Submitted:
11 March 2025
Posted:
11 March 2025
You are already at the latest version
Abstract
Keywords:
1. Hypoxia-Inducible Factors (HIFs) and the Cellular Oxygen Sensor
2. Cis-Regulatory Elements and the 3D Genome
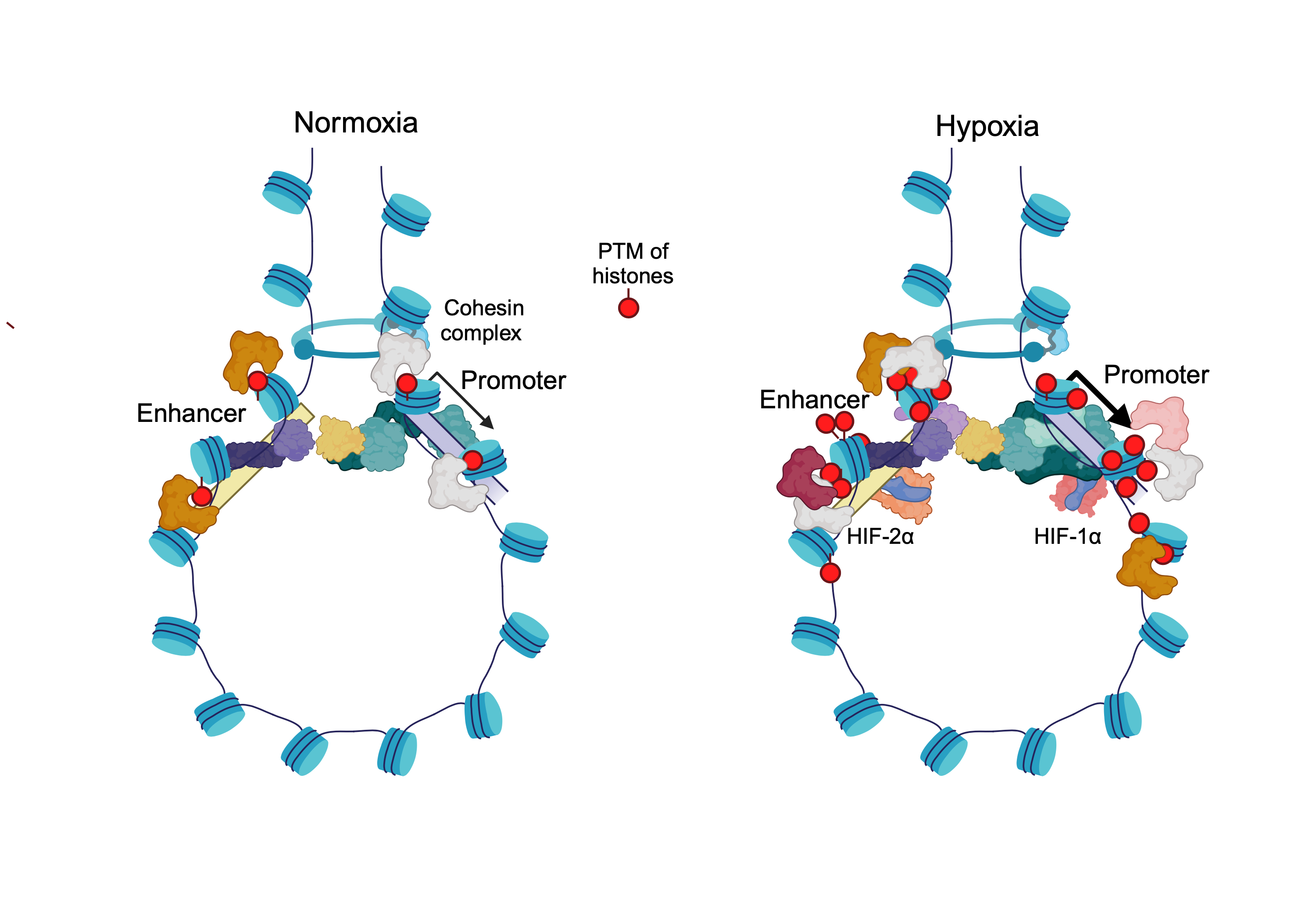
3. Distal cis-Regulatory Elements and the Hypoxia Response
4. The 3D Genome Under Hypoxia
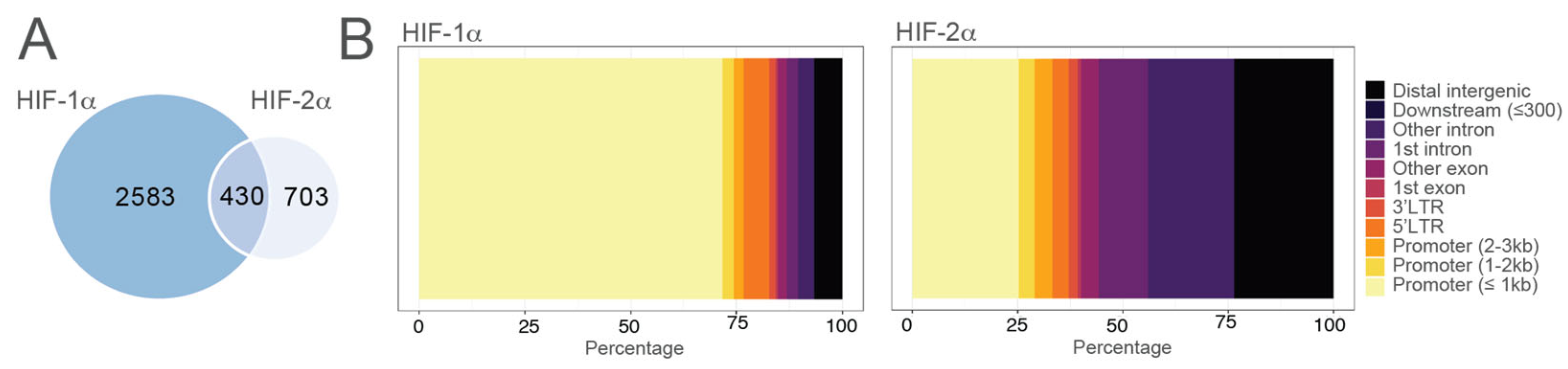
5. The Landscape of HIFs Genomic Binding
6. Chromatin Accessibility Under Hypoxia
7. Epigenetic Modifications
7.1. DNA Methylation
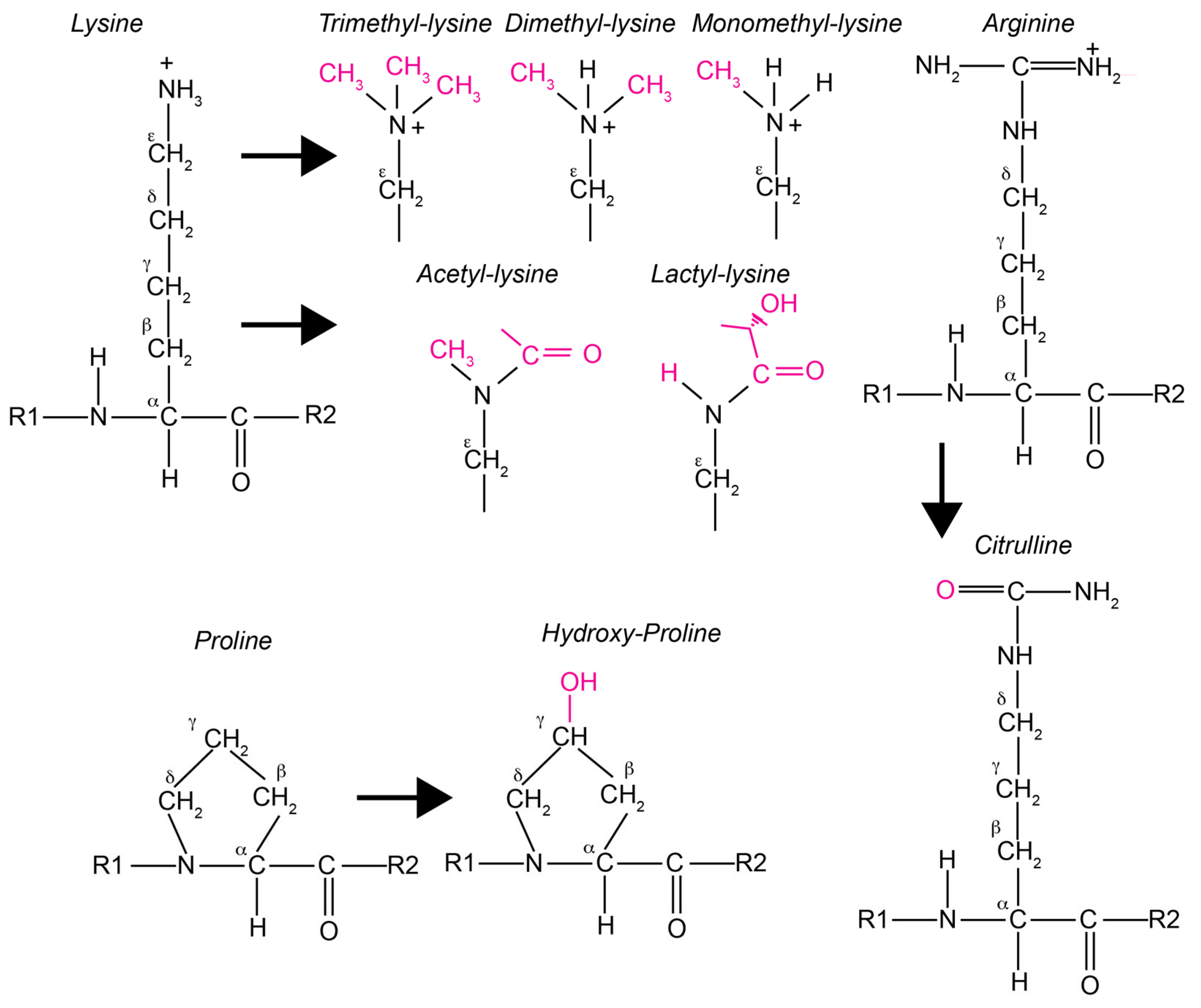
7.2. The Epigenetic of Histones
7.3. Histone Acetylation Under Hypoxia
7.4. Histone Methylation Under Hypoxia
7.5. Other PTMs of Histones and Hypoxia
7.5.1. Histone Phosphorylation
7.5.2. Histone Ubiquitylation
7.5.3. Histone Hydroxylation
7.5.4. Histone Citrullination
7.5.5. Histone Lactylation
| Histone modification |
Cell line/Tissue | Hypoxia | Reference | GEO ID | |
|---|---|---|---|---|---|
| H3K27ac | HUVECs | 1% | 146 | GSE38555 GSE50144 |
|
| H3K27ac | MCF-7 | 0.5% | 49, 145 | GSE78113 | |
| H3K27ac | hMSMCs | 3% | 149 | HRI266643 (GSA repository) |
|
| H3K27ac | PANC-1 | pO2=1% | 143 | GSE93982 GSE93989 |
|
| H3K27ac | EA.Hy926 | 1% | 148 | GSE120527 | |
| H3K27ac | FaDu | 1% | 81 | GSE260872 | |
| H3K27ac | ccRCC | VHL inactivation | 40 | GSE86095 | |
| H3K27ac, H3ac, H4ac |
HUVECs | 1% | 62 | GSE35932 | |
| H3ac | DLD-1, TIG-3 | 1% | 63 | DRA000285-000288 DRA000293-000296 (DDBJ database) |
|
| H3K4ac | FaDu | 1% | 153 | GSE80218 | |
| H3K27ac | Human placenta | FGR | 146 | N.A.* |
| Histone modifications |
Cell line Tissue |
Hypoxia | Reference | GEO ID | |
|---|---|---|---|---|---|
| H3K4me3, H3K36me3 | HeLa cells | 1% | 169 | GSE120339 | |
| H3K4me3 H3K4me1 | FaDu | 1% | 81 | GSE260872 | |
| H3K4me3 | ESF, DSC | 1% | 183 | GSE167946 | |
| H3K4me3, H3K4me1 | HUVEC | 1% | 62 | GSE39089 | |
| H3K4me3, H3K27me3 | MCF7 | <0.02% | 168 | GSE71031 | |
| H3K4me3 H3K27me3 | MCF7 | <0.02% | 167 | GSE71031 | |
| H3K4me3 | MCF-7, Human PTCs | 1% | 145 | GSE78113 | |
| H3K4me3 H3K4me1 | MCF7, RCC4, SK-MEL-28, A549 | 0.5% | 105 | GSE85352 | |
| H3K4me3 | HepG2, U87 | 0.5% | 172 | GSE18505 | |
| H3K27me3 | MCF7, HMLER | <0.02 to 1.0% | 174 | GSE61740 | |
| H3K27me3 | HCC1806 cells | 1% | 176 | GSE253833 |
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Pugh, C. W.; Ratcliffe, P. J. New Horizons in Hypoxia Signaling Pathways. Exp. Cell Res. 2017, 356 (2), 116–121.
- Choudhry, H.; Harris, A. L. Advances in Hypoxia-Inducible Factor Biology. Cell Metab. 2018, 27 (2), 281–298. [CrossRef]
- Semenza, G. L. HIF-1 Mediates Metabolic Responses to Intratumoral Hypoxia and Oncogenic Mutations. J. Clin. Invest. 2013, 123 (9), 3664–3671. [CrossRef]
- Kaelin, W. G. Von Hippel–Lindau Disease: Insights into Oxygen Sensing, Protein Degradation, and Cancer. J. Clin. Invest. 2022, 132 (18).
- Minisini, M.; Cricchi, E.; Brancolini, C. Acetylation and Phosphorylation in the Regulation of Hypoxia-Inducible Factor Activities: Additional Options to Modulate Adaptations to Changes in Oxygen Levels. Life 2024, 14 (1), 20. [CrossRef]
- Islam, M. S.; Leissing, T. M.; Chowdhury, R.; Hopkinson, R. J.; Schofield, C. J. 2-Oxoglutarate-Dependent Oxygenases. Annu. Rev. Biochem. 2018, 87 (Volume 87, 2018), 585–620.
- Whyte, W. A.; Orlando, D. A.; Hnisz, D.; Abraham, B. J.; Lin, C. Y.; Kagey, M. H.; Rahl, P. B.; Lee, T. I.; Young, R. A. Master Transcription Factors and Mediator Establish Super-Enhancers at Key Cell Identity Genes. Cell 2013, 153 (2), 307–319. [CrossRef]
- Hnisz, D.; Abraham, B. J.; Lee, T. I.; Lau, A.; Saint-André, V.; Sigova, A. A.; Hoke, H. A.; Young, R. A. Super-Enhancers in the Control of Cell Identity and Disease. Cell 2013, 155 (4), 934–947. [CrossRef]
- Tang, S. C.; Vijayakumar, U.; Zhang, Y.; Fullwood, M. J. Super-Enhancers, Phase-Separated Condensates, and 3D Genome Organization in Cancer. Cancers 2022, 14 (12), 2866. [CrossRef]
- Di Giorgio, E.; Benetti, R.; Kerschbamer, E.; Xodo, L.; Brancolini, C. Chapter Three - Super-Enhancer Landscape Rewiring in Cancer: The Epigenetic Control at Distal Sites. In International Review of Cell and Molecular Biology; Spada, S., Galluzzi, L., Eds.; Epigenetic Regulation of Cancer - Part A; Academic Press, 2023; Vol. 380, pp 97–148.
- Blayney, J. W.; Francis, H.; Rampasekova, A.; Camellato, B.; Mitchell, L.; Stolper, R.; Cornell, L.; Babbs, C.; Boeke, J. D.; Higgs, D. R.; Kassouf, M. Super-Enhancers Include Classical Enhancers and Facilitators to Fully Activate Gene Expression. Cell 2023, 186 (26), 5826-5839.e18. [CrossRef]
- Galouzis, C. C.; Furlong, E. E. M. Regulating Specificity in Enhancer–Promoter Communication. Curr. Opin. Cell Biol. 2022, 75, 102065. [CrossRef]
- Rao, S. S. P.; Huang, S.-C.; Hilaire, B. G. S.; Engreitz, J. M.; Perez, E. M.; Kieffer-Kwon, K.-R.; Sanborn, A. L.; Johnstone, S. E.; Bascom, G. D.; Bochkov, I. D.; Huang, X.; Shamim, M. S.; Shin, J.; Turner, D.; Ye, Z.; Omer, A. D.; Robinson, J. T.; Schlick, T.; Bernstein, B. E.; Casellas, R.; Lander, E. S.; Aiden, E. L. Cohesin Loss Eliminates All Loop Domains. Cell 2017, 171 (2), 305-320.e24. [CrossRef]
- Fursova, N. A.; Larson, D. R. Transcriptional Machinery as an Architect of Genome Structure. Curr. Opin. Struct. Biol. 2024, 89, 102920. [CrossRef]
- Barshad, G.; Lewis, J. J.; Chivu, A. G.; Abuhashem, A.; Krietenstein, N.; Rice, E. J.; Ma, Y.; Wang, Z.; Rando, O. J.; Hadjantonakis, A.-K.; Danko, C. G. RNA Polymerase II Dynamics Shape Enhancer–Promoter Interactions. Nat. Genet. 2023, 55 (8), 1370–1380. [CrossRef]
- Galbraith, M. D.; Allen, M. A.; Bensard, C. L.; Wang, X.; Schwinn, M. K.; Qin, B.; Long, H. W.; Daniels, D. L.; Hahn, W. C.; Dowell, R. D.; Espinosa, J. M. HIF1A Employs CDK8-Mediator to Stimulate RNAPII Elongation in Response to Hypoxia. Cell 2013, 153 (6), 1327–1339. [CrossRef]
- Choudhry, H.; Schödel, J.; Oikonomopoulos, S.; Camps, C.; Grampp, S.; Harris, A. L.; Ratcliffe, P. J.; Ragoussis, J.; Mole, D. R. Extensive Regulation of the Non-coding Transcriptome by Hypoxia: Role of HIF in Releasing Paused RNApol2. EMBO Rep. 2014, 15 (1), 70–76. [CrossRef]
- Chen, Y.; Zhang, B.; Bao, L.; Jin, L.; Yang, M.; Peng, Y.; Kumar, A.; Wang, J. E.; Wang, C.; Zou, X.; Xing, C.; Wang, Y.; Luo, W. ZMYND8 Acetylation Mediates HIF-Dependent Breast Cancer Progression and Metastasis. J. Clin. Invest. 2018, 128 (5), 1937–1955. [CrossRef]
- Stadhouders, R.; Vidal, E.; Serra, F.; Di Stefano, B.; Le Dily, F.; Quilez, J.; Gomez, A.; Collombet, S.; Berenguer, C.; Cuartero, Y.; Hecht, J.; Filion, G. J.; Beato, M.; Marti-Renom, M. A.; Graf, T. Transcription Factors Orchestrate Dynamic Interplay between Genome Topology and Gene Regulation during Cell Reprogramming. Nat. Genet. 2018, 50 (2), 238–249. [CrossRef]
- Weintraub, A. S.; Li, C. H.; Zamudio, A. V.; Sigova, A. A.; Hannett, N. M.; Day, D. S.; Abraham, B. J.; Cohen, M. A.; Nabet, B.; Buckley, D. L.; Guo, Y. E.; Hnisz, D.; Jaenisch, R.; Bradner, J. E.; Gray, N. S.; Young, R. A. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 2017, 171 (7), 1573-1588.e28.
- Barajas-Mora, E. M.; Kleiman, E.; Xu, J.; Carrico, N. C.; Lu, H.; Oltz, E. M.; Murre, C.; Feeney, A. J. A B-Cell-Specific Enhancer Orchestrates Nuclear Architecture to Generate a Diverse Antigen Receptor Repertoire. Mol. Cell 2019, 73 (1), 48-60.e5. [CrossRef]
- Aboreden, N. G.; Lam, J. C.; Goel, V. Y.; Wang, S.; Wang, X.; Midla, S. C.; Quijano, A.; Keller, C. A.; Giardine, B. M.; Hardison, R. C.; Zhang, H.; Hansen, A. S.; Blobel, G. A. LDB1 Establishes Multi-Enhancer Networks to Regulate Gene Expression. Mol. Cell 2025, 85 (2), 376-393.e9. [CrossRef]
- Blanchard, K. L.; Acquaviva, A. M.; Galson, D. L.; Franklin Bunn, H. Hypoxic Induction of the Human Erythropoietin Gene: Cooperation between the Promoter and Enhancer, Each of Which Contains Steroid Receptor Response Elements. Mol. Cell. Biol. 1992, 12 (12), 5373–5385.
- Semenza, G. L.; Wang, G. L. A Nuclear Factor Induced by Hypoxia via De Novo Protein Synthesis Binds to the Human Erythropoietin Gene Enhancer at a Site Required for Transcriptional Activation. Mol. Cell. Biol. 1992, 12 (12), 5447–5454. [CrossRef]
- Madan, A.; Curtin, P. T. A 24-Base-Pair Sequence 3’ to the Human Erythropoietin Gene Contains a Hypoxia-Responsive Transcriptional Enhancer. Proc. Natl. Acad. Sci. 1993, 90 (9), 3928–3932. [CrossRef]
- Liu, Y.; Cox, S. R.; Morita, T.; Kourembanas, S. Hypoxia Regulates Vascular Endothelial Growth Factor Gene Expression in Endothelial Cells. Circ. Res. 1995, 77 (3), 638–643.
- Liang, Y.; Li, X.-Y.; Rebar, E. J.; Li, P.; Zhou, Y.; Chen, B.; Wolffe, A. P.; Case, C. C. Activation of Vascular Endothelial Growth Factor A Transcription in Tumorigenic Glioblastoma Cell Lines by an Enhancer with Cell Type-Specific DNase I Accessibility *. J. Biol. Chem. 2002, 277 (22), 20087–20094. [CrossRef]
- Behrooz, A.; Ismail-Beigi, F. Dual Control of Glut1 Glucose Transporter Gene Expression by Hypoxia and by Inhibition of Oxidative Phosphorylation *. J. Biol. Chem. 1997, 272 (9), 5555–5562. [CrossRef]
- Kambe, T.; Tada, J.; Chikuma, M.; Masuda, S.; Nagao, M.; Tsuchiya, T.; Ratcliffe, P. J.; Sasaki, R. Embryonal Carcinoma P19 Cells Produce Erythropoietin Constitutively But Express Lactate Dehydrogenase in an Oxygen-Dependent Manner. Blood 1998, 91 (4), 1185–1195. [CrossRef]
- Graven, K. K.; Yu, Q.; Pan, D.; Roncarati, J. S.; Farber, H. W. Identification of an Oxygen Responsive Enhancer Element in the Glyceraldehyde-3-Phosphate Dehydrogenase Gene. Biochim. Biophys. Acta BBA - Gene Struct. Expr. 1999, 1447 (2), 208–218.
- Fukasawa, M.; Tsuchiya, T.; Takayama, E.; Shinomiya, N.; Uyeda, K.; Sakakibara, R.; Seki, S. Identification and Characterization of the Hypoxia-Responsive Element of the Human Placental 6-Phosphofructo-2-Kinase/Fructose-2,6-Bisphosphatase Gene. J. Biochem. (Tokyo) 2004, 136 (3), 273–277. [CrossRef]
- Stancevic, B.; Varda-Bloom, N.; Cheng, J.; Fuller, J. D.; Rotolo, J. A.; García-Barros, M.; Feldman, R.; Rao, S.; Weichselbaum, R. R.; Harats, D.; Haimovitz-Friedman, A.; Fuks, Z.; Sadelain, M.; Kolesnick, R. Adenoviral Transduction of Human Acid Sphingomyelinase into Neo-Angiogenic Endothelium Radiosensitizes Tumor Cure. PLOS ONE 2013, 8 (8), e69025. [CrossRef]
- Romney, S. J.; Newman, B. S.; Thacker, C.; Leibold, E. A. HIF-1 Regulates Iron Homeostasis in Caenorhabditis Elegans by Activation and Inhibition of Genes Involved in Iron Uptake and Storage. PLOS Genet. 2011, 7 (12), e1002394. [CrossRef]
- Wang, X.; Brea, L.; Lu, X.; Gritsina, G.; Park, S. H.; Xie, W.; Zhao, J. C.; Yu, J. FOXA1 Inhibits Hypoxia Programs through Transcriptional Repression of HIF1A. Oncogene 2022, 41 (37), 4259–4270. [CrossRef]
- Wenger, R. H.; Stiehl, D. P.; Camenisch, G. Integration of Oxygen Signaling at the Consensus HRE. Sci. STKE 2005, 2005 (306), re12–re12.
- Wang, F.; Zhang, R.; Beischlag, T. V.; Muchardt, C.; Yaniv, M.; Hankinson, O. Roles of Brahma and Brahma/SWI2-Related Gene 1 in Hypoxic Induction of the Erythropoietin Gene *. J. Biol. Chem. 2004, 279 (45), 46733–46741. [CrossRef]
- Makita, T.; Duncan, S. A.; Sucov, H. M. Retinoic Acid, Hypoxia, and GATA Factors Cooperatively Control the Onset of Fetal Liver Erythropoietin Expression and Erythropoietic Differentiation. Dev. Biol. 2005, 280 (1), 59–72. [CrossRef]
- Kobayashi, M.; Morinibu, A.; Koyasu, S.; Goto, Y.; Hiraoka, M.; Harada, H. A Circadian Clock Gene, PER2, Activates HIF-1 as an Effector Molecule for Recruitment of HIF-1α to Promoter Regions of Its Downstream Genes. FEBS J. 2017, 284 (22), 3804–3816.
- Rioja, P.; Rey-Cardenas, M.; Velasco, G. D. Targeting HIF-2α and Anemia: A Therapeutic Breakthrough for Clear-Cell Renal Cell Carcinoma. Cancer Treat. Rev. 2024, 129. [CrossRef]
- Yao, X.; Tan, J.; Lim, K. J.; Koh, J.; Ooi, W. F.; Li, Z.; Huang, D.; Xing, M.; Chan, Y. S.; Qu, J. Z.; Tay, S. T.; Wijaya, G.; Lam, Y. N.; Hong, J. H.; Lee-Lim, A. P.; Guan, P.; Ng, M. S. W.; He, C. Z.; Lin, J. S.; Nandi, T.; Qamra, A.; Xu, C.; Myint, S. S.; Davies, J. O. J.; Goh, J. Y.; Loh, G.; Tan, B. C.; Rozen, S. G.; Yu, Q.; Tan, I. B. H.; Cheng, C. W. S.; Li, S.; Chang, K. T. E.; Tan, P. H.; Silver, D. L.; Lezhava, A.; Steger, G.; Hughes, J. R.; Teh, B. T.; Tan, P. VHL Deficiency Drives Enhancer Activation of Oncogenes in Clear Cell Renal Cell Carcinoma. Cancer Discov. 2017, 7 (11), 1284–1305. [CrossRef]
- Ricketts, C. J.; Linehan, W. M. Insights into Epigenetic Remodeling in VHL-Deficient Clear Cell Renal Cell Carcinoma. Cancer Discov. 2017, 7 (11), 1221–1223.
- Patel, S. A.; Hirosue, S.; Rodrigues, P.; Vojtasova, E.; Richardson, E. K.; Ge, J.; Syafruddin, S. E.; Speed, A.; Papachristou, E. K.; Baker, D.; Clarke, D.; Purvis, S.; Wesolowski, L.; Dyas, A.; Castillon, L.; Caraffini, V.; Bihary, D.; Yong, C.; Harrison, D. J.; Stewart, G. D.; Machiela, M. J.; Purdue, M. P.; Chanock, S. J.; Warren, A. Y.; Samarajiwa, S. A.; Carroll, J. S.; Vanharanta, S. The Renal Lineage Factor PAX8 Controls Oncogenic Signalling in Kidney Cancer. Nature 2022, 606 (7916), 999–1006. [CrossRef]
- Protze, J.; Naas, S.; Krüger, R.; Stöhr, C.; Kraus, A.; Grampp, S.; Wiesener, M.; Schiffer, M.; Hartmann, A.; Wullich, B.; Schödel, J. The Renal Cancer Risk Allele at 14q24.2 Activates a Novel Hypoxia-Inducible Transcription Factor-Binding Enhancer of DPF3 Expression. J. Biol. Chem. 2022, 298 (3). [CrossRef]
- Grampp, S.; Platt, J. L.; Lauer, V.; Salama, R.; Kranz, F.; Neumann, V. K.; Wach, S.; Stöhr, C.; Hartmann, A.; Eckardt, K.-U.; Ratcliffe, P. J.; Mole, D. R.; Schödel, J. Genetic Variation at the 8q24.21 Renal Cancer Susceptibility Locus Affects HIF Binding to a MYC Enhancer. Nat. Commun. 2016, 7 (1), 13183. [CrossRef]
- Wang, X.; Guan, X.; Zhu, X.; Zhang, L.; Ma, C.; He, S.; Bai, J.; Mei, J.; Li, Q.; Sun, N.; Wu, B.; Zhu, D. CircNAP1L4 Regulates Pulmonary Artery Smooth Muscle Cell Proliferation via the NAP1L4-Mediated Super-Enhancer-Driven Glycolysis Gene Hexokinase II (HK II) in Pulmonary Hypertension. FASEB J. 2024, 38 (15), e23868. [CrossRef]
- Shi, J.; Jia, Z.; Sun, J.; Wang, X.; Zhao, X.; Zhao, C.; Liang, F.; Song, X.; Guan, J.; Jia, X.; Yang, J.; Chen, Q.; Yu, K.; Jia, Q.; Wu, J.; Wang, D.; Xiao, Y.; Xu, X.; Liu, Y.; Wu, S.; Zhong, Q.; Wu, J.; Cui, S.; Bo, X.; Wu, Z.; Park, M.; Kellis, M.; He, K. Structural Variants Involved in High-Altitude Adaptation Detected Using Single-Molecule Long-Read Sequencing. Nat. Commun. 2023, 14 (1), 8282.
- Fu, X.; Pereira, R.; De Angelis, C.; Veeraraghavan, J.; Nanda, S.; Qin, L.; Cataldo, M. L.; Sethunath, V.; Mehravaran, S.; Gutierrez, C.; Chamness, G. C.; Feng, Q.; O’Malley, B. W.; Selenica, P.; Weigelt, B.; Reis-Filho, J. S.; Cohen, O.; Wagle, N.; Nardone, A.; Jeselsohn, R.; Brown, M.; Rimawi, M. F.; Osborne, C. K.; Schiff, R. FOXA1 Upregulation Promotes Enhancer and Transcriptional Reprogramming in Endocrine-Resistant Breast Cancer. Proc. Natl. Acad. Sci. 2019, 116 (52), 26823–26834. [CrossRef]
- Moreau, P. R.; Örd, T.; Downes, N. L.; Niskanen, H.; Bouvy-Liivrand, M.; Aavik, E.; Ylä-Herttuala, S.; Kaikkonen, M. U. Transcriptional Profiling of Hypoxia-Regulated Non-Coding RNAs in Human Primary Endothelial Cells. Front. Cardiovasc. Med. 2018, 5. [CrossRef]
- Platt, J. L.; Salama, R.; Smythies, J.; Choudhry, H.; Davies, J. O.; Hughes, J. R.; Ratcliffe, P. J.; Mole, D. R. Capture-C Reveals Preformed Chromatin Interactions between HIF-binding Sites and Distant Promoters. EMBO Rep. 2016, 17 (10), 1410–1421. [CrossRef]
- Niskanen, H.; Tuszynska, I.; Zaborowski, R.; Heinäniemi, M.; Ylä-Herttuala, S.; Wilczynski, B.; Kaikkonen, M. U. Endothelial Cell Differentiation Is Encompassed by Changes in Long Range Interactions between Inactive Chromatin Regions. Nucleic Acids Res. 2018, 46 (4), 1724–1740. [CrossRef]
- Krueger, K.; Catanese, L.; Sciesielski, L. K.; Kirschner, K. M.; Scholz, H. Deletion of an Intronic HIF-2α Binding Site Suppresses Hypoxia-Induced WT1 Expression. Biochim. Biophys. Acta BBA - Gene Regul. Mech. 2019, 1862 (1), 71–83. [CrossRef]
- Schörg, A.; Santambrogio, S.; Platt, J. L.; Schödel, J.; Lindenmeyer, M. T.; Cohen, C. D.; Schrödter, K.; Mole, D. R.; Wenger, R. H.; Hoogewijs, D. Destruction of a Distal Hypoxia Response Element Abolishes Trans-Activation of the PAG1 Gene Mediated by HIF-Independent Chromatin Looping. Nucleic Acids Res. 2015, 43 (12), 5810–5823. [CrossRef]
- Nakayama, K.; Shachar, S.; Finn, E. H.; Sato, H.; Hirakawa, A.; Misteli, T. Large-Scale Mapping of Positional Changes of Hypoxia-Responsive Genes upon Activation. Mol. Biol. Cell 2022, 33 (8), ar72. [CrossRef]
- Kakani, P.; Dhamdhere, S. G.; Pant, D.; Joshi, R.; Mishra, S.; Pandey, A.; Notani, D.; Shukla, S. Hypoxia-Induced CTCF Mediates Alternative Splicing via Coupling Chromatin Looping and RNA Pol II Pause to Promote EMT in Breast Cancer. Cell Rep. 2025, 44 (2). [CrossRef]
- Kindrick, J. D.; Mole, D. R. Hypoxic Regulation of Gene Transcription and Chromatin: Cause and Effect. Int. J. Mol. Sci. 2020, 21 (21), 8320. [CrossRef]
- Wiesener, M. S.; Jürgensen, J. S.; Rosenberger, C.; Scholze, C.; Hörstrup, J. H.; Warnecke, C.; Mandriota, S.; Bechmann, I.; Frei, U. A.; Pugh, C. W.; Ratcliffe, P. J.; Bachmann, S.; Maxwell, P. H.; Eckardt, K.-U. Widespread, Hypoxia-Inducible Expression of HIF-2α in Distinct Cell Populations of Different Organs. FASEB J. 2003, 17 (2), 271–273.
- Shi, Y.; Gilkes, D. M. HIF-1 and HIF-2 in Cancer: Structure, Regulation, and Therapeutic Prospects. Cell. Mol. Life Sci. 2025, 82 (1), 44.
- Loboda, A.; Jozkowicz, A.; Dulak, J. HIF-1 and HIF-2 Transcription Factors - Similar but Not Identical. Mol. Cells 2010, 29 (5), 435–442. [CrossRef]
- Lombardi, O.; Li, R.; Halim, S.; Choudhry, H.; Ratcliffe, P. J.; Mole, D. R. Pan-Cancer Analysis of Tissue and Single-Cell HIF-Pathway Activation Using a Conserved Gene Signature. Cell Rep. 2022, 41 (7). [CrossRef]
- Schödel, J.; Oikonomopoulos, S.; Ragoussis, J.; Pugh, C. W.; Ratcliffe, P. J.; Mole, D. R. High-Resolution Genome-Wide Mapping of HIF-Binding Sites by ChIP-Seq. Blood 2011, 117 (23), e207–e217.
- Smythies, J. A.; Sun, M.; Masson, N.; Salama, R.; Simpson, P. D.; Murray, E.; Neumann, V.; Cockman, M. E.; Choudhry, H.; Ratcliffe, P. J.; Mole, D. R. Inherent DNA-binding Specificities of the HIF-1α and HIF-2α Transcription Factors in Chromatin. EMBO Rep. 2019, 20 (1), e46401.
- Mimura, I.; Nangaku, M.; Kanki, Y.; Tsutsumi, S.; Inoue, T.; Kohro, T.; Yamamoto, S.; Fujita, T.; Shimamura, T.; Suehiro, J.; Taguchi, A.; Kobayashi, M.; Tanimura, K.; Inagaki, T.; Tanaka, T.; Hamakubo, T.; Sakai, J.; Aburatani, H.; Kodama, T.; Wada, Y. Dynamic Change of Chromatin Conformation in Response to Hypoxia Enhances the Expression of GLUT3 (SLC2A3) by Cooperative Interaction of Hypoxia-Inducible Factor 1 and KDM3A. Mol. Cell. Biol. 2012, 32 (15), 3018–3032. [CrossRef]
- Tanimoto, K.; Tsuchihara, K.; Kanai, A.; Arauchi, T.; Esumi, H.; Suzuki, Y.; Sugano, S. Genome-Wide Identification and Annotation of HIF-1α Binding Sites in Two Cell Lines Using Massively Parallel Sequencing. HUGO J. 2010, 4 (1), 35–48. [CrossRef]
- Loftus, S. K.; Baxter, L. L.; Cronin, J. C.; Fufa, T. D.; Program, N. C. S.; Pavan, W. J. Hypoxia-Induced HIF1α Targets in Melanocytes Reveal a Molecular Profile Associated with Poor Melanoma Prognosis. Pigment Cell Melanoma Res. 2017, 30 (3), 339–352. [CrossRef]
- Tran, M. G. B.; Bibby, B. A. S.; Yang, L.; Lo, F.; Warren, A. Y.; Shukla, D.; Osborne, M.; Hadfield, J.; Carroll, T.; Stark, R.; Scott, H.; Ramos-Montoya, A.; Massie, C.; Maxwell, P.; West, C. M. L.; Mills, I. G.; Neal, D. E. Independence of HIF1a and Androgen Signaling Pathways in Prostate Cancer. BMC Cancer 2020, 20 (1), 469. [CrossRef]
- Martí, J. M.; Garcia-Diaz, A.; Delgado-Bellido, D.; O’Valle, F.; González-Flores, A.; Carlevaris, O.; Rodríguez-Vargas, J. M.; Amé, J. C.; Dantzer, F.; King, G. L.; Dziedzic, K.; Berra, E.; de Álava, E.; Amaral, A. T.; Hammond, E. M.; Oliver, F. J. Selective Modulation by PARP-1 of HIF-1α-Recruitment to Chromatin during Hypoxia Is Required for Tumor Adaptation to Hypoxic Conditions. Redox Biol. 2021, 41, 101885. [CrossRef]
- Feng, D.; Qu, L.; Powell-Coffman, J. A. Whole Genome Profiling of Short-Term Hypoxia Induced Genes and Identification of HIF-1 Binding Sites Provide Insights into HIF-1 Function in Caenorhabditis Elegans. PLOS ONE 2024, 19 (5), e0295094. [CrossRef]
- Hu, C.-J.; Wang, L.-Y.; Chodosh, L. A.; Keith, B.; Simon, M. C. Differential Roles of Hypoxia-Inducible Factor 1α (HIF-1α) and HIF-2α in Hypoxic Gene Regulation. Mol. Cell. Biol. 2003, 23 (24), 9361–9374.
- Raval, R. R.; Lau, K. W.; Tran, M. G. B.; Sowter, H. M.; Mandriota, S. J.; Li, J.-L.; Pugh, C. W.; Maxwell, P. H.; Harris, A. L.; Ratcliffe, P. J. Contrasting Properties of Hypoxia-Inducible Factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-Associated Renal Cell Carcinoma. Mol. Cell. Biol. 2005, 25 (13), 5675–5686.
- Scortegagna, M.; Ding, K.; Zhang, Q.; Oktay, Y.; Bennett, M. J.; Bennett, M.; Shelton, J. M.; Richardson, J. A.; Moe, O.; Garcia, J. A. HIF-2α Regulates Murine Hematopoietic Development in an Erythropoietin-Dependent Manner. Blood 2005, 105 (8), 3133–3140. [CrossRef]
- Warnecke, C.; Zaborowska, Z.; Kurreck, J.; Erdmann, V. A.; Frei, U.; Wiesener, M.; Eckardt, K.-U. Differentiating the Functional Role of Hypoxia-Inducible Factor (HIF)-1α and HIF-2α (EPAS-1) by the Use of RNA Interference: Erythropoietin Is a HIF-2α Target Gene in Hep3B and Kelly Cells. FASEB J. 2004, 18 (12), 1462–1464.
- Rankin, E. B.; Biju, M. P.; Liu, Q.; Unger, T. L.; Rha, J.; Johnson, R. S.; Simon, M. C.; Keith, B.; Haase, V. H. Hypoxia-Inducible Factor–2 (HIF-2) Regulates Hepatic Erythropoietin in Vivo. J. Clin. Invest. 2007, 117 (4), 1068–1077. [CrossRef]
- Covello, K. L.; Kehler, J.; Yu, H.; Gordan, J. D.; Arsham, A. M.; Hu, C.-J.; Labosky, P. A.; Simon, M. C.; Keith, B. HIF-2α Regulates Oct-4: Effects of Hypoxia on Stem Cell Function, Embryonic Development, and Tumor Growth. Genes Dev. 2006, 20 (5), 557–570.
- Sowter, H. M.; Raval, R.; Moore, J.; Ratcliffe, P. J.; Harris, A. L. Predominant Role of Hypoxia-Inducible Transcription Factor (Hif)-1α versus Hif-2α in Regulation of the Transcriptional Response to Hypoxia1. Cancer Res. 2003, 63 (19), 6130–6134.
- Turner, K. J.; Moore, J. W.; Jones, A.; Taylor, C. F.; Cuthbert-Heavens, D.; Han, C.; Leek, R. D.; Gatter, K. C.; Maxwell, P. H.; Ratcliffe, P. J.; Cranston, D.; Harris, A. L. Expression of Hypoxia-Inducible Factors in Human Renal Cancer: Relationship to Angiogenesis and to the von Hippel-Lindau Gene Mutation. Cancer Res. 2002, 62 (10), 2957–2961.
- Gordan, J. D.; Bertout, J. A.; Hu, C.-J.; Diehl, J. A.; Simon, M. C. HIF-2α Promotes Hypoxic Cell Proliferation by Enhancing c-Myc Transcriptional Activity. Cancer Cell 2007, 11 (4), 335–347. [CrossRef]
- Chen, X.; Litzenburger, U. M.; Wei, Y.; Schep, A. N.; LaGory, E. L.; Choudhry, H.; Giaccia, A. J.; Greenleaf, W. J.; Chang, H. Y. Joint Single-Cell DNA Accessibility and Protein Epitope Profiling Reveals Environmental Regulation of Epigenomic Heterogeneity. Nat. Commun. 2018, 9 (1), 4590. [CrossRef]
- Xin, J.; Zhang, H.; He, Y.; Duren, Z.; Bai, C.; Chen, L.; Luo, X.; Yan, D.-S.; Zhang, C.; Zhu, X.; Yuan, Q.; Feng, Z.; Cui, C.; Qi, X.; Ouzhuluobu; Wong, W. H.; Wang, Y.; Su, B. Chromatin Accessibility Landscape and Regulatory Network of High-Altitude Hypoxia Adaptation. Nat. Commun. 2020, 11 (1), 4928. [CrossRef]
- Gualandi, N.; Minisini, M.; Bertozzo, A.; Brancolini, C. Dissecting Transposable Elements and Endogenous Retroviruses Upregulation by HDAC Inhibitors in Leiomyosarcoma Cells: Implications for the Interferon Response. Genomics 2024, 116 (5), 110909. [CrossRef]
- Siebenthall, K. T.; Miller, C. P.; Vierstra, J. D.; Mathieu, J.; Tretiakova, M.; Reynolds, A.; Sandstrom, R.; Rynes, E.; Haugen, E.; Johnson, A.; Nelson, J.; Bates, D.; Diegel, M.; Dunn, D.; Frerker, M.; Buckley, M.; Kaul, R.; Zheng, Y.; Himmelfarb, J.; Ruohola-Baker, H.; Akilesh, S. Integrated Epigenomic Profiling Reveals Endogenous Retrovirus Reactivation in Renal Cell Carcinoma. EBioMedicine 2019, 41, 427–442. [CrossRef]
- Lai, J. C.-Y.; Hsu, K.-W.; Wu, K.-J. Interrogation of the Interplay between DNA N6-Methyladenosine (6mA) and Hypoxia-Induced Chromatin Accessibility by a Randomized Empirical Model (EnrichShuf). Nucleic Acids Res. 2024, 52 (22), 13605–13624. [CrossRef]
- Batie, M.; Frost, J.; Shakir, D.; Rocha, S. Regulation of Chromatin Accessibility by Hypoxia and HIF. Biochem. J. 2022, 479 (6), 767–786. [CrossRef]
- Miar, A.; Arnaiz, E.; Bridges, E.; Beedie, S.; Cribbs, A. P.; Downes, D. J.; Beagrie, R. A.; Rehwinkel, J.; Harris, A. L. Hypoxia Induces Transcriptional and Translational Downregulation of the Type I IFN Pathway in Multiple Cancer Cell Types. Cancer Res. 2020, 80 (23), 5245–5256. [CrossRef]
- Li, Y.; Gruber, J. J.; Litzenburger, U. M.; Zhou, Y.; Miao, Y. R.; LaGory, E. L.; Li, A. M.; Hu, Z.; Yip, M.; Hart, L. S.; Maris, J. M.; Chang, H. Y.; Giaccia, A. J.; Ye, J. Acetate Supplementation Restores Chromatin Accessibility and Promotes Tumor Cell Differentiation under Hypoxia. Cell Death Dis. 2020, 11 (2), 1–17. [CrossRef]
- Leszczynska, K. B.; Dzwigonska, M.; Estephan, H.; Moehlenbrink, J.; Bowler, E.; Giaccia, A. J.; Mieczkowski, J.; Kaminska, B.; Hammond, E. M. Hypoxia-Mediated Regulation of DDX5 through Decreased Chromatin Accessibility and Post-Translational Targeting Restricts R-Loop Accumulation. Mol. Oncol. 2023, 17 (7), 1173–1191. [CrossRef]
- Eustermann, S.; Patel, A. B.; Hopfner, K.-P.; He, Y.; Korber, P. Energy-Driven Genome Regulation by ATP-Dependent Chromatin Remodellers. Nat. Rev. Mol. Cell Biol. 2024, 25 (4), 309–332. [CrossRef]
- Li, Z.; Zhao, J.; Tang, Y. Advances in the Role of SWI/SNF Complexes in Tumours. J. Cell. Mol. Med. 2023, 27 (8), 1023–1031. [CrossRef]
- Kang, J.-S.; Kim, D.; Rhee, J.; Seo, J.-Y.; Park, I.; Kim, J.-H.; Lee, D.; Lee, W.; Kim, Y. L.; Yoo, K.; Bae, S.; Chung, J.; Seong, R. H.; Kong, Y.-Y. Baf155 Regulates Skeletal Muscle Metabolism via HIF-1a Signaling. PLOS Biol. 2023, 21 (7), e3002192. [CrossRef]
- Sena, J. A.; Wang, L.; Hu, C.-J. BRG1 and BRM Chromatin-Remodeling Complexes Regulate the Hypoxia Response by Acting as Coactivators for a Subset of Hypoxia-Inducible Transcription Factor Target Genes. Mol. Cell. Biol. 2013, 33 (19), 3849–3863. [CrossRef]
- Gao, W.; Li, W.; Xiao, T.; Liu, X. S.; Kaelin, W. G. Inactivation of the PBRM1 Tumor Suppressor Gene Amplifies the HIF-Response in VHL−/− Clear Cell Renal Carcinoma. Proc. Natl. Acad. Sci. 2017, 114 (5), 1027–1032.
- Nargund, A. M.; Pham, C. G.; Dong, Y.; Wang, P. I.; Osmangeyoglu, H. U.; Xie, Y.; Aras, O.; Han, S.; Oyama, T.; Takeda, S.; Ray, C. E.; Dong, Z.; Berge, M.; Hakimi, A. A.; Monette, S.; Lekaye, C. L.; Koutcher, J. A.; Leslie, C. S.; Creighton, C. J.; Weinhold, N.; Lee, W.; Tickoo, S. K.; Wang, Z.; Cheng, E. H.; Hsieh, J. J. The SWI/SNF Protein PBRM1 Restrains VHL-Loss-Driven Clear Cell Renal Cell Carcinoma. Cell Rep. 2017, 18 (12), 2893–2906. [CrossRef]
- Liu, X.; Li, Z.; Wang, Z.; Liu, F.; Zhang, L.; Ke, J.; Xu, X.; Zhang, Y.; Yuan, Y.; Wei, T.; Shan, Q.; Chen, Y.; Huang, W.; Gao, J.; Wu, N.; Chen, F.; Sun, L.; Qiu, Z.; Deng, Y.; Wang, X. Chromatin Remodeling Induced by ARID1A Loss in Lung Cancer Promotes Glycolysis and Confers JQ1 Vulnerability. Cancer Res. 2022, 82 (5), 791–804. [CrossRef]
- Wei, A.; Wu, H. Mammalian DNA Methylome Dynamics: Mechanisms, Functions and New Frontiers. Development 2022, 149 (24), dev182683. [CrossRef]
- Greenberg, M. V. C.; Bourc’his, D. The Diverse Roles of DNA Methylation in Mammalian Development and Disease. Nat. Rev. Mol. Cell Biol. 2019, 20 (10), 590–607.
- Honer, M. A.; Ferman, B. I.; Gray, Z. H.; Bondarenko, E. A.; Whetstine, J. R. Epigenetic Modulators Provide a Path to Understanding Disease and Therapeutic Opportunity. Genes Dev. 2024, 38 (11–12), 473–503. [CrossRef]
- Wenger, R. H.; Kvietikova, I.; Rolfs, A.; Camenisch, G.; Gassmann, M. Oxygen-Regulated Erythropoietin Gene Expression Is Dependent on a CpG Methylation-Free Hypoxia-Inducible Factor-1 DNA-Binding Site. Eur. J. Biochem. 1998, 253 (3), 771–777.
- Okami, J.; Simeone, D. M.; Logsdon, C. D. Silencing of the Hypoxia-Inducible Cell Death Protein BNIP3 in Pancreatic Cancer. Cancer Res. 2004, 64 (15), 5338–5346. [CrossRef]
- Lee, S. H.; Kim, J.; Kim, W.-H.; Lee, Y. M. Hypoxic Silencing of Tumor Suppressor RUNX3 by Histone Modification in Gastric Cancer Cells. Oncogene 2009, 28 (2), 184–194. [CrossRef]
- Lu, Y.; Chu, A.; Turker, M. S.; Glazer, P. M. Hypoxia-Induced Epigenetic Regulation and Silencing of the BRCA1 Promoter. Mol. Cell. Biol. 2011, 31 (16), 3339–3350. [CrossRef]
- Lee, J. S.; Kim, Y.; Bhin, J.; Shin, H.-J. R.; Nam, H. J.; Lee, S. H.; Yoon, J.-B.; Binda, O.; Gozani, O.; Hwang, D.; Baek, S. H. Hypoxia-Induced Methylation of a Pontin Chromatin Remodeling Factor. Proc. Natl. Acad. Sci. 2011, 108 (33), 13510–13515. [CrossRef]
- Lachance, G.; Uniacke, J.; Audas, T. E.; Holterman, C. E.; Franovic, A.; Payette, J.; Lee, S. DNMT3a Epigenetic Program Regulates the HIF-2α Oxygen-Sensing Pathway and the Cellular Response to Hypoxia. Proc. Natl. Acad. Sci. 2014, 111 (21), 7783–7788. [CrossRef]
- Liu, Q.; Liu, L.; Zhao, Y.; Zhang, J.; Wang, D.; Chen, J.; He, Y.; Wu, J.; Zhang, Z.; Liu, Z. Hypoxia Induces Genomic DNA Demethylation through the Activation of HIF-1α and Transcriptional Upregulation of MAT2A in Hepatoma Cells. Mol. Cancer Ther. 2011, 10 (6), 1113–1123. [CrossRef]
- Pal, A.; Srivastava, T.; Sharma, M. K.; Mehndiratta, M.; Das, P.; Sinha, S.; Chattopadhyay, P. Aberrant Methylation and Associated Transcriptional Mobilization of Alu Elements Contributes to Genomic Instability in Hypoxia. J. Cell. Mol. Med. 2010, 14 (11), 2646–2654. [CrossRef]
- Cherkasova, E.; Malinzak, E.; Rao, S.; Takahashi, Y.; Senchenko, V. N.; Kudryavtseva, A. V.; Nickerson, M. L.; Merino, M.; Hong, J. A.; Schrump, D. S.; Srinivasan, R.; Linehan, W. M.; Tian, X.; Lerman, M. I.; Childs, R. W. Inactivation of the von Hippel–Lindau Tumor Suppressor Leads to Selective Expression of a Human Endogenous Retrovirus in Kidney Cancer. Oncogene 2011, 30 (47), 4697–4706. [CrossRef]
- D’Anna, F.; Van Dyck, L.; Xiong, J.; Zhao, H.; Berrens, R. V.; Qian, J.; Bieniasz-Krzywiec, P.; Chandra, V.; Schoonjans, L.; Matthews, J.; De Smedt, J.; Minnoye, L.; Amorim, R.; Khorasanizadeh, S.; Yu, Q.; Zhao, L.; De Borre, M.; Savvides, S. N.; Simon, M. C.; Carmeliet, P.; Reik, W.; Rastinejad, F.; Mazzone, M.; Thienpont, B.; Lambrechts, D. DNA Methylation Repels Binding of Hypoxia-Inducible Transcription Factors to Maintain Tumor Immunotolerance. Genome Biol. 2020, 21 (1), 182.
- Hu, C. Y.; Mohtat, D.; Yu, Y.; Ko, Y.-A.; Shenoy, N.; Bhattacharya, S.; Izquierdo, M. C.; Park, A. S. D.; Giricz, O.; Vallumsetla, N.; Gundabolu, K.; Ware, K.; Bhagat, T. D.; Suzuki, M.; Pullman, J.; Liu, X. S.; Greally, J. M.; Susztak, K.; Verma, A. Kidney Cancer Is Characterized by Aberrant Methylation of Tissue-Specific Enhancers That Are Prognostic for Overall Survival. Clin. Cancer Res. 2014, 20 (16), 4349–4360. [CrossRef]
- Childebayeva, A.; Jones, T. R.; Goodrich, J. M.; Leon-Velarde, F.; Rivera-Chira, M.; Kiyamu, M.; Brutsaert, T. D.; Dolinoy, D. C.; Bigham, A. W. LINE-1 and EPAS1 DNA Methylation Associations with High-Altitude Exposure. Epigenetics 2019, 14 (1), 1–15. [CrossRef]
- Belmonte, K. C. D.; Harman, J. C.; Lanson, N. A.; Gidday, J. M. Intra- and Intergenerational Changes in the Cortical DNA Methylome in Response to Therapeutic Intermittent Hypoxia in Mice. Physiol. Genomics 2020, 52 (1), 20–34. [CrossRef]
- Childebayeva, A.; Harman, T.; Weinstein, J.; Day, T.; Brutsaert, T. D.; Bigham, A. W. Genome-Wide DNA Methylation Changes Associated With High-Altitude Acclimatization During an Everest Base Camp Trek. Front. Physiol. 2021, 12. [CrossRef]
- Basang, Z.; Zhang, S.; Yang, L.; Quzong, D.; Li, Y.; Ma, Y.; Hao, M.; Pu, W.; Liu, X.; Xie, H.; Liang, M.; Wang, J.; Danzeng, Q. Correlation of DNA Methylation Patterns to the Phenotypic Features of Tibetan Elite Alpinists in Extreme Hypoxia. J. Genet. Genomics 2021, 48 (10), 928–935. [CrossRef]
- Yadav, P.; Pandey, A.; Kakani, P.; Mutnuru, S. A.; Samaiya, A.; Mishra, J.; Shukla, S. Hypoxia-Induced Loss of SRSF2-Dependent DNA Methylation Promotes CTCF-Mediated Alternative Splicing of VEGFA in Breast Cancer. iScience 2023, 26 (6), 106804. [CrossRef]
- Parry, A.; Rulands, S.; Reik, W. Active Turnover of DNA Methylation during Cell Fate Decisions. Nat. Rev. Genet. 2021, 22 (1), 59–66. [CrossRef]
- Mariani, C. J.; Vasanthakumar, A.; Madzo, J.; Yesilkanal, A.; Bhagat, T.; Yu, Y.; Bhattacharyya, S.; Wenger, R. H.; Cohn, S. L.; Nanduri, J.; Verma, A.; Prabhakar, N. R.; Godley, L. A. TET1-Mediated Hydroxymethylation Facilitates Hypoxic Gene Induction in Neuroblastoma. Cell Rep. 2014, 7 (5), 1343–1352. [CrossRef]
- Zhu, J.; Wang, K.; Li, T.; Chen, J.; Xie, D.; Chang, X.; Yao, J.; Wu, J.; Zhou, Q.; Jia, Y.; Duan, T. Hypoxia-Induced TET1 Facilitates Trophoblast Cell Migration and Invasion through HIF1α Signaling Pathway. Sci. Rep. 2017, 7 (1), 8077. [CrossRef]
- Hains, A. E.; Uppal, S.; Cao, J. Z.; Salwen, H. R.; Applebaum, M. A.; Cohn, S. L.; Godley, L. A. MYCN and HIF-1 Directly Regulate TET1 Expression to Control 5-hmC Gains and Enhance Neuroblastoma Cell Migration in Hypoxia. Epigenetics 2022, 17 (13), 2056–2074. [CrossRef]
- Tsai, Y.-P.; Chen, H.-F.; Chen, S.-Y.; Cheng, W.-C.; Wang, H.-W.; Shen, Z.-J.; Song, C.; Teng, S.-C.; He, C.; Wu, K.-J. TET1 Regulates Hypoxia-Induced Epithelial-Mesenchymal Transition by Acting as a Co-Activator. Genome Biol. 2014, 15 (12), 513. [CrossRef]
- Cao, J. Z.; Liu, H.; Wickrema, A.; Godley, L. A. HIF-1 Directly Induces TET3 Expression to Enhance 5-hmC Density and Induce Erythroid Gene Expression in Hypoxia. Blood Adv. 2020, 4 (13), 3053–3062. [CrossRef]
- Wu, M.-Z.; Chen, S.-F.; Nieh, S.; Benner, C.; Ger, L.-P.; Jan, C.-I.; Ma, L.; Chen, C.-H.; Hishida, T.; Chang, H.-T.; Lin, Y.-S.; Montserrat, N.; Gascon, P.; Sancho-Martinez, I.; Izpisua Belmonte, J. C. Hypoxia Drives Breast Tumor Malignancy through a TET–TNFα–P38–MAPK Signaling Axis. Cancer Res. 2015, 75 (18), 3912–3924.
- Burr, S.; Caldwell, A.; Chong, M.; Beretta, M.; Metcalf, S.; Hancock, M.; Arno, M.; Balu, S.; Kropf, V. L.; Mistry, R. K.; Shah, A. M.; Mann, G. E.; Brewer, A. C. Oxygen Gradients Can Determine Epigenetic Asymmetry and Cellular Differentiation via Differential Regulation of Tet Activity in Embryonic Stem Cells. Nucleic Acids Res. 2018, 46 (3), 1210–1226. [CrossRef]
- de la Calle-Fabregat, C.; Calafell-Segura, J.; Gardet, M.; Dunsmore, G.; Mulder, K.; Ciudad, L.; Silvin, A.; Moreno-Càceres, J.; Corbí, Á. L.; Muñoz-Pinedo, C.; Michels, J.; Gouy, S.; Dutertre, C.-A.; Rodríguez-Ubreva, J.; Ginhoux, F.; Ballestar, E. NF-κB and TET2 Promote Macrophage Reprogramming in Hypoxia That Overrides the Immunosuppressive Effects of the Tumor Microenvironment. Sci. Adv. 2024, 10 (38), eadq5226. [CrossRef]
- Prasad, P.; Mittal, S. A.; Chongtham, J.; Mohanty, S.; Srivastava, T. Hypoxia-Mediated Epigenetic Regulation of Stemness in Brain Tumor Cells. Stem Cells 2017, 35 (6), 1468–1478. [CrossRef]
- Nosyreva, Elena. D.; Thompson, D.; Syeda, R. Identification and Functional Characterization of the Piezo1 Channel Pore Domain. J. Biol. Chem. 2021, 296, 100225. [CrossRef]
- Thienpont, B.; Steinbacher, J.; Zhao, H.; D’Anna, F.; Kuchnio, A.; Ploumakis, A.; Ghesquière, B.; Van Dyck, L.; Boeckx, B.; Schoonjans, L.; Hermans, E.; Amant, F.; Kristensen, V. N.; Koh, K. P.; Mazzone, M.; Coleman, M. L.; Carell, T.; Carmeliet, P.; Lambrechts, D. Tumour Hypoxia Causes DNA Hypermethylation by Reducing TET Activity. Nature 2016, 537 (7618), 63–68. [CrossRef]
- Hao, Z.; Wu, T.; Cui, X.; Zhu, P.; Tan, C.; Dou, X.; Hsu, K.-W.; Lin, Y.-T.; Peng, P.-H.; Zhang, L.-S.; Gao, Y.; Hu, L.; Sun, H.-L.; Zhu, A.; Liu, J.; Wu, K.-J.; He, C. N6-Deoxyadenosine Methylation in Mammalian Mitochondrial DNA. Mol. Cell 2020, 78 (3), 382-395.e8. [CrossRef]
- Hsu, K.-W.; Lai, J. C.-Y.; Chang, J.-S.; Peng, P.-H.; Huang, C.-H.; Lee, D.-Y.; Tsai, Y.-C.; Chung, C.-J.; Chang, H.; Chang, C.-H.; Chen, J.-L.; Pang, S.-T.; Hao, Z.; Cui, X.-L.; He, C.; Wu, K.-J. METTL4-Mediated Nuclear N6-Deoxyadenosine Methylation Promotes Metastasis through Activating Multiple Metastasis-Inducing Targets. Genome Biol. 2022, 23 (1), 249. [CrossRef]
- Stillman, B. Histone Modifications: Insights into Their Influence on Gene Expression. Cell 2018, 175 (1), 6–9. [CrossRef]
- Millán-Zambrano, G.; Burton, A.; Bannister, A. J.; Schneider, R. Histone Post-Translational Modifications — Cause and Consequence of Genome Function. Nat. Rev. Genet. 2022, 23 (9), 563–580. [CrossRef]
- Jash, E.; Csankovszki, G. Chromatin Organization during C. Elegans Early Development. DNA 2024, 4 (1), 64–83. [CrossRef]
- Luo, W.; Chang, R.; Zhong, J.; Pandey, A.; Semenza, G. L. Histone Demethylase JMJD2C Is a Coactivator for Hypoxia-Inducible Factor 1 That Is Required for Breast Cancer Progression. Proc. Natl. Acad. Sci. 2012, 109 (49), E3367–E3376. [CrossRef]
- Lyu, Y.; Yang, Y.; Talwar, V.; Lu, H.; Chen, C.; Salman, S.; Wicks, E. E.; Huang, T. Y.-T.; Drehmer, D.; Wang, Y.; Zuo, Q.; Datan, E.; Jackson, W.; Dordai, D.; Wang, R.; Semenza, G. L. Hypoxia-Inducible Factor 1 Recruits FACT and RNF20/40 to Mediate Histone Ubiquitination and Transcriptional Activation of Target Genes. Cell Rep. 2024, 43 (4), 113972.
- Perez-Perri, J. I.; Dengler, V. L.; Audetat, K. A.; Pandey, A.; Bonner, E. A.; Urh, M.; Mendez, J.; Daniels, D. L.; Wappner, P.; Galbraith, M. D.; Espinosa, J. M. The TIP60 Complex Is a Conserved Coactivator of HIF1A. Cell Rep. 2016, 16 (1), 37–47. [CrossRef]
- Wang, Y.; Lyu, Y.; Tu, K.; Xu, Q.; Yang, Y.; Salman, S.; Le, N.; Lu, H.; Chen, C.; Zhu, Y.; Wang, R.; Liu, Q.; Semenza, G. L. Histone Citrullination by PADI4 Is Required for HIF-Dependent Transcriptional Responses to Hypoxia and Tumor Vascularization. Sci. Adv. 2021, 7 (35), eabe3771. [CrossRef]
- Qu, M.; Long, Y.; Wang, Y.; Yin, N.; Zhang, X.; Zhang, J. Hypoxia Increases ATX Expression by Histone Crotonylation in a HIF-2α-Dependent Manner. Int. J. Mol. Sci. 2023, 24 (8), 7031. [CrossRef]
- Karagiota, A.; Kanoura, A.; Paraskeva, E.; Simos, G.; Chachami, G. Pyruvate Dehydrogenase Phosphatase 1 (PDP1) Stimulates HIF Activity by Supporting Histone Acetylation under Hypoxia. FEBS J. 2023, 290 (8), 2165–2179. [CrossRef]
- Collier, H.; Albanese, A.; Kwok, C.-S.; Kou, J.; Rocha, S. Functional Crosstalk between Chromatin and Hypoxia Signalling. Cell. Signal. 2023, 106, 110660. [CrossRef]
- Brancolini, C.; Gagliano, T.; Minisini, M. HDACs and the Epigenetic Plasticity of Cancer Cells: Target the Complexity. Pharmacol. Ther. 2022, 238, 108190. [CrossRef]
- Charidemou, E.; Kirmizis, A. A Two-Way Relationship between Histone Acetylation and Metabolism. Trends Biochem. Sci. 2024, 49 (12), 1046–1062. [CrossRef]
- Sabari, B. R.; Zhang, D.; Allis, C. D.; Zhao, Y. Metabolic Regulation of Gene Expression through Histone Acylations. Nat. Rev. Mol. Cell Biol. 2017, 18 (2), 90–101.
- Drazic, A.; Myklebust, L. M.; Ree, R.; Arnesen, T. The World of Protein Acetylation. Biochim. Biophys. Acta BBA - Proteins Proteomics 2016, 1864 (10), 1372–1401.
- Kamphorst, J. J.; Chung, M. K.; Fan, J.; Rabinowitz, J. D. Quantitative Analysis of Acetyl-CoA Production in Hypoxic Cancer Cells Reveals Substantial Contribution from Acetate. Cancer Metab. 2014, 2 (1), 23. [CrossRef]
- Schug, Z. T.; Peck, B.; Jones, D. T.; Zhang, Q.; Grosskurth, S.; Alam, I. S.; Goodwin, L. M.; Smethurst, E.; Mason, S.; Blyth, K.; McGarry, L.; James, D.; Shanks, E.; Kalna, G.; Saunders, R. E.; Jiang, M.; Howell, M.; Lassailly, F.; Thin, M. Z.; Spencer-Dene, B.; Stamp, G.; van den Broek, N. J. F.; Mackay, G.; Bulusu, V.; Kamphorst, J. J.; Tardito, S.; Strachan, D.; Harris, A. L.; Aboagye, E. O.; Critchlow, S. E.; Wakelam, M. J. O.; Schulze, A.; Gottlieb, E. Acetyl-CoA Synthetase 2 Promotes Acetate Utilization and Maintains Cancer Cell Growth under Metabolic Stress. Cancer Cell 2015, 27 (1), 57–71.
- Gao, X.; Lin, S.-H.; Ren, F.; Li, J.-T.; Chen, J.-J.; Yao, C.-B.; Yang, H.-B.; Jiang, S.-X.; Yan, G.-Q.; Wang, D.; Wang, Y.; Liu, Y.; Cai, Z.; Xu, Y.-Y.; Chen, J.; Yu, W.; Yang, P.-Y.; Lei, Q.-Y. Acetate Functions as an Epigenetic Metabolite to Promote Lipid Synthesis under Hypoxia. Nat. Commun. 2016, 7 (1), 11960. [CrossRef]
- Kondo, A.; Yamamoto, S.; Nakaki, R.; Shimamura, T.; Hamakubo, T.; Sakai, J.; Kodama, T.; Yoshida, T.; Aburatani, H.; Osawa, T. Extracellular Acidic pH Activates the Sterol Regulatory Element-Binding Protein 2 to Promote Tumor Progression. Cell Rep. 2017, 18 (9), 2228–2242. [CrossRef]
- Quevedo, M.; Meert, L.; Dekker, M. R.; Dekkers, D. H. W.; Brandsma, J. H.; van den Berg, D. L. C.; Ozgür, Z.; van IJcken, W. F. J.; Demmers, J.; Fornerod, M.; Poot, R. A. Mediator Complex Interaction Partners Organize the Transcriptional Network That Defines Neural Stem Cells. Nat. Commun. 2019, 10 (1), 2669. [CrossRef]
- Lauer, V.; Grampp, S.; Platt, J.; Lafleur, V.; Lombardi, O.; Choudhry, H.; Kranz, F.; Hartmann, A.; Wullich, B.; Yamamoto, A.; Coleman, M. L.; Ratcliffe, P. J.; Mole, D. R.; Schödel, J. Hypoxia Drives Glucose Transporter 3 Expression through Hypoxia-Inducible Transcription Factor (HIF)–Mediated Induction of the Long Noncoding RNA NICI. J. Biol. Chem. 2020, 295 (13), 4065–4078. [CrossRef]
- Inoue, T.; Kohro, T.; Tanaka, T.; Kanki, Y.; Li, G.; Poh, H.-M.; Mimura, I.; Kobayashi, M.; Taguchi, A.; Maejima, T.; Suehiro, J.; Sugiyama, A.; Kaneki, K.; Aruga, H.; Dong, S.; Stevens, J. F.; Yamamoto, S.; Tsutsumi, S.; Fujita, T.; Ruan, X.; Aburatani, H.; Nangaku, M.; Ruan, Y.; Kodama, T.; Wada, Y. Cross-Enhancement of ANGPTL4 Transcription by HIF1 Alpha and PPAR Beta/Delta Is the Result of the Conformational Proximity of Two Response Elements. Genome Biol. 2014, 15 (4), R63. [CrossRef]
- Paauw, N. D.; Lely, A. T.; Joles, J. A.; Franx, A.; Nikkels, P. G.; Mokry, M.; van Rijn, B. B. H3K27 Acetylation and Gene Expression Analysis Reveals Differences in Placental Chromatin Activity in Fetal Growth Restriction. Clin. Epigenetics 2018, 10 (1), 85. [CrossRef]
- Wu, Z.; Zhang, W.; Kang, Y. J. Copper Affects the Binding of HIF-1α to the Critical Motifs of Its Target Genes†. Metallomics 2019, 11 (2), 429–438. [CrossRef]
- Wen, B.; Zheng, Z.; Wang, L.; Qian, X.; Wang, X.; Chen, Y.; Bao, J.; Jiang, Y.; Ji, K.; Liu, H. HIF-1α Is Essential for the Augmentation of Myometrial Contractility during Labor†. Biol. Reprod. 2022, 107 (6), 1540–1550. [CrossRef]
- Kasper, L. H.; Qu, C.; Obenauer, J. C.; McGoldrick, D. J.; Brindle, P. K. Genome-Wide and Single-Cell Analyses Reveal a Context Dependent Relationship between CBP Recruitment and Gene Expression. Nucleic Acids Res. 2014, 42 (18), 11363–11382. [CrossRef]
- Luo, W.; Wang, Y. Epigenetic Regulators: Multifunctional Proteins Modulating Hypoxia-Inducible Factor-α Protein Stability and Activity. Cell. Mol. Life Sci. 2018, 75 (6), 1043–1056. [CrossRef]
- Kang, J.; Kang, Y.; Kim, A. Histone H3K4ac, as a Marker of Active Transcription Start Sites and Enhancers, Plays Roles in Histone Eviction and RNA Transcription. Biochim. Biophys. Acta BBA - Gene Regul. Mech. 2024, 1867 (2), 195021. [CrossRef]
- Wang, J.-Q.; Yan, F.-Q.; Wang, L.-H.; Yin, W.-J.; Chang, T.-Y.; Liu, J.-P.; Wu, K.-J. Identification of New Hypoxia-Regulated Epithelial-Mesenchymal Transition Marker Genes Labeled by H3K4 Acetylation. Genes. Chromosomes Cancer 2020, 59 (2), 73–83. [CrossRef]
- Mirtschink, P.; Bischof, C.; Pham, M.-D.; Sharma, R.; Khadayate, S.; Rossi, G.; Fankhauser, N.; Traub, S.; Sossalla, S.; Hagag, E.; Berthonneche, C.; Sarre, A.; Stehr, Sebastian. N.; Grote, P.; Pedrazzini, T.; Dimmeler, S.; Krek, W.; Krishnan, J. Inhibition of the Hypoxia-Inducible Factor 1α–Induced Cardiospecific HERNA1 Enhance-Templated RNA Protects From Heart Disease. Circulation 2019, 139 (24), 2778–2792. [CrossRef]
- Tiana, M.; Acosta-Iborra, B.; Puente-Santamaría, L.; Hernansanz-Agustin, P.; Worsley-Hunt, R.; Masson, N.; García-Rio, F.; Mole, D.; Ratcliffe, P.; Wasserman, W. W.; Jimenez, B.; del Peso, L. The SIN3A Histone Deacetylase Complex Is Required for a Complete Transcriptional Response to Hypoxia. Nucleic Acids Res. 2018, 46 (1), 120–133. [CrossRef]
- Musselman, C. A.; Lalonde, M.-E.; Côté, J.; Kutateladze, T. G. Perceiving the Epigenetic Landscape through Histone Readers. Nat. Struct. Mol. Biol. 2012, 19 (12), 1218–1227. [CrossRef]
- Bae, A. A.; Zheng, Y. G. Hetero-Oligomeric Interaction as a New Regulatory Mechanism for Protein Arginine Methyltransferases. Biochem. Soc. Trans. 2024, 52 (5), 2193–2201. [CrossRef]
- Wesche, J.; Kühn, S.; Kessler, B. M.; Salton, M.; Wolf, A. Protein Arginine Methylation: A Prominent Modification and Its Demethylation. Cell. Mol. Life Sci. 2017, 74 (18), 3305–3315. [CrossRef]
- Ortmann, B. M.; Burrows, N.; Lobb, I. T.; Arnaiz, E.; Wit, N.; Bailey, P. S. J.; Jordon, L. H.; Lombardi, O.; Peñalver, A.; McCaffrey, J.; Seear, R.; Mole, D. R.; Ratcliffe, P. J.; Maxwell, P. H.; Nathan, J. A. The HIF Complex Recruits the Histone Methyltransferase SET1B to Activate Specific Hypoxia-Inducible Genes. Nat. Genet. 2021, 53 (7), 1022–1035.
- Kim, J.; Lee, H.; Yi, S.-J.; Kim, K. Gene Regulation by Histone-Modifying Enzymes under Hypoxic Conditions: A Focus on Histone Methylation and Acetylation. Exp. Mol. Med. 2022, 54 (7), 878–889. [CrossRef]
- Chen, H.; Yan, Y.; Davidson, T. L.; Shinkai, Y.; Costa, M. Hypoxic Stress Induces Dimethylated Histone H3 Lysine 9 through Histone Methyltransferase G9a in Mammalian Cells. Cancer Res. 2006, 66 (18), 9009–9016. [CrossRef]
- Hancock, R. L.; Masson, N.; Dunne, K.; Flashman, E.; Kawamura, A. The Activity of JmjC Histone Lysine Demethylase KDM4A Is Highly Sensitive to Oxygen Concentrations. ACS Chem. Biol. 2017, 12 (4), 1011–1019. [CrossRef]
- Fiorini, G.; Schofield, C. J. Biochemistry of the Hypoxia-Inducible Factor Hydroxylases. Curr. Opin. Chem. Biol. 2024, 79, 102428. [CrossRef]
- Chakraborty, A. A.; Laukka, T.; Myllykoski, M.; Ringel, A. E.; Booker, M. A.; Tolstorukov, M. Y.; Meng, Y. J.; Meier, S. R.; Jennings, R. B.; Creech, A. L.; Herbert, Z. T.; McBrayer, S. K.; Olenchock, B. A.; Jaffe, J. D.; Haigis, M. C.; Beroukhim, R.; Signoretti, S.; Koivunen, P.; Kaelin, W. G. Histone Demethylase KDM6A Directly Senses Oxygen to Control Chromatin and Cell Fate. Science 2019, 363 (6432), 1217–1222.
- Qian, X.; Li, X.; Shi, Z.; Bai, X.; Xia, Y.; Zheng, Y.; Xu, D.; Chen, F.; You, Y.; Fang, J.; Hu, Z.; Zhou, Q.; Lu, Z. KDM3A Senses Oxygen Availability to Regulate PGC-1α-Mediated Mitochondrial Biogenesis. Mol. Cell 2019, 76 (6), 885-895.e7. [CrossRef]
- Ortmann, B. M.; Taylor, C. T.; Rocha, S. Hypoxia Research, Where to Now? Trends Biochem. Sci. 2024, 49 (7), 573–582.
- Adriaens, M. E.; Prickaerts, P.; Chan-Seng-Yue, M.; van den Beucken, T.; Dahlmans, V. E. H.; Eijssen, L. M.; Beck, T.; Wouters, B. G.; Voncken, J. W.; Evelo, C. T. A. Quantitative Analysis of ChIP-Seq Data Uncovers Dynamic and Sustained H3K4me3 and H3K27me3 Modulation in Cancer Cells under Hypoxia. Epigenetics Chromatin 2016, 9 (1), 48. [CrossRef]
- Prickaerts, P.; Adriaens, M. E.; Beucken, T. van den; Koch, E.; Dubois, L.; Dahlmans, V. E. H.; Gits, C.; Evelo, C. T. A.; Chan-Seng-Yue, M.; Wouters, B. G.; Voncken, J. W. Hypoxia Increases Genome-Wide Bivalent Epigenetic Marking by Specific Gain of H3K27me3. Epigenetics Chromatin 2016, 9 (1), 46. [CrossRef]
- Batie, M.; Frost, J.; Frost, M.; Wilson, J. W.; Schofield, P.; Rocha, S. Hypoxia Induces Rapid Changes to Histone Methylation and Reprograms Chromatin. Science 2019, 363 (6432), 1222–1226.
- Semenza, G. L. Targeting HIF-1 for Cancer Therapy. Nat. Rev. Cancer 2003, 3 (10), 721–732.
- Ivan, M.; Kaelin, W. G. The EGLN-HIF O2-Sensing System: Multiple Inputs and Feedbacks. Mol. Cell 2017, 66 (6), 772–779. [CrossRef]
- Xia, X.; Kung, A. L. Preferential Binding of HIF-1 to Transcriptionally Active Loci Determines Cell-Type Specific Response to Hypoxia. Genome Biol. 2009, 10 (10), R113. [CrossRef]
- Macrae, T. A.; Fothergill-Robinson, J.; Ramalho-Santos, M. Regulation, Functions and Transmission of Bivalent Chromatin during Mammalian Development. Nat. Rev. Mol. Cell Biol. 2023, 24 (1), 6–26. [CrossRef]
- van den Beucken, T.; Koch, E.; Chu, K.; Rupaimoole, R.; Prickaerts, P.; Adriaens, M.; Voncken, J. W.; Harris, A. L.; Buffa, F. M.; Haider, S.; Starmans, M. H. W.; Yao, C. Q.; Ivan, M.; Ivan, C.; Pecot, C. V.; Boutros, P. C.; Sood, A. K.; Koritzinsky, M.; Wouters, B. G. Hypoxia Promotes Stem Cell Phenotypes and Poor Prognosis through Epigenetic Regulation of DICER. Nat. Commun. 2014, 5 (1), 5203. [CrossRef]
- Ho, J. J. D.; Metcalf, J. L.; Yan, M. S.; Turgeon, P. J.; Wang, J. J.; Chalsev, M.; Petruzziello-Pellegrini, T. N.; Tsui, A. K. Y.; He, J. Z.; Dhamko, H.; Man, H. S. J.; Robb, G. B.; Teh, B. T.; Ohh, M.; Marsden, P. A. Functional Importance of Dicer Protein in the Adaptive Cellular Response to Hypoxia*. J. Biol. Chem. 2012, 287 (34), 29003–29020. [CrossRef]
- Chen, H.; Yu, S.; Ma, R.; Deng, L.; Yi, Y.; Niu, M.; Xu, C.; Xiao, Z.-X. J. Hypoxia-Activated XBP1s Recruits HDAC2-EZH2 to Engage Epigenetic Suppression of ΔNp63α Expression and Promote Breast Cancer Metastasis Independent of HIF1α. Cell Death Differ. 2024, 31 (4), 447–459. [CrossRef]
- Duncan, E. M.; Muratore-Schroeder, T. L.; Cook, R. G.; Garcia, B. A.; Shabanowitz, J.; Hunt, D. F.; Allis, C. D. Cathepsin L Proteolytically Processes Histone H3 During Mouse Embryonic Stem Cell Differentiation. Cell 2008, 135 (2), 284–294. [CrossRef]
- Duarte, L. F.; Young, A. R. J.; Wang, Z.; Wu, H.-A.; Panda, T.; Kou, Y.; Kapoor, A.; Hasson, D.; Mills, N. R.; Ma’ayan, A.; Narita, M.; Bernstein, E. Histone H3.3 and Its Proteolytically Processed Form Drive a Cellular Senescence Programme. Nat. Commun. 2014, 5 (1), 5210.
- Ferrari, K. J.; Amato, S.; Noberini, R.; Toscani, C.; Fernández-Pérez, D.; Rossi, A.; Conforti, P.; Zanotti, M.; Bonaldi, T.; Tamburri, S.; Pasini, D. Intestinal Differentiation Involves Cleavage of Histone H3 N-Terminal Tails by Multiple Proteases. Nucleic Acids Res. 2021, 49 (2), 791–804. [CrossRef]
- Di Giorgio, E.; Paluvai, H.; Dalla, E.; Ranzino, L.; Renzini, A.; Moresi, V.; Minisini, M.; Picco, R.; Brancolini, C. HDAC4 Degradation during Senescence Unleashes an Epigenetic Program Driven by AP-1/P300 at Selected Enhancers and Super-Enhancers. Genome Biol. 2021, 22 (1), 129.
- Otero-Albiol, D.; Carnero, A. Cellular Senescence or Stemness: Hypoxia Flips the Coin. J. Exp. Clin. Cancer Res. 2021, 40 (1), 243. [CrossRef]
- Chang, S.; Moon, R.; Nam, D.; Lee, S.-W.; Yoon, I.; Lee, D.-S.; Choi, S.; Paek, E.; Hwang, D.; Hur, J. K.; Nam, Y.; Chang, R.; Park, H. Hypoxia Increases Methylated Histones to Prevent Histone Clipping and Heterochromatin Redistribution during Raf-Induced Senescence. Nucleic Acids Res. 2025, 53 (3), gkae1210. [CrossRef]
- Rytkönen, K. T.; Faux, T.; Mahmoudian, M.; Heinosalo, T.; Nnamani, M. C.; Perheentupa, A.; Poutanen, M.; Elo, L. L.; Wagner, G. P. Histone H3K4me3 Breadth in Hypoxia Reveals Endometrial Core Functions and Stress Adaptation Linked to Endometriosis. iScience 2022, 25 (5), 104235. [CrossRef]
- Rogakou, E. P.; Pilch, D. R.; Orr, A. H.; Ivanova, V. S.; Bonner, W. M. DNA Double-Stranded Breaks Induce Histone H2AX Phosphorylation on Serine 139 *. J. Biol. Chem. 1998, 273 (10), 5858–5868. [CrossRef]
- Burma, S.; Chen, B. P.; Murphy, M.; Kurimasa, A.; Chen, D. J. ATM Phosphorylates Histone H2AX in Response to DNA Double-Strand Breaks *. J. Biol. Chem. 2001, 276 (45), 42462–42467. [CrossRef]
- Ward, I. M.; Chen, J. Histone H2AX Is Phosphorylated in an ATR-Dependent Manner in Response to Replicational Stress *. J. Biol. Chem. 2001, 276 (51), 47759–47762. [CrossRef]
- Matsuoka, S.; Ballif, B. A.; Smogorzewska, A.; McDonald, E. R.; Hurov, K. E.; Luo, J.; Bakalarski, C. E.; Zhao, Z.; Solimini, N.; Lerenthal, Y.; Shiloh, Y.; Gygi, S. P.; Elledge, S. J. ATM and ATR Substrate Analysis Reveals Extensive Protein Networks Responsive to DNA Damage. Science 2007, 316 (5828), 1160–1166. [CrossRef]
- Economopoulou, M.; Langer, H. F.; Celeste, A.; Orlova, V. V.; Choi, E. Y.; Ma, M.; Vassilopoulos, A.; Callen, E.; Deng, C.; Bassing, C. H.; Boehm, M.; Nussenzweig, A.; Chavakis, T. Histone H2AX Is Integral to Hypoxia-Driven Neovascularization. Nat. Med. 2009, 15 (5), 553–558. [CrossRef]
- Lyu, X.; Chastain, M.; Chai, W. Genome-Wide Mapping and Profiling of γH2AX Binding Hotspots in Response to Different Replication Stress Inducers. BMC Genomics 2019, 20 (1), 579. [CrossRef]
- Di Giorgio, E.; Dalla, E.; Tolotto, V.; D’Este, F.; Paluvai, H.; Ranzino, L.; Brancolini, C. HDAC4 Influences the DNA Damage Response and Counteracts Senescence by Assembling with HDAC1/HDAC2 to Control H2BK120 Acetylation and Homology-Directed Repair. Nucleic Acids Res. 2024, 52 (14), 8218–8240.
- Rezaeian, A.-H.; Li, C.-F.; Wu, C.-Y.; Zhang, X.; Delacerda, J.; You, M. J.; Han, F.; Cai, Z.; Jeong, Y. S.; Jin, G.; Phan, L.; Chou, P.-C.; Lee, M.-H.; Hung, M.-C.; Sarbassov, D.; Lin, H.-K. A Hypoxia-Responsive TRAF6–ATM–H2AX Signalling Axis Promotes HIF1α Activation, Tumorigenesis and Metastasis. Nat. Cell Biol. 2017, 19 (1), 38–51.
- Pan, M.-R.; Peng, G.; Hung, W.-C.; Lin, S.-Y. Monoubiquitination of H2AX Protein Regulates DNA Damage Response Signaling. J. Biol. Chem. 2011, 286 (32), 28599–28607.
- Formosa, T.; Winston, F. The Role of FACT in Managing Chromatin: Disruption, Assembly, or Repair? Nucleic Acids Res. 2020, 48 (21), 11929–11941. [CrossRef]
- Fields, J. K.; Hicks, C. W.; Wolberger, C. Diverse Modes of Regulating Methyltransferase Activity by Histone Ubiquitination. Curr. Opin. Struct. Biol. 2023, 82, 102649. [CrossRef]
- Liu, X.; Wang, J.; Boyer, J. A.; Gong, W.; Zhao, S.; Xie, L.; Wu, Q.; Zhang, C.; Jain, K.; Guo, Y.; Rodriguez, J.; Li, M.; Uryu, H.; Liao, C.; Hu, L.; Zhou, J.; Shi, X.; Tsai, Y.-H.; Yan, Q.; Luo, W.; Chen, X.; Strahl, B. D.; von Kriegsheim, A.; Zhang, Q.; Wang, G. G.; Baldwin, A. S.; Zhang, Q. Histone H3 Proline 16 Hydroxylation Regulates Mammalian Gene Expression. Nat. Genet. 2022, 54 (11), 1721–1735. [CrossRef]
- Christophorou, M. A. The Virtues and Vices of Protein Citrullination. R. Soc. Open Sci. 2022, 9 (6), 220125. [CrossRef]
- Cuthbert, G. L.; Daujat, S.; Snowden, A. W.; Erdjument-Bromage, H.; Hagiwara, T.; Yamada, M.; Schneider, R.; Gregory, P. D.; Tempst, P.; Bannister, A. J.; Kouzarides, T. Histone Deimination Antagonizes Arginine Methylation. Cell 2004, 118 (5), 545–553. [CrossRef]
- Darrah, E.; Rosen, A.; Giles, J. T.; Andrade, F. Peptidylarginine Deiminase 2, 3 and 4 Have Distinct Specificities against Cellular Substrates: Novel Insights into Autoantigen Selection in Rheumatoid Arthritis. Ann. Rheum. Dis. 2012, 71 (1), 92–98. [CrossRef]
- Sharma, P.; Azebi, S.; England, P.; Christensen, T.; Møller-Larsen, A.; Petersen, T.; Batsché, E.; Muchardt, C. Citrullination of Histone H3 Interferes with HP1-Mediated Transcriptional Repression. PLOS Genet. 2012, 8 (9), e1002934. [CrossRef]
- Guertin, M. J.; Zhang, X.; Anguish, L.; Kim, S.; Varticovski, L.; Lis, J. T.; Hager, G. L.; Coonrod, S. A. Targeted H3R26 Deimination Specifically Facilitates Estrogen Receptor Binding by Modifying Nucleosome Structure. PLOS Genet. 2014, 10 (9), e1004613. [CrossRef]
- Missiaen, R.; Lesner, N. P.; Simon, M. C. HIF: A Master Regulator of Nutrient Availability and Metabolic Cross-talk in the Tumor Microenvironment. EMBO J. 2023, 42 (6), e112067. [CrossRef]
- Gao, J.; Liu, R.; Huang, K.; Li, Z.; Sheng, X.; Chakraborty, K.; Han, C.; Zhang, D.; Becker, L.; Zhao, Y. Dynamic Investigation of Hypoxia-Induced L-Lactylation. Proc. Natl. Acad. Sci. 2025, 122 (10), e2404899122. [CrossRef]
- Pérez-Tomás, R.; Pérez-Guillén, I. Lactate in the Tumor Microenvironment: An Essential Molecule in Cancer Progression and Treatment. Cancers 2020, 12 (11), 3244. [CrossRef]
- Zhang, Z.; Zhang, L.; Zhou, Y.; Li, L.; Zhao, J.; Qin, W.; Jin, Z.; Liu, W. Increase in HDAC9 Suppresses Myoblast Differentiation via Epigenetic Regulation of Autophagy in Hypoxia. Cell Death Dis. 2019, 10 (8), 552.
- Moreno-Yruela, C.; Zhang, D.; Wei, W.; Bæk, M.; Liu, W.; Gao, J.; Danková, D.; Nielsen, A. L.; Bolding, J. E.; Yang, L.; Jameson, S. T.; Wong, J.; Olsen, C. A.; Zhao, Y. Class I Histone Deacetylases (HDAC1–3) Are Histone Lysine Delactylases. Sci. Adv. 2022, 8 (3), eabi6696. [CrossRef]
- Galle, E.; Wong, C.-W.; Ghosh, A.; Desgeorges, T.; Melrose, K.; Hinte, L. C.; Castellano-Castillo, D.; Engl, M.; de Sousa, J. A.; Ruiz-Ojeda, F. J.; De Bock, K.; Ruiz, J. R.; von Meyenn, F. H3K18 Lactylation Marks Tissue-Specific Active Enhancers. Genome Biol. 2022, 23 (1), 207. [CrossRef]
- Chen, J.; Zhang, M.; Liu, Y.; Zhao, S.; Wang, Y.; Wang, M.; Niu, W.; Jin, F.; Li, Z. Histone Lactylation Driven by mROS-Mediated Glycolytic Shift Promotes Hypoxic Pulmonary Hypertension. J. Mol. Cell Biol. 2022, 14 (12), mjac073. [CrossRef]
- Xu, Y.; Meng, W.; Dai, Y.; Xu, L.; Ding, N.; Zhang, J.; Zhuang, X. Anaerobic Metabolism Promotes Breast Cancer Survival via Histone-3 Lysine-18 Lactylation Mediating PPARD Axis. Cell Death Discov. 2025, 11 (1), 1–16. [CrossRef]
- Zang, Y.; Wang, A.; Zhang, J.; Xia, M.; Jiang, Z.; Jia, B.; Lu, C.; Chen, C.; Wang, S.; Zhang, Y.; Wang, C.; Cao, X.; Niu, Z.; He, C.; Bai, X.; Tian, S.; Zhai, G.; Cao, H.; Chen, Y.; Zhang, K. Hypoxia Promotes Histone H3K9 Lactylation to Enhance LAMC2 Transcription in Esophageal Squamous Cell Carcinoma. iScience 2024, 27 (7), 110188. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





