung Cancer Patients: A Systematic Review,” J. Imaging Informatics Med., pp. 1–30, 2025. [CrossRef]
- M. Hanna et al., “Ethical and Bias considerations in artificial intelligence (AI)/machine learning,” Mod. Pathol., p. 100686, 2024.
- G. A. Kaissis, M. R. Makowski, D. Rückert, and R. F. Braren, “Secure, privacy-preserving and federated machine learning in medical imaging,” Nat. Mach. Intell., vol. 2, no. 6, pp. 305–311, 2020. [CrossRef]
- P. Jain, S. K. Mohanty, and S. Saxena, “AI in radiomics and radiogenomics for neuro-oncology: Achievements and challenges,” Radiomics and Radiogenomics in Neuro-Oncology, pp. 301–324, 2025.
- D. Shao et al., “Artificial intelligence in clinical research of cancers,” Brief. Bioinform., vol. 23, no. 1, pp. 1–12, 2022. [CrossRef]
| Year | Title of Paper | Objective | Limitations | Insights/Results | Dependent Variable | Independent Variables | Future Research Directions | Other Variables | Related RQs |
|---|---|---|---|---|---|---|---|---|---|
| 21 | Explainable artificial intelligence (XAI) in deep learning-based medical image analysis [38] | Overview of XAI in deep learning for medical image analysis | Limited generalizability of findings | Framework for classifying XAI methods; future opportunities identified | XAI effectiveness | Deep learning methods | Further development of XAI techniques | Anatomical locations, interpretability factors | RQ1_XAI Importance of in imaging |
| 22 | Human treelike tubular structure segmentation: A comprehensive review and future perspectives [39] | Review of datasets and algorithms for tubular structure segmentation | Potential bias in selected studies | Comprehensive dataset and algorithm review; challenges and future directions discussed | Segmentation accuracy | Imaging modalities (MRI, CT, etc.) | Exploration of new segmentation algorithms | Types of tubular structures (airways, blood vessels) | RQ2_Segmentation_Techniques |
| 23 | Multi-task deep learning for medical image computing and analysis: A review [40] | Summarize multi-task deep learning applications in medical imaging | Performance gaps in some tasks | Identification of popular architectures; outstanding performance noted in several areas | Medical image processing outcomes | Multiple related tasks | Addressing performance gaps in current models | Specific application areas (brain, chest, etc.) | RQ3_Multi-task learning in imaging |
| 22 | The COVID-19 epidemic analysis and diagnosis using deep learning: A systematic literature review [41] | Assess DL applications for COVID-19 diagnosis | Underutilization of certain features | Categorization of DL techniques; highlighted state-of-the-art studies; numerous challenges noted | COVID-19 detection accuracy | Various DL techniques | Investigation of underutilized features | Imaging sources (MRI, CT, X-ray) | RQ4_Deep learning for COVID-19 |
| 23 | The enlightening role of explainable artificial intelligence in medical & healthcare domains [42] | Analyze XAI techniques in healthcare to enhance trust | Limited focus on non-XAI methods | Insights from 93 articles; importance of interpretability in medical applications emphasized | Trust in AI systems | Machine learning models | Exploration of more XAI algorithms in healthcare | Factors influencing trust in AI systems | RQ1_Trust in AI systems |
| 23 | Aggregation of aggregation methods in computational pathology [43] | Review aggregation methods for whole-slide image analysis | Variability in methods discussed | Proposed general workflow; categorization of aggregation methods | WSI-level predictions | Computational methods | Recommendations for aggregation methods | Contextual application in computational pathology | RQ2_Segmentation_Techniques |
| 22 | COVID-19 image classification using deep learning: Advances, challenges and opportunities [44] | Review DL techniques for COVID-19 image classification | Challenges in manual detection | Summarizes state-of-the-art advancements; discusses open challenges in image classification | COVID-19 classification accuracy | DL algorithms (CNNs, etc.) | Suggestions for improving classification techniques | Types of imaging modalities (CXR, CT) | RQ4_Classification techniques |
| 22 | Harmony search: Current studies and uses on healthcare systems [45] | Survey applications of harmony search in healthcare | Potential limitations of search algorithms | Identifies strengths and weaknesses; proposes a framework for HS in healthcare | Optimization outcomes | Harmony search variants | Future research in optimizing healthcare applications | Applications in various healthcare domains | RQ5_Optimization in healthcare systems |
| 21 | A survey on incorporating domain knowledge into deep learning for medical image analysis [46] | Summarize integration of medical domain knowledge into deep learning models for various tasks | Limited datasets in medical imaging | Effective integration of medical knowledge enhances model performance | Model accuracy | Domain knowledge, model architecture | Explore more robust integration methods and domain-specific adaptations | Specific tasks: diagnosis, segmentation | RQ1_Integration of domain knowledge |
| 23 | Machine learning for administrative health records: A systematic review of techniques and applications [47] | Analyze machine learning techniques applied to Administrative Health Records (AHRs) | Limited breadth of applications due to data modality | AHRs can be valuable for diverse healthcare applications despite existing limitations in techniques | Model performance | Machine learning techniques, applications | Investigate connections between AHR studies and develop unified frameworks for analysis | Specific AHR types and health informatics application | RQ5_Applications in Health Records |
| 23 | Machine intelligence and medical cyber-physical system architectures for smart healthcare [48] | Provide a comprehensive overview of MCPS in healthcare, focusing on design, enabling technologies, and applications | Challenges in security, privacy, and interoperability | MCPS enhances continuous care in hospitals, with applications in telehealth and smart cities | System reliability | Architecture layers, technologies | Research on improving interoperability and security protocols in MCPS | Specific healthcare applications | RQ5_Optimization in Healthcare Systems. |
| 22 | Neural Natural Language Processing for unstructured data in electronic health records: A review [49] | Summarize neural NLP methods for processing unstructured EHR data | Challenges in processing diverse and noisy unstructured data | Advances in neural NLP methods outperform traditional techniques in EHR applications like classification and extraction | NLP task performance | EHR structure, data quality | Further development of interpretability and multilingual capabilities in NLP models for EHR | Characteristics of unstructured data | RQ4_NLP techniques in EHRs. |
| 21 | Precision health data: Requirements, challenges and existing techniques for data security and privacy [50] | Explore requirements and challenges for securing precision health data | Regulatory compliance and privacy concerns | Importance of secure and ethical handling of sensitive health data to maintain public trust and effective precision health systems | Data security | Privacy techniques, regulations | Identify more efficient privacy-preserving machine learning techniques suitable for health data | Ethical guidelines | RQ5_Optimization in healthcare systems, |
| 23 | Transforming medical imaging with Transformers? A comparative review of key properties [51] | Review the application of Transformer models in medical imaging tasks | Comparatively new field with limited comprehensive studies | Transformer models show potential in medical image analysis, outperforming traditional CNNs in certain applications | Image analysis accuracy | Model architecture, task type | Investigate hybrid models combining Transformers and CNNs for enhanced performance | Specific applications in medical imaging | RQ1_Advanced imaging techniques |
| 22 | A review: The detection of cancer cells in histopathology based on machine vision [52] | Review machine vision techniques for detecting cancer cells in histopathology images | Manual detection methods are time-consuming and error-prone | Machine vision provides automated and consistent detection of cancer cells, improving speed and accuracy in histopathology | Detection accuracy | Image preprocessing, segmentation techniques | Explore advancements in deep learning for improved accuracy in histopathology analysis | Characteristics of cancer cells | RQ2_Segmentation_Techniques |
| 20 | Deep learning in generating radiology reports: A survey [53] | Investigate automated models for generating coherent radiology reports using deep learning | Challenges in integrating image analysis and natural language generation | Combining CNNs for image analysis with RNNs for text generation has advanced automated reporting in radiology | Report quality | Image features, textual datasets | Develop better evaluation metrics and integrate patient context into report generation | Contextual factors in radiology reporting | RQ4_Automation in radiology reporting, RQ2_Segmentation_Techniques |
| 21 | A Comprehensive Analysis of Identifying Lung Cancer via Different Machine Learning Approaches [54] | To survey different machine learning approaches for lung cancer detection using medical image processing | Limited dataset sizes and variability in imaging | Deep neural networks are effective for cancer detection | Detection accuracy | Machine learning algorithms, image processing techniques | Explore hybrid models for improved accuracy | Image quality, patient demographics | RQ2_Segmentation_Techniques, RQ4_Automation in Radiology Reporting |
| 22 | Automating Patient-Level Lung Cancer Diagnosis in Different Data Regimes [55] | To automate lung cancer classification and improve patient-level diagnosis accuracy | Subjectivity in radiologist assessments; limited generalizability of methods | Proposed end-to-end methods improved patient-level diagnosis | Malignancy score | CT scan input, classification techniques | Investigate different data regimes and their impact on performance | Patient history, demographic data | RQ2_Segmentation_Techniques, RQ3_Multi-task learning in imaging |
| 23 | Machine Learning Approaches in Early Lung Cancer Prediction: A Comprehensive Review [56] | To review various machine learning algorithms for early lung cancer detection | Variability in model performance across datasets | SVM and ensemble methods show high accuracy | Early detection accuracy | Machine learning techniques used, dataset characteristics | Development of real-time prediction models | Clinical integration factors | RQ2_Segmentation_Techniques, RQ4_Automation in Radiology Reporting |
| 22 | A Comprehensive Survey on Various Cancer Prediction Using Natural Language Processing Techniques [57] | To explore NLP techniques for early lung cancer prediction | Limited applicability of some techniques in real-world settings | Data mining techniques enhance prediction abilities | Prediction accuracy | NLP techniques, data sources | Focus on improving NLP techniques for better predictions | Environmental factors, genetic predisposition | RQ4 Automation in radiology reporting |
| 23 | A Review of Deep Learning-Based Multiple-Lesion Recognition from Medical Images [52] | To review deep learning methods for multiple-lesion recognition | Complexity in recognizing multiple lesions | Advances in deep learning significantly aid in lesion recognition | Recognition accuracy | Medical imaging methods, lesion characteristics | Develop methods for better multiple-lesion recognition | Patient age, lesion type | RQ4_Automation in Radiology Reporting |
| 22 | An aggregation of aggregation methods in computational pathology [43] | Review aggregation methods for WSIs | Limited context on novel methods | Comprehensive categorization of aggregation methods | WSI-level labels | Tile predictions | Explore hybrid aggregation techniques | CPath use cases | RQ2_Segmentation_Techniques, RQ5_Optimization in Healthcare Systems. |
| 33 | Data mining and machine learning in heart disease prediction [58] | Survey ML and data mining techniques for heart disease prediction | Potential overfitting in small datasets | Several ML techniques yield promising predictive performance | Prediction accuracy | Data sources, features | Investigate integration of diverse data types | Health metrics | RQ3_Multi-task Learning in Imaging |
| 21 | The role of AI in precision medicine: Applications and challenges [59] | Analyze applications of AI in precision medicine | Ethical concerns regarding bias | AI can optimize treatment strategies and improve patient outcomes | Treatment effectiveness | Patient data types, AI algorithms | Future studies to address bias and enhance model transparency | Clinical settings | RQ5_Optimization in Healthcare Systems |
| 23 | Advances in medical image analysis: A comprehensive survey [60] | Comprehensive review of recent advances in medical image analysis techniques | Limitations in the scope of reviewed studies | Highlights the importance of advanced techniques like DL in medical imaging | Image analysis outcomes | Imaging methods | Integration of Imaging and Genomic Data in Cancer Detection Using AI Models | RQ3_Multi-task learning in imaging2023 |
| Title of Paper | Objective | Limitations | Insights/Results | Dependent Variable | Independent Variables | Future Research Directions | Related RQs | Year |
|---|---|---|---|---|---|---|---|---|
| Hybrid AI-Radiomics Models for Lung Cancer Diagnosis | Evaluate the effectiveness of hybrid models combining deep learning and radiomics | Limited studies on hybrid models | Hybrid models improve diagnostic accuracy, interpretability, and robustness | Diagnostic accuracy | Deep learning, radiomics features | Explore integration of radiomics with advanced deep learning architectures | RQ1, RQ3 | 2023 |
| Challenges in Clinical Validation of AI Models for Lung Cancer Diagnosis | Identify barriers to clinical validation and propose a framework | Lack of standardized datasets and evaluation metrics | Multi-center trials and diverse datasets are essential for real-world implementation | Clinical adoption | Dataset diversity, evaluation metrics | Develop a framework emphasizing diverse datasets, multi-center trials, and standardized metrics | RQ2, RQ4 | 2023 |
| Explainable AI in Medical Imaging | Overview of XAI in deep learning for medical image analysis | Limited generalizability of findings | Framework for classifying XAI methods; future opportunities identified | XAI effectiveness | Deep learning methods | Further development of XAI techniques | RQ1, RQ5 | 2021 |
| Human Treelike Tubular Structure Segmentation | Review of datasets and algorithms for tubular structure segmentation | Potential bias in selected studies | Comprehensive dataset and algorithm review; challenges and future directions discussed | Segmentation accuracy | Imaging modalities (MRI, CT, etc.) | Exploration of new segmentation algorithms | RQ2 | 2022 |
| Multi-task Deep Learning in Medical Imaging | Summarize multi-task deep learning applications in medical imaging | Performance gaps in some tasks | Identification of popular architectures; outstanding performance noted in several areas | Medical image processing outcomes | Multiple related tasks | Addressing performance gaps in current models | RQ3 | 2023 |
| Deep Learning for Chest X-ray Analysis | Review DL applications in chest X-ray analysis | Varied quality and methodologies in studies | Categorization of tasks and datasets used in chest X-ray analysis | X-ray analysis accuracy | DL methods, types of tasks | Address gaps in dataset utilization and model applicability | RQ1, RQ2 | 2021 |
| Transformers in Medical Imaging | Review applications of Transformers in medical imaging | Complexity of implementation and adaptation from NLP | Highlights advantages of Transformers over CNNs in capturing global context for medical imaging tasks | Imaging performance | Transformer architectures, medical tasks | Address challenges in adaptation and optimization of Transformers | RQ1, RQ3 | 2023 |
| Uncertainty Quantification in AI for Healthcare | Review uncertainty techniques in AI models for healthcare | Scarcity of studies on physiological signals | Highlights the importance of uncertainty quantification for reliable medical predictions and decisions | Prediction accuracy | AI models (Bayesian, Fuzzy, etc.) | Investigate uncertainty quantification in physiological signals | RQ4, RQ5 | 2023 |
| Graph Neural Networks in Computational Histopathology | Examine the use of graph neural networks in histopathological analysis | Limited understanding of contextual feature extraction | Summarizes clinical applications and proposes improved graph construction methods | Diagnostic accuracy | Graph neural networks | Further research on model generalization in histopathology | RQ3, RQ4 | 2023 |
| Recent Advances in Deep Learning for Medical Imaging | Summarize recent advances in deep learning for medical imaging tasks | Lack of large annotated datasets | Reviews the effectiveness of deep learning techniques in various medical imaging applications | Imaging performance | Deep learning models | Address dataset limitations and enhance model robustness | RQ1, RQ2 | 202 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





