Submitted:
15 March 2025
Posted:
19 March 2025
Read the latest preprint version here
Abstract
Keywords:
1. Introduction
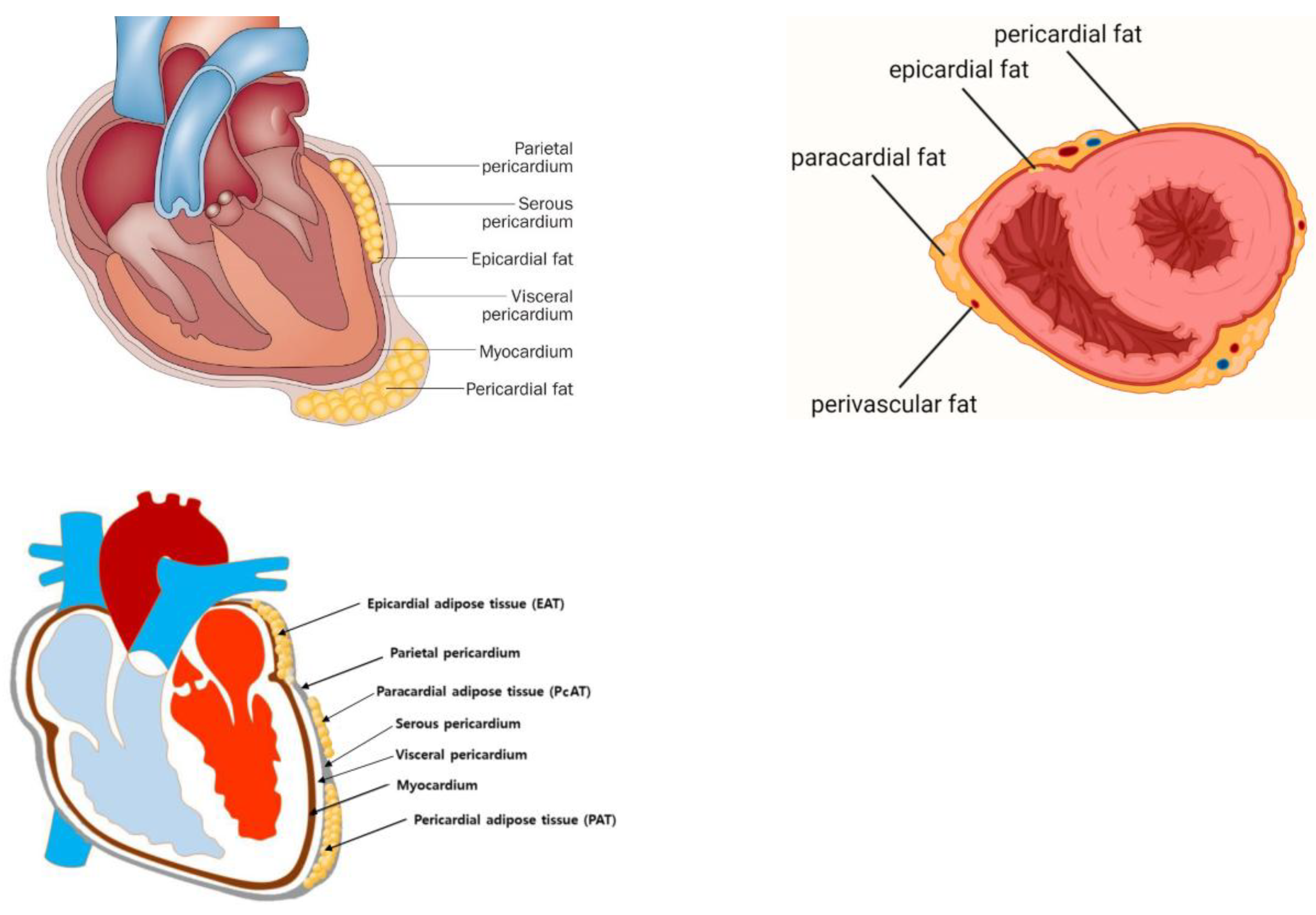
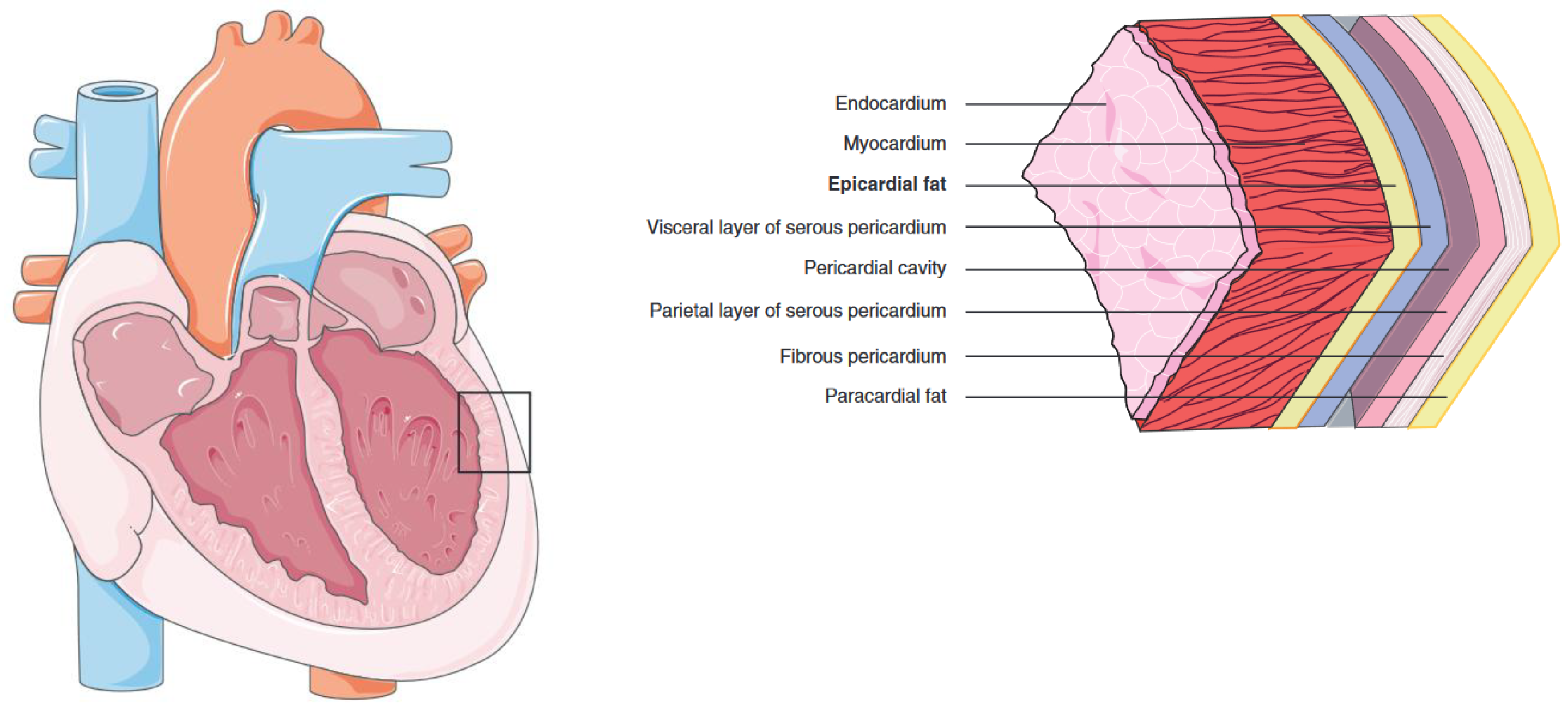
2. Cardiac Tissue Layers and Adipose Depots
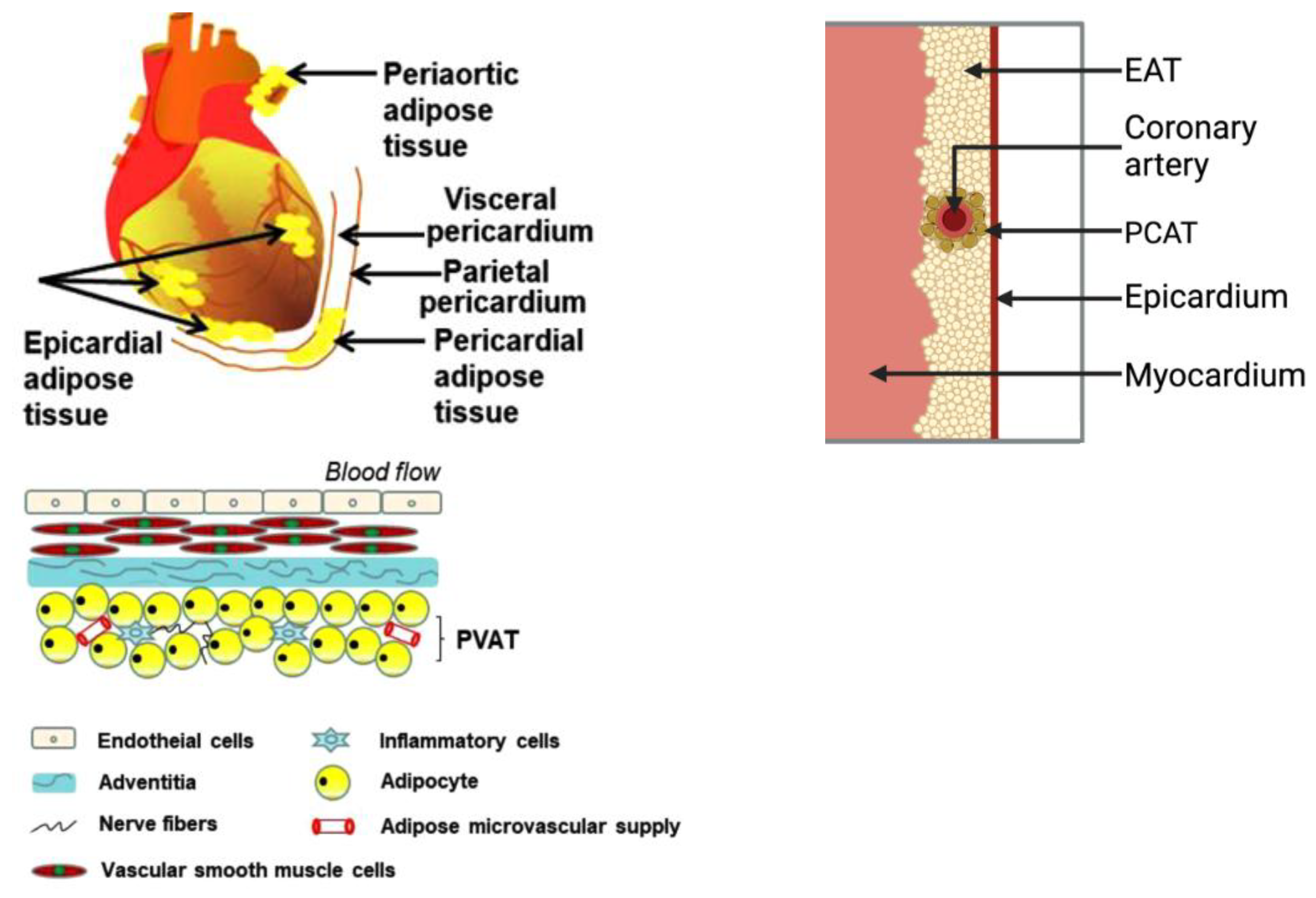
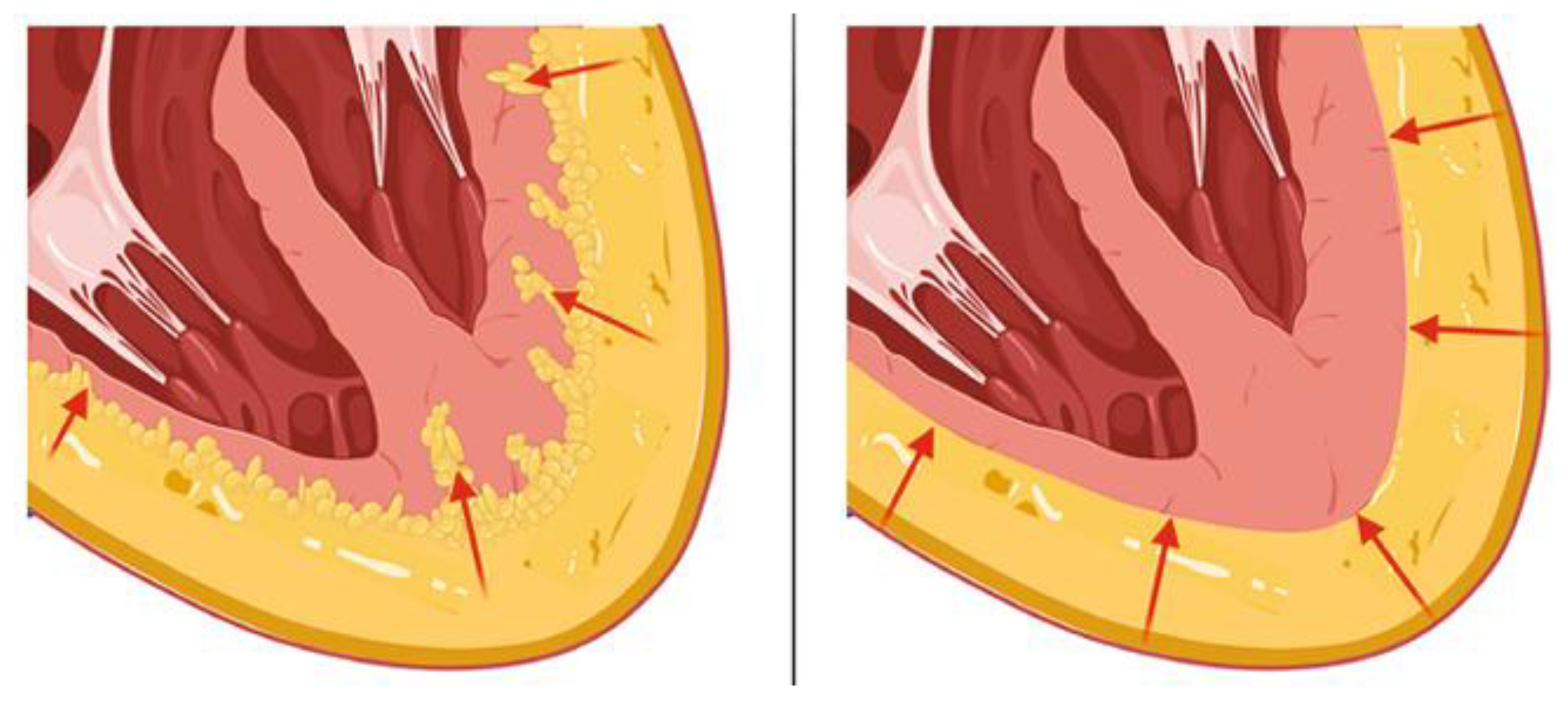
2.1. Epicardial Adipose Tissue
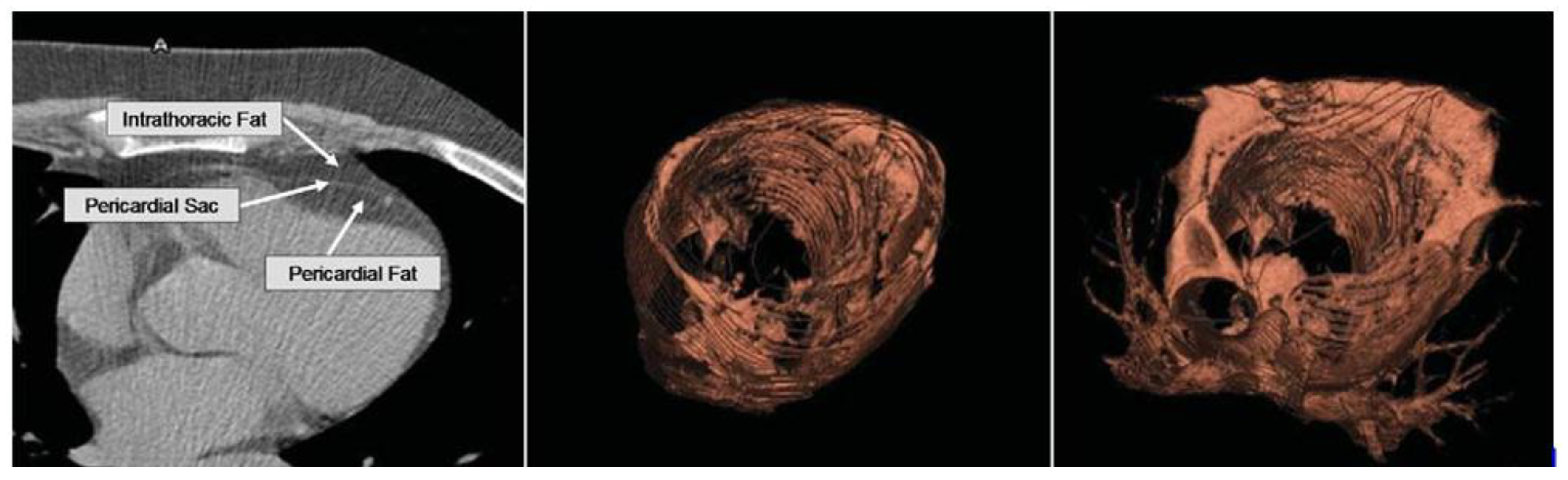
2.2. Pericardial Adipose Tissue
2.3. Paracardial Adipose Tissue
2.4. Perivascular Adipose Tissue
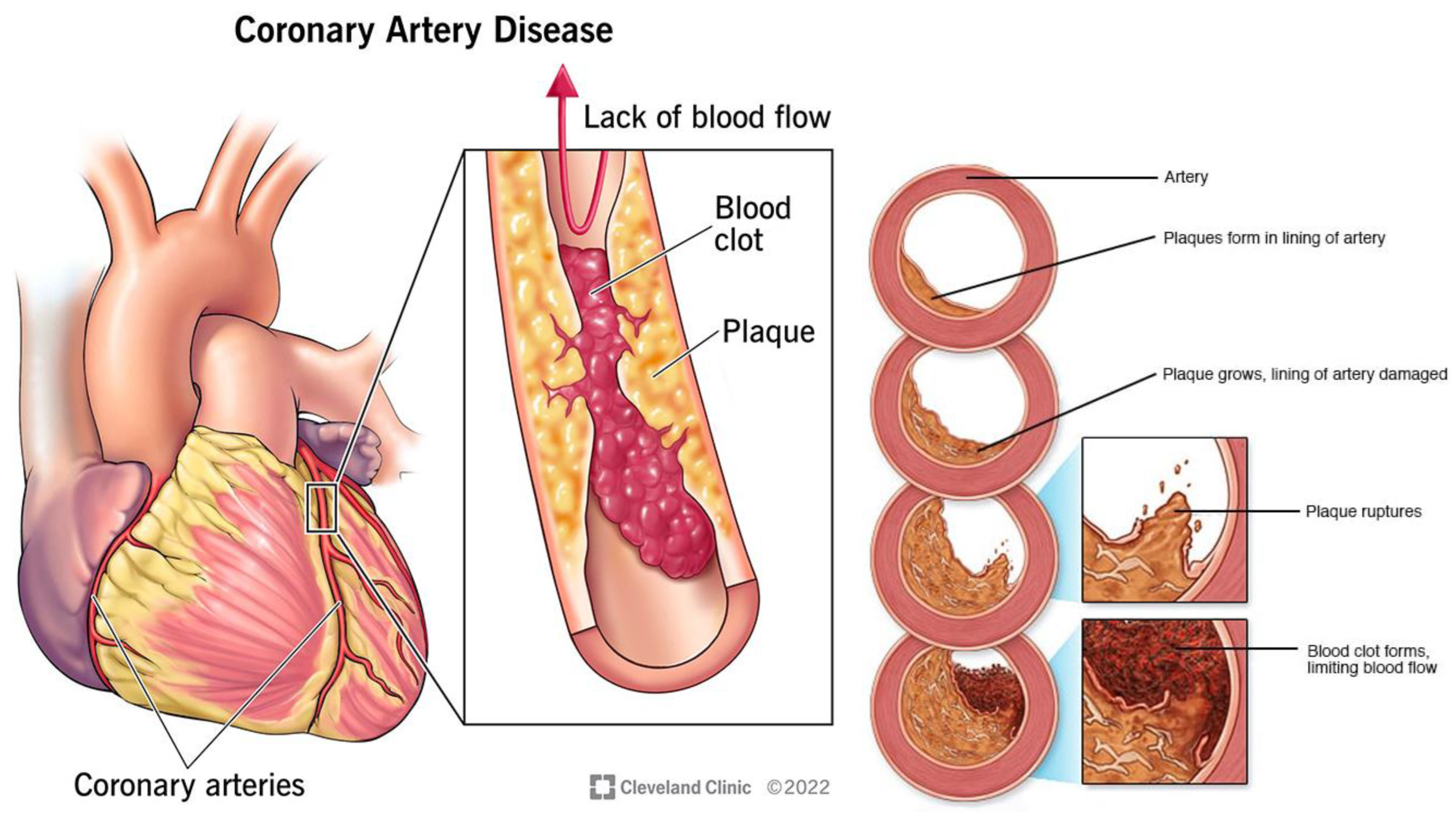
3. Cardiac Fat and Cardiovascular Disease
3.1. Coronary Artery Disease
3.2. Heart Failure (HF)
3.3. Atrial Fibrillation
3.4. Ischemic Heart Disease
3.5. Heart Valve Stenosis
3.6. Cardiac Steatosis
3.7. Cardiac Fibrosis
3.8. Cardiac Lipoma
4. Imaging Modalities to Assess CAT
4.1. Assessment Metrics
4.2. Cardiac Magnetic Resonance (CMR)
4.2.1. Imaging Techniques
4.2.2. CAT Segmentation
4.2.3. CAT Quantification
4.2.4. Limitations
4.3. Computed Tomography (CT scan)
4.3.1. Imaging Techniques
4.3.2. CAT Segmentation
4.3.3. CAT Quantification
4.3.4. Limitations
4.4. Echocardiography or Ultrasound (US)
4.4.1. Imaging Techniques
4.4.2. CAT Segmentation
4.4.3. CAT Quantification
4.4.4. Limitations
5. Conclusions
Funding
Conflicts of Interest
References
- Kim, I.K.; Song, B.W.; Lim, S.; Kim, S.W.; Lee, S. The Role of Epicardial Adipose Tissue-Derived MicroRNAs in the Regulation of Cardiovascular Disease: A Narrative Review. Biology. 2023, 12, 498. [Google Scholar] [CrossRef]
- Adipose Tissue: What Is It, Location, Function, and More | Osmosis. Available online: https://www.osmosis.org/answers/adipose-tissue (accessed on 6 June 2024).
- Sacks, H.; Symonds, M.E. Anatomical Locations of Human Brown Adipose Tissue: Functional Relevance and Implications in Obesity and Type 2 Diabetes. Diabetes. 2013, 62, 1783–1790. [Google Scholar] [CrossRef]
- Farese, R.V.; Walther, T.C. Lipid Droplets Finally Get a Little R-E-S-P-E-C-T. Cell. 2009, 139, 855–860. [Google Scholar] [CrossRef]
- Gaborit, B.; Sengenes, C.; Ancel, P.; Jacquier, A.; Dutour, A. Role of Epicardial Adipose Tissue in Health and Disease: A Matter of Fat? Comprehensive Physiology. 2017, 7, 1051–1082. [Google Scholar] [CrossRef]
- Iacobellis, G. Local and systemic effects of the multifaceted epicardial adipose tissue depot. Nat Rev Endocrinol. 2015, 11, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Cheładze, P.; Martuszewski, A.; Poręba, R.; Gać, P. The Importance of the Assessment of Epicardial Adipose Tissue in Scientific Research. J Clin Med. 2022, 11, 5621. [Google Scholar] [CrossRef]
- Konwerski, M.; Gąsecka, A.; Opolski, G.; Grabowski, M.; Mazurek, T. Role of Epicardial Adipose Tissue in Cardiovascular Diseases: A Review. Biology. 2022, 11, 355. [Google Scholar] [CrossRef] [PubMed]
- Mazurek, T.; Zhang, L.; Zalewski, A.; et al. Human Epicardial Adipose Tissue Is a Source of Inflammatory Mediators. Circulation. 2003, 108, 2460–2466. [Google Scholar] [CrossRef]
- Chhabra, L.; Gurukripa Kowlgi, N. Cardiac adipose tissue: Distinction between epicardial and pericardial fat remains important! International Journal of Cardiology. 2015, 201, 274–275. [Google Scholar] [CrossRef]
- Krauz, K.; Kempiński, M.; Jańczak, P.; et al. The Role of Epicardial Adipose Tissue in Acute Coronary Syndromes, Post-Infarct Remodeling and Cardiac Regeneration. International Journal of Molecular Sciences. 2024, 25, 3583. [Google Scholar] [CrossRef]
- Silaghi, A.; Piercecchi-Marti, M.D.; Grino, M.; et al. Epicardial adipose tissue extent: Relationship with age, body fat distribution, and coronaropathy. Obesity (Silver Spring). 2008, 16, 2424–2430. [Google Scholar] [CrossRef] [PubMed]
- Rhee, T.M.; Lee, J.H.; Choi, E.K.; et al. Increased Risk of Atrial Fibrillation and Thromboembolism in Patients with Severe Psoriasis: A Nationwide Population-based Study. Sci Rep. 2017, 7, 9973. [Google Scholar] [CrossRef]
- Gaeta, M.; Bandera, F.; Tassinari, F.; et al. Is epicardial fat depot associated with atrial fibrillation? A systematic review and meta-analysis. Europace. 2017, 19, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Wang, H.; Chen, J.; Zhao, L. Epicardial adipose tissue and atrial fibrillation: Possible mechanisms, potential therapies, and future directions. Pacing Clin Electrophysiol. 2020, 43, 133–145. [Google Scholar] [CrossRef]
- Xiao, J.; Lu, Y.; Yang, X. Ultrasound Detection of Epicardial Adipose Tissue Combined With Ischemic Modified Albumin in the Diagnosis of Coronary Heart Disease. The Heart Surgery Forum. 2020, 23, E461–E464. [Google Scholar] [CrossRef] [PubMed]
- Abdulkareem, M.; Brahier, M.S.; Zou, F.; et al. Quantification of Epicardial Adipose Tissue Volume and Attenuation for Cardiac CT Scans Using Deep Learning in a Single Multi-Task Framework. Rev Cardiovasc Med. 2022, 23, 412. [Google Scholar] [CrossRef]
- Iacobellis, G. Epicardial adipose tissue in contemporary cardiology. Nat Rev Cardiol. 2022, 19, 593–606. [Google Scholar] [CrossRef]
- Mahmoud, I.; Dykun, I.; Kärner, L.; et al. Epicardial adipose tissue differentiates in patients with and without coronary microvascular dysfunction. Int J Obes. 2021, 45, 2058–2063. [Google Scholar] [CrossRef]
- Song, Y.; Tan, Y.; Deng, M.; et al. Epicardial adipose tissue, metabolic disorders, and cardiovascular diseases: Recent advances classified by research methodologies. MedComm. 2023, 4, e413. [Google Scholar] [CrossRef]
- van Woerden, G.; van Veldhuisen, D.J.; Westenbrink, B.D.; de Boer, R.A.; Rienstra, M.; Gorter, T.M. Connecting epicardial adipose tissue and heart failure with preserved ejection fraction: Mechanisms, management and modern perspectives. European Journal of Heart Failure. 2022, 24, 2238–2250. [Google Scholar] [CrossRef]
- Si, Y.; Feng, Z.; Liu, Y.; et al. Inflammatory biomarkers, angiogenesis and lymphangiogenesis in epicardial adipose tissue correlate with coronary artery disease. Sci Rep. 2023, 13, 2831. [Google Scholar] [CrossRef]
- Drake, R.; Vogl, A.W.; Mitchell, A.W.M. Gray’s Anatomy for Students E-Book. Elsevier Health Sciences; 2014.
- Stauffer, C.M.; Meshida, K.; Bernor, R.L.; Granite, G.E.; Boaz, N.T. Anatomy, Thorax, Pericardiacophrenic Vessels. In: StatPearls. StatPearls Publishing; 2024. Available online: http://www.ncbi.nlm.nih.gov/books/NBK559242/ (accessed on 20 June 2024).
- Ding, J.; Kritchevsky, S.B.; Harris, T.B.; et al. The Association of Pericardial Fat With Calcified Coronary Plaque. Obesity. 2008, 16, 1914–1919. [Google Scholar] [CrossRef] [PubMed]
- Pericardial Rather Than Epicardial Fat is a Cardiometabolic Risk Marker: An MRI vs Echo Study - ClinicalKey. Available online: https://www.clinicalkey.com/#!/content/playContent/1-s2.0-S0894731711004718?returnurl=https:%2F%2Flinkinghub.elsevier.com%2Fretrieve%2Fpii%2FS0894731711004718%3Fshowall%3Dtrue&referrer= (accessed on 19 June 2024).
- Yamaguchi, Y.; Cavallero, S.; Patterson, M.; et al. Adipogenesis and epicardial adipose tissue: A novel fate of the epicardium induced by mesenchymal transformation and PPARγ activation. Proceedings of the National Academy of Sciences. 2015, 112, 2070–2075. [Google Scholar] [CrossRef]
- Mahabadi, A.A.; Massaro, J.M.; Rosito, G.A.; et al. Association of pericardial fat, intrathoracic fat, and visceral abdominal fat with cardiovascular disease burden: The Framingham Heart Study. Eur Heart, J. 2009, 30, 850–856. [Google Scholar] [CrossRef]
- Rodrigues, É.O. On the Automated Segmentation of Epicardial and Mediastinal Cardiac Adipose Tissues Using Classification Algorithms.
- Zhang, P.; Konja, D.; Wang, Y. Adipose tissue secretory profile and cardiometabolic risk in obesity. Endocrine and Metabolic Science. 2020, 1, 100061. [Google Scholar] [CrossRef]
- Grigoras, A.; Amalinei, C.; Balan, R.A.; Giusca, S.E.; Caruntu, I.D. Perivascular adipose tissue in cardiovascular diseases-an update. Anatol J Cardiol. 2019, 22, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Tran, K.V.; Fitzgibbons, T.; Min, S.Y.; DeSouza, T.; Corvera, S. Distinct adipocyte progenitor cells are associated with regional phenotypes of perivascular aortic fat in mice. Molecular Metabolism. 2018, 9, 199–206. [Google Scholar] [CrossRef]
- van Dam, A.D.; Boon, M.R.; Berbée, J.F.P.; Rensen, P.C.N.; van Harmelen, V. Targeting white, brown and perivascular adipose tissue in atherosclerosis development. European Journal of Pharmacology. 2017, 816, 82–92. [Google Scholar] [CrossRef]
- Virdis, A.; Duranti, E.; Rossi, C.; et al. Tumour necrosis factor-alpha participates on the endothelin-1/nitric oxide imbalance in small arteries from obese patients: Role of perivascular adipose tissue. Eur Heart J. 2015, 36, 784–794. [Google Scholar] [CrossRef]
- Withers, S.B.; Bussey, C.E.; Saxton, S.N.; Melrose, H.M.; Watkins, A.E.; Heagerty, A.M. Mechanisms of adiponectin-associated perivascular function in vascular disease. Arterioscler Thromb Vasc Biol. 2014, 34, 1637–1642. [Google Scholar] [CrossRef]
- Association, A.H.; American Stroke Association. Cardiovascular Disease: A Costly Burden for America Projections Through 2035. Available online: chrome-extension://efaidnbmnnnibpcajpcglclefindmkaj/https://www.heart.org/-/media/Files/About-Us/Policy-Research/Fact-Sheets/Public-Health-Advocacy-and-Research/CVD-A-Costly-Burden-for-America-Projections-Through-2035.pdf.
- Milewicz, D.M.; Seidman, C.E. Genetics of Cardiovascular Disease. Circulation. 2000, 102 (Suppl. S4), Iv-103. [Google Scholar] [CrossRef]
- Nerlekar, N.; Thakur, U.; Lin, A.; et al. The Natural history of Epicardial Adipose Tissue Volume and Attenuation: A long-term prospective cohort follow-up study. Sci Rep. 2020, 10, 7109. [Google Scholar] [CrossRef] [PubMed]
- What’s Draggin’ Your Heart Down? Cleveland Clinic. Available online: https://my.clevelandclinic.org/health/diseases/16898-coronary-artery-disease (accessed on 29 May 2024).
- Coronary artery disease - Symptoms and causes. Mayo Clinic. Available online: https://www.mayoclinic.org/diseases-conditions/coronary-artery-disease/symptoms-causes/syc-20350613 (accessed on 29 May 2024).
- Mancio, J.; Azevedo, D.; Saraiva, F.; et al. Epicardial adipose tissue volume assessed by computed tomography and coronary artery disease: A systematic review and meta-analysis. European Heart Journal - Cardiovascular Imaging. 2018, 19, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Hsu, F.C.; Harris, T.B.; et al. The association of pericardial fat with incident coronary heart disease: The Multi-Ethnic Study of Atherosclerosis (MESA). Am J Clin Nutr. 2009, 90, 499–504. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.Z.; Huang, Y.L.; Wang, Y.C.; et al. Impact of location of epicardial adipose tissue, measured by coronary artery calcium-scoring computed tomography on obstructive coronary artery disease. Am J Cardiol. 2013, 112, 943–949. [Google Scholar] [CrossRef]
- Hirata, Y.; Yamada, H.; Kusunose, K.; et al. Clinical Utility of Measuring Epicardial Adipose Tissue Thickness with Echocardiography Using a High-Frequency Linear Probe in Patients with Coronary Artery Disease. J Am Soc Echocardiogr. 2015, 28, 1240–1246.e1. [Google Scholar] [CrossRef]
- Bachar, G.N.; Dicker, D.; Kornowski, R.; Atar, E. Epicardial adipose tissue as a predictor of coronary artery disease in asymptomatic subjects. Am J Cardiol. 2012, 110, 534–538. [Google Scholar] [CrossRef]
- Heart failure - Symptoms and causes. Mayo Clinic. Available online: https://www.mayoclinic.org/diseases-conditions/heart-failure/symptoms-causes/syc-20373142 (accessed on 19 June 2024).
- Malik, A.; Brito, D.; Vaqar, S.; Chhabra, L. Congestive Heart Failure. In StatPearls. StatPearls Publishing; 2024. Available online: http://www.ncbi.nlm.nih.gov/books/NBK430873/ (accessed on 19 June 2024).
- Pugliese, N.R.; Paneni, F.; Mazzola, M.; et al. Impact of epicardial adipose tissue on cardiovascular haemodynamics, metabolic profile, and prognosis in heart failure. Eur J Heart Fail. 2021, 23, 1858–1871. [Google Scholar] [CrossRef]
- Tromp, J.; Bryant, J.A.; Jin, X.; et al. Epicardial fat in heart failure with reduced versus preserved ejection fraction. Eur J Heart Fail. 2021, 23, 835–838. [Google Scholar] [CrossRef]
- Li, C.; Liu, X.; Adhikari, B.K.; et al. The role of epicardial adipose tissue dysfunction in cardiovascular diseases: An overview of pathophysiology, evaluation, and management. Front Endocrinol. 2023, 14. [Google Scholar] [CrossRef]
- Large, S.R.; Hosseinpour, A.R.; Wisbey, C.; Wells, F.C. Spontaneous cardioversion and mitral valve repair: A role for surgical cardioversion (Cox-maze)? Eur J Cardiothorac Surg. 1997, 11, 76–80. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.P.; Hua, T.A.; Böhm, M.; Wachtell, K.; Kjeldsen, S.E.; Schmieder, R.E. Prevention of atrial fibrillation by Renin-Angiotensin system inhibition a meta-analysis. J Am Coll Cardiol. 2010, 55, 2299–2307. [Google Scholar] [CrossRef] [PubMed]
- Tsao, H.M.; Hu, W.C.; Wu, M.H.; et al. Quantitative analysis of quantity and distribution of epicardial adipose tissue surrounding the left atrium in patients with atrial fibrillation and effect of recurrence after ablation. Am J Cardiol. 2011, 107, 1498–1503. [Google Scholar] [CrossRef]
- Nakanishi, K.; Fukuda, S.; Tanaka, A.; et al. Peri-atrial epicardial adipose tissue is associated with new-onset nonvalvular atrial fibrillation. Circ, J. 2012, 76, 2748–2754. [Google Scholar] [CrossRef]
- Nagashima, K.; Okumura, Y.; Watanabe, I.; et al. Association between epicardial adipose tissue volumes on 3-dimensional reconstructed CT images and recurrence of atrial fibrillation after catheter ablation. Circ, J. 2011, 75, 2559–2565. [Google Scholar] [CrossRef]
- Steenbergen, C.; Frangogiannis, N.G. Chapter 36—Ischemic Heart Disease. In: Hill, J.A.; Olson, E.N.; eds. Muscle. Academic Press; 2012; pp. 495–521. [CrossRef]
- Gul, Z.; Shams, P.; Makaryus, A.N. Silent Myocardial Ischemia. In: StatPearls. StatPearls Publishing; 2024. Available online: http://www.ncbi.nlm.nih.gov/books/NBK536915/ (accessed on 18 June 2024).
- Tamarappoo, B.; Dey, D.; Shmilovich, H.; et al. Increased pericardial fat volume measured from noncontrast CT predicts myocardial ischemia by SPECT. JACC Cardiovasc Imaging. 2010, 3, 1104–1112. [Google Scholar] [CrossRef] [PubMed]
- Hell, M.M.; Ding, X.; Rubeaux, M.; et al. Epicardial adipose tissue volume but not density is an independent predictor for myocardial ischemia. J Cardiovasc Comput Tomogr. 2016, 10, 141–149. [Google Scholar] [CrossRef]
- Mitral valve stenosis - Symptoms and causes. Mayo Clinic. Available online: https://www.mayoclinic.org/diseases-conditions/mitral-valve-stenosis/symptoms-causes/syc-20353159 (accessed on 29 May 2024).
- Mahabadi, A.A.; Kahlert, H.A.; Dykun, I.; Balcer, B.; Kahlert, P.; Rassaf, T. Epicardial Adipose Tissue Thickness Independently Predicts Severe Aortic Valve Stenosis. J Heart Valve Dis. 2017, 26, 262–267. [Google Scholar]
- Nabati, M.; Salehi, A.; Hatami, G.; Dabirian, M.; Yazdani, J.; Parsaee, H. Epicardial adipose tissue and its association with cardiovascular risk factors and mitral annular calcium deposits. Ultrasound. 2019, 27, 217–224. [Google Scholar] [CrossRef]
- Aortic valve stenosis - Symptoms and causes. Mayo Clinic. Available online: https://www.mayoclinic.org/diseases-conditions/aortic-stenosis/symptoms-causes/syc-20353139 (accessed on 29 May 2024).
- Liu, C.Y.; Redheuil, A.; Ouwerkerk, R.; Lima, J.A.C.; Bluemke, D.A. Myocardial Fat Quantification in Humans: Evaluation by Two-Point Water-Fat Imaging and Localized Proton Spectroscopy. Magn Reson Med. 2010, 63, 892–901. [Google Scholar] [CrossRef]
- Wu, C.K.; Lee, J.K.; Hsu, J.C.; et al. Myocardial adipose deposition and the development of heart failure with preserved ejection fraction. European Journal of Heart Failure. 2020, 22, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Schimmel, K.; Ichimura, K.; Reddy, S.; Haddad, F.; Spiekerkoetter, E. Cardiac Fibrosis in the Pressure Overloaded Left and Right Ventricle as a Therapeutic Target. Frontiers in Cardiovascular Medicine. 2022, 9, 886553. [Google Scholar] [CrossRef]
- Wu, C.K.; Tsai, H.Y.; Su, M.Y.M.; et al. Evolutional change in epicardial fat and its correlation with myocardial diffuse fibrosis in heart failure patients. Journal of Clinical Lipidology. 2017, 11, 1421–1431. [Google Scholar] [CrossRef] [PubMed]
- Ismail, I.; Al-Khafaji, K.; Mutyala, M.; et al. Cardiac lipoma. J Community Hosp Intern Med Perspect. 2015, 5, 10.3402–jchimp.v5. [Google Scholar] [CrossRef]
- Radswiki, T. Cardiac lipoma | Radiology Reference Article | Radiopaedia.org. Radiopaedia. [CrossRef]
- Weerakkody, Y. Epicardial lipomatosis | Radiology Reference Article | Radiopaedia.org. Radiopaedia. [CrossRef]
- Myerson, S.G.; Roberts, R.; Moat, N.; Pennell, D.J. Tamponade caused by cardiac lipomatous hypertrophy. J Cardiovasc Magn Reson. 2004, 6, 565–568. [Google Scholar] [CrossRef]
- Miller, C.A.; Schmitt, M. Epicardial Lipomatous Hypertrophy Mimicking Pericardial Effusion. Circulation: Cardiovascular Imaging. 2011, 4, 77–78. [Google Scholar] [CrossRef]
- Gaillard, F. Lipomatous hypertrophy of the interatrial septum | Radiology Reference Article | Radiopaedia.org. Radiopaedia. [CrossRef]
- Antonopoulos, A.S.; Antoniades, C. Cardiac Magnetic Resonance Imaging of Epicardial and Intramyocardial Adiposity as an Early Sign of Myocardial Disease. Circulation: Cardiovascular Imaging. 2018, 11, e008083. [Google Scholar] [CrossRef]
- Marwan, M.; Koenig, S.; Schreiber, K.; et al. Quantification of epicardial adipose tissue by cardiac CT: Influence of acquisition parameters and contrast enhancement. European Journal of Radiology. 2019, 121, 108732. [Google Scholar] [CrossRef]
- Iacobellis, G.; Willens, H.J. Echocardiographic Epicardial Fat: A Review of Research and Clinical Applications. Journal of the American Society of Echocardiography. 2009, 22, 1311–1319. [Google Scholar] [CrossRef]
- Pohost, G.M. The History of Cardiovascular Magnetic Resonance. JACC: Cardiovascular Imaging. 2008, 1, 672–678. [Google Scholar] [CrossRef]
- Guglielmo, M.; Lin, A.; Dey, D.; et al. Epicardial fat and coronary artery disease: Role of cardiac imaging. Atherosclerosis. 2021, 321, 30–38. [Google Scholar] [CrossRef]
- Bertaso, A.G.; Bertol, D.; Duncan, B.B.; Foppa, M. Epicardial Fat: Definition, Measurements and Systematic Review of Main Outcomes. Arquivos Brasileiros de Cardiologia. Published online 2013. [CrossRef]
- Salerno, M.; Sharif, B.; Arheden, H.; et al. Recent Advances in Cardiovascular Magnetic Resonance: Techniques and Applications. Circ: Cardiovascular Imaging. 2017, 10, e003951. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Carrick, D.; Layland, J.; Oldroyd, K.G.; Berry, C. The Role of Cardiac Magnetic Resonance Imaging (MRI) in Acute Myocardial Infarction (AMI). Heart, Lung and Circulation. 2013, 22, 243–255. [Google Scholar] [CrossRef]
- Curtis, A.D.; Cheng, H.M. Primer and Historical Review on Rapid Cardiac CINE MRI. Magnetic Resonance Imaging. 2022, 55, 373–388. [Google Scholar] [CrossRef]
- Homsi, R.; Meier-Schroers, M.; Gieseke, J.; et al. 3D-Dixon MRI based volumetry of peri- and epicardial fat. Int J Cardiovasc Imaging. 2016, 32, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Militello, C.; Rundo, L.; Toia, P.; et al. A semi-automatic approach for epicardial adipose tissue segmentation and quantification on cardiac CT scans. Computers in Biology and Medicine. 2019, 114, 103424. [Google Scholar] [CrossRef]
- Malavazos, A.E.; Di Leo, G.; Secchi, F.; et al. Relation of Echocardiographic Epicardial Fat Thickness and Myocardial Fat. The American Journal of Cardiology. 2010, 105, 1831–1835. [Google Scholar] [CrossRef]
- Chen, S.; An, D.; Feng, C.; Bian, Z.; Wu, L.M. Segmentation of Pericardial Adipose Tissue in CMR Images: A Benchmark Dataset MRPEAT and a Triple-Stage Network 3SUnet. IEEE Trans Med Imaging. 2023, 42, 2386–2399. [Google Scholar] [CrossRef]
- Feng, F.; Carlhäll, C.J.; Tan, Y.; et al. FM-Net: A Fully Automatic Deep Learning Pipeline for Epicardial Adipose Tissue Segmentation. In Statistical Atlases and Computational Models of the Heart. Regular and CMRxRecon Challenge Papers; Notes in Computer Science; Camara, O., Puyol-Antón, E., Sermesant, M., et al., Eds.; Switzerland: Springer Nature, 2024; Volume 14507, pp. 88–97. [Google Scholar] [CrossRef]
- Daudé, P.; Ancel, P.; Confort Gouny, S.; et al. Deep-Learning Segmentation of Epicardial Adipose Tissue Using Four-Chamber Cardiac Magnetic Resonance Imaging. Diagnostics. 2022, 12, 126. [Google Scholar] [CrossRef]
- Kulasekara, M.; Dinh, V.Q.; Fernandez-del-Valle, M.; Klingensmith, J.D. Comparison of two-dimensional and three-dimensional U-Net architectures for segmentation of adipose tissue in cardiac magnetic resonance images. Med Biol Eng Comput. 2022, 60, 2291–2306. [Google Scholar] [CrossRef]
- Fulton, M.R.; Givan, A.H.; Fernandez-del-Valle, M.; Klingensmith, J.D. Segmentation of epicardial adipose tissue in cardiac MRI using deep learning. In: Gimi, B.S.; Krol, A.; eds. Medical Imaging 2020: Biomedical Applications in Molecular, Structural, and Functional Imaging. SPIE; 2020, 25. [CrossRef]
- Requena-Ibáñez, J.A.; Santos-Gallego, C.G.; Rodriguez Cordero, A.J.; et al. Not only how much, but also how to, when measuring epicardial adipose tissue. Magnetic Resonance Imaging. 2022, 86, 149–151. [Google Scholar] [CrossRef] [PubMed]
- Henningsson, M.; Brundin, M.; Scheffel, T.; Edin, C.; Viola, F.; Carlhäll, C.J. Quantification of epicardial fat using 3D cine Dixon, M.R.I. BMC Med Imaging. 2020, 20, 80. [Google Scholar] [CrossRef] [PubMed]
- Guglielmo, M.; Penso, M.; Carerj, M.L.; et al. DEep LearnIng-based QuaNtification of epicardial adipose tissue predicts MACE in patients undergoing stress, C.M.R. Atherosclerosis 2024, 117549. [Google Scholar] [CrossRef] [PubMed]
- Secchi, F.; Asteria, C.; Monti, C.B.; et al. Quantification of epicardial adipose tissue in obese patients using an open-bore MR scanner. Eur Radiol Exp. 2022, 6, 25. [Google Scholar] [CrossRef]
- Meloni, A.; Frijia, F.; Panetta, D.; et al. Photon-Counting Computed Tomography (PCCT): Technical Background and Cardio-Vascular Applications. Diagnostics. 2023, 13, 645. [Google Scholar] [CrossRef]
- Flohr, T.; Schmidt, B.; Ulzheimer, S.; Alkadhi, H. Cardiac imaging with photon counting, C.T. The British Journal of Radiology. 2023, 96, 20230407. [Google Scholar] [CrossRef]
- Benčević, M.; Galić, I.; Habijan, M.; Pižurica, A. Recent Progress in Epicardial and Pericardial Adipose Tissue Segmentation and Quantification Based on Deep Learning: A Systematic Review. Applied Sciences. 2022, 12, 5217. [Google Scholar] [CrossRef]
- Greco, F.; Salgado, R.; Van Hecke, W.; Del Buono, R.; Parizel, P.M.; Mallio, C.A. Epicardial and pericardial fat analysis on CT images and artificial intelligence: A literature review. Quant Imaging Med Surg. 2022, 12, 2075–2089. [Google Scholar] [CrossRef]
- La Grutta, L.; Toia, P.; Farruggia, A.; et al. Quantification of epicardial adipose tissue in coronary calcium score and CT coronary angiography image data sets: Comparison of attenuation values, thickness and volumes. BJR. 2016, 89, 20150773. [Google Scholar] [CrossRef]
- Marwan, M.; Achenbach, S. Quantification of epicardial fat by computed tomography: Why, when and how? Journal of Cardiovascular Computed Tomography. 2013, 7, 3–10. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Guo, B.; Lei, Y.; et al. Automatic epicardial fat segmentation in cardiac CT imaging using 3D deep attention U-Net. In: Landman, B.A.; Išgum, I.; eds. Medical Imaging 2020: Image Processing. SPIE; 2020:84. [CrossRef]
- Bencevic, M.; Habijan, M.; Galic, I. Epicardial Adipose Tissue Segmentation from CT Images with A Semi-3D Neural Network. In: 2021 International Symposium ELMAR. IEEE; 2021:87-90. [CrossRef]
- Commandeur, F.; Goeller, M.; Razipour, A.; et al. Fully Automated CT Quantification of Epicardial Adipose Tissue by Deep Learning: A Multicenter Study. Radiology: Artificial Intelligence. 2019, 1, e190045. [Google Scholar] [CrossRef]
- Li, X.; Sun, Y.; Xu, L.; et al. Automatic quantification of epicardial adipose tissue volume. Medical Physics. 2021, 48, 4279–4290. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhou, J.; Zhang, B.; Jia, W.; Wu, E. Automatic Epicardial Fat Segmentation and Quantification of CT Scans Using Dual U-Nets With a Morphological Processing Layer. IEEE Access. 2020, 8, 128032–128041. [Google Scholar] [CrossRef]
- Priya, C.; Sudha, S. Adaptive Fruitfly Based Modified Region Growing Algorithm for Cardiac Fat Segmentation Using Optimal Neural Network. J Med Syst. 2019, 43, 104. [Google Scholar] [CrossRef]
- Norlén, A.; Alvén, J.; Molnar, D.; et al. Automatic pericardium segmentation and quantification of epicardial fat from computed tomography angiography. J Med Imaging (Bellingham). 2016, 3, 034003. [Google Scholar] [CrossRef]
- Rodrigues, É.O.; Morais, F.F.C.; Morais, N.A.O.S.; Conci, L.S.; Neto, L.V.; Conci, A. A novel approach for the automated segmentation and volume quantification of cardiac fats on computed tomography. Computer Methods and Programs in Biomedicine. 2016, 123, 109–128. [Google Scholar] [CrossRef] [PubMed]
- Vajihi, Z.; Rosado-Mendez, I.; Hall, T.J.; Rivaz, H. L1 And L2 Norm Depth-Regularized Estimation Of The Acoustic Attenuation And Backscatter Coefficients Using Dynamic Programming. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019); 2019; pp. 1749–1752. [Google Scholar] [CrossRef]
- Hoori, A.; Hu, T.; Lee, J.; Al-Kindi, S.; Rajagopalan, S.; Wilson, D.L. Deep learning segmentation and quantification method for assessing epicardial adipose tissue in CT calcium score scans. Sci Rep. 2022, 12, 2276. [Google Scholar] [CrossRef] [PubMed]
- Commandeur, F.; Goeller, M.; Betancur, J.; et al. Deep Learning for Quantification of Epicardial and Thoracic Adipose Tissue From Non-Contrast, C.T. IEEE Trans Med Imaging. 2018, 37, 1835–1846. [Google Scholar] [CrossRef]
- D’Errico, L.; Salituri, F.; Ciardetti, M.; et al. Quantitative analysis of epicardial fat volume: Effects of scanning protocol and reproducibility of measurements in non-contrast cardiac CT vs. coronary CT angiography. Quant Imaging Med Surg. 2017, 7, 326–335. [Google Scholar] [CrossRef]
- Insana, M. Handbook of Physics in Medicine and Biology - Ultrasonic Imaging. 0 ed. (Splinter, R.; ed.). CRC Press; 2010. [CrossRef]
- Finel, V. 3D ultrafast echocardiography: Doctoral Thesis. Sorbonne; 2018.
- Standard Transthoracic Echocardiogram: Complete Imaging Protocol. Cardiovascular Education. Available online: https://ecgwaves.com/topic/the-standard-adult-transthoracic-echocardiogram-a-protocol-to-obtain-a-complete-study/ (accessed on 8 July 2024).
- My medical illustrations. Patrick Lynch. January 22, 2017. Available online: https://coastfieldguides.com/my-medical-illustrations/ (accessed on 8 July 2024).
- Chen, C.; Qin, C.; Qiu, H.; et al. Deep Learning for Cardiac Image Segmentation: A Review. Front Cardiovasc Med. 2020, 7. [Google Scholar] [CrossRef]
- Leclerc, S.; Smistad, E.; Pedrosa, J.; et al. Deep Learning for Segmentation Using an Open Large-Scale Dataset in 2D Echocardiography. IEEE Trans Med Imaging. 2019, 38, 2198–2210. [Google Scholar] [CrossRef]
- MoosaviTayebi, R. Echocardiography Image Segmentation: A Survey. IEEE. Available online: https://www.academia.edu/7479069/Echocardiography_Image_Segmentation_A_Survey (accessed on 18 April 2024).
- Painchaud, N.; Duchateau, N.; Bernard, O.; Jodoin, P.M. Echocardiography Segmentation With Enforced Temporal Consistency. IEEE Trans Med Imaging. 2022, 41, 2867–2878. [Google Scholar] [CrossRef] [PubMed]
- Zyuzin, V.; Mukhtarov, A.; Neustroev, D.; Chumarnaya, T. Segmentation of 2D Echocardiography Images using Residual Blocks in U-Net Architectures. In: 2020 Ural Symposium on Biomedical Engineering, Radioelectronics and Information Technology (USBEREIT). ; 2020:499-502. [CrossRef]
- Cuellar, J.R.; Gillette, L.; Dinh, V.; Woodard, P.; Burri, M.; Klingensmith, J.D. Echocardiogram image segmentation and cardiac adipose tissue estimation using spectral analysis and deep learning.
- Klingensmith, J.D.; Karlapalem, A.; Kulasekara, M.M.; Fernandez-del-Valle, M. Spectral analysis of ultrasound radiofrequency backscatter for the identification of epicardial adipose tissue. J Med Imag. [CrossRef]
- Iacobellis, G.; Assael, F.; Ribaudo, M.C.; et al. Epicardial Fat from Echocardiography: A New Method for Visceral Adipose Tissue Prediction. Obesity Research. 2003, 11, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Schejbal, V. [Epicardial fatty tissue of the right ventricle--morphology, morphometry and functional significance]. Pneumologie. 1989, 43, 490–499. [Google Scholar] [PubMed]
- Nesti, L.; Pugliese, N.R.; Chiriacò, M.; Trico, D.; Baldi, S.; Natali, A. Epicardial adipose tissue thickness is associated with reduced peak oxygen consumption and systolic reserve in patients with type 2 diabetes and normal heart function. Diabetes Obes Metab. 2023, 25, 177–188. [Google Scholar] [CrossRef]
- Wang, Q.; Chi, J.; Wang, C.; Yang, Y.; Tian, R.; Chen, X. Epicardial Adipose Tissue in Patients with Coronary Artery Disease: A Meta-Analysis. J Cardiovasc Dev Dis. 2022, 9, 253. [Google Scholar] [CrossRef]
- Eren, H.; Omar, M.B.; Öcal, L. Epicardial fat tissue may predict new-onset atrial fibrillation in patients with non-ST-segment elevation myocardial infarction. Archives of the Turkish Society of Cardiology. 2021, 49, 430–438. [Google Scholar] [CrossRef]
- Parisi, V.; Petraglia, L.; Formisano, R.; et al. Validation of the echocardiographic assessment of epicardial adipose tissue thickness at the Rindfleisch fold for the prediction of coronary artery disease. Nutrition, Metabolism and Cardiovascular Diseases. 2020, 30, 99–105. [Google Scholar] [CrossRef]
- Meenakshi, K.; Rajendran, M.; Srikumar, S.; Chidambaram, S. Epicardial fat thickness: A surrogate marker of coronary artery disease – Assessment by echocardiography. Indian Heart, J. 2016, 68, 336–341. [Google Scholar] [CrossRef]
- Nerlekar, N.; Baey, Y.W.; Brown, A.J.; et al. Poor Correlation, Reproducibility, and Agreement Between Volumetric Versus Linear Epicardial Adipose Tissue Measurement: A 3D Computed Tomography Versus 2D Echocardiography Comparison. JACC: Cardiovascular Imaging. 2018, 11, 1035–1036. [Google Scholar] [CrossRef] [PubMed]
- Iacobellis, G.; Willens, H.J.; Barbaro, G.; Sharma, A.M. Threshold values of high-risk echocardiographic epicardial fat thickness. Obesity (Silver Spring). 2008, 16, 887–892. [Google Scholar] [CrossRef] [PubMed]
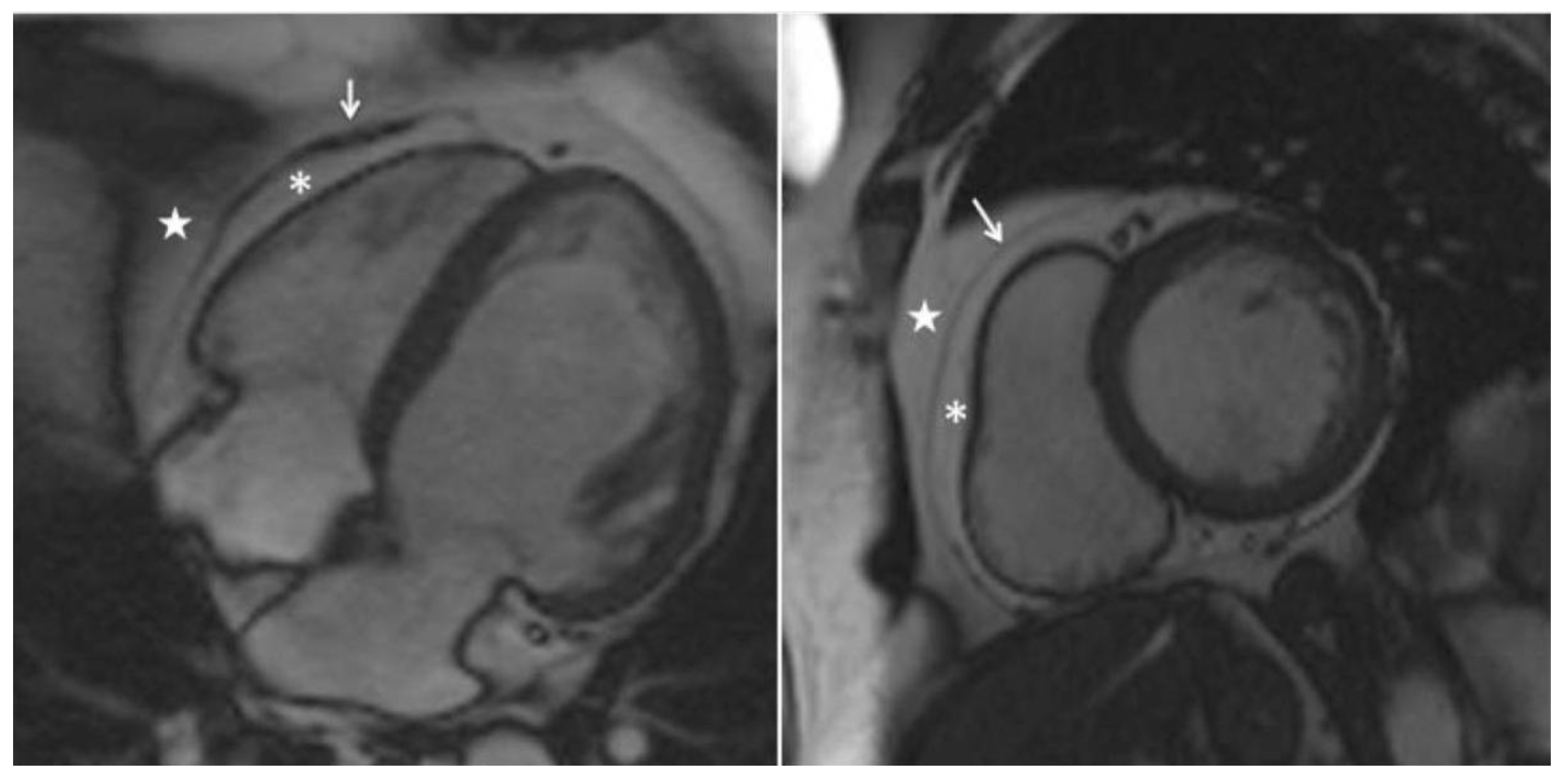
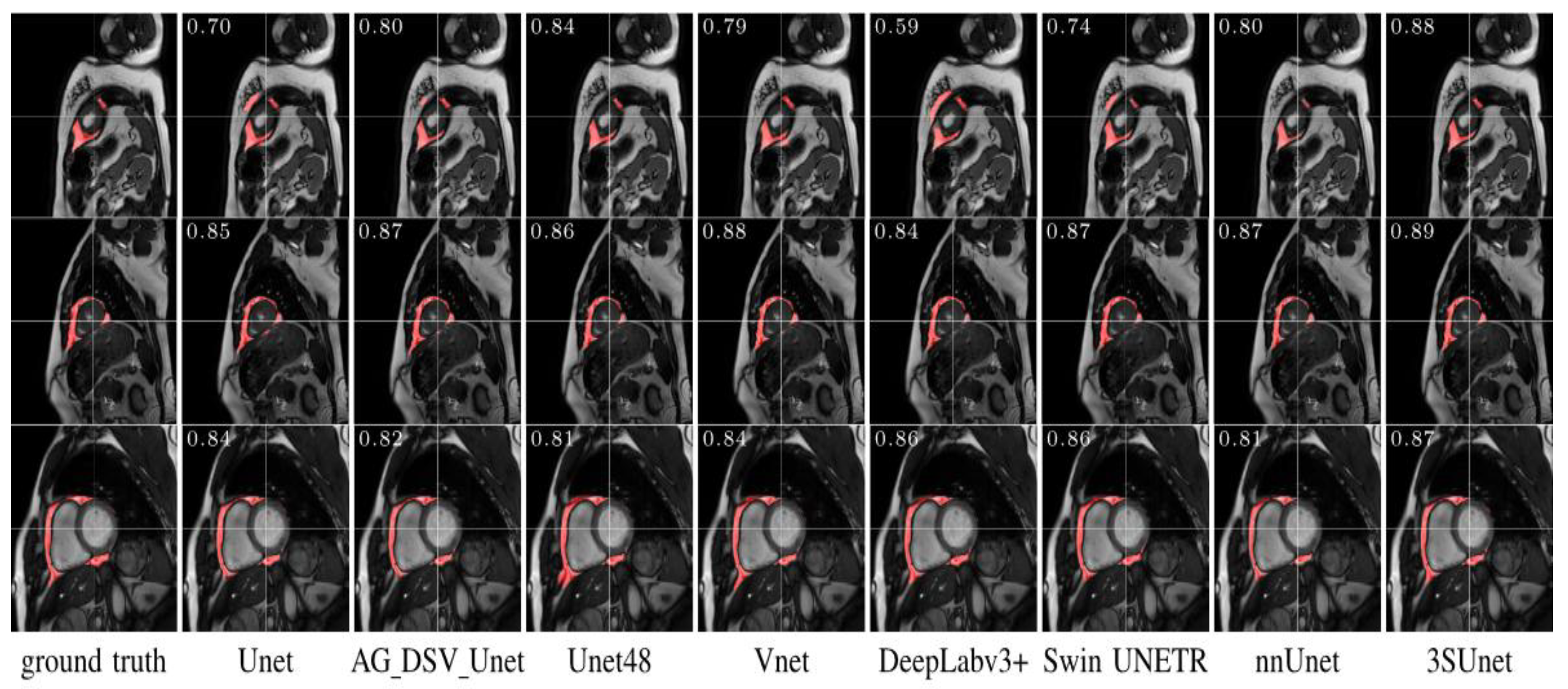
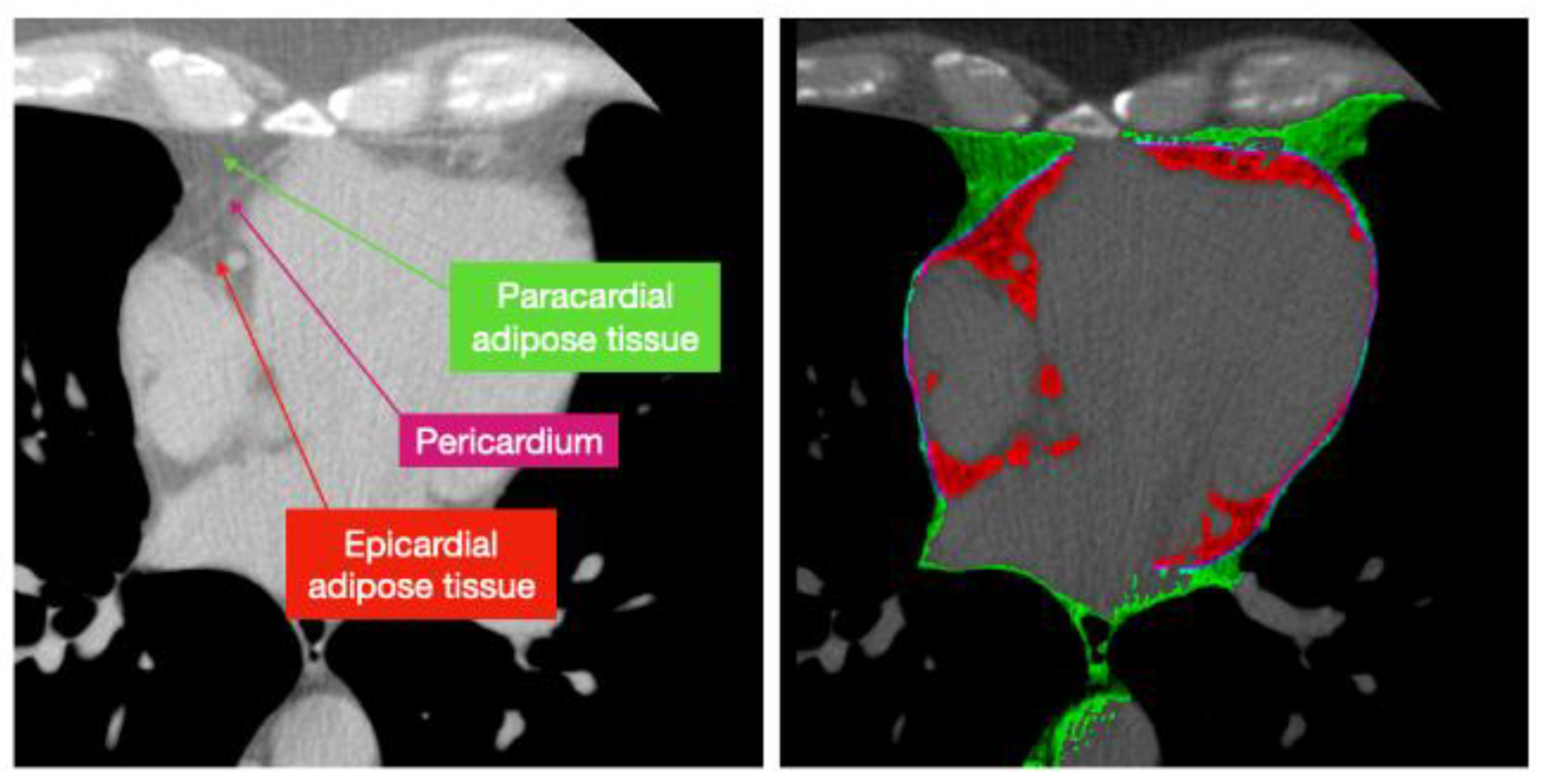
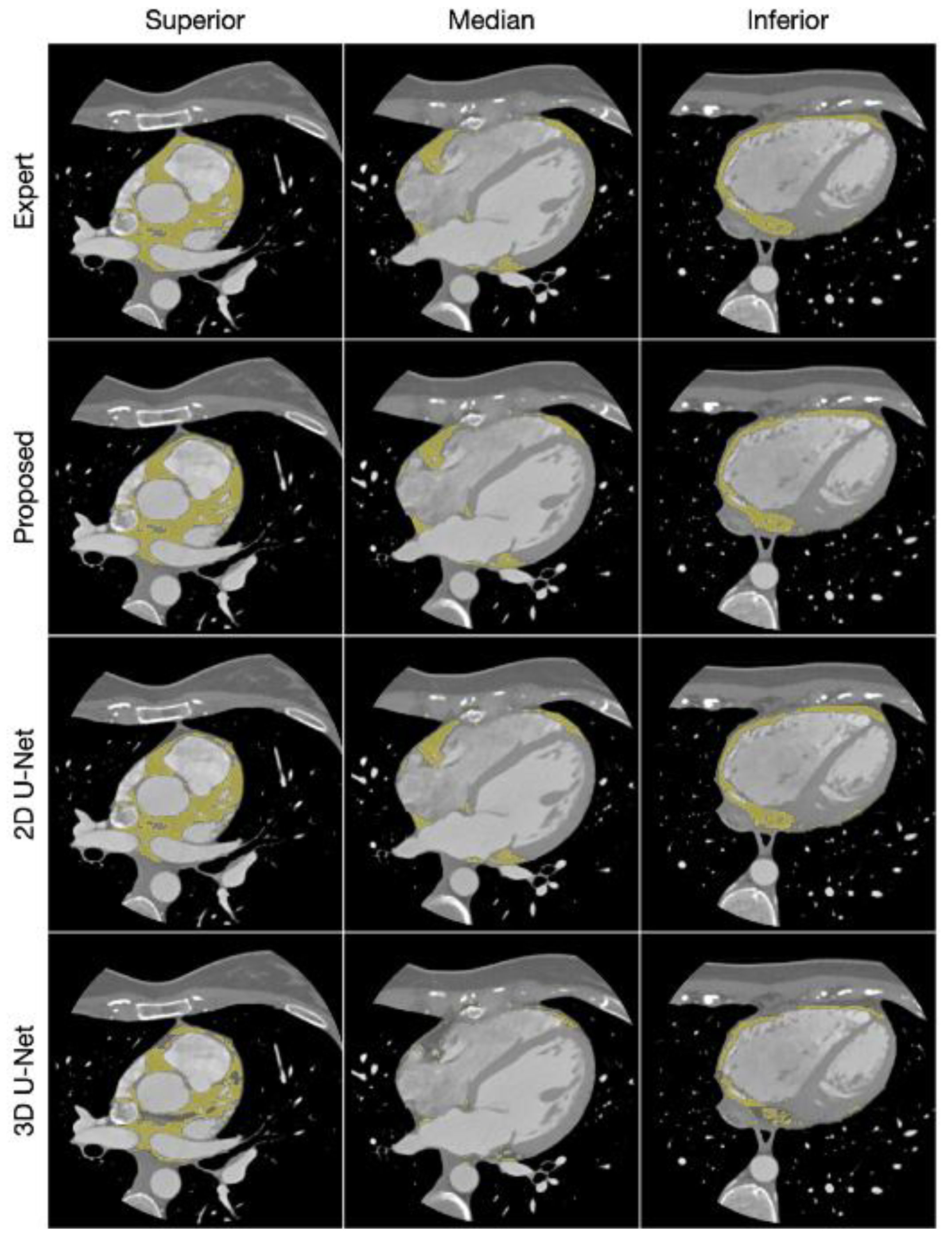
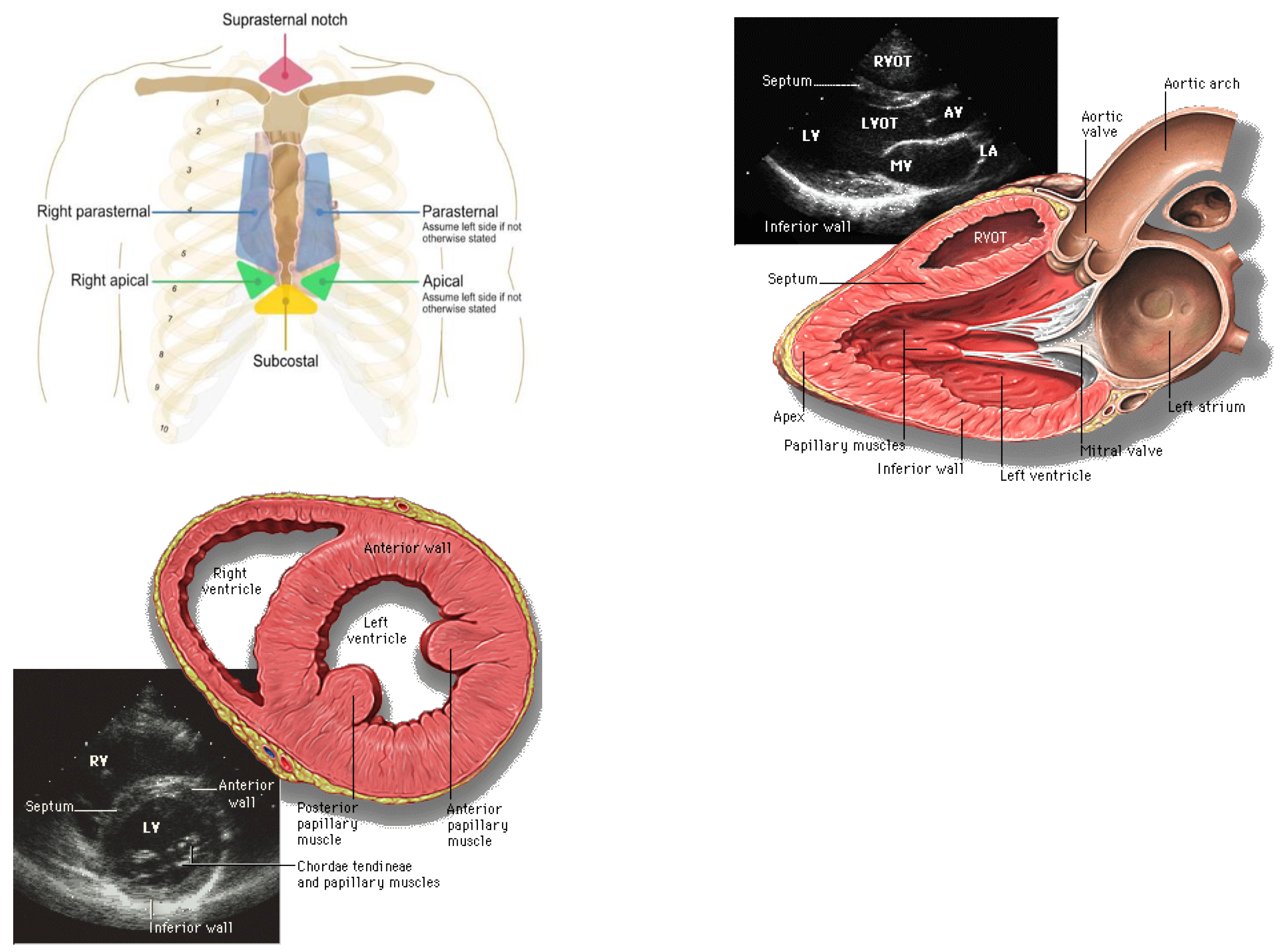
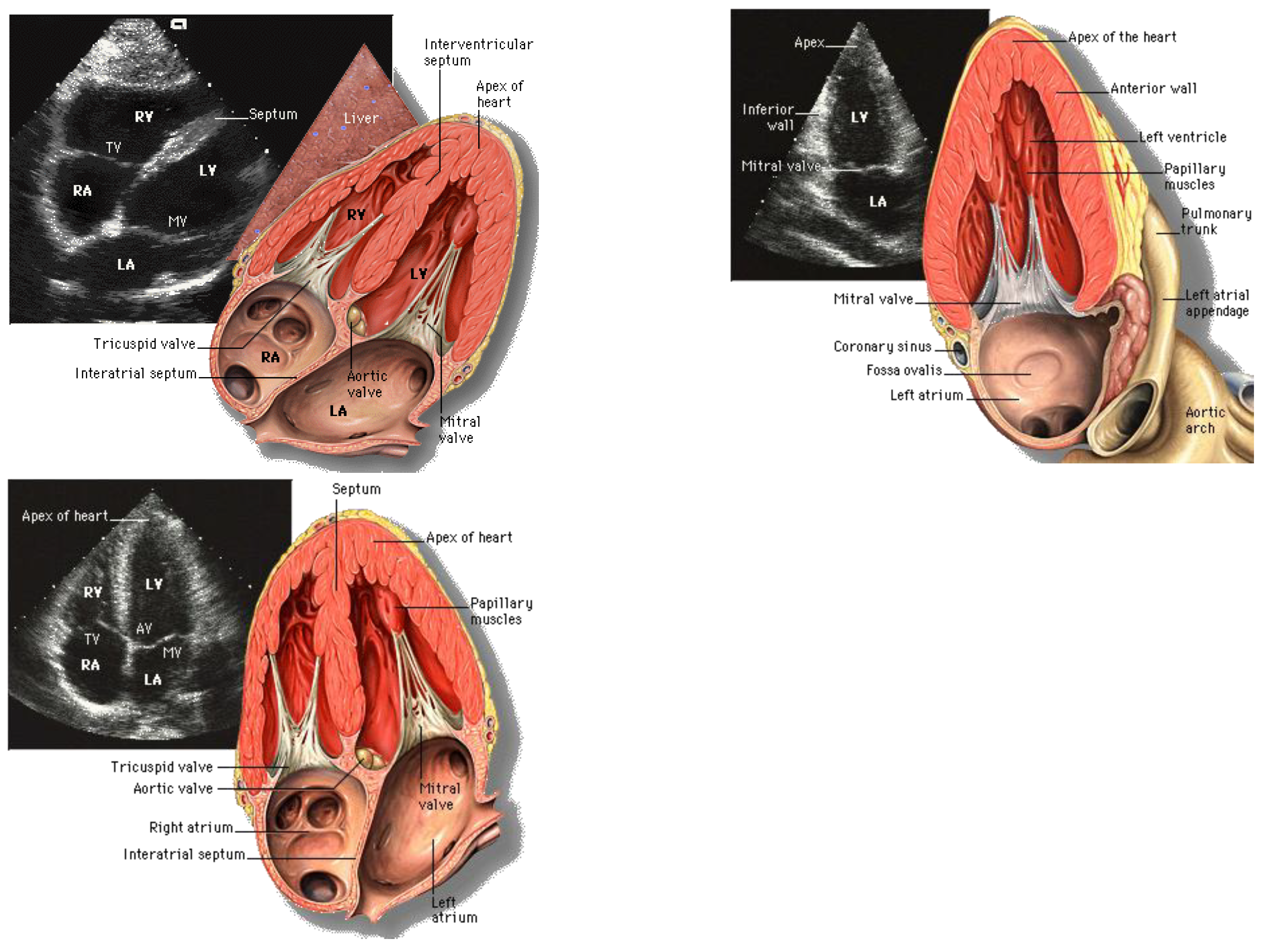
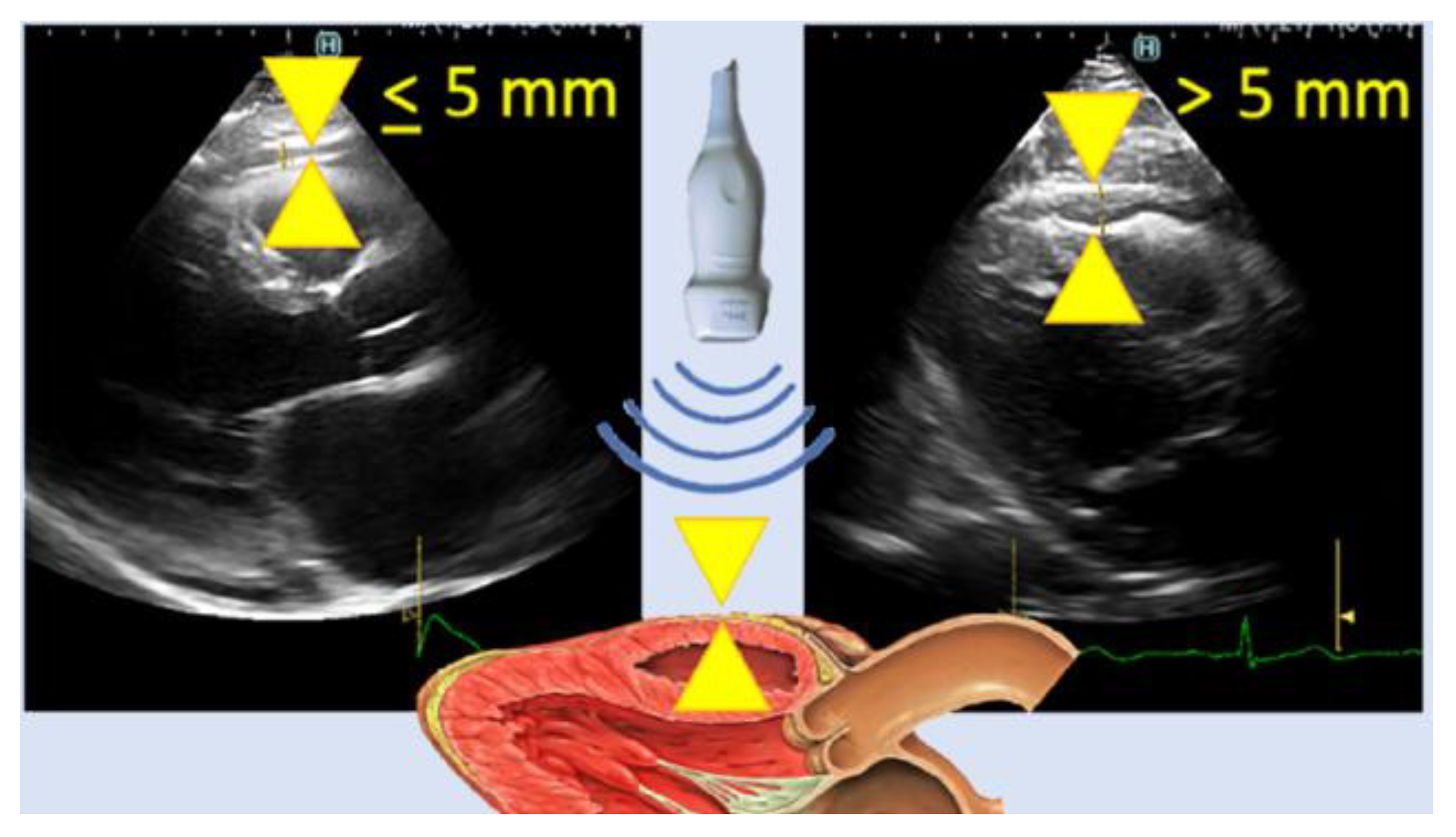

| Author | Tissue | Study | Metrics | Values |
|---|---|---|---|---|
| Feng et al.[87] - 2024 | EAT | Automatic double Res-Unet CNN based on fat maps, Dixon MRI | DSC | 0.8630 |
| Chen et al.[86] - 2023 | PAT | Automatic triple-stage 3SUnet, 2D SA MRI | Precision Recall |
0.766±0.152 0.831±0.126 |
| Daude et al.[88] - 2022 | PAT EAT |
Automatic four-chamber FCNs, cine MRI | DSC MSD (mm) DSC MSD (mm) |
0.7700 1.71 0.8000 2.38 |
| Kulasekara et al.[89] - 2022 | CAT | Automatic 3D U-Net, cine MRI | DSC | 0.7170 |
| Fulton et al.[90] - 2020 | EAT | Automatic double NN, cine MRI | DSC | 0.56±0.12 |
| Author | Tissue | Study | Units | Values | Correlation |
|---|---|---|---|---|---|
| Guglielmo et al.[93] - 2024 | EAT | Automated deep learning volume measurement | mL | 43.5±9.0 | p < 0.001 |
| Secchi et al.[94] - 2022 | EAT | Manual volume measurement using open-bore MR, cine MRI | Systole cm3 Diastole cm3 |
88.25 87.00 |
p < 0.124 p < 0.551 |
| Henningsson et al.[92] - 2020 | EAT | Manual volume measurement using cine Dixon technique, 3D Dixon MRI | mL | 145±90 | p < 0.01 |
| Author | Tissue | Study | Metrics | Values | Correlation |
|---|---|---|---|---|---|
| Zhang et al.[105] - 2020 | EAT | Automatic dual U-Nets, CT | DSC | 0.9119 | 0.9304 |
| He et al.[101] -2020 | EAT | Automatic 3D deep attention U-Net, CCTA | DSC Precision Recall |
0.8550 0.8640 0.8950 |
NA |
| Militello et al.[84] - 2019 | EAT | Semi-automatic image analysis, CS and CCTA | DSC MAD |
0.9374, 0.9248 2.18, 2.87 |
(Pearson) 0.9591 0.9513 |
| Priya et al.[106] - 2019 | EAT PAT |
Adaptive Region Growing Algorithm, NC CT | Accuracy DSC Accuracy DSC |
0.9850 0.9870 0.9640 0.9530 |
NA |
| Norlén et al.[107] - 2016 | Automatic supervised, CCTA | DSC | 0.9900 | 0.9900 | |
| Rodrigues et al.[108] - 2016 | EAT PAT |
Automatic supervised, CT | DSC Accuracy |
0.9810 0.9850 |
NA |
| Author | Tissue | Study | Units | Values | Correlation |
|---|---|---|---|---|---|
| Hoori et al.[110] -2022 | EATd | Automatic DeepFat, NC low-dose CS CT | cm3 | 100.2 138.6 |
R = 0.9833 R = 0.9852 |
| Abdulkareem et al.[17] - 2022 | EATv | Automatic Single Multi-task framework, ECG-gated CT | mL | 101.16 | R = 0.9300 |
| Commandeur et al.[103] - 2019 | EATd | Automatic CNN, NC CS CT |
cm3 | 86.75 | R = 0.9740, p < 0.001 |
| Commandeur et al.[111] - 2018 | EATd TATd |
Automatic dual ConvNet, NC CCTA |
cm3 | 130.35 130.94 |
R = 0.945 p < 0.001 |
| D’Errico et al.[112] - 2017 | T–EATd RV–EATd LV–EATd |
Manual volume analysis, NC CCTA |
cm3 | 103.62, 94.96 67.23, 57.41 38.01, 35.27 |
ICC = 0.9900 |
| Author | Tissue | Study | Units | Values | Correlation |
|---|---|---|---|---|---|
| Average from metanalysis in[127] - 2022 | EAT | Metanalysis of EAT in patients with CAD and Non-CAD groups |
mm | 5.68 avg 3.61 avg |
NA |
| Eren et al.[128] - 2021 | EAT | EAT for atrial fibrillation prediction univariate, multivariate regression ROC EAT > 6.5 mm |
mm mm Sensitivity Specificity |
8.300, 6.100 5.850, 3.521 0.720 0.770 |
p < 0.001 |
| Xiao et al.[16] - 2020 | EAT | EAT thickness and heart disease (control) Coronary heart disease Single vessel disease Double vessel disease Multi vessel disease |
mm | 4.88 6.51 5.66 6.24 6.86 |
p < 0.01 vs control group |
| Parisi et al.[129] - 2020 | EAT | Validation of EAT thickness assessment for predicting CAD | mm | 11.00 (median) 1.00 mm (min) 29.00 mm (max) |
p < 0.001 |
| Meenakshi et al.[130] - 2016 | EAT | EAT thickness as CAD marker | mm | 0.9 min 13.5 max 5.56 avg (men) 5.97 avg (women) |
p (CAD) = 0.0001 p (BMI) = 0.08 |
| Iacobellis et al.[124] - 2003 | RV – EAT |
Epicardial Fat from Echocardiography, thickness | mm | 1.90 min 15.70 max 7.30 avg (men) 6.84 avg (women) |
r (VAT) = 0.798 r (WC) = 0.74 |
| Feature | MRI | CT | Echocardiography |
|---|---|---|---|
| Radiation Exposure | None | High radiation dose | Minimal or no radiation |
| Examination time | Longer scan time | Fast acquisition time | Real-time imaging |
| Cost | High | Moderate | Low |
| Image Quality | High spatial and temporal resolution, Multiplanar imaging capabilities, High soft tissue contrast, excellent for fat quantification | Good spatial resolution, accurate fat attenuation, good for calcium scoring | Lower resolution, limited depth penetration |
| CAT Assessment | Accurate quantification, can differentiate fat types | Good for assessing fat distribution, but less accurate for quantification | Limited ability to quantify fat, primarily qualitative assessment, free wall of RV thickness |
| CAT quantification | Volume, thickness | Volume, thickness | Thickness |
| Contraindications | Contraindicated for patients with certain metal implants | Can be performed on patients with pacemakers/defibrillators | None |
| Constraints | Difficult for claustrophobic and robust patients, requires specialized cardiac MRI protocols | Motion artifacts can affect image quality, may require contrast agents | Difficulty in imaging obese patients, limited field of view, operator-dependent |
| Availability | Widely available | Widely available | Widely available |
| Infrastructure | Large, requires a shielded room | Moderate size room | Minimal, portable |
| Other Applications | Cardiac function, tissue characterization, perfusion imaging | Chest imaging, vascular imaging | Cardiac function, valve assessment, cardiac chamber dimensions |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).