1. Introduction
It is known that metabolic-reprograming is a tumour hallmark that is required to support the rapid energy needs and biosynthetic demands of growing tumors[
1,
2].
MYC is a key transcription factor that governs numerous cellular processes, including metabolism, growth, and differentiation [
3,
4,
5,
6]. In cancer, MYC is often deregulated and overexpressed, acting as a master regulator of metabolic reprogramming that sustains tumor survival and proliferation. A key hallmark of MYC metabolic function is its pivotal role in driving the Warburg effect by enhancing glycolysis, where it increases the expression of glucose transporters and enzymes involved in the glycolytic pathway, thus accelerating the conversion of glucose to lactate even in the presence of oxygen [
7,
8,
9].
Despite the wealth of reviews addressing MYC's metabolic functions [
7,
8,
9] there remains a gap in comprehensive analyses that describes the mechanisms and factors influencing MYC's metabolic roles. A deeper understanding of these mechanisms is crucial for mapping how various factors impact MYC’s activity and metabolic regulation,uncovering thus potential molecular targets that regulate or modulate MYC's metabolic function.
A key area that has emerged is the role of non-coding RNAs (ncRNAs), which have been shown to modulate MYC's function including metabolic functions through a variety of mechanisms . With the recent advancements in RNA-based therapies—such as RNA interference ,small molecules, and antisense oligonucleotides— which have demonstrated the potential to modulate ncRNA activity [
10,
11] , a better understanding of the role of ncRNAs in the context of MYC metaboism would be of great benifit opening avenues for discovering synthetic lethality targets of MYC related functions in cancer .
While some reviews have discussed ncRNAs that regulate MYC expression across different tissues[
12,
13], this review specifically aims to explore ncRNAs that directly influence MYC’s metabolic functions.
While MYC plays a central role in regulating glycolysis, it is not always the case that any ncRNA affecting MYC stability will directly impact glycolytic activity. The relationship between MYC and metabolism is complex and context-dependent, influenced by factors such as cell type, metabolic state, and compensatory mechanisms. Additionally, MYC's effects on glycolysis can be modulated by other pathways, including those involving hypoxia and other oncogenes and tumour suppressors such as p53 and AMPK.
Therefore, this review specifically focuses on ncRNAs that have shown clear evidence of modulating MYC-driven metabolic pathways,specifically the warburg effect, aiming to examining the mechanisms through which these interactions occurs. The review also aims to explore the interactions between various types of ncRNAs, shedding light on how they may collaborate or influence each other’s functions in the regulation of MYC’s metabolic function.
By shedding light on these complex relationships, our goal is to deepen the understanding of MYC’s role in metabolism and highlight how ncRNAs contribute to this process. By narrowing the scope to MYC-related metabolic regulation this offers a more targeted approach to therapeutic development, potentially leading to the identification of biomarkers and synthetic lethality targets to disrupt MYC-driven metabolic reprogramming in cancer.
2. Overview on MYC Metabolic Function
MYC's involvement in glycolysis is considered the hallmark of MYC’s metabolic reprograming in vitro and in vivo [
7,
8,
9] .MYC activates the transcription of most glycolytic enzymes, either directly by binding to classical E-box sequences or indirectly through other mechanisms. It is known to enhances the transcription and thus the expression of key glycolytic enzymes and proteins such as glucose transporter 1( GLUT1) ,hexokinase 2(HK2), lactate dehydrogenase A (LDHA), and alpha enolase (Eno1) [
7,
8,
9] a feature that is also observed in MYC
high patient samples [
14]
The further direct link between glycolysis and MYC was confirmed in vivo in inducible MYC-driven malignancies analysed by hyperpolarized 13C pyruvate Magnetic Resonance Spectroscopic Imaging, in which an increase flux of pyruvate to lactate is seen in primary tumour compared to normal or pre-tumour nodules, and which was also reversible upon MYC inhibition [
15,
16] .In addition, further supporting glycolysis, MYC was shown to increase the expression of monocarboxylate transporters MCT1 and MCT2 which are known to be over expressed in tumours and can mediate lactate efflux thus maintaining glycolytic rates [
17,
18].This was further confirmed in vivo in Eμ-Myc transgenic mouse model of human B lymphoma, in addition to various MYC driven malignancies in human data sets [
17]. MYC influence on phosphofructokinase muscle isoform 2 (PFKM2), the enzyme responsible for the final step of glycolysis and which in contrast to PFKM1 favors glycolysis over oxidative phosphorylation, was also observed . MYC was found to activate the transcription of splicing factors, promoting the production of PKM2 [
19]. Another suggested mechanism was through the activation of the nuclear RNA helicase MTR4 transcription by MYC in hepatocellular carcinoma [
20]. MTR4 plays a significant role in RNA metabolism and stability and found to significantly activate glycolytic enzymes expression mainly through alternative splicing regulation in this study. Further confirming the role of MYC in glycolysis were results from a small cell lung carcinoma (SCLC) xenografts model, where it has bee shown that MYC
high tumors are more sensitive to glycolysis inhibition [
14] .
3. Non-Coding RNAs and their mechanisms in MYC-mediated metabolic Regulation
Non-coding RNAs (ncRNAs) are diverse RNA molecules that do not encode proteins but regulate gene expression and various cellular processes. Among the most studied types are long non-coding RNAs (lncRNAs), circular RNAs (circRNAs), and microRNAs (miRNAs), each playing significant roles in cancer progression and cellular metabolism. LncRNAs, typically a heterogeneous class of >500 nucleotide transcripts , are involved in chromatin remodeling, transcriptional regulation, and post-transcriptional control. It is now clear that lncRNA can play a signficant role in regulating MYC expression and its downstream target genes[
21] , with some being themselves under MYC regulation. In the recent year lncRNA are being strongly studies as potential biomarkers for cancer progression and/or targets for theraputic intervention through its interactions with MYC [
22,
23].
CircRNAs, which are formed through back-splicing of pre-mRNA into a closed loop are extremely stable molecules, and have also been closely associated with MYC. MYC driven circRNA were found to be upregulated in tumours such as triple negative breast cancer (TNBC) and SCLC , and can promote tumour progression in these cells[
24,
25]. CircRNA can also act as miRNAs sponges modulating gene expression including MYC itself [
26].
MiRNAs, small RNAs of 20-24 nucleotides, regulate gene expression by binding to mRNA and inhibiting translation or promoting degradation. Dysregulated miRNAs are common in cancers, and they can either function as oncogenes or tumor suppressors, and similar to the previous ncRNAs, miRNAs can both target and be regulated by MYC[
27,
28]. Thus in summary ncRNAs has been implemented in MYC-mediated functions and offer potential therapeutic avenues for MYC-driven cancers .
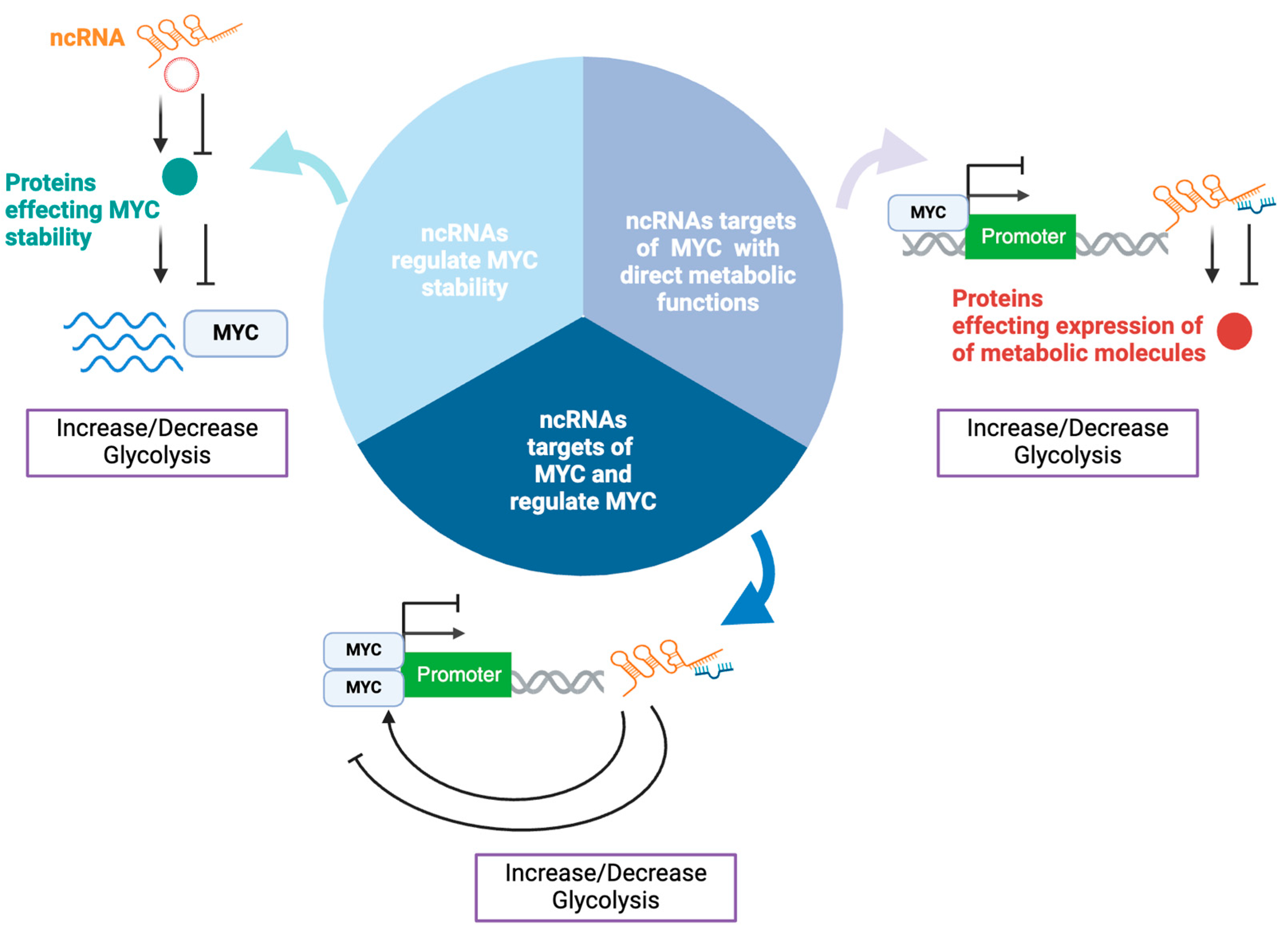
In a similar manner ncRNAs influence MYC-mediated metabolic regulation through three key mechanisms (
Figure 1): 1)
ncRNAs regulate MYC stability, affecting its degradation or accumulation, and thus its ability to control metabolic pathways; 2)
ncRNAs with direct metabolic functions and are targets of MYC, in which MYC controls the expression of certain ncRNAs which can modulate metabolic pathways, such as glycolysis and oxidative phosphorylation; 3)
ncRNAs that are target of MYC and regulate MYC expression in a feedback loop, where MYC influences the expression of ncRNAs, while these ncRNAs also affect MYC’s stability and activity, creating a dynamic regulatory cycle.
The following sections describe in details the ncRNA involved in these mechanisms which have shown to modulate MYC metabolic functions specifically its glycolytic effect
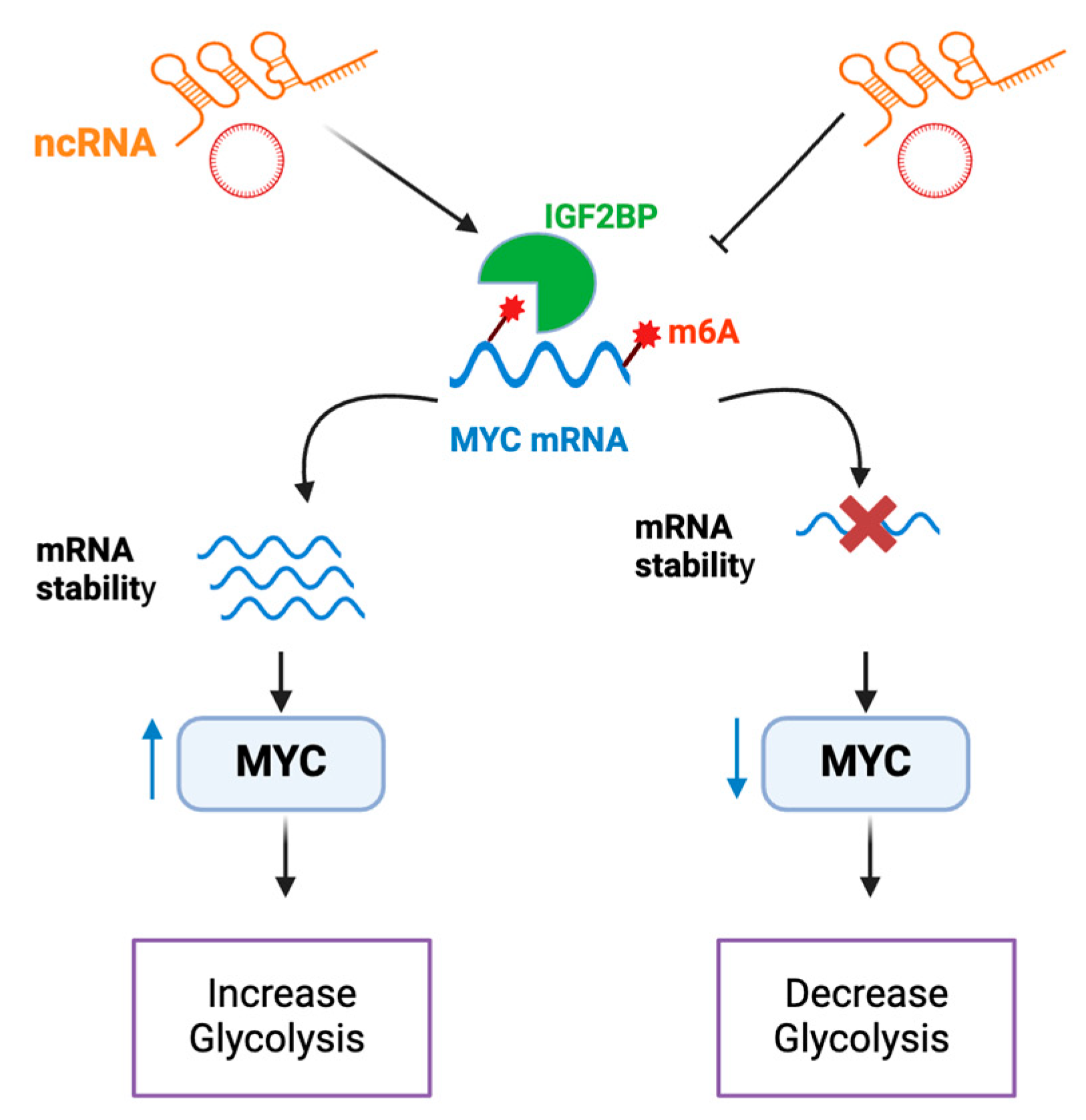
3.1. ncRNAs that affect metabolism by regulating MYC Stability
The role of lncRNA in glucose metabolism through mechanisms linked to MYC have been introduced recently in several tumours. One mechanism being their interaction with insulin-like growth factor mRNA-binding proteins (IGFBPs) (
Figure 2). N6-methyladenosine (m
6A)is a posttranscriptional RNA modification which plays a role in mRNA metabolism, splicing , stability and translation [
29] .IGFBPs have been recently described as a N
6-methyladenosine (m
6A) readers that can maintain the stability of m
6A modified mRNAs [
30], and increase the translation of target transcripts such as MYC [
31,
32,
33]. In colorectal cancer cells (CRC), LINRIS (Long Intergenic Noncoding RNA for IGFBP2 Stability) was found to block ubiquitination and thus stop degradation of IGFBP2[
34]. LINRIS knock down in CRC was associated with a decrease in expression of MYC and glycolytic enzymes with an overall attenuation of glycolysis which was rescued to some extent with over expression IGF2BP2, indicating a role for LINRIS
-IGF2BP2-MYC in glycolysis and cancer proliferation [
34]. In the same mechanism, however with opposite effects, LncRNA FGF13-AS1 in breast cancer exhibited a tumour suppressor function reducing MYC stability by disrupting MYC- IGF2BP2 binding. Interestingly, the lncRNA was able to attenuate glycolysis and stemness in cells [
35].
It is important to note that other lncRNA , such as gastric carcinoma high expressed transcript 1 (lncRNA GHET1) for example, were found to enhances the interaction between MYC mRNA and IGF2BP1 thus increasing MYC stability , however a direct contribution on MYC metabolic function was not confirmed [
36]
Emphasizing the role of circRNAs in MYC glycolytic function, a study was conducted in prostate cancer (PCa) [
37]. The circARHGAP29, a circRNA generated from the circularization of the Rho GTPase Activating Protein 29 (
ARHGAP29) gene, promoted docetaxel resistance in PCa through promoting aerobic glycolysis . In a similar manner, circARHGAP29 was able to stabilize MYC through interacting with IGF2BP2 . Interestingly this circRNA was found to also directly bind to MYC protein, thus stabilizing MYC mRNA and protein levels . Inhibition of circARHGAP29 decreased LDHA expression and extracellular glucose consumption in vitro and in vivo , which could be rescued by MYC over expression.
Another lncRNA which was shown to stabilize MYC is the lncRNA GLCC1, a glycolysis associated lnRNA which was found to be overexpressed in CRC tissue compared to adjacent tissue and correlated with tumour progression [
38]. Through knockdown and in-gain of function expression of GLCC1in vitro, its expression was induced by glucose starvation and resulted in glycolysis. Interestingly, GLCC1-induced glycolytic metabolism appeared to be mediated by its interaction with HSP90 in glucose starvation condition. HSP90 protein chaperone is essential for the stability of many proteins, and mediate MYC stability in CRC cells, [
38]. Thus GLCC1 stabilizes MYC through its direct interaction with HSP90.
CircPDK1 was found to be highly expressed in hypoxic-induced exosomes in pancreatic cancer , and was associated with advanced pathology and poor prognosis [
39]. In a recent study, circPDK1 played a significant role in tumour growth and in promoting glycolysis in vitro and in vivo in a pancreatic subcutaneous tumour mice model through a MYC related pathway [
39]. circPDK1 served as a competing endogenous RNA (ceRNA) sponging miR-628-3p thereby releasing Bromodomain and PHD finger-containing transcription factor (BPTF) from the inhibiting effects of miR-628-3p. BPTF proteins can regulated the expression of various oncogenes and tumor suppressors through their chromatin modulating activity. Notably, BPTF is essential for MYC binding of target genes and its overall transcriptional activity[
40]. Thus the study provides insight for the role of hypoxia and hypoxia induced factor 1 alpha (HIF1 α) in inducing circular RNA which can promote MYC mediated glycolytic effect [
39].
Using casteration-resistance prostate cancer cells, miR-644a was found to be a potent tumour suppressor that can inhibit glycolytic activity and the key glycolytic enzyme glyceraldehyde 3-phosphate dehydrogenase( GAPDH) expression in vitro and in xenograft models [
41]. Interestingly MYC was found to be a direct target of miR-644a, suggesting the possible mechanism by which miR-644 suppress the Warburg effect, however given the broad range of effect of miR-644a downregulating various driver molecules [
41], further experiments might be required to establish the exact mechanism. In miR-155ko/ko isolated from miR-155 deficient breast cancer mouse model, the loss of miR-155 was associated with a decrease in glucose transporters and main glycolytic enzymes expression which was linked to a decrease in MYC expression [
42]. MiR-155 was found to indirectly target FOXO3a which is known to destabilize MYC. MiR-155 metabolic effect was further confirmed in human breast cancer cell lines, in addition miR-155 high tumours displayed higher glucose uptake in patient samples seen through PET images, with miR-155 correlating negatively and positively with FOXO3a and MYC respectively.
3.2. ncRNAs with direct metabolic functions and regulated by MYC
In addition to their role in stabilizng MYC, another important mechanism by which ncRNAs influence MYC metabolism is through their direct regulation of glycolysis. These ncRNAs are transcriptional targets of MYC, and can modulate key metabolic pathways that MYC is involved in, further shaping the cellular metabolic landscape
The pan-cancer lncRNA Motor neuron and pancreas homeobox 1-antisense RNA1 (MNX1-AS1) MNX1-AS1 which acts as an oncogene, promoting the Warburg effect, was found to be a direct target of MYC, in HCC [
43]. The lncRNA was found to be induced by MYC following epidermal growth factor signaling and facilitates the nuclear transportation of PKM2 making use of its non-glycolytic function as a coactivator of the transcription of LDHA, PDK1, and GLUT1 genes . Importantly, using MYC depleted HepG2, silencing of MNX1-AS1 decreased the RNA and protein levels of glycolytic enzymes which could not be rescued by ectopic expression of MYC , indicating that this could be an essential mechanism by which MYC can mediate its metabolic function . The correlation between MNX1-AS1, nuclear PKM2, and C-MYC was further confirmed in patient tumour samples [
43].
MYC can also regulate glycolysis through repressing the expression of ncRNA expression. MYC , for example, was shown to mediate the repression of the lncRNA IDH1 antisense RNA1 in various cell lines [
44]. lncRNA IDH1-AS1 effected MYC- glycolytic function, since over expression of this lncRNA partially inhibited the increase of glucose uptake and lactate production associated with MYC over expression , while silencing lncRNA IDH1 showed opposite effects. The mechanism of action of this lncRNA was linked to an isocitrate dehydrogenase 1-alpha ketoglutarate - hypoxia-inducible factor 1α (IDH1-α-KG-HIF1α) axis under normoxic conditions only. The lncRNA increases the activity of IDH1 leading to an increase of α-KG which can suppress HIF1a, limiting its glycolytic function, thus demonstrating an example of how lncRNA can link MYC and HIF1 α functions in the Warburg effect
3.3. ncRNAs That Regulate MYC and Are Targets of MYC
Another mechanism that links both previous mechanisms is MYC’s ability to create feedback loops with ncRNA molecules that can induce or suppress glycolysis [
45,
46,
47]. These ncRNA not only impact MYC stability but are also direct transcriptional targets of MYC, thus creating a dynamic feedback loop by which MYC can sustain its metabolic activity through a self-perpetuating positive feedback mechanism. However, in some cases, these ncRNA were shown to possess other MYC-independent metabolic regulations that can enhance MYC metabolic effect. An example is the sophisticated work done with the novel lncRNA glycoLINC (gLINC) induced by MYC [
47]. gLINC played a crucial role in forming a complex network with enzymes involved in the glycolytic pathway, including Phosphoglycerate Kinase 1(PGK1), Phosphoglycerate Mutase 1(PGAM1), ENO1, PKM2, and LDHA. The assembled complex, known as a metabolon, enhances glycolysis, increases ATP production, and allows cancer cells to survive during serine deprivation in vitro and in vivo . Interestingly the lncRNA promoted MYC expression suggesting a positive feedback loop. It was also shown that gLINC silencing, although partially, was able to reverse MYC’s overexpression glycolytic effect , indicating that gLINC can enhance MYC-mediated glycolysis. MYC -gLINC axis therefore shows a mechanism by which cancer cells sustain cell survival during serine deprivation through enhancing glycolysis and sustaining ATP production .
Further support of the importance of m
6A modifications in cancer metabolism , and the cross talk between m
6A modification and lncRNA, was seen in a study showing the reciprocal regulation between the FTO intronic Transcript 1 lncRNA (FTO-IT1) and MYC [
45]. Through stabilizing its gene product, the FTO demethylase, this lncRNA was associated with increasing glycolysis in hepatocellular carcinoma (HCC) cells and an increase of GLUT1 and PKM2 stability through FTO mediated demethylation of m
6A. The expression of this lncRNA increased by MYC in hypoglycemic conditions on the other hand MYC was in itself regulated by FTO via m
6A modification. FTO-IT1 was also associated with tumour growth and glycolysis in a subcutaneous tumour model [
45]. These results differ from previous studies that have shown m
6A modification to be important for stability of MYC, indicating that m
6A modification can have context-dependent effects on MYC stability .
Another example was LINC01123 which was upregulated in non small cell lung cancer ( NSCLC )patients with high
18F-FDG uptake on PET/CT scans, and correlated with poor survival [
46]. A novel function was introduced for this lncRNA with its ability to increase
18F-FDG uptake, lactate production and expression of glycolytic enzymes in vitro and in vivo in xenograft model. MYC regulated the transcription of LINC01123, interestingly however , LINC01123 was also found to increase MYC expression through competitively decoying miR-199a-5p which can bind to MYC and have an inhibitory effect . It is worth noting that studies have also identified miR-199a-5p as a tumor suppressor across various cancer types, with evidence suggesting it can inhibit the Warburg effect by targeting other molecules such as HIF-1α[
48].
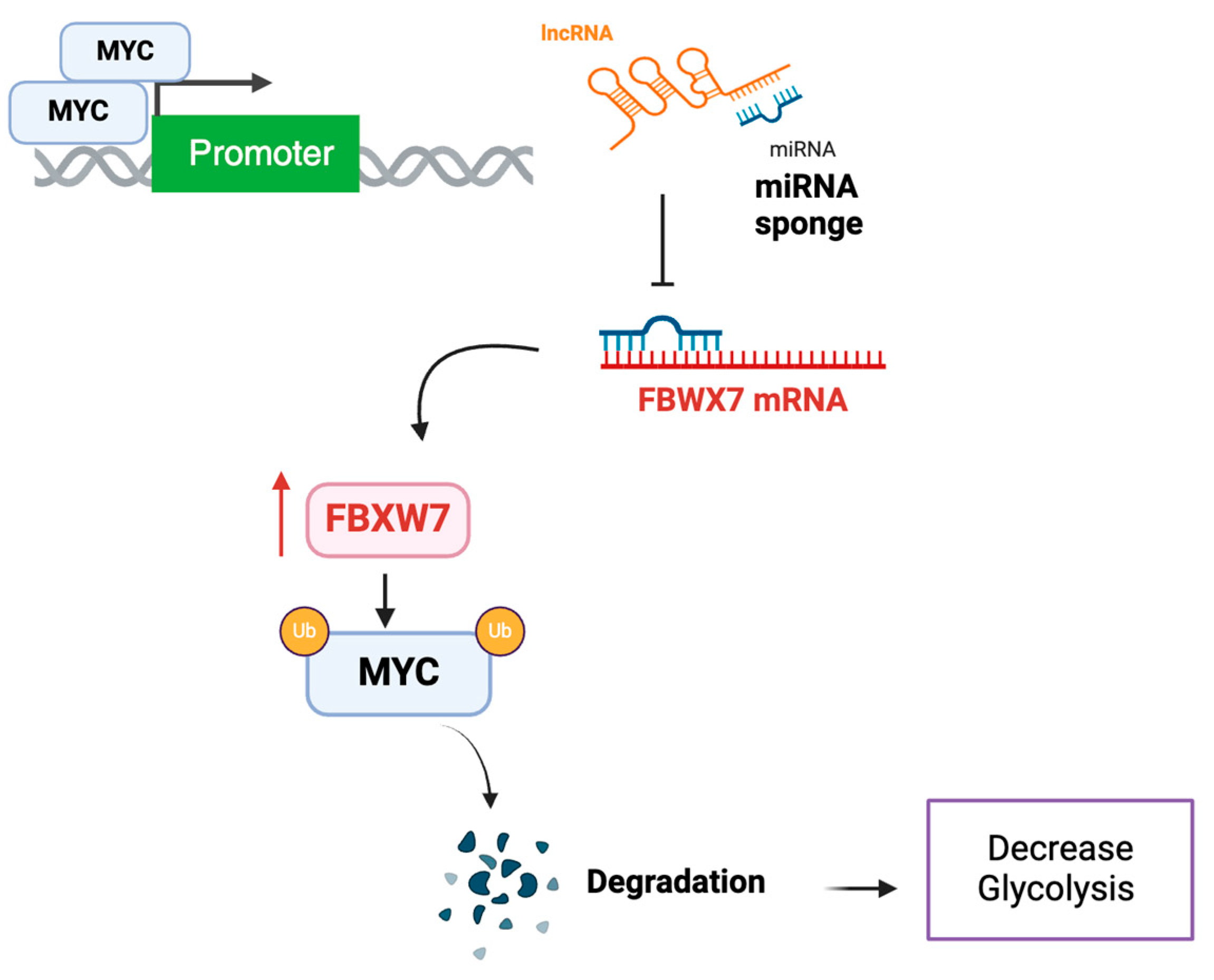
Another intresting ncRNA is lncRNA Myc inhibitory factor (LncRNA-MIF) which is identified as a direct transcript of MYC but has shown to regulate MYC protein stability in various cell lines, including HeLa and HCT116 [
49]. It functions as a competing endogenous RNA (ceRNA) for miR-586, which modulates the expression of F-box and WD repeat domain containingg 7 (FBWX7), a member of the F-box protein family, acting as a substrate recognition component of the an E3 ubiquitin ligase involved in MYC degradation (
Figure 3). Consequently, lncRNA-MIF is associated with reduced MYC levels. Knockdown of lncRNA-MIF increases the expression of MYC target genes related to glycolysis, such as GLUT1, LDHA, PKM2, and HK2 while over-expression of the lncRNA was associated with a decrease in the expression of those genes and a decrease in lactate production confirming its role in regulating glycolysis [
49].
This study further confirms other studies that show the importance of FBXW7 in decreasing glycolysis and tumour progression in vitro and in vivo[
50], underscoring the importance of exploring this axis. The complexity further lies on how FBXW7 in tumours is regulated through various means on the transcriptional and epigenetic level. For example, MYC itself was found to negatively or positively regulate FBXW7 expression through miRNA and lncRNA expression [
51].
Overall these studies show that lncRNAs not only mediate MYC stability, therefore regulating its metabolic function, but MYC can also regulate the expression of lncRNA, which in turn have metabolic functions, creating a complex regulatory network.
3. Clinical Relevance of the ncRNAs Discussed
The majority of the described ncRNAs are still in the early stages of research, with much yet to be explored in terms of their biological functions and mechanisms and clinical signficance. However, we provide an overview of the current understanding of their clinical correlations, highlighting their potential implications in various diseases and therapeutic contexts.
In a cohort of 95 CRC cases, LncRNA GLCC1 expression was found to be positively correlated with tumor size and invasion depth. Higher levels of GLCC1 expression were associated with more aggressive tumor characteristics and shorter survival times. Univariate and multivariate regression analyses showed that GLCC1 expression was an independent predictor of poor clinical outcomes,indicating its potential use as a prognostic marker [
38]. Similary, LINRIS expression was associated with predicting poor survival in a sample of 62 NSCLC ptainets [
52].
CircPDK1 expression also significantly correlated with tumor size, TNM stage, tumor recurrence, and paclitaxel resistance in 40 patients of NSLC [
53]
The low expression of miR-644a on the other hand was found to correlate with TNM stage, metastasis ,and low overall survival rate in 80 patients with NSCLS [
54]. Interestingly, and further demonstrating the complex netwrok of ncRNA interactions, the circRNA CircGLIS3 was shown to function as an oncogene in this study via sponging multiple tumor-suppressive miRNAs including miR-644a [
54].
Among miRNA-based therapies, miR-155 inhibition has emerged as one of the advanced in clinical development. Cobomarsen, an oligonucleotide miR-155 inhibitor, has progressed to phase 1/2 clinical trials for the treatment of relapsed and refractory lymphoma, showing promising potential in targeting this oncogenic miRNA[
55] .
Being highly explored and showing growing interest is the clincail impact of LncRNA MNX1-AS1. Aberrant expression of MNX1-AS1 has been shown in various tumour including and not limited to; gastric cancer,lung , osteosarcoma, hepatocellular carcinoma, and ovarian with its upregulation associated with clinicopathological parameters such as lymphatic metastasis, tumor size, stage and poor survival [
56].
FTO-IT1 was shown to be upregulated in HCC samples , and PCa,and correlated with decreased overall survival , and disease free survival [
45,
57] . Similarly LINC01123 was found to be upregulated and associated with poor prognosis in CRC [
58], NSLC [
46], and oral squamous cell carcinoma[
59].
Despite being previously described as undruggable , recent advances in MYC theraputics have been promosing [
60]. For example small molecules and peptide-based approaches like the Omomyc miniprotein, which disrupts MYC/MAX dimerization and MYC-DNA binding, have shown promising preclinical results, and has successfully completed Phase 1 clinical trials for advanced solid tumors[
60,
61]. With recent advances in MYC targeting, combining MYC inhibitors with ncRNA therapeutics could provide a powerful new strategy for treating MYC-driven cancers.
4. Conclusion
In summary , ncRNAs can regulate MYC metabolic and glycolytic activity. These RNA classes are known to interact together and create complex networks that is tissue-specific and context-specific. Currently as studied those ncRNA can effect MYC metabolic function by either; affecting MYC stability , or by being a target of MYC but having metabolic functions, or through both mechansisms combined. One of the limitations is that the studied ncRNAs are still in their early research phases, and much remains unknown about their full roles and mechanisms. As further studies are conducted, it is likely that new mechanisms will emerge, such as some ncRNAs currently classified in the second category potentially being reclassified into the third category, or entirely new mechanisms being discovered to explain their functions. Another potential limitation is that findings from in vitro studies may not always fully reflect the in vivo mechanisms, particularly in the context of MYC-mediated metabolic regulation. The complexity of MYC’s role in metabolism is influenced by a variety of external and internal factors, such as hypoxia, and cell-specific conditions, which are difficult to replicate in simplified in vitro models. These additional factors can significantly alter MYC’s impact on metabolism and MYC’s interaction with ncRNA. It is therefore crucial to investigate the role of these ncRNAs in vivo and in patient samples to better understand their true impact on MYC-driven metabolic regulation.
Funding
This research received no external funding.
Data Availability Statement
Not applicable
Conflicts of Interest
The authors declare no conflicts of interest.”
Abbreviations
The following abbreviations are used in this manuscript:
| ncRNA GLUT1 |
Non-coding RNAglucose transporter 1 |
| HK2 |
hexokinase 2 |
| LDHA |
lactate dehydrogenase A |
| Eno1 |
alpha enolase |
| MCT |
Monocarboxylate transporters |
| PFKM |
phosphofructokinase muscle isoform |
| SCLC |
Small cell lung carcinoma |
| NSCLC |
Non small cell lung cancer |
| LncRNA |
Long non-coding RNA |
| CircRNA |
Circular RNA |
| MiRNA |
MicroRNA |
| TNBC |
Triple negative breast cancer |
| IGFBP |
insulin-like growth factor mRNA-binding proteins |
| M6A |
N6-methyladenosine |
| CRC |
Colorectal cancer cells |
| HCC |
Hepatocellular carcinoma |
| PCa |
Prostata cancer |
| BPTF |
Bromodomain and PHD finger-containing transcription factor |
| HIF1α |
Hypxia inducible factor 1 alpha |
| FBXW7 |
F-box and WD repeat domain containingg 7 |
References
- Martínez-Reyes, I. and N.S. Chandel, Cancer metabolism: looking forward. Nat Rev Cancer, 2021, 21, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J. and C.B. Thompson, Metabolic regulation of cell growth and proliferation. Nat Rev Mol Cell Biol, 2019, 20, 436–450. [Google Scholar] [CrossRef] [PubMed]
- Dhanasekaran, R. , et al., The MYC oncogene - the grand orchestrator of cancer growth and immune evasion. Nat Rev Clin Oncol, 2022, 19, 23–36. [Google Scholar] [CrossRef]
- Das, S.K., B. A. Lewis, and D. Levens, MYC: a complex problem. Trends Cell Biol, 2023, 33, 235–246. [Google Scholar] [CrossRef]
- Gabay, M., Y. Li, and D.W. Felsher, MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med, 2014. 4(6).
- Chen, H., H. Liu, and G. Qing, Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct Target Ther, 2018. 3: p. 5.
- Dong, Y. , et al., Regulation of cancer cell metabolism: oncogenic MYC in the driver's seat. Signal Transduct Target Ther, 2020, 5, 124. [Google Scholar] [CrossRef]
- Dejure, F.R. and M. Eilers, MYC and tumor metabolism: chicken and egg. Embo j, 2017, 36, 3409–3420. [Google Scholar] [CrossRef]
- Stine, Z.E. , et al., MYC, Metabolism, and Cancer. Cancer Discov, 2015, 5, 1024–1039. [Google Scholar] [CrossRef] [PubMed]
- Winkle, M. , et al., Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discov, 2021, 20, 629–651. [Google Scholar] [CrossRef]
- Coan, M. , et al., Targeting and engineering long non-coding RNAs for cancer therapy. Nat Rev Genet, 2024, 25, 578–595. [Google Scholar] [CrossRef]
- Stasevich, E.M. , et al., The Role of Non-Coding RNAs in the Regulation of the Proto-Oncogene MYC in Different Types of Cancer. Biomedicines, 2021. 9(8).
- Jahangiri, L. , et al., The Contribution of Autophagy and LncRNAs to MYC-Driven Gene Regulatory Networks in Cancers. Int J Mol Sci, 2021. 22(16).
- Cargill, K.R. , et al., Targeting MYC-enhanced glycolysis for the treatment of small cell lung cancer. Cancer Metab, 2021, 9, 33. [Google Scholar] [CrossRef]
- Hu, S. , et al., 13C-pyruvate imaging reveals alterations in glycolysis that precede c-Myc-induced tumor formation and regression. Cell Metab, 2011, 14, 131–42. [Google Scholar] [CrossRef] [PubMed]
- Shin, P.J. , et al., Cancer recurrence monitoring using hyperpolarized [1-(13)C]pyruvate metabolic imaging in murine breast cancer model. Magn Reson Imaging, 2017, 43, 105–109. [Google Scholar] [CrossRef]
- Doherty, J.R. , et al., Blocking lactate export by inhibiting the Myc target MCT1 Disables glycolysis and glutathione synthesis. Cancer Res, 2014, 74, 908–20. [Google Scholar] [CrossRef]
- Gan, L. , et al., Metabolic targeting of oncogene MYC by selective activation of the proton-coupled monocarboxylate family of transporters. Oncogene, 2016, 35, 3037–48. [Google Scholar] [CrossRef] [PubMed]
- David, C.J. , et al., HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature, 2010, 463, 364–8. [Google Scholar] [CrossRef]
- Yu, L. , et al., MTR4 drives liver tumorigenesis by promoting cancer metabolic switch through alternative splicing. Nat Commun, 2020, 11, 708. [Google Scholar] [CrossRef]
- Lei, Z. , et al., Reciprocal interactions between lncRNAs and MYC in colorectal cancer: partners in crime. Cell Death Dis, 2024, 15, 539. [Google Scholar] [CrossRef] [PubMed]
- Chao, C. , et al., Oncogenic roles and related mechanisms of the long non-coding RNA MINCR in human cancers. Front Cell Dev Biol, 2023, 11, 1087337. [Google Scholar] [CrossRef]
- Benetatos, L., A. Benetatou, and G. Vartholomatos, Long non-coding RNAs and MYC association in hematological malignancies. Ann Hematol, 2020, 99, 2231–2242. [Google Scholar] [CrossRef]
- Wang, S. , et al., Myc derived circRNA promotes triple-negative breast cancer progression via reprogramming fatty acid metabolism. Discov Oncol, 2023, 14, 67. [Google Scholar] [CrossRef]
- Yang, X. , et al., CircMYC promotes proliferation, migration, invasion and inhibits apoptosis of small cell lung cancer by targeting miR-145/ Matrix Metallopeptidase 2 axis. Bioengineered, 2022. 13(4): p. 10552-10563.
- Wang, J. , et al., Circular RNA circ-LRP6 facilitates Myc-driven tumorigenesis in esophageal squamous cell cancer. Bioengineered, 2020. 11(1): p. 932-938.
- Kim, J.W., S. Mori, and J.R. Nevins, Myc-induced microRNAs integrate Myc-mediated cell proliferation and cell fate. Cancer Res, 2010. 70(12): p. 4820-8.
- Li, Y., Y. Zhu, and E.V. Prochownik, MicroRNA-based screens for synthetic lethal interactions with c-Myc. RNA Dis, 2016. 3(3).
- Zhao, B.S., I. A. Roundtree, and C. He, Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol, 2017. 18(1): p. 31-42.
- Huang, H. , et al., Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol, 2018. 20(3): p. 285-295.
- Samuels, T.J. , et al., Imp/IGF2BP levels modulate individual neural stem cell growth and division through myc mRNA stability. Elife, 2020. 9.
- Gao, Y. , et al., A novel lncRNA MTAR1 promotes cancer development through IGF2BPs mediated post-transcriptional regulation of c-MYC. Oncogene, 2022, 41, 4736–4753. [Google Scholar] [CrossRef]
- Zhu, P. , et al. , A novel hypoxic long noncoding RNA KB-1980E6.3 maintains breast cancer stem cell stemness via interacting with IGF2BP1 to facilitate c-Myc mRNA stability. Oncogene, 2021, 40, 1609–1627. [Google Scholar] [PubMed]
- Wang, Y. , et al., LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol Cancer, 2019. 18(1): p. 174.
- Ma, F. , et al., Long non-coding RNA FGF13-AS1 inhibits glycolysis and stemness properties of breast cancer cells through FGF13-AS1/IGF2BPs/Myc feedback loop. Cancer Lett, 2019. 450: p. 63-75.
- Yang, F. , et al., Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. Febs j, 2014. 281(3): p. 802-13.
- Jiang, X. , et al., EIF4A3-Induced circARHGAP29 Promotes Aerobic Glycolysis in Docetaxel-Resistant Prostate Cancer through IGF2BP2/c-Myc/LDHA Signaling. Cancer Res, 2022, 82, 831–845. [Google Scholar] [CrossRef] [PubMed]
- Tang, J. , et al., LncRNA GLCC1 promotes colorectal carcinogenesis and glucose metabolism by stabilizing c-Myc. Nat Commun, 2019. 10(1): p. 3499.
- Lin, J. , et al., Hypoxia-induced exosomal circPDK1 promotes pancreatic cancer glycolysis via c-myc activation by modulating miR-628-3p/BPTF axis and degrading BIN1. J Hematol Oncol, 2022. 15(1): p. 128.
- Richart, L. , et al., BPTF is required for c-MYC transcriptional activity and in vivo tumorigenesis. Nat Commun, 2016. 7: p. 10153.
- Ebron, J.S. , et al., MiR-644a Disrupts Oncogenic Transformation and Warburg Effect by Direct Modulation of Multiple Genes of Tumor-Promoting Pathways. Cancer Res, 2019. 79(8): p. 1844-1856.
- Kim, S. , et al., microRNA-155 positively regulates glucose metabolism via PIK3R1-FOXO3a-cMYC axis in breast cancer. Oncogene, 2018, 37, 2982–2991. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y. , et al., MNX1-AS1, a c-Myc induced lncRNA, promotes the Warburg effect by regulating PKM2 nuclear translocation. J Exp Clin Cancer Res, 2022, 41, 337. [Google Scholar] [CrossRef] [PubMed]
- Xiang, S. , et al., LncRNA IDH1-AS1 links the functions of c-Myc and HIF1α via IDH1 to regulate the Warburg effect. Proc Natl Acad Sci U S A, 2018. 115(7): p. E1465-e1474.
- Wang, F. , et al., LncRNA FTO-IT1 promotes glycolysis and progression of hepatocellular carcinoma through modulating FTO-mediated N6-methyladenosine modification on GLUT1 and PKM2. J Exp Clin Cancer Res, 2023. 42(1): p. 267.
- Hua, Q. , et al., LINC01123, a c-Myc-activated long non-coding RNA, promotes proliferation and aerobic glycolysis of non-small cell lung cancer through miR-199a-5p/c-Myc axis. J Hematol Oncol, 2019. 12(1): p. 91.
- Zhu, Y. , et al., The long noncoding RNA glycoLINC assembles a lower glycolytic metabolon to promote glycolysis. Mol Cell, 2022. 82(3): p. 542-554.e6.
- Li, B. , et al., Mutual Regulation of MiR-199a-5p and HIF-1α Modulates the Warburg Effect in Hepatocellular Carcinoma. J Cancer, 2017, 8, 940–949. [Google Scholar] [CrossRef]
- Zhang, P. , et al., LncRNA-MIF, a c-Myc-activated long non-coding RNA, suppresses glycolysis by promoting Fbxw7-mediated c-Myc degradation. EMBO Rep, 2016, 17, 1204–20. [Google Scholar] [CrossRef]
- Ji, S. , et al., FBW7 (F-box and WD Repeat Domain-Containing 7) Negatively Regulates Glucose Metabolism by Targeting the c-Myc/TXNIP (Thioredoxin-Binding Protein) Axis in Pancreatic Cancer. Clin Cancer Res, 2016. 22(15): p. 3950-60.
- Fan, J. , et al., Clinical significance of FBXW7 loss of function in human cancers. Mol Cancer, 2022. 21(1): p. 87.
- Zhu, Y. , et al., Long non-coding RNA LINRIS is upregulated in non-small cell lung cancer and its silencing inhibits cell proliferation by suppressing microRNA-10a maturation. Bioengineered, 2022. 13(2): p. 4340-4346.
- Feng, Y., T. Zhang, and H. Liu, circPDK1 competitively binds miR-4731-5p to mediate GIGYF1 expression and increase paclitaxel sensitivity in non-small cell lung cancer. Discov Oncol, 2024. 15(1): p. 157.
- Wu, Z. , et al., A circGLIS3/miR-644a/PTBP1 positive feedback loop promotes the malignant biological progressions of non-small cell lung cancer. Am J Cancer Res, 2021. 11(1): p. 108-122.
- Anastasiadou, E. , et al., Cobomarsen, an Oligonucleotide Inhibitor of miR-155, Slows DLBCL Tumor Cell Growth In Vitro and In Vivo. Clin Cancer Res, 2021, 27, 1139–1149. [Google Scholar] [CrossRef]
- Li, T. , et al., LncRNA MNX1-AS1: A novel oncogenic propellant in cancers. Biomed Pharmacother, 2022. 149: p. 112801.
- Zhang, J. , et al., A lncRNA from the FTO locus acts as a suppressor of the m(6)A writer complex and p53 tumor suppression signaling. Mol Cell, 2023. 83(15): p. 2692-2708.e7.
- Liu, Z. , et al., Long non-coding RNA LINC01123 promotes cell proliferation, migration and invasion via interacting with SRSF7 in colorectal cancer. Pathol Res Pract, 2022. 232: p. 153843.
- Qin, H., C. Wang, and Y. Hua, LINC01123 is associated with prognosis of oral squamous cell carcinoma and involved in tumor progression by sponging miR-34a-5p. Oral Surg Oral Med Oral Pathol Oral Radiol, 2022. 133(1): p. 50-59.
- Atibalentja, D.F., A. Deutzmann, and D.W. Felsher, A big step for MYC-targeted therapies. Trends Cancer, 2024, 10, 383–385. [Google Scholar] [CrossRef]
- Garralda, E. , et al., MYC targeting by OMO-103 in solid tumors: a phase 1 trial. Nat Med, 2024, 30, 762–771. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).