Submitted:
10 February 2025
Posted:
13 February 2025
You are already at the latest version
Abstract
African swine fever (ASF), a highly contagious and lethal viral disease, continues to devastate the global swine industry. The emergence of ASF virus (ASFV) variants with varying genomic deletions poses significant challenges to ASF control. This study presents a novel, sensitive, and reliable quadplex real-time PCR assay for detecting ASFV variants lacking key genes (I177L, EP402R, and MGF360-14L), either individually or in combination. The assay targets conserved regions within these genes, ensuring broad coverage of diverse ASFV genotypes. A porcine beta-actin (ACTB) internal control was incorporated to minimize false-negative results. Optimization and evaluation using spike-in tests demonstrated high sensitivity with a limit of detection (LOD) ranging from 1-10 plasmid copies or 0.1 TCID50 of ASFV isolates per reaction. No cross-reactivity was observed when testing serum samples from pigs infected with other common swine viruses. Further validation across a diverse panel of samples, comprising those from naturally ASFV-infected field pigs (n=54), experimentally ASFV-infected pigs (n=50), PBS-inoculated pigs (n=50), ASFV-free field pigs (n=100), and feral pigs (n=6), confirmed 100% specificity. This robust assay provides a valuable tool for rapid and accurate ASF surveillance and control efforts, facilitating the timely detection and mitigation of outbreaks caused by emerging ASFV variants.
Keywords:
1. Introduction
2. Materials and Methods
2.1. Viruses and Porcine Serum Samples
- (i)
- ASFV negative pig sera: serum samples from pigs inoculated with phosphate-buffered saline (PBS, pH7.4, Thermo Scientific, Bridgewater, NJ, USA) (n=50).
- (ii)
- Experimentally ASFV infected pig sera: serum samples from pigs infected with the ASFV VNUA-ASFV-05L1 which confirmed ASFV positive when tested by standard ASFV RT-PCR [41] (n=50).
- (iii)
- Naturally ASFV-infected pig sera: serum samples from naturally ASFV infected field domestic pigs in ASFV epidemic country (The Philippines) (n=54).
- (iv)
- ASFV-free field domestic pig sera: serum samples from pigs in local farms in Kansas, USA (n=100).
- (v)
- Feral pig sera: serum samples from feral pigs caught in Kansas (collaboration with USDA APHIS Wildlife Services, Kansas Wildlife Services, USA) (n=6).
- (vi)
- Other common swine virus infected pig sera: serum samples from pigs infected with CSFV (n=50), PRRSV (n=50), PRV(n=10), and Bovine viral diarrhea virus (BVDV, n=4).
2.2. Construction Databases, Sequence Analysis, and Design of Primer and Probes
2.3. Preparation of Standard Plasmid and Optimization of Amplification Conditions
2.4. Analytical Sensitivity and Specificity Evaluation by Spiking Experiments
2.5. Validate the Quadplex RT-PCR with Experimental and Field Samples
2.6. Statistical Analysis
3. Results
3.1. Database for Sequence Alignment and the Design of Primers and Probes
3.2. Optimization of Quadplex RT-PCR Through Standard Plasmid Spiked Experiments Using Various Multiplex Reaction Buffers
3.3. Analytical Sensitivity and Specificity of Quadplex RT-PCR in Virus Spiked Samples
3.4. Diagnostic Sensitivity and Specificity of Quadplex RT-PCR in Clinical Samples from Experimental Infected Pigs
3.5. Performance of Quadplex RT-PCR on Field Clinical Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dixon, L.K.; Stahl, K.; Jori, F.; Vial, L.; Pfeiffer, D.U. African swine fever epidemiology and control. Annu. Rev. Anim. Biosci. 2020, 8, 221–246. [Google Scholar] [CrossRef]
- Montgomery, R.E. On a form of swine fever occurring in British East Africa (Kenya Colony). J. Comp. Pathol. Ther. 1921, 34, 159–191. [Google Scholar] [CrossRef]
- Li, Z.; Chen, W.; Qiu, Z.; Li, Y.; Fan, J.; Wu, K.; Li, X.; Zhao, M.; Ding, H.; Fan, S.; Chen, J. African Swine Fever Virus: A Review. Life (Basel) 2024, 14(3), 381. [Google Scholar] [CrossRef]
- Wang, L.; Ganges, L.; Dixon, L.K.; Bu, Z.; Zhao, D.; Truong, Q.L.; Richt, J.A.; Jin, M.; Netherton, C.L.; Benarafa, C.; Summerfield, A.; Weng, C.; Peng, G.; Reis, A.L.; Han, J.; Penrith, M. L.; Mo, Y.; Su, Z.; Vu Hoang, D.; Pogranichniy, R.M.; Balaban-Oglan, D.A.; Li, Y.; Wang, K.; Cai, X.; Shi, J. International African Swine Fever Workshop: Critical Issues That Need to Be Addressed for ASF Control. Viruses 2023, 16(1), 4. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wang, L.; McVey, D.S. Of pigs and men: The best-laid plans for prevention and control of swine fevers. Anim. Front. 2021, 11, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Ruedas-Torres, I.; Thi To Nga, B.; Salguero, F.J. Pathogenicity and virulence of African swine fever virus. Virulence 2024, 15, 2375550. [Google Scholar] [CrossRef]
- Karger, A.; Pérez-Núñez, D.; Urquiza, J.; Hinojar, P.; Alonso, C.; Freitas, F. B.; Revilla, Y.; Le Potier, M. F.; Montoya, M. An Update on African Swine Fever Virology. Viruses 2019, 11(9), 864. [Google Scholar] [CrossRef]
- Sánchez-Vizcaíno, J.M.; Mur, L.; Martínez-López, B. African swine fever: an epidemiological update. Transbound. Emerg. Dis. 2012, 59 Suppl 1, 27–35. [Google Scholar] [CrossRef]
- Vu, H.L.X.; McVey, D.S. Recent progress on gene-deleted live-attenuated African swine fever virus vaccines. NPJ Vaccines. 2024, 9(1), 60. [Google Scholar] [CrossRef]
- Post, J.; Weesendorp, E.; Montoya, M.; Loeffen, W.L. Influence of Age and Dose of African Swine Fever Virus Infections on Clinical Outcome and Blood Parameters in Pigs. Viral Immunol. 2017, 30(1), 58–69. [Google Scholar] [CrossRef]
- Dixon, L.K.; Sun, H.; Roberts, H. African swine fever. Antiviral Res. 2019, 165, 34–41. [Google Scholar] [CrossRef]
- Alejo, A.; Matamoros, T.; Guerra, M.; Andrés, G. A Proteomic Atlas of the African Swine Fever Virus Particle. J. Virol. 2018, 92(23), e01293–18. [Google Scholar] [CrossRef]
- Quembo, C. J.; Jori, F.; Vosloo, W.; Heath, L. Genetic characterization of African swine fever virus isolates from soft ticks at the wildlife/domestic interface in Mozambique and identification of a novel genotype. Transbound Emerg. Dis. 2018, 65(2), 420–431. [Google Scholar] [CrossRef] [PubMed]
- Arias, M.; de la Torre, A.; Dixon, L.; Gallardo, C.; Jori, F.; Laddomada, A.; Martins, C.; Parkhouse, R.M.; Revilla, Y.; Rodriguez, F.A.J.; Sanchez-Vizcaino. Approaches and Perspectives for Development of African Swine Fever Virus Vaccines. Vaccines (Basel) 2017, 5(4), 35. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Vizcaíno, J. M.; Mur, L.; Martínez-López, B. African swine fever (ASF): five years around Europe. Vet. Microbiol. 2013, 165(1-2), 45–50. [CrossRef]
- Ge, S.; Li, J.; Fan, X.; Liu, F.; Li, L.; Wang, Q.; Ren, W; Bao, J; Liu, C; Wang, H; Liu, Y.; Zhang, Y.; Xu, T.; Wu, X.; Wang, Z. Molecular characterization of African swine fever virus, China, 2018. Emerg. Infect. Dis. 2018, 24(11), 2131–2133. [CrossRef]
- Cwynar, P.; Stojkov, J.; Wlazlak, K. African swine fever status in Europe. Viruses 2019, 11(4), 310. [Google Scholar] [CrossRef]
- Ruiz-Saenz, J.; Diaz, A.; Bonilla-Aldana, D. K.; Rodríguez-Morales, A. J.; Martinez-Gutierrez, M.; Aguilar, P. V. African swine fever virus: A re-emerging threat to the swine industry and food security in the Americas. Front. Microbiol. 2022, 13, 1011891. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Sun, E.; Huang, L.; Ding, L.; Zhu, Y.; Zhang, J.; Shen, D.; Zhang, X.; Zhang, Z.; Ren, T.; Wang, W.; Li, F.; He, X.; Bu, Z. Highly lethal genotype I and II recombinant African swine fever viruses detected in pigs. Nat. Commun. 2023, 14(1), 3096. [Google Scholar] [CrossRef]
- Le, V.P.; Nguyen, V.T.; Le, T.B.; Mai, N.T.A.; Nguyen, V.D.; Than, T.T.; Lai, T.N.H.; Cho, K.H.; Hong, S.K.; Kim, Y.H.; Bui, T.A.D.; Nguyen, T.L.; Song, D.; Ambagala, A. Detection of Recombinant African Swine Fever Virus Strains of p72 Genotypes I and II in Domestic Pigs, Vietnam, 2023. Emerg. Infect. Dis. 2024, 30(5), 991–994. [Google Scholar] [CrossRef]
- Escribano, J.M.; Galindo, I.; Alonso, C. Antibody-mediated neutralization of African swine fever virus: myths and facts. Virus Res. 2013, 173(1), 101–109. [Google Scholar] [CrossRef]
- Pikalo, J.; Porfiri, L.; Akimkin, V.; Roszyk, H.; Pannhorst, K.; Kangethe, R.T.; Wijewardana, V.; Sehl-Ewert, J.; Beer, M.; Cattoli, G.; Blome, S. Vaccination with a Gamma Irradiation-Inactivated African Swine Fever Virus Is Safe but Does Not Protect Against a Challenge. Front Immunol. 2022, 13, 832264. [Google Scholar] [CrossRef]
- Rock, D.L. Thoughts on African Swine Fever Vaccines. Viruses 2021, 13(5), 943. [Google Scholar] [CrossRef] [PubMed]
- Gaudreault, N.N.; Richt, J.A. Subunit Vaccine Approaches for African Swine Fever Virus. Vaccines (Basel) 2019, 7(2), 56. [Google Scholar] [CrossRef]
- O'Donnell, V.; Holinka, L.G.; Gladue, D.P.; Sanford, B.; Krug, P.W.; Lu, X.; Arzt, J.; Reese, B.; Carrillo, C.; Risatti, G.R.; Borca, M.V. African Swine Fever Virus Georgia Isolate Harboring Deletions of MGF360 and MGF505 Genes Is Attenuated in Swine and Confers Protection against Challenge with Virulent Parental Virus. J Virol. 2015, 89(11), 6048–56. [Google Scholar] [CrossRef] [PubMed]
- Borca, M.V.; Ramirez-Medina, E.; Silva, E.; Vuono, E.; Rai, A.; Pruitt, S.; Holinka, L.G.; Velazquez-Salinas, L.; Zhu, J.; Gladue, D.P. Development of a Highly Effective African Swine Fever Virus Vaccine by Deletion of the I177L Gene Results in Sterile Immunity against the Current Epidemic Eurasia Strain. J Virol. 2020, 94(7), e02017–19. [Google Scholar] [CrossRef]
- Tran, X.H.; Le, T.T.P.; Nguyen, Q.H.; Do, T.T.; Nguyen, V.D.; Gay, C.G.; Borca, M.V.; Gladue, D.P. African swine fever virus vaccine candidate ASFV-G-ΔI177L efficiently protects European and native pig breeds against circulating Vietnamese field strain. Transbound Emerg Dis. 2022, 69(4), e497–e504. [Google Scholar] [CrossRef]
- Borca, M.V.; Rai, A.; Ramirez-Medina, E.; Silva, E.; Velazquez-Salinas, L.; Vuono, E.; Pruitt, S.; Espinoza, N.; Gladue, D.P. A Cell Culture-Adapted Vaccine Virus against the Current African Swine Fever Virus Pandemic Strain. J Virol. 2021, 95(14), e0012321. [Google Scholar] [CrossRef]
- Xie, Z.; Liu, Y.; Di, D.; Liu, J.; Gong, L.; Chen, Z.; Li, Y.; Yu, W.; Lv, L.; Zhong, Q.; Song, Y.; Liao, X.; Song, Q.; Wang, H.; Chen, H. Protection Evaluation of a Five-Gene-Deleted African Swine Fever Virus Vaccine Candidate Against Homologous Challenge. Front Microbiol. 2022, 13, 902932. [Google Scholar] [CrossRef] [PubMed]
- Petrovan, V.; Rathakrishnan, A.; Islam, M.; Goatley, L.C.; Moffat, K.; Sanchez-Cordon, P.J.; Reis, A.L.; Dixon, L.K. Role of African Swine Fever Virus Proteins EP153R and EP402R in Reducing Viral Persistence in Blood and Virulence in Pigs Infected with BeninΔDP148R. J Virol. 2022, 96(1), e0134021. [Google Scholar] [CrossRef]
- Teklue, T.; Wang, T.; Luo, Y.; Hu, R.; Sun, Y.; Qiu, H.J. Generation and Evaluation of an African Swine Fever Virus Mutant with Deletion of the CD2v and UK Genes. Vaccines (Basel) 2020, 8(4), 763. [Google Scholar] [CrossRef]
- Chen, W.; Zhao, D.; He, X.; Liu, R.; Wang, Z.; Zhang, X.; Li, F.; Shan, D.; Chen, H.; Zhang, J.; Wang, L.; Wen, Z.; Wang, X.; Guan, Y.; Liu, J.; Bu, Z. A seven-gene-deleted African swine fever virus is safe and effective as a live attenuated vaccine in pigs. Sci. China Life Sci. 2020, 63(5), 623–634. [Google Scholar] [CrossRef]
- Wang, L.; Fu, D.; Tesfagaber, W.; Li, F.; Chen, W.; Zhu, Y.; Sun, E.; Wang, W.; He, X.; Guo, Y.; Bu, Z.; Zhao, D. Development of an ELISA Method to Differentiate Animals Infected with Wild-Type African Swine Fever Viruses and Attenuated HLJ/18-7GD Vaccine Candidate. Viruses 2022, 14(8), 1731. [Google Scholar] [CrossRef]
- Sun, E.; Huang, L.; Zhang, X.; Zhang, J.; Shen, D.; Zhang, Z.; Wang, Z.; Huo, H.; Wang, W.; Huangfu, H.; Wang, W.; Li, F.; Liu, R.; Sun, J.; Tian, Z.; Xia, W.; Guan, Y.; He, X.; Zhu, Y.; Zhao, D.; Bu, Z. Genotype I African swine fever viruses emerged in domestic pigs in China and caused chronic infection. Emerg. Microbes Infect. 2021, 10(1), 2183–2193. [Google Scholar] [CrossRef] [PubMed]
- Leitão, A.; Cartaxeiro, C.; Coelho, R.; Cruz, B.; Parkhouse, R.M.E.; Portugal, F.C.; Vigário, J.D.; Martins, C.L.V. The non-haemadsorbing African swine fever virus isolate ASFV/NH/P68 provides a model for defining the protective anti-virus immune response. J. Gen. Virol. 2001, 82(Pt 3), 513–523. [Google Scholar] [CrossRef]
- Gallardo, C.; Soler, A.; Rodze, I.; Nieto, R.; Cano-Gómez, C.; Fernandez-Pinero, J.; Arias, M. Attenuated and non-haemadsorbing (non-HAD) genotype II African swine fever virus (ASFV) isolated in Europe, Latvia 2017. Transbound Emerg. Dis. 2019, 66(3), 1399–1404. [Google Scholar] [CrossRef] [PubMed]
- Boinas, F.S.; Hutchings, G.H.; Dixon, L.K.; Wilkinson, P. J. Characterization of pathogenic and non-pathogenic African swine fever virus isolates from Ornithodoros erraticus inhabiting pig premises in Portugal. J. Gen. Virol. 2004, 85(Pt 8), 2177–2187. [Google Scholar] [CrossRef]
- World Organisation for Animal Health. Available at: https://www.Woah.Org/En/What-We-Do/Standards/Codes-and-Manuals/Terrestrial-Manual-Online-Access/ (Accessed January 25, 2025).
- Oura, C.A.; Edwards, L.; Batten, C.A. Virological diagnosis of African swine fever--comparative study of available tests. Virus Res. 2013, 173(1), 150–158. [Google Scholar] [CrossRef]
- Hu, L.; Lin, X.Y.; Yang, Z.X.; Yao, X.P.; Li, G.L.; Peng, S.Z.; Wang, Y. A multiplex PCR for simultaneous detection of classical swine fever virus, African swine fever virus, highly pathogenic porcine reproductive and respiratory syndrome virus, porcine reproductive and respiratory syndrome virus and pseudorabies in swine. Pol. J. Vet. Sci. 2015, 18(4), 715–23. [Google Scholar] [CrossRef]
- Shi, X.; Liu, X.; Wang, Q.; Das, A.; Ma, G.; Xu, L.; Sun, Q.; Peddireddi, L.; Jia, W.; Liu, Y.; Anderson, G.; Bai, J.; Shi, J. A multiplex real-time PCR panel assay for simultaneous detection and differentiation of 12 common swine viruses. J. Virol. Methods 2016, 236, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25(24), 4876–82. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol. 2021, 38(7), 3022–3027. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, L.; Noll, L.; Stoy, C.; Porter, E.; Fu, J.; Feng, Y.; Peddireddi, L.; Liu, X.; Dodd, K.A.; Jia, W.; Bai, J. Development of a real-time PCR assay for detection of African swine fever virus with an endogenous internal control. Transbound Emerg Dis. 2020, 67(6), 2446–2454. [Google Scholar] [CrossRef] [PubMed]
- Truong, Q. L.; Nguyen, T. L.; Nguyen, T. H.; Shi, J.; Vu, H. L. X.; Lai, T. L. H.; Nguyen, V. G. Genome Sequence of a Virulent African Swine Fever Virus Isolated in 2020 from a Domestic Pig in Northern Vietnam. Microbiol Resour Announc. 2021, 10(19), e00193–21. [Google Scholar] [CrossRef]
- Hu, Z.; Tian, X.; Lai, R.; Wang, X.; Li, X. Current detection methods of African swine fever virus. Front Vet Sci. 2023, 10, 1289676. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhang, M.; Jie, Z.; Guo, S.; Zhu, Z.; Tao, S.C. Strategic nucleic acid detection approaches for diagnosing African swine fever (ASF): navigating disease dynamics. Vet Res. 2024, 55(1), 131. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Hu, L.; Shi, K.; Wei, H.; Shi, Y.; Hu, X.; Zhou, Q.; Feng, S.; Long, F.; Mo, S.; Li, Z. Development of a triplex real-time quantitative PCR for detection and differentiation of genotypes I and II African swine fever virus. Front Vet Sci. 2023, 10, 1278714. [Google Scholar] [CrossRef]
- Chen, Y.; Shi, K.; Liu, H.; Yin, Y.; Zhao, J.; Long, F.; Lu, W.; Si, H. Development of a multiplex qRT-PCR assay for detection of African swine fever virus, classical swine fever virus and porcine reproductive and respiratory syndrome virus. J. Vet. Sci. 2021, 22(6), e87. [Google Scholar] [CrossRef]
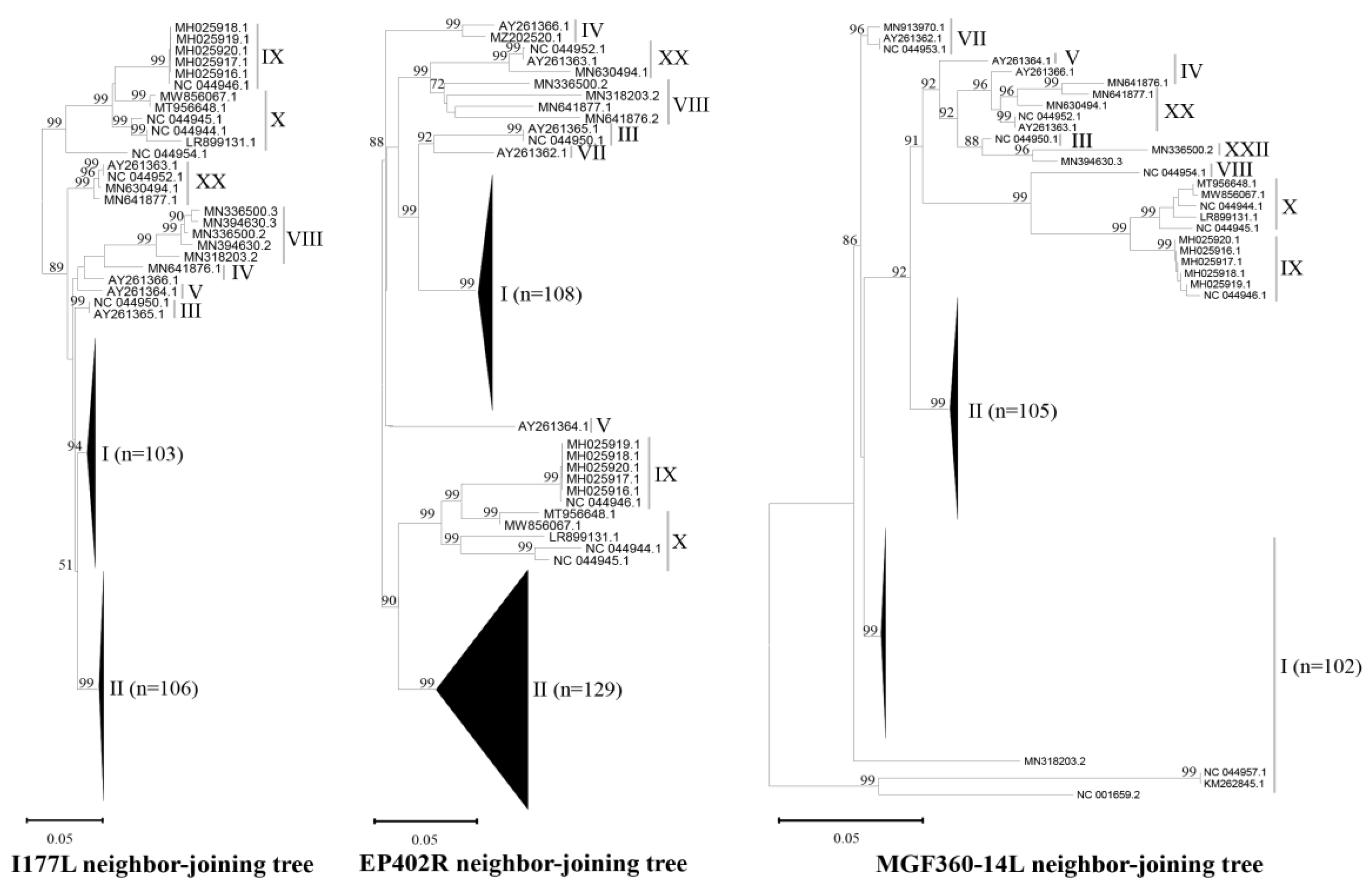
| Target Gene (product size) | Primer/Probe | Sequence (5'-3') | Genotypes/sequence numbers (n=) and coverages (%) | Total | ||||||||||
| I | II | III | IV | V | VII | VIII | IX | X | XX | XXII | ||||
| I177L (147 bp) | n=103 | n=106 | n=1 | n=2 | n=1 | n=3 | n=2 | n=6 | n=5 | n=5 | n=1 | n=235 | ||
| Forward1 | TGTACTGGAAAAAACTTTATCGG | 100% | 100% | 1% | ||||||||||
| Forward2 | TGAACTGGAAAAAACTTTAACGG | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 94% | |||||
| Forward3 | TGAACTGATATAAATCCTTAACGG | 100% | 100% | 5% | ||||||||||
| Reverse1 | AATGTGGAAAGATAATGAACAGG | 100% | 100% | 1% | ||||||||||
| Reverse2 | AATGTGGAAAGTTAATGATCAGG | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 96% | ||||
| Reverse3 | AATGTGGAAAATTGATGATAAGG | 100% | 3% | |||||||||||
| Probe | GAAGGGGGATCCGTATAAAATCCTAGCTTG | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | |
| EP402R (145 bp) | n=108 | n=129 | n=1 | n=2 | n=1 | n=3 | n=2 | n=6 | n=5 | n=5 | n=1 | n=263 | ||
| Forward1 | ACATGTTGAAGAAATAGAAAGTC | 100% | 100% | 100% | 50% | 100% | 100% | 100% | 100% | 100% | 60% | 98% | ||
| Forward2 | CATGTTGCAGAAATACAAAGTCC | 50% | 40% | 100% | 2% | |||||||||
| Reverse1 | AGGTGTATTATATTGATAACGACT | 60% | 60% | 2% | ||||||||||
| Reverse2 | AGGTGTATTATACTGATAACGACT | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 40% | 40% | 100% | 98% | |
| Probe | TCTCCCAGAGAACCATTACTTCCTAAGCC | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | |
|
MGF 360-14L (101 bp) |
n=102 | n=105 | n=1 | n=2 | n=1 | n=3 | n=5 | n=6 | n=5 | n=5 | n=1 | n=236 | ||
| Forward1 | AGAAGACGGGGTTCGGATACAG | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 93% | ||||
| Forward2 | AGAAGACGAGATTCGGAGACAG | 100% | 100% | 100% | 7% | |||||||||
| Reverse1 | GCAAATCCTGAATATGGGCTTATACG | 100% | 100% | 100% | 80% | 40% | 100% | 100% | ||||||
| Reverse2 | GCAAATCCTGAATATGGACTTATACG | 100% | 100% | 100% | 20% | 100% | 100% | 60% | ||||||
| Probe1 | CCTCCCAGTTCCGCACACAGCCG | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | ||||
| Probe2 | CCTCCTAGTTCCGTGCACAGCCG | 100% | 100% | 100% | ||||||||||
| Viruses | Probes with different dyes | |||
| VIC labeled EP402R probe |
FAM labeled I177L probe |
Texas Red labeled MGF360-14L probe | Cy5 labeled ACTB probe | |
| Wild-type ASFVs | + | + | + | + |
| ASFVΔEP402R | - | + | + | + |
| ASFVΔI177L | + | - | + | + |
| ASFVΔMGF360-14L | + | + | - | + |
| Target | Quadplex RT-PCR with different reaction buffer | |||||
| B1 | B2 | B3 | B4 | B5 | ||
| I177L | R2 | 0.99 | 0.96 | 0.96 | 0.94 | 0.97 |
| E | 105% | 109% | 91% | 100% | 95% | |
| LOD | 1 | 100 | 100 | 10 | 100 | |
| EP402R | R2 | 0.99 | 0.96 | 0.94 | 0.98 | 0.98 |
| E | 105% | 102% | 89% | 90% | 81% | |
| LOD | 10 | 10 | 100 | 100 | 100 | |
| MGF360-14L | R2 | 0.98 | 0.98 | 0.98 | 0.99 | 0.96 |
| E | 104% | 108% | 80% | 101% | 92% | |
| LOD | 1 | 10 | 10 | 10 | 10 | |
| Viruses | Quantity (TCID50) spiked | Quadplex RT-PCR |
Standard singular RT-PCR |
|||
| Sensitivity | Specificity | Sensitivity | Specificity | |||
| ASFV | OURT88/1 (GI) | Ten-fold serial dilution from 105 |
LOD=0.1 | + | LOD=0.1 | + |
| VNUA-ASFV-05L1 (GII) | LOD=0.1 | + | LOD=0.1 | + | ||
| Georgia strain (GII) | LOD=0.1 | + | LOD=0.1 | + | ||
| CSFV | Alfort strain | 105 | UD | - | UD | - |
| C-strain | UD | - | UD | - | ||
| PRRSV | VR-2332 | 105 | UD | - | UD | - |
| NADC-20 | UD | - | UD | - | ||
| JXA1-R | UD | - | UD | - | ||
| 1-4-4L1C | UD | - | UD | - | ||
| PCV | PCV2b | 105 | UD | - | UD | - |
| PRV | Bartha-K61 | 105 | UD | - | UD | - |
| Pig serum samples | Number of samples | Standard singular ASFV RT-PCR | Quadplex RT-PCR | Positive | Negative | Specificity | |||
| I177L | EP402R | MGF | ACTB | ||||||
| ASFV infected | 50 | + (Ct 19-40) |
+ (Ct 19-40) |
+ (Ct 19-40) |
+ (Ct 19-40) |
+ (Ct 28) |
50/50 | 0/50 | 100% |
| PBS injected | 50 | - | - | - | - | + | 0/50 | 50/50 | 100% |
| CSFV-infected | 50 | - | - | - | - | + | 0/50 | 50/50 | 100% |
| PRRSV-infected | 50 | - | - | - | - | + | 0/50 | 50/50 | 100% |
| PRV-infected | 10 | - | - | - | - | + | 0/10 | 10/10 | 100% |
| BVDV-infected | 4 | - | - | - | - | + | 0/4 | 4/4 | 100% |
| Samples | Number of samples | Standard singular ASFV RT-PCR | Quadplex RT-PCR | Positive | Negative | Specificity | |||
| I177L | EP402R | MGF | ACTB | ||||||
| Naturally ASFV-infected pig sera | 54 | + | + | + | + | + | 54/54 | 0/54 | 100% |
| ASFV free pig sera | 100 | - | - | - | - | + | 0/100 | 100/100 | 100% |
| Feral pig sera | 6 | - | - | - | - | + | 0/6 | 6/6 | 100% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





