Submitted:
11 March 2025
Posted:
12 March 2025
You are already at the latest version
Abstract
Keywords:
HIGHLIGHTS
- Expression efficiency in distinct cell lineages.
- Immunogenicity and efficacy as chimeric nanovaccine.
- Capacity as drug carriers.
1. Introduction
2. Overview of basic concepts
2.1. Virus-like Particle Vaccines and Platforms for Vaccine Development
2.2. Uses of VLPs
- Preventive Vaccines: VLPs have been used in vaccines against viruses such as HPV (human papillomavirus) and hepatitis B. They induce an effective immune response, helping to prevent infections.
- Oncology Therapies: In addition to vaccines, VLPs are being explored as nanocarriers for cancer therapies, allowing targeted delivery of drugs directly to tumor cells.
- Personalized Vaccines: VLPs' versatility allows them to be adapted to create personalized vaccines for different pathogens or virus variants, responding quickly to emerging outbreaks.
- Delivery Platforms: VLPs can deliver antigens from different pathogens, facilitating the creation of combination vaccines.
2.3. Advantages of VLPs Compared to Other Vaccine Approaches
- Safety: Because VLPs do not contain viral genetic material, they cannot cause infections, making them a safe option for vaccination.
- Immunogenicity: VLPs are highly immunogenic and can induce a strong and long-lasting immune response, often with fewer doses than other vaccines.
- Versatility: The ability to modify VLPs to include different antigens allows them to be used in a wide range of diseases, from viral infections to cancer.
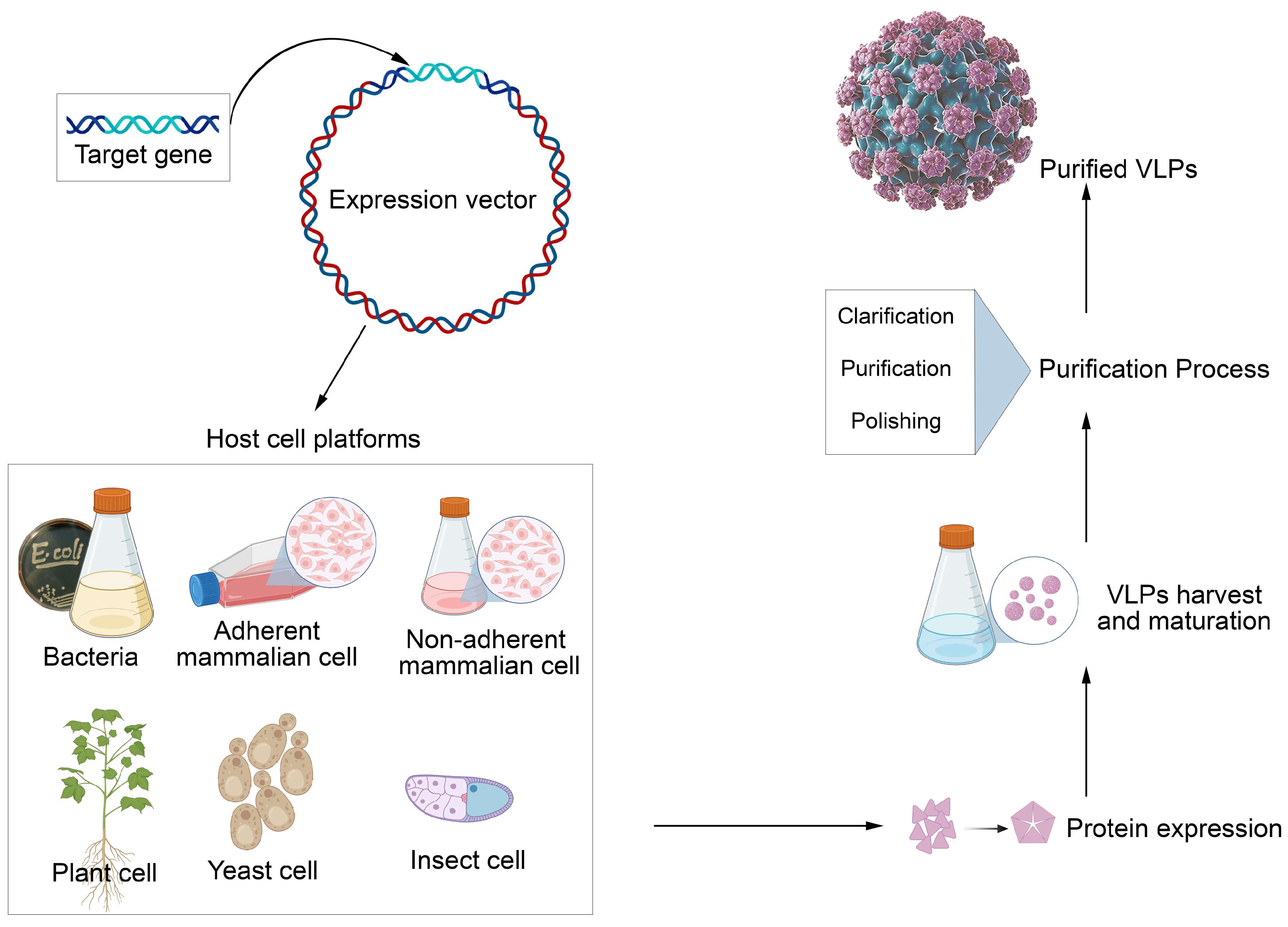
- Efficient Production: VLPs can be produced in mammalian cells, such as HEK cell lines, which are ideal for large-scale production and can quickly be adapted to various antigens.
- Fewer Side Effects: The non-infectious nature of VLPs makes them less likely to cause side effects than the attenuated virus vaccines.
2.4. Structural basis of viral function
2.4.1. Basic Structure of Viruses
- I.
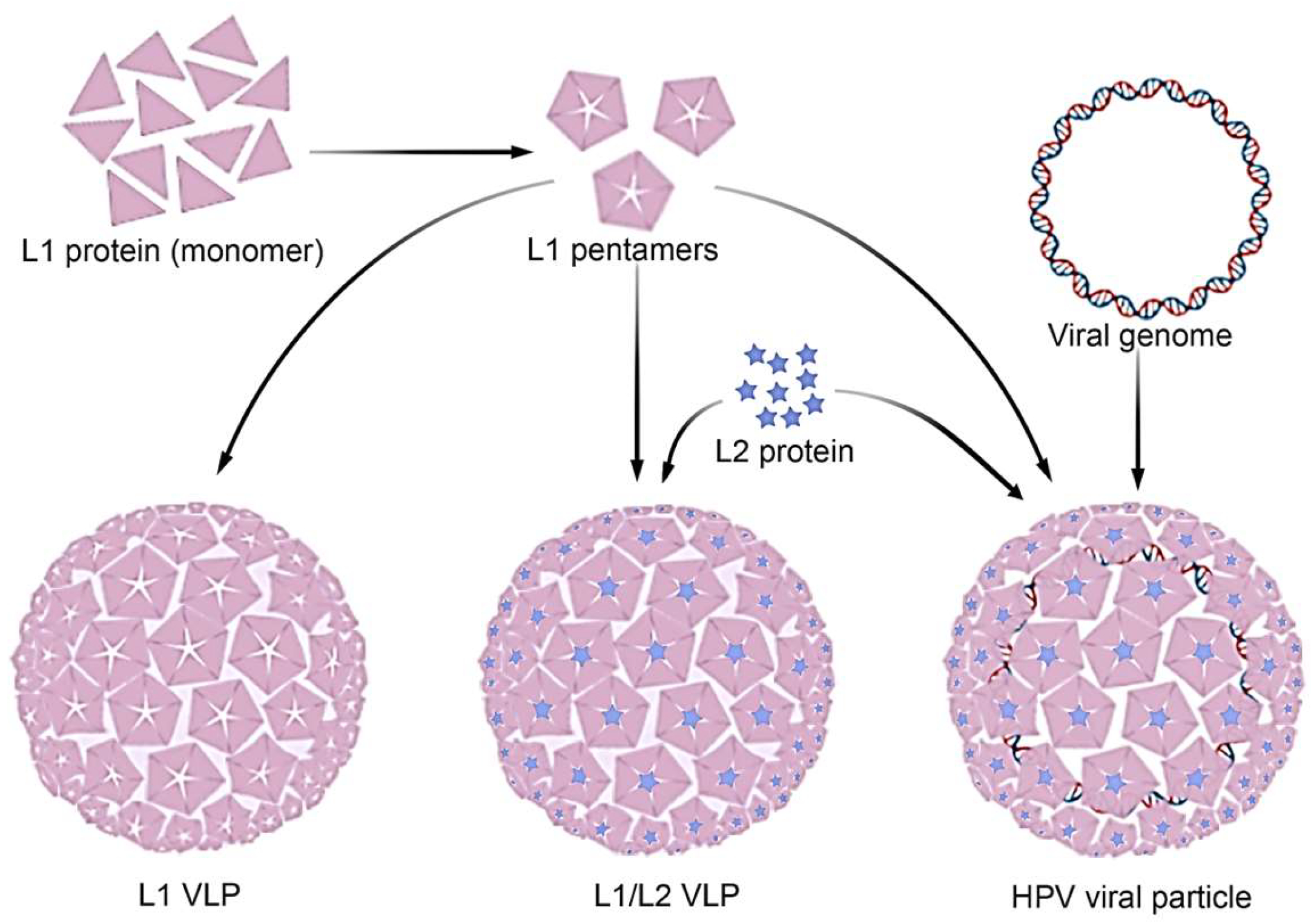
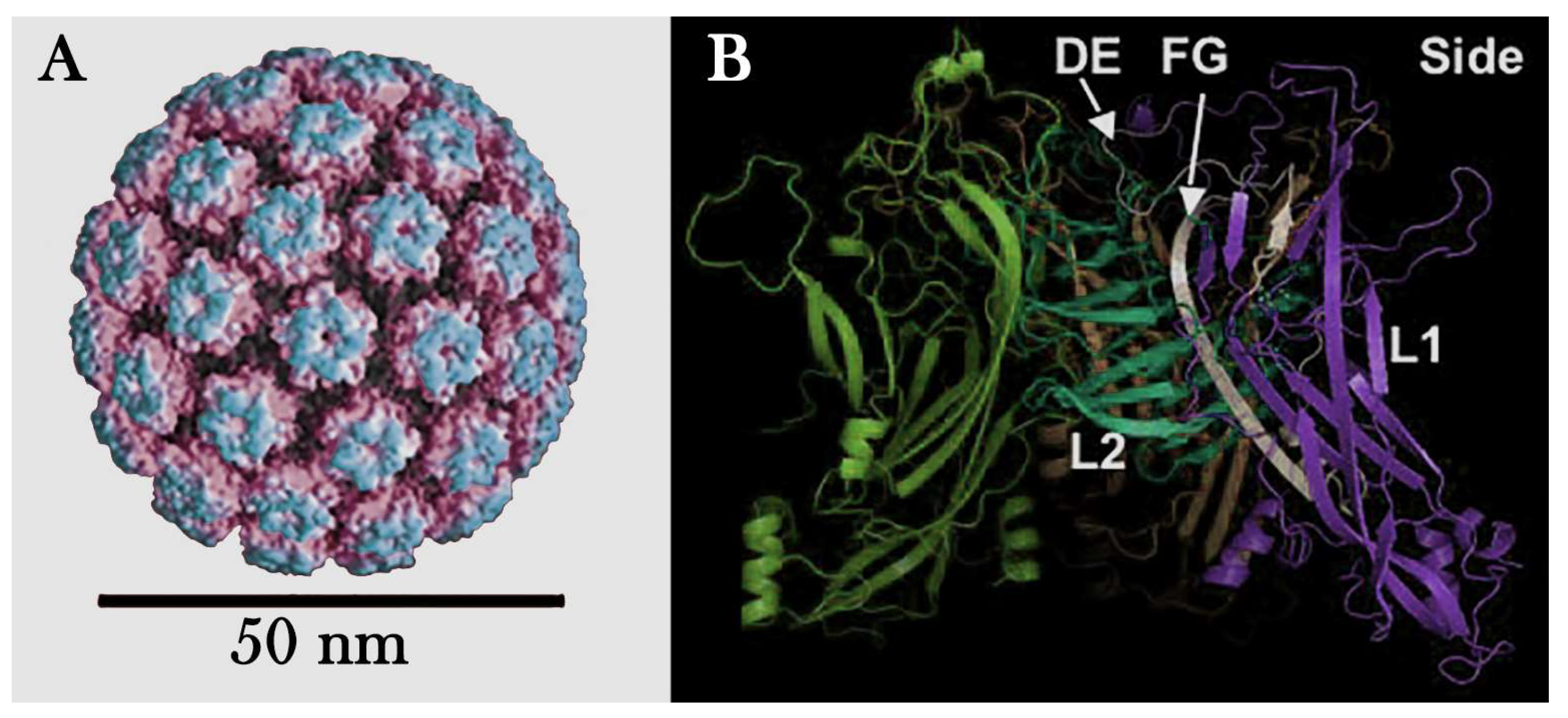
- Capsid: The capsid is a protein coat that surrounds and protects the virus's genetic material. It is composed of proteins called capsomeres, which are arranged in specific patterns. The shape of the capsid can be icosahedral, helical, or complex, depending on the type of virus. For example, the capsid of HPV is composed of two structural proteins, L1 and L2, which assemble to form an icosahedral structure. The major capsid protein, L1, organizes into 72 pentameric capsomeres, arranged in a T=7 symmetric lattice (Figure 3.A). These capsomeres create the outer shell of the capsid, providing the virus with its characteristic spherical shape. The minor capsid protein, L2, is located internally and plays a role in stabilizing the structure and facilitating viral genome packaging. Together, L1 and L2 form a robust and highly organized capsid that protects the viral DNA and mediates host cell entry.
- II.
- Genetic Material: Viruses contain genetic material, either DNA or RNA, which can be single- or double-stranded. This material allows the virus to hijack host cell machinery, replicate, and produce new viral particles. For example, HPV has a double-stranded DNA genome that encodes proteins for replication and assembly, enabling it to infect and spread within host cells. This genetic diversity is key to viruses' ability to infect a wide range of hosts and cause disease.
- III.
- Viral Envelope: Some viruses have a lipid envelope surrounding the capsid. This envelope is derived from the host cell membrane and contains viral proteins that help the virus adhere to and enter the cells. Enveloped viruses, such as HIV and influenza viruses, are generally more sensitive to disinfectants and environmental conditions.
2.4.2. Viral Function
- I.
- Attachment and Entry: Proteins on the surface of the viral capsid or envelope bind to specific receptors on the host cell surface. For example, in HPV, the major capsid protein L1 interacts with heparan sulfate proteoglycans (HSPGs) on the cell surface. This binding trigger structural changes in the virus, exposing other proteins essential for internalization. In HPV, the minor capsid protein L2 becomes exposed after the L1-HSPG interaction. The exposed L2 protein then interacts with additional cell receptors, such as integrins, facilitating clathrin- or caveolin-mediated endocytosis. This process enables the virus to enter the host cell and initiate infection.
- II.
- Replication: Once inside the cell, the virus's genetic material is released and uses the host cell machinery to replicate itself. This may involve transcription and translation of viral genetic material to produce new viral proteins.
- III.
- Assembly: After replication, new viral particles (viruses) are assembled from the genetic material and proteins produced. Depending on the type of virus, this may occur in the nucleus or cytoplasm of the cell.
- IV.
- Release: Finally, the new viruses are released from the host cell. In enveloped viruses, this usually occurs by budding, where the virus acquires its envelope from the host cell membrane. In non-enveloped viruses, the cell may rupture, releasing viral particles.
3. Overview of Specific Concepts
3.1. Prophylactic and Therapeutic DNA Vaccine against HPV and Virus-Associated Cancers
3.2. Peptide-Based Vaccine: Current Landscape in the Therapeutic HPV-Related Cancers
3.3. In Silico Peptides Evaluation Directed to HPV
3.5. Chimeras Designed for New HPV Vaccines
4. Viral Structure and Physical Properties
4.1. Imaging Methods
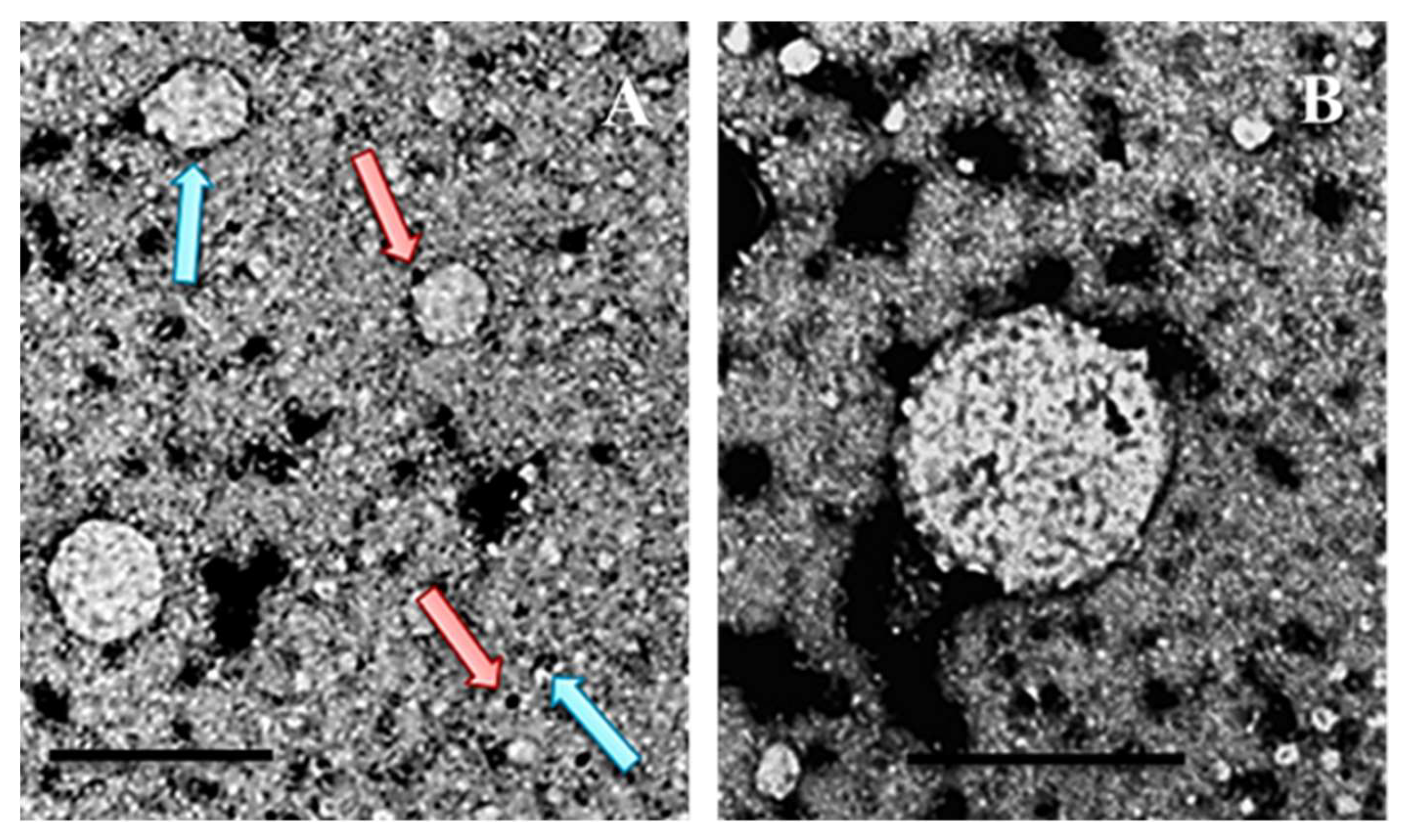
- Electron Microscopy (EM) is one of the main techniques for visualizing viral morphology. It allows the observation of viruses at high resolution, revealing details about the shape, size, and organization of the capsid and envelope, if present (Figure 4.A and B).
- X-ray Crystallography allows the determination of the three-dimensional structure of viral proteins at the atomic level, providing detailed information about the arrangement of proteins in the capsid.
4.2. Spectroscopy
- Nuclear Magnetic Resonance Spectroscopy (NMR) can be used to study the structure of viral proteins in solutions, helping to understand how proteins interact with each other and with the genetic material.
- Mass Spectrometry (MS) can be used to analyze the protein composition of viruses, identifying the present proteins and their post-translational modifications.
4.3. Biochemical Analysis
- Virus Purification Studies allow the analysis of their physical properties, such as size, shape, and stability. Methods such as density gradient centrifugation help separate viruses from other cellular components.
- Stability Assays Tests under different conditions (pH, temperature, presence of solvents) help determine viruses' physical properties, such as their resistance and sensitivity to disinfectants.
4.4. Computational Modeling
- Molecular Simulations, performed in computational models, can predict the structure and interactions of viral proteins, helping to understand how the structure influences the virus's function and pathogenicity (Figure 3.A and B).
4.5. Genomics and Proteomics
- Virus Genetic Sequencing provides information about the organization and composition of the viral genome, which is essential to understanding the structure and function of the encoded proteins.
- Proteomic Analysis is a fundamental approach to identifying and quantifying viral proteins, improving our understanding of viral biology and interactions with host cells. The following topic discusses how proteomic analysis is crucial in viral studies.
4.6. Highlights in Proteomic Analysis
4.6.1. Viral Protein Identification
4.6.2. Quantification of Viral Proteins
4.6.3. Functional Analysis
Acknowledgments
References
- Ahmels, M.; Mariz, F.C.; Braspenning-Wesch, I.; Stephan, S.; Huber, B.; Schmidt, G.; Cao, R.; Müller, M.; Kirnbauer, R.; Rösl, F.; et al. Next Generation L2-Based HPV Vaccines Cross-Protect against Cutaneous Papillomavirus Infection and Tumor Development. Front. Immunol. 2022, 13, 1010790. [CrossRef]
- Gardella, B.; Pasquali, M.F.; La Verde, M.; Cianci, S.; Torella, M.; Dominoni, M. The Complex Interplay between Vaginal Microbiota, HPV Infection, and Immunological Microenvironment in Cervical Intraepithelial Neoplasia: A Literature Review. IJMS 2022, 23, 7174. [CrossRef]
- Mohsen, M.O.; Bachmann, M.F. Virus-like Particle Vaccinology, from Bench to Bedside. Cell Mol Immunol 2022, 19, 993–1011. [CrossRef]
- Yan, D.; Wei, Y.-Q.; Guo, H.-C.; Sun, S.-Q. The Application of Virus-like Particles as Vaccines and Biological Vehicles. Appl Microbiol Biotechnol 2015, 99, 10415–10432. [CrossRef]
- Kheirvari, M.; Liu, H.; Tumban, E. Virus-like Particle Vaccines and Platforms for Vaccine Development. Viruses 2023, 15, 1109. [CrossRef]
- Crisci, E.; Bárcena, J.; Montoya, M. Virus-like Particle-Based Vaccines for Animal Viral Infections. Inmunología 2013, 32, 102–116. [CrossRef]
- Bárcena, J.; Blanco, E. Design of Novel Vaccines Based on Virus-Like Particles or Chimeric Virions. In Structure and Physics of Viruses; Mateu, M.G., Ed.; Subcellular Biochemistry; Springer Netherlands: Dordrecht, 2013; Vol. 68, pp. 631–665 ISBN 978-94-007-6551-1.
- González-Domínguez, I.; Lorenzo, E.; Bernier, A.; Cervera, L.; Gòdia, F.; Kamen, A. A Four-Step Purification Process for Gag VLPs: From Culture Supernatant to High-Purity Lyophilized Particles. Vaccines 2021, 9, 1154. [CrossRef]
- Boxus, M.; Fochesato, M.; Miseur, A.; Mertens, E.; Dendouga, N.; Brendle, S.; Balogh, K.K.; Christensen, N.D.; Giannini, S.L. Broad Cross-Protection Is Induced in Preclinical Models by a Human Papillomavirus Vaccine Composed of L1/L2 Chimeric Virus-Like Particles. J Virol 2016, 90, 6314–6325. [CrossRef]
- Nooraei, S.; Bahrulolum, H.; Hoseini, Z.S.; Katalani, C.; Hajizade, A.; Easton, A.J.; Ahmadian, G. Virus-like Particles: Preparation, Immunogenicity and Their Roles as Nanovaccines and Drug Nanocarriers. J Nanobiotechnol 2021, 19, 59. [CrossRef]
- Petrov, G.V.; Galkina, D.A.; Koldina, A.M.; Grebennikova, T.V.; Eliseeva, O.V.; Chernoryzh, Y.Yu.; Lebedeva, V.V.; Syroeshkin, A.V. Controlling the Quality of Nanodrugs According to Their New Property—Radiothermal Emission. Pharmaceutics 2024, 16, 180. [CrossRef]
- Structure and Physics of Viruses: An Integrated Guide; Mateu, M.G., Ed.; Subcellular Biochemistry; Springer Nature Switzerland: Cham, 2024; Vol. 105; ISBN 978-3-031-65186-1.
- Sakauchi, D.; Sasaki, E.A.K.; Anni, F.O.B.; Cianciarullo, A.M. Production of HPV16 Capsid Proteins in Suspension Cultures of Human Epithelial Cells. World J. Adv. Res. Rev. 2021, 9, 258–268. [CrossRef]
- Bei, L.; Gao, S.; Zhao, D.; Kou, Y.; Liang, S.; Wu, Y.; Zhang, X.; Meng, D.; Lu, J.; Luo, C.; et al. Immunogenicity Assessment of a 14-Valent Human Papillomavirus Vaccine Candidate in Mice. Vaccines 2024, 12, 1262. [CrossRef]
- Wang, R.; Huang, H.; Yu, C.; Li, X.; Wang, Y.; Xie, L. Current Status and Future Directions for the Development of Human Papillomavirus Vaccines. Front. Immunol. 2024, 15, 1362770. [CrossRef]
- Gupta, R.; Arora, K.; Roy, S.S.; Joseph, A.; Rastogi, R.; Arora, N.M.; Kundu, P.K. Platforms, Advances, and Technical Challenges in Virus-like Particles-Based Vaccines. Front. Immunol. 2023, 14, 1123805. [CrossRef]
- Nierengarten, M.B. Men Face Substantial Lifelong Risk of Oral HPV Infection: A Study Provides Information on Rates of Newly Acquired Oral HPV Infections and Associated Risk Factors for Acquiring HPV Infections in a Multinational Cohort of More than 3000 Men. Cancer 2025, 131, e35714. [CrossRef]
- Zhao, Q.; Modis, Y.; High, K.; Towne, V.; Meng, Y.; Wang, Y.; Alexandroff, J.; Brown, M.; Carragher, B.; Potter, C.S.; et al. Disassembly and Reassembly of Human Papillomavirus Virus-like Particles Produces More Virion-like Antibody Reactivity. Virol J 2012, 9, 52. [CrossRef]
- Lowe, J.; Panda, D.; Rose, S.; Jensen, T.; Hughes, W.A.; Tso, F.Y.; Angeletti, P.C. Evolutionary and Structural Analyses of Alpha-Papillomavirus Capsid Proteins Yields Novel Insights into L2 Structure and Interaction with L1. Virol J 2008, 5, 150. [CrossRef]
- Zella, D.; Gallo, R.C. Viruses and Bacteria Associated with Cancer: An Overview. Viruses 2021, 13, 1039. [CrossRef]
- Singh, D.; Vignat, J.; Lorenzoni, V.; Eslahi, M.; Ginsburg, O.; Lauby-Secretan, B.; Arbyn, M.; Basu, P.; Bray, F.; Vaccarella, S. Global Estimates of Incidence and Mortality of Cervical Cancer in 2020: A Baseline Analysis of the WHO Global Cervical Cancer Elimination Initiative. The Lancet Global Health 2023, 11, e197–e206. [CrossRef]
- Adelstein, D.J.; Rodriguez, C.P. Human Papillomavirus: Changing Paradigms in Oropharyngeal Cancer. Curr Oncol Rep 2010, 12, 115–120. [CrossRef]
- Pickard, R.K.L.; Xiao, W.; Broutian, T.R.; He, X.; Gillison, M.L. The Prevalence and Incidence of Oral Human Papillomavirus Infection Among Young Men and Women, Aged 18–30 Years. Sexually Transmitted Diseases 2012, 39, 559–566. [CrossRef]
- Andersen, A.S.; Koldjær Sølling, A.S.; Ovesen, T.; Rusan, M. The Interplay between HPV and Host Immunity in Head and Neck Squamous Cell Carcinoma. Intl Journal of Cancer 2014, 134, 2755–2763. [CrossRef]
- De Vuyst, H.; Clifford, G.M.; Nascimento, M.C.; Madeleine, M.M.; Franceschi, S. Prevalence and Type Distribution of Human Papillomavirus in Carcinoma and Intraepithelial Neoplasia of the Vulva, Vagina and Anus: A Meta-analysis. Intl Journal of Cancer 2009, 124, 1626–1636. [CrossRef]
- De Martel, C.; Plummer, M.; Vignat, J.; Franceschi, S. Worldwide Burden of Cancer Attributable to HPV by Site, Country and HPV Type. Intl Journal of Cancer 2017, 141, 664–670. [CrossRef]
- Accardi, R.; Gheit, T. Cutaneous HPV and Skin Cancer. La Presse Médicale 2014, 43, e435–e443. [CrossRef]
- Paolini, F.; Cota, C.; Amantea, A.; Curzio, G.; Venuti, A. Mucosal Alpha-Papillomavirus (HPV89) in a Rare Skin Lesion. Virol J 2015, 12, 105. [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J Clinicians 2024, 74, 229–263. [CrossRef]
- International Agency for Reasearch on Cancer (IARC) Cancer Today.
- Brazil Healthy Ministry NOTA TÉCNICA No 41/2024-CGICI/DPNI/SVSA/MS 2024.
- Cianciarullo, A.M. Revista SODEBRAS. 2014, pp. 134–141.
- INCA, I.N. de C. Prevenção do câncer do colo do útero Available online: https://www.gov.br/inca/pt-br/assuntos/gestor-e-profissional-de-saude/controle-do-cancer-do-colo-do-utero/acoes/prevencao (accessed on 18 January 2025).
- Herrero, R.; González, P.; Markowitz, L.E. Present Status of Human Papillomavirus Vaccine Development and Implementation. The Lancet Oncology 2015, 16, e206–e216. [CrossRef]
- Wang, D.; Li, Z.; Xiao, J.; Wang, J.; Zhang, L.; Liu, Y.; Fan, F.; Xin, L.; Wei, M.; Kong, Z.; et al. Identification of Broad-Genotype HPV L2 Neutralization Site for Pan-HPV Vaccine Development by a Cross-Neutralizing Antibody. PLoS ONE 2015, 10, e0123944. [CrossRef]
- Vande Pol, S.B.; Klingelhutz, A.J. Papillomavirus E6 Oncoproteins. Virology 2013, 445, 115–137. [CrossRef]
- Cianciarullo, A.M.; Sasaki, E.A.K. Process of Producing an Immunological Composition of Prophylatic and Therapeutic DNA against HPV and Cancers Associated with the Virus, Hybrid Protein, Expression Vector, Immunologial Composition, and Its Use. Patent 2022.
- Cianciarullo, A.M.; Sasaki, E.A.K. PROCESSO DE PRODUÇÃO DE UMA COMPOSIÇÃO IMUNOLÓGICA DE DNA PROFILÁTICA E TERAPÊUTICA CONTRA HPV E CÂNCERES ASSOCIADOS AO VÍRUS, PROTEÍNA HÍBRIDA, VETOR DE EXPRESSÃO, COMPOSIÇÃO IMUNOLÓGICA E SEUS USOS. Patent 2019.
- Hancock, G.; Hellner, K.; Dorrell, L. Therapeutic HPV Vaccines. Best Practice & Research Clinical Obstetrics & Gynaecology 2018, 47, 59–72. [CrossRef]
- Hamley, I.W. Peptides for Vaccine Development. ACS Appl. Bio Mater. 2022, 5, 905–944. [CrossRef]
- Jaradat, D. Thirteen Decades of Peptide Synthesis: Key Developments in Solid Phase Peptide Synthesis and Amide Bond Formation Utilized in Peptide Ligation. Amino Acids 2018, 50, 1–30. [CrossRef]
- Merrifield, B. [1] Concept and Early Development of Solid-Phase Peptide Synthesis. In Methods in Enzymology; Elsevier, 1997; Vol. 289, pp. 3–13 ISBN 978-0-12-182190-6.
- El-Baky, N.A.; Elkhawaga, M.A.; Abdelkhalek, E.S.; Sharaf, M.M.; Redwan, E.M.; Kholef, H.R. De Novo Expression and Antibacterial Potential of Four Lactoferricin Peptides in Cell-Free Protein Synthesis System. Biotechnology Reports 2021, 29, e00583. [CrossRef]
- Torres, M.D.T.; Cao, J.; Franco, O.L.; Lu, T.K.; De La Fuente-Nunez, C. Synthetic Biology and Computer-Based Frameworks for Antimicrobial Peptide Discovery. ACS Nano 2021, 15, 2143–2164. [CrossRef]
- Tîrziu, A.; Avram, S.; Mada, L.; Crișan-Vida, M.; Popovici, C.; Popovici, D.; Faur, C.; Duda-Seiman, C.; Păunescu, V.; Vernic, C. Design of a Synthetic Long Peptide Vaccine Targeting HPV-16 and -18 Using Immunoinformatic Methods. Pharmaceutics 2023, 15, 1798. [CrossRef]
- Zhai, L.; Peabody, J.; Pang, Y.-Y.S.; Schiller, J.; Chackerian, B.; Tumban, E. A Novel Candidate HPV Vaccine: MS2 Phage VLP Displaying a Tandem HPV L2 Peptide Offers Similar Protection in Mice to Gardasil-9. Antiviral Research 2017, 147, 116–123. [CrossRef]
- Namvar, A.; Bolhassani, A.; Javadi, G.; Noormohammadi, Z. In Silico/In Vivo Analysis of High-Risk Papillomavirus L1 and L2 Conserved Sequences for Development of Cross-Subtype Prophylactic Vaccine. Sci Rep 2019, 9, 15225. [CrossRef]
- Ronchi, V.P.; Klein, J.M.; Edwards, D.J.; Haas, A.L. The Active Form of E6-Associated Protein (E6AP)/UBE3A Ubiquitin Ligase Is an Oligomer. Journal of Biological Chemistry 2014, 289, 1033–1048. [CrossRef]
- Pal, A.; Kundu, R. Human Papillomavirus E6 and E7: The Cervical Cancer Hallmarks and Targets for Therapy. Front. Microbiol. 2020, 10, 3116. [CrossRef]
- Greenfield, W.W.; Stratton, S.L.; Myrick, R.S.; Vaughn, R.; Donnalley, L.M.; Coleman, H.N.; Mercado, M.; Moerman-Herzog, A.M.; Spencer, H.J.; Andrews-Collins, N.R.; et al. A Phase I Dose-Escalation Clinical Trial of a Peptide-Based Human Papillomavirus Therapeutic Vaccine with Candida Skin Test Reagent as a Novel Vaccine Adjuvant for Treating Women with Biopsy-Proven Cervical Intraepithelial Neoplasia 2/3. OncoImmunology 2015, 4, e1031439. [CrossRef]
- Van Poelgeest, M.I.E.; Welters, M.J.P.; Van Esch, E.M.G.; Stynenbosch, L.F.M.; Kerpershoek, G.; Van Persijn Van Meerten, E.L.; Van Den Hende, M.; Löwik, M.J.G.; Berends-van Der Meer, D.M.A.; Fathers, L.M.; et al. HPV16 Synthetic Long Peptide (HPV16-SLP) Vaccination Therapy of Patients with Advanced or Recurrent HPV16-Induced Gynecological Carcinoma, a Phase II Trial. J Transl Med 2013, 11, 88. [CrossRef]
- De Vos Van Steenwijk, P.J.; Van Poelgeest, M.I.E.; Ramwadhdoebe, T.H.; Löwik, M.J.G.; Berends-van Der Meer, D.M.A.; Van Der Minne, C.E.; Loof, N.M.; Stynenbosch, L.F.M.; Fathers, L.M.; Valentijn, A.R.P.M.; et al. The Long-Term Immune Response after HPV16 Peptide Vaccination in Women with Low-Grade Pre-Malignant Disorders of the Uterine Cervix: A Placebo-Controlled Phase II Study. Cancer Immunol Immunother 2014, 63, 147–160. [CrossRef]
- Gomes, D.; Silvestre, S.; Duarte, A.P.; Venuti, A.; Soares, C.P.; Passarinha, L.; Sousa, Â. In Silico Approaches: A Way to Unveil Novel Therapeutic Drugs for Cervical Cancer Management. Pharmaceuticals 2021, 14, 741. [CrossRef]
- Matos, A.S.; Invenção, M.D.C.V.; Moura, I.A.D.; Freitas, A.C.D.; Batista, M.V.D.A. Immunoinformatics Applications in the Development of Therapeutic Vaccines against Human Papillomavirus-related Infections and Cervical Cancer. Reviews in Medical Virology 2023, 33, e2463. [CrossRef]
- Yang, A.; Farmer, E.; Wu, T.C.; Hung, C.-F. Perspectives for Therapeutic HPV Vaccine Development. J Biomed Sci 2016, 23, 75. [CrossRef]
- Yang, A.; Jeang, J.; Cheng, K.; Cheng, T.; Yang, B.; Wu, T.-C.; Hung, C.-F. Current State in the Development of Candidate Therapeutic HPV Vaccines. Expert Review of Vaccines 2016, 15, 989–1007. [CrossRef]
- Kayyal, M.; Bolhassani, A.; Noormohammadi, Z.; Sadeghizadeh, M. In Silico Design and Immunological Studies of Two Novel Multiepitope DNA-Based Vaccine Candidates Against High-Risk Human Papillomaviruses. Mol Biotechnol 2021, 63, 1192–1222. [CrossRef]
- Kaufmann, A.M.; Nieland, J.D.; Jochmus, I.; Baur, S.; Friese, K.; Gabelsberger, J.; Gieseking, F.; Gissmann, L.; Glasschröder, B.; Grubert, T.; et al. Vaccination Trial with HPV16 L1E7 Chimeric Virus-like Particles in Women Suffering from High Grade Cervical Intraepithelial Neoplasia (CIN 2/3). Intl Journal of Cancer 2007, 121, 2794–2800. [CrossRef]
- Garcia, F.; Petry, K.U.; Muderspach, L.; Gold, M.A.; Braly, P.; Crum, C.P.; Magill, M.; Silverman, M.; Urban, R.G.; Hedley, M.L.; et al. ZYC101a for Treatment of High-Grade Cervical Intraepithelial Neoplasia: A Randomized Controlled Trial. Obstetrics & Gynecology 2004, 103, 317–326. [CrossRef]
- Gaillard, S.; Deery, A.; Fader, A.N.; Huh, W.; Liang, M.; Straughn, M.; Arend, R.; Wu, T.C.; Leath, C.; Roden, R. Safety and Feasibility of an HPV Therapeutic Vaccine (TA-CIN) in Patients with HPV16 Associated Cervical Cancer (458). Gynecologic Oncology 2022, 166, S228–S229. [CrossRef]
- Kaufmann, A.M.; Stern, P.L.; Rankin, E.M.; Sommer, H.; Nuessler, V.; Schneider, A.; Adams, M.; Onon, T.S.; Bauknecht, T.; Wagner, U.; et al. Safety and Immunogenicity of TA-HPV, a Recombinant Vaccinia Virus Expressing Modified Human Papillomavirus (HPV)-16 and HPV-18 E6 and E7 Genes, in Women with Progressive Cervical Cancer. Clin Cancer Res 2002, 8, 3676–3685.
- Roman, L.D.; Wilczynski, S.; Muderspach, L.I.; Burnett, A.F.; O’Meara, A.; Brinkman, J.A.; Kast, W.M.; Facio, G.; Felix, J.C.; Aldana, M.; et al. A Phase II Study of Hsp-7 (SGN-00101) in Women with High-Grade Cervical Intraepithelial Neoplasia. Gynecologic Oncology 2007, 106, 558–566. [CrossRef]
- Kayyal, M.; Bolhassani, A.; Noormohammadi, Z.; Sadeghizadeh, M. Immunological Responses and Anti-Tumor Effects of HPV16/18 L1-L2-E7 Multiepitope Fusion Construct along with Curcumin and Nanocurcumin in C57BL/6 Mouse Model. Life Sciences 2021, 285, 119945. [CrossRef]
- Bahmani, B.; Amini-bayat, Z.; Ranjbar, M.M.; Bakhtiari, N.; Zarnani, A.-H. HPV16-E7 Protein T Cell Epitope Prediction and Global Therapeutic Peptide Vaccine Design Based on Human Leukocyte Antigen Frequency: An In-Silico Study. Int J Pept Res Ther 2021, 27, 365–378. [CrossRef]
- Gupta, S.K.; Singh, A.; Srivastava, M.; Gupta, S.K.; Akhoon, B.A. In Silico DNA Vaccine Designing against Human Papillomavirus (HPV) Causing Cervical Cancer. Vaccine 2009, 28, 120–131. [CrossRef]
- Zhu, F.-C.; Zhong, G.-H.; Huang, W.-J.; Chu, K.; Zhang, L.; Bi, Z.-F.; Zhu, K.-X.; Chen, Q.; Zheng, T.-Q.; Zhang, M.-L.; et al. Head-to-Head Immunogenicity Comparison of an Escherichia Coli-Produced 9-Valent Human Papillomavirus Vaccine and Gardasil 9 in Women Aged 18–26 Years in China: A Randomised Blinded Clinical Trial. The Lancet Infectious Diseases 2023, 23, 1313–1322. [CrossRef]
- Gambhira, R.; Karanam, B.; Jagu, S.; Roberts, J.N.; Buck, C.B.; Bossis, I.; Alphs, H.; Culp, T.; Christensen, N.D.; Roden, R.B.S. A Protective and Broadly Cross-Neutralizing Epitope of Human Papillomavirus L2. J Virol 2007, 81, 13927–13931. [CrossRef]
- Zhou, X.; Lian, H.; Li, H.; Fan, M.; Xu, W.; Jin, Y. Nanotechnology in Cervical Cancer Immunotherapy: Therapeutic Vaccines and Adoptive Cell Therapy. Front. Pharmacol. 2022, 13, 1065793. [CrossRef]
- Mukerjee, N.; Maitra, S.; Gorai, S.; Ghosh, A.; Alexiou, A.; Thorat, N.D. Revolutionizing Human Papillomavirus (HPV)-related Cancer Therapies: Unveiling the Promise of Proteolysis Targeting Chimeras (PROTACs) and Proteolysis Targeting Antibodies (PROTABs) in Cancer Nano-vaccines. Journal of Medical Virology 2023, 95, e29135. [CrossRef]
- Structure and Physics of Viruses: An Integrated Textbook; Mateu, M.G., Ed.; Subcellular Biochemistry; Springer Netherlands: Dordrecht, 2013; Vol. 68; ISBN 978-94-007-6551-1.
- Johnson, J.E.; Speir, J.A. Virus Particle Structure: Principles. In Encyclopedia of Virology; Elsevier, 2008; pp. 393–401 ISBN 978-0-12-374410-4.
- Johnson, J.E. Multi-Disciplinary Studies of Viruses: The Role of Structure in Shaping the Questions and Answers. Journal of Structural Biology 2008, 163, 246–253. [CrossRef]
- Cianciarullo A.M.; Oliveira C.S.; Vicente P.I.; Sakauchi D.; Sasaki E.A.K. Prophylactic and Therapeutic Vaccines against HPV and Associated Cancers; 1st ed.; CRV Publisher, 2024; ISBN 978-65-251-5296-7.
- Lum, K.K.; Cristea, I.M. Proteomic Approaches to Uncovering Virus–Host Protein Interactions during the Progression of Viral Infection. Expert Review of Proteomics 2016, 13, 325–340. [CrossRef]
- Zheng, J.; Tan, B.H.; Sugrue, R.; Tang, K. Current Approaches on Viral Infection: Proteomics and Functional Validations. Front. Microbio. 2012, 3. [CrossRef]
- Balvers, M.; Gordijn, I.F.; Voskamp-Visser, I.A.I.; Schelling, M.F.A.; Schuurman, R.; Heikens, E.; Braakman, R.; Stingl, C.; Van Leeuwen, H.C.; Luider, T.M.; et al. Proteome2virus: Shotgun Mass Spectrometry Data Analysis Pipeline for Virus Identification. Journal of Clinical Virology Plus 2023, 3, 100147. [CrossRef]
- Saha, D.; Iannuccelli, M.; Brun, C.; Zanzoni, A.; Licata, L. The Intricacy of the Viral-Human Protein Interaction Networks: Resources, Data, and Analyses. Front. Microbiol. 2022, 13, 849781. [CrossRef]
- Kocjan, B.J.; Seme, K.; Poljak, M. Detection and Differentiation of Human Papillomavirus Genotypes HPV-6 and HPV-11 by FRET-Based Real-Time PCR. Journal of Virological Methods 2008, 153, 245–249. [CrossRef]
- Castelletto, S.; Boretti, A. Viral Particle Imaging by Super-Resolution Fluorescence Microscopy. Chemical Physics Impact 2021, 2, 100013. [CrossRef]
- Holmes, E.C.; Krammer, F.; Goodrum, F.D. Virology—The next Fifty Years. Cell 2024, 187, 5128–5145. [CrossRef]
| Bivalent Cervarix® |
Bivalent Cecolin® |
Bivalent WalrinVax® |
Quadrivalent Gardasil® |
Quadrivalent Cervavac-4* |
Nonavalent Gardasil-9® |
14-valent SCT-1000 |
|
|---|---|---|---|---|---|---|---|
| Approval | 2009 | 2019 | 2022 | 2006 | 2022 | 2014 | Phase III |
| Manufacturer | GlaxoSmith Kline plc |
Xiamen Innovax Co., Ltd | Walvax Co., Ltd |
Merck & Co., Inc | Serum Institute of India | Merck & Co., Inc | SinoCelltech Group |
| VLP HPV Type (protein dose) | HPV-16 (20 µg) HPV-18 (20 µg) |
HPV-16 (40 µg) HPV-18 (20 µg) |
HPV-16 (40 µg) HPV-18 (20 µg) |
HPV-6, 18 (20 µg each) HPV-11, 16 (40 µg each) |
HPV-6, 18 (20 µg each) HPV-11, 16 (40 µg each) |
HPV-6 (30 mg) HPV-16 (60 mg) HPV-11, 18 (40 mg each) HPV-31, 33, 45, 52, 58 (20 mg each) |
HPV-11, 18 (40 µg each) HPV-6 (30 µg) HPV-16 (60 µg) HPV-31, 33, 35, 39, 45, 51, 52, 56, 58, 59 (20 µg each) |
| Expression system | Trichoplusia ni insect cell line | Escherichia coli | Pichia pastoris | Saccharomyces cerevisiae | Hansenula polymorpha | Saccharomyces cerevisiae | SF9 insect cells, transfected with Baculovirus |
| Adjuvant | AS04 | Aluminum hydroxide | Aluminum phosphate | AAHS | Aluminum hydroxide | AAHS | Aluminum phosphate |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





