Submitted:
18 March 2025
Posted:
20 March 2025
Read the latest preprint version here
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. DNA Synthesis and Engineering
2.2. Strains and Vectors
2.3. Enzymatic Activities
3. Results
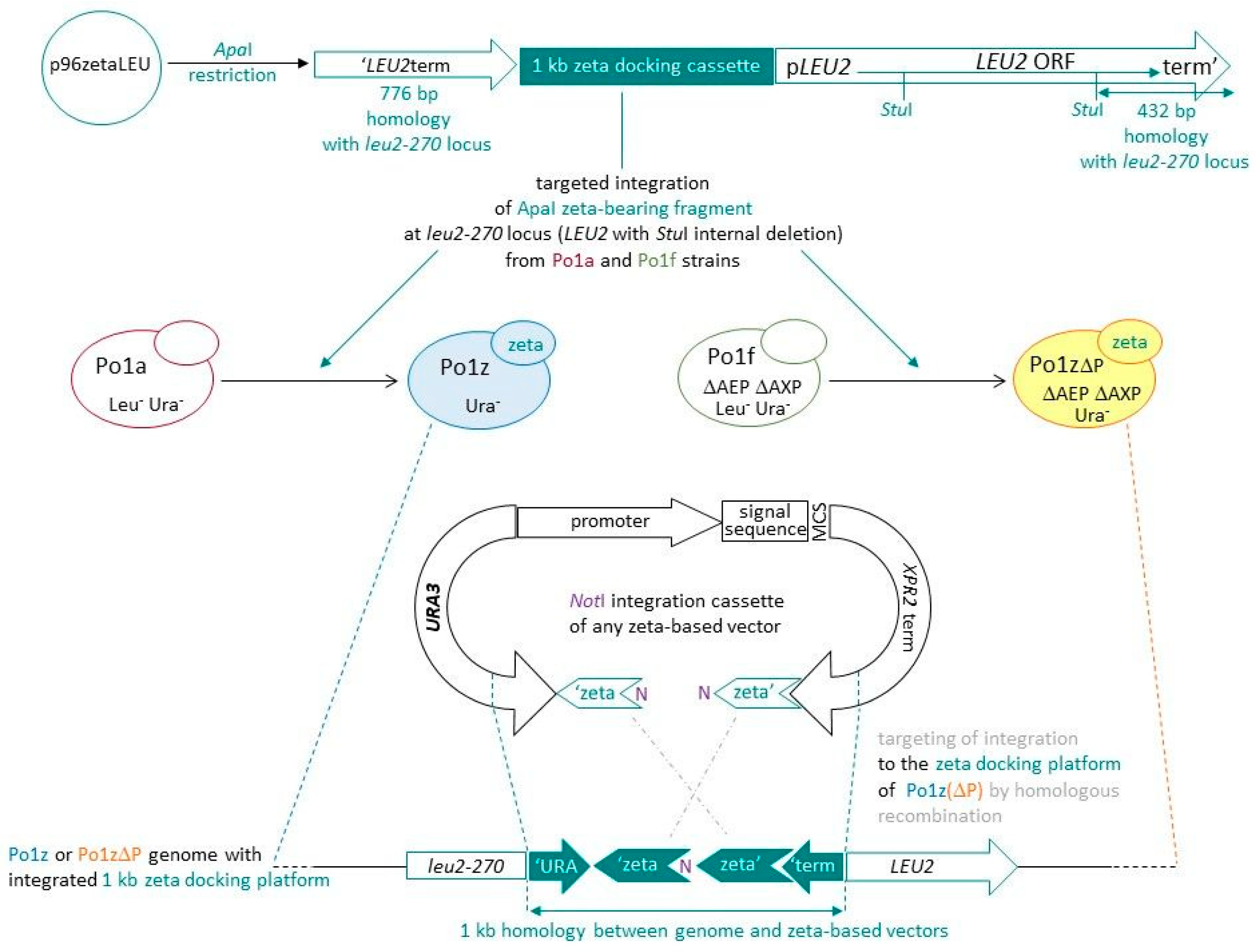
3.1. Design of Strains Equipped with a Zeta Docking Platform
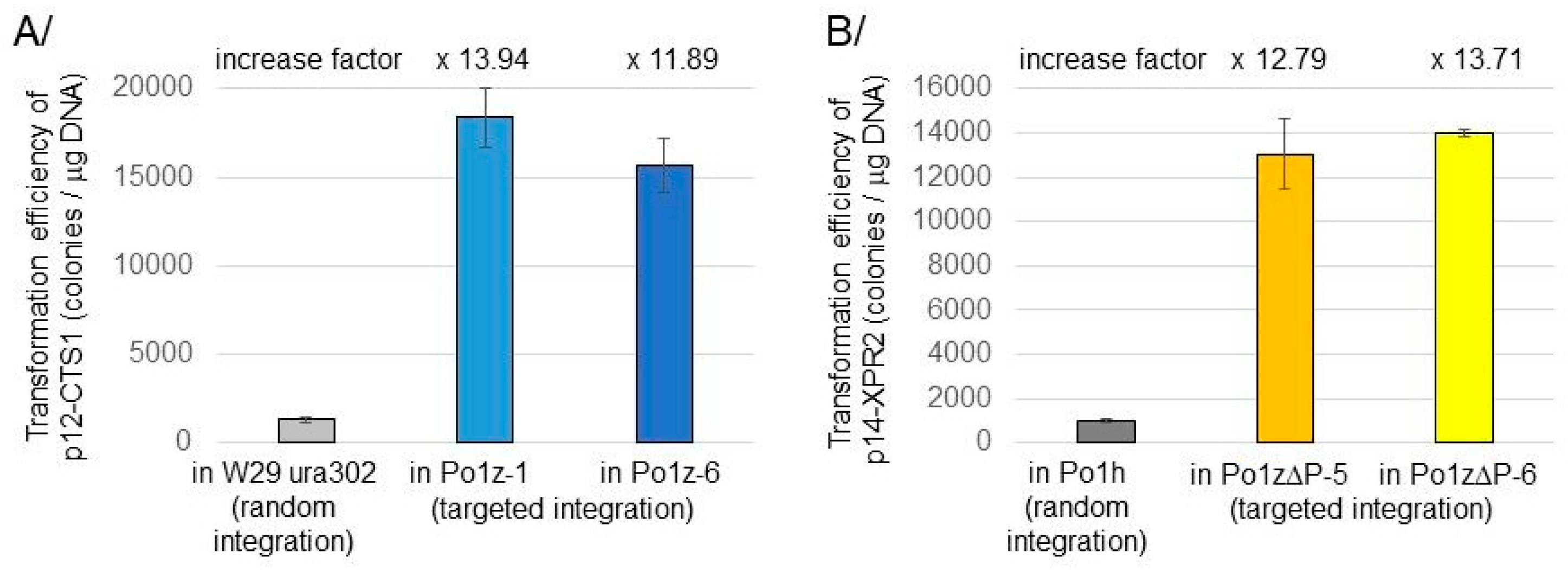
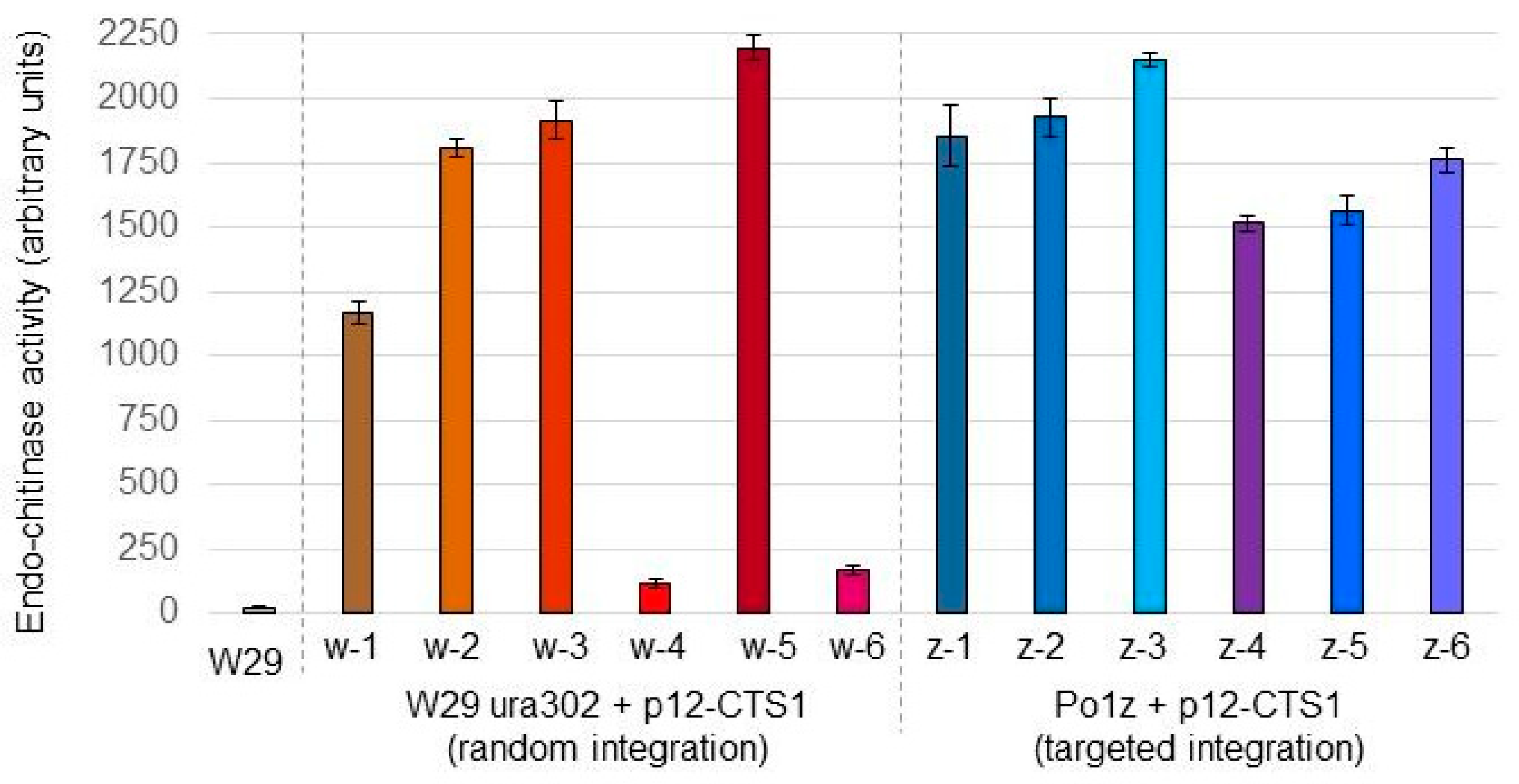
3.2. Validation of New Po1z and Po1zΔP Strains
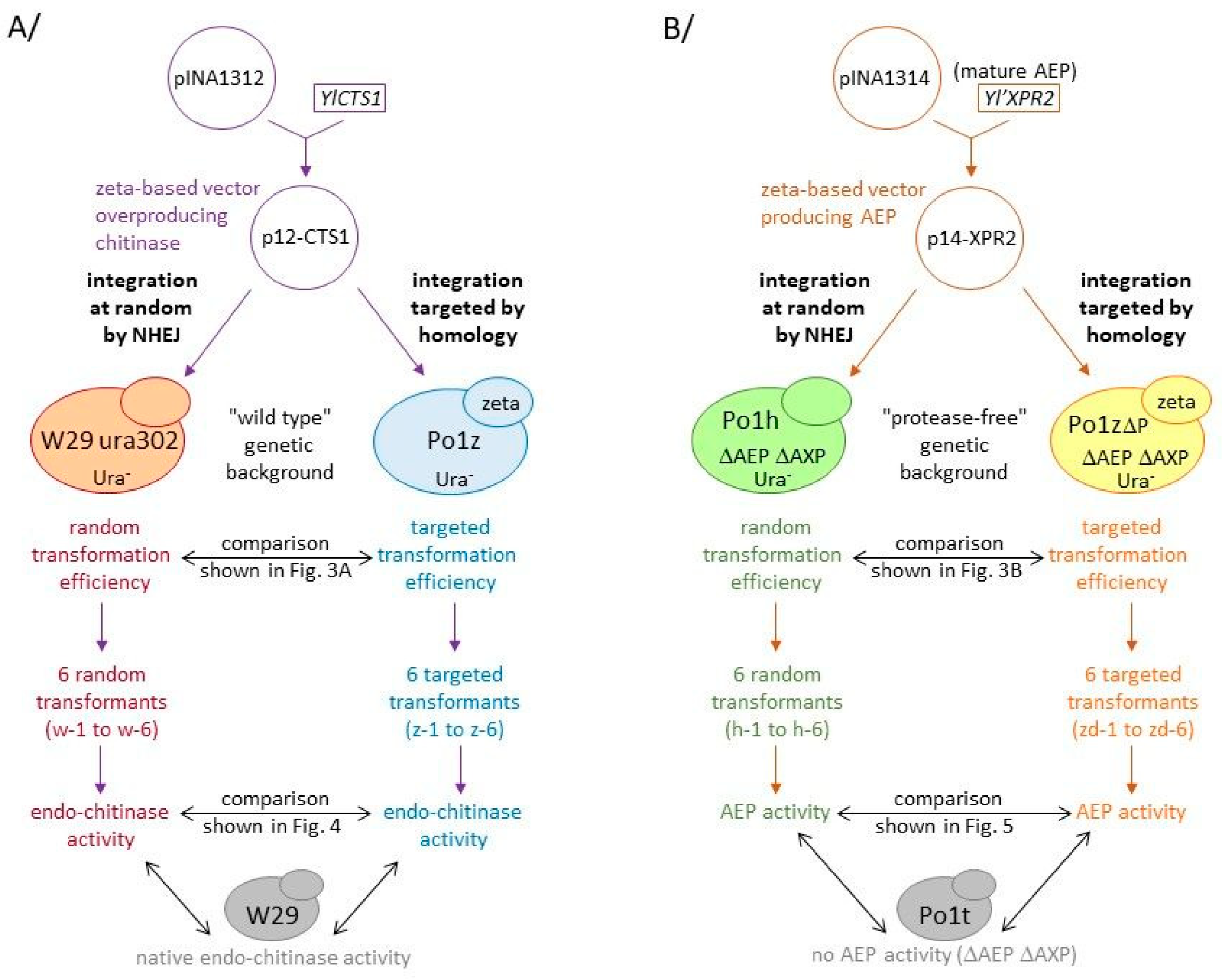
3.3. Design of New Zeta-Based Vectors
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 4MU AEP CRISPR C-term. DNA FDA GM GRAS |
4-Methylumbelliferyl β-D-N,N',N''-triacetylchitotriose Alkaline Extracellular Protease Clustered Regularly Interspaced Short Palindromic Repeats C-terminal extremity of the corresponding protein Deoxyribonucleic acid Food and Drug Administration Genetically Modified Generally Regarded as Safe |
| LTR | Long Terminal Repeats |
| MCS | Multiple Cloning Site |
| NHEJ ORF p…. term. UAS |
Non Homologous End Joining Open reading Frame Promoter of the corresponding gene Terminator Upstream Activating Sequence |
References
- Groenewald, M.; Boekhout, T.; Neuvéglise, C.; Gaillardin, C.; van Dijck, P.W.; Wyss, M. Yarrowia lipolytica: safety assessment of an oleaginous yeast with a great industrial potential. Crit. Rev. Microbiol. 2014, 40, 187–206. [Google Scholar] [CrossRef] [PubMed]
- Madzak, C. Yarrowia lipolytica Strains and Their Biotechnological Applications: How Natural Biodiversity and Metabolic Engineering Could Contribute to Cell Factories Improvement. J. Fungi (Basel) 2021, 7, 548. [Google Scholar] [CrossRef] [PubMed]
- Bankar, A.V.; Kumar, A.R.; Zinjarde, S.S. Environmental and industrial applications of Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2009, 84, 847–865. [Google Scholar] [CrossRef]
- Liu, H.H.; Ji, X.J.; Huang, H. Biotechnological applications of Yarrowia lipolytica: Past, present and future. Biotechnol. Adv. 2015, 33, 1522–1546. [Google Scholar] [CrossRef]
- Zhu, Q.; Jackson, E.N. Metabolic engineering of Yarrowia lipolytica for industrial applications. Curr. Opin. Biotechnol. 2015, 36, 65–72. [Google Scholar] [CrossRef]
- Madzak, C. Engineering Yarrowia lipolytica for Use in Biotechnological Applications: A Review of Major Achievements and Recent Innovations. Mol. Biotechnol. 2018, 60, 621–635. [Google Scholar] [CrossRef]
- Miller, K.K.; Alper, H.S. Yarrowia lipolytica: more than an oleaginous workhorse. Appl. Microbiol. Biotechnol. 2019, 103, 9251–9262. [Google Scholar] [CrossRef]
- Park, Y.K.; Ledesma-Amaro, R. What makes Yarrowia lipolytica well suited for industry? Trends Biotechnol. 2023, 41, 242–254. [Google Scholar] [CrossRef]
- Sun, M.L.; Gao, X.; Lin, L.; Yang, J.; Ledesma-Amaro, R.; Ji, X.J. Building Yarrowia lipolytica Cell Factories for Advanced Biomanufacturing: Challenges and Solutions. J. Agric. Food Chem. 2024, 72, 94–107. [Google Scholar] [CrossRef]
- Dujon, B.; Sherman, D.; Fischer, G.; Durrens, P.; Casaregola, S.; Lafontaine, I.; De Montigny, J.; Marck, C.; Neuveglise, C.; Talla, E.; et al. Genome evolution in yeasts. Nature 2004, 430, 35–44. [Google Scholar] [CrossRef]
- Bigey, F.; Pasteur, E.; Połomska, X.; Thomas, S.; Crutz-Le Coq, A.M.; Devillers, H.; Neuvéglise, C. Insights into the Genomic and Phenotypic Landscape of the Oleaginous Yeast Yarrowia lipolytica. J. Fungi (Basel). 2023, 9, 76. [Google Scholar] [CrossRef] [PubMed]
- Magnan, C.; Yu, J.; Chang, I.; Jahn, E.; Kanomata, Y.; Wu, J.; Zeller, M.; Oakes, M.; Baldi, P.; Sandmeyer, S. Sequence Assembly of Yarrowia lipolytica Strain W29/CLIB89 Shows Transposable Element Diversity. PLoS ONE 2016, 11, e0162363. [Google Scholar] [CrossRef]
- Liu, L.; Alper, H.S. Draft Genome Sequence of the Oleaginous Yeast Yarrowia lipolytica PO1f, a Commonly Used Metabolic Engineering Host. Genome Announc. 2014, 2, e00652-14. [Google Scholar] [CrossRef]
- Madzak, C. Yarrowia lipolytica engineering as a source of microbial cell factories. In Microbial Cell Factories Engineering for Production of Biomolecules; Singh, V., Ed.; Academic Press: Cambridge, MA, USA, 2021; pp. 345–380. [Google Scholar]
- Georgiadis, I.; Tsiligkaki, C.; Patavou, V.; Orfanidou, M.; Tsoureki, A.; Andreadelli, A.; Theodosiou, E.; Makris, A.M. Identification and Construction of Strong Promoters in Yarrowia lipolytica Suitable for Glycerol-Based Bioprocesses. Microorganisms. 2023, 11, 1152. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, S.; Lu, Z.; Zhao, B.; Wang, S.; Zhang, C.; Xiao, D.; Foo, J.L.; Yu, A. Hybrid promoter engineering strategies in Yarrowia lipolytica: isoamyl alcohol production as a test study. Biotechnol. Biofuels. 2021, 14, 149. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Zheng, H.; Zhang, J.; Jiang, Z.; Zhu, Z.; Liu, X.; Qi, Q.; Hou, J. A CRISPR/Cas9-Mediated, Homology-Independent Tool Developed for Targeted Genome Integration in Yarrowia lipolytica. Appl. Environ. Microbiol. 2021, 87, e02666-20. [Google Scholar] [CrossRef]
- Liu, X.; Cui, Z.; Su, T.; Lu, X.; Hou, J.; Qi, Q. Identification of genome integration sites for developing a CRISPR-based gene expression toolkit in Yarrowia lipolytica. Microb. Biotechnol. 2022, 15, 2223–2234. [Google Scholar] [CrossRef]
- Li, Y.W.; Yang, C.L.; Shen, Q.; Peng, Q.Q.; Guo, Q.; Nie, Z.K.; Sun, X.M.; Shi, T.Q.; Ji, X.J.; Huang, H. YALIcloneNHEJ: An Efficient Modular Cloning Toolkit for NHEJ Integration of Multigene Pathway and Terpenoid Production in Yarrowia lipolytica. Front. Bioeng. Biotechnol. 2022, 9, 816980. [Google Scholar] [CrossRef]
- Shen, Q.; Yan, F.; Li, Y.W.; Wang, J.; Ji, J.; Yan, W.X.; He, D.C.; Song, P.; Shi, T.Q. Expansion of YALIcloneHR toolkit for Yarrowia lipolytica combined with Golden Gate and CRISPR technology. Biotechnol. Lett. 2024, 46, 37–46. [Google Scholar] [CrossRef]
- Luo, Z.; Shi, J.T.; Chen, X.L.; Chen, J.; Liu, F.; Wei, L.J.; Hua, Q. Iterative gene integration mediated by 26S rDNA and non-homologous end joining for the efficient production of lycopene in Yarrowia lipolytica. Bioresour. Bioprocess. 2023, 10, 83. [Google Scholar] [CrossRef]
- Zhang, B.; Cao, J. Improving and Streamlining Gene Editing in Yarrowia lipolytica via Integration of Engineered Cas9 Protein. J. Fungi (Basel). 2024, 10, 63. [Google Scholar] [CrossRef]
- Gorczyca, M.; Białas, W.; Nicaud, J.M.; Celińska, E. 'Mother(Nature) knows best' - hijacking nature-designed transcriptional programs for enhancing stress resistance and protein production in Yarrowia lipolytica; presentation of YaliFunTome database. Microb. Cell Fact. 2024, 23, 26. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Su, L.; Liu, Q.; Zhu, Y.; Dai, Z.; Wang, Q. Dissecting carbon metabolism of Yarrowia lipolytica type strain W29 using genome-scale metabolic modelling. Comput. Struct. Biotechnol. J. 2022, 20, 2503–2511. [Google Scholar] [CrossRef] [PubMed]
- Madzak, C.; Tréton, B.; Blanchin-Roland, S. Strong hybrid promoters and integrative expression/secretion vectors for quasiconstitutive expression of heterologous proteins in the yeast Yarrowia lipolytica. J. Mol. Microbiol. Biotechnol. 2000, 2, 207–216. [Google Scholar]
- Nicaud, J.M.; Madzak, C.; van den Broek, P.; Gysler, C.; Duboc, P.; Niederberger, P.; Gaillardin, C. Protein expression and secretion in the yeast Yarrowia lipolytica. FEMS Yeast Res. 2002, 2, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Madzak, C. New tools for heterologous protein production in the yeast Yarrowia lipolytica. In Recent Research Developments in Microbiology; Pandalai, S.G., Ed.; Research Signpost: Trivandrum, India, 2003; Volume 7, pp. 453–479. [Google Scholar]
- Yue, L.; Chi, Z.; Wang, L.; Liu, J.; Madzak, C.; Li, J.; Wang, X. Construction of a new plasmid for surface display on cells of Yarrowia lipolytica. J. Microbiol. Methods 2008, 72, 116–123. [Google Scholar] [CrossRef]
- Kopecný, D.; Pethe, C.; Sebela, M.; Houba-Hérin, N.; Madzak, C.; Majira, A.; Laloue, M. High-level expression and characterization of Zea mays cytokinin oxidase/dehydrogenase in Yarrowia lipolytica. Biochimie 2005, 87, 1011–1022. [Google Scholar] [CrossRef]
- Madzak, C.; Mimmi, M.C.; Caminade, E.; Brault, A.; Baumberger, S.; Briozzo, P.; Mougin, C.; Jolivalt, C. Shifting the optimal pH of activity for a laccase from the fungus Trametes versicolor by structure-based mutagenesis. Protein Eng. Des. Sel. 2006, 9, 77–84. [Google Scholar] [CrossRef]
- Galli, C.; Gentili, P.; Jolivalt, C.; Madzak, C.; Vadalà, R. How is the reactivity of laccase affected by single-point mutations? Engineering laccase for improved activity towards sterically demanding substrates. Appl. Microbiol. Biotechnol. 2011, 91, 123–131. [Google Scholar] [CrossRef]
- Nicaud, J.M.; Fabre, E.; Gaillardin, C. Expression of invertase activity in Yarrowia lipolytica and its use as a selective marker. Curr. Genet. 1989, 16, 253–260. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Meth. 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Blazeck, J.; Liu, L.; Redden, H.; Alper, H. Tuning gene expression in Yarrowia lipolytica by a hybrid promoter approach. Appl. Environ. Microbiol. 2011, 77, 7905–7914. [Google Scholar] [CrossRef] [PubMed]
- Blazeck, J.; Reed, B.; Garg, R.; Gerstner, R.; Pan, A.; Agarwala, V.; Alper, H.S. Generalizing a hybrid synthetic promoter approach in Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2013, 97, 3037–3052. [Google Scholar] [CrossRef] [PubMed]
- Dulermo, R.; Brunel, F.; Dulermo, T.; Ledesma-Amaro, R.; Vion, J.; Trassaert, M.; Thomas, S.; Nicaud, J.M.; Leplat, C. Using a vector pool containing variable-strength promoters to optimize protein production in Yarrowia lipolytica. Microb. Cell Fact. 2017, 16, 31. [Google Scholar] [CrossRef]

|
Strain name * Origin |
Genotype Phenotype |
Reference number at CIRM-Levures (and other microorganism collections) |
| W29 French natural isolate [33] |
MatA Wild type |
CLIB 89 (ATCC 20460, CBS 7504, CICC 1778, NBRC 113670, NRLL Y-3178, VKPM Y-3178) |
| W29_ura302 GM from W29 [2] |
MatA, ura3-302 *§ Ura-, Suc+ |
CLIB 141 |
| Po1a GM from W29_ura302 [2] |
MatA, leu2-270, ura3-302 Leu-, Ura-, Suc+ |
CLIB 140 |
|
Po1z GM from Po1a |
MatA, leu2-270::LEU2-zeta, ura3-302 Ura-, Suc+, zeta docking platform |
CLIB 4231 |
| Po1f GM from Po1a [25] |
MatA, leu2-270, ura3-302, xpr2-322, axp1-2 Leu-, Ura-, Suc+, ΔAEP, ΔAXP |
CLIB 724 (ATCC MYA2613, VKPM Y-3155) |
|
Po1zΔP GM from Po1f |
MatA, leu2-270::LEU2-zeta, ura3-302, xpr2-322, axp1-2 Ura-, Suc+, ΔAEP, ΔAXP, zeta docking platform |
CLIB 4232 |
| Po1h GM from Po1f [27] |
MatA, ura3-302, xpr2-322, axp1-2 Ura-, Suc+, ΔAEP, ΔAXP |
CLIB 882 |
| Po1t GM from Po1f [27] |
MatA, leu2-270, LEU2, ura3-302::URA3, xpr2-322, axp1-2 Suc+, ΔAEP, ΔAXP |
CLIB 883 |
| Vector * |
Promoter Secretion signal (Surface display signal) |
Upstream / Downstream cloning sites |
Terminator |
CIRM-Levures reference number |
| pINA1311 [26] | hp4d § no signal |
PmlI (blunt) / BamHI, KpnI, AvrII |
LIP2 term. | 1222032 |
| pINA1312 [26] | hp4d no signal |
PmlI (blunt) / BamHI, KpnI |
XPR2 term. | 1222033 |
|
pINA1312-8UASs aka pINA2008 |
hp8d no signal |
PmlI (blunt) / BamHI, KpnI |
XPR2 term. | 1223001 |
|
pINA1312-12UASs aka pINA2012 |
hp12d no signal |
PmlI (blunt) / BamHI, KpnI |
XPR2 term. | 1223002 |
| pINA1313 [26] | hp4d LIP2 prepro |
XmnI (in pro) / BamHI, KpnI, AvrII |
LIP2 term | 1222034 |
| pINA1314 [26] | hp4d XPR2 prepro |
SfiI (in pro) / KpnI |
XPR2 term. | 1222035 |
| pINA1317 [26] | hp4d XPR2 pre |
SfiI (in pre) / AvrII, BamHI, KpnI |
XPR2 term. | 1222036 |
|
pINA1317-8UASs aka pINA7008 |
hp8d XPR2 pre |
SfiI (in pre) / AvrII, BamHI, KpnI |
XPR2 term. | 1223003 |
|
pINA1317-12UASs aka pINA7012 |
hp12d XPR2 pre |
SfiI (in pre) / AvrII, BamHI, KpnI |
XPR2 term. | 1223004 |
| pINA1317-YlCWP110 aka pINA1710, pSD1 [28] |
hp4d XPR2 pre (CWP1 C-term. 110 bp) |
SfiI (in pre) / KpnI |
XPR2 term. | 1222256 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





