Submitted:
07 February 2025
Posted:
10 February 2025
You are already at the latest version
Abstract
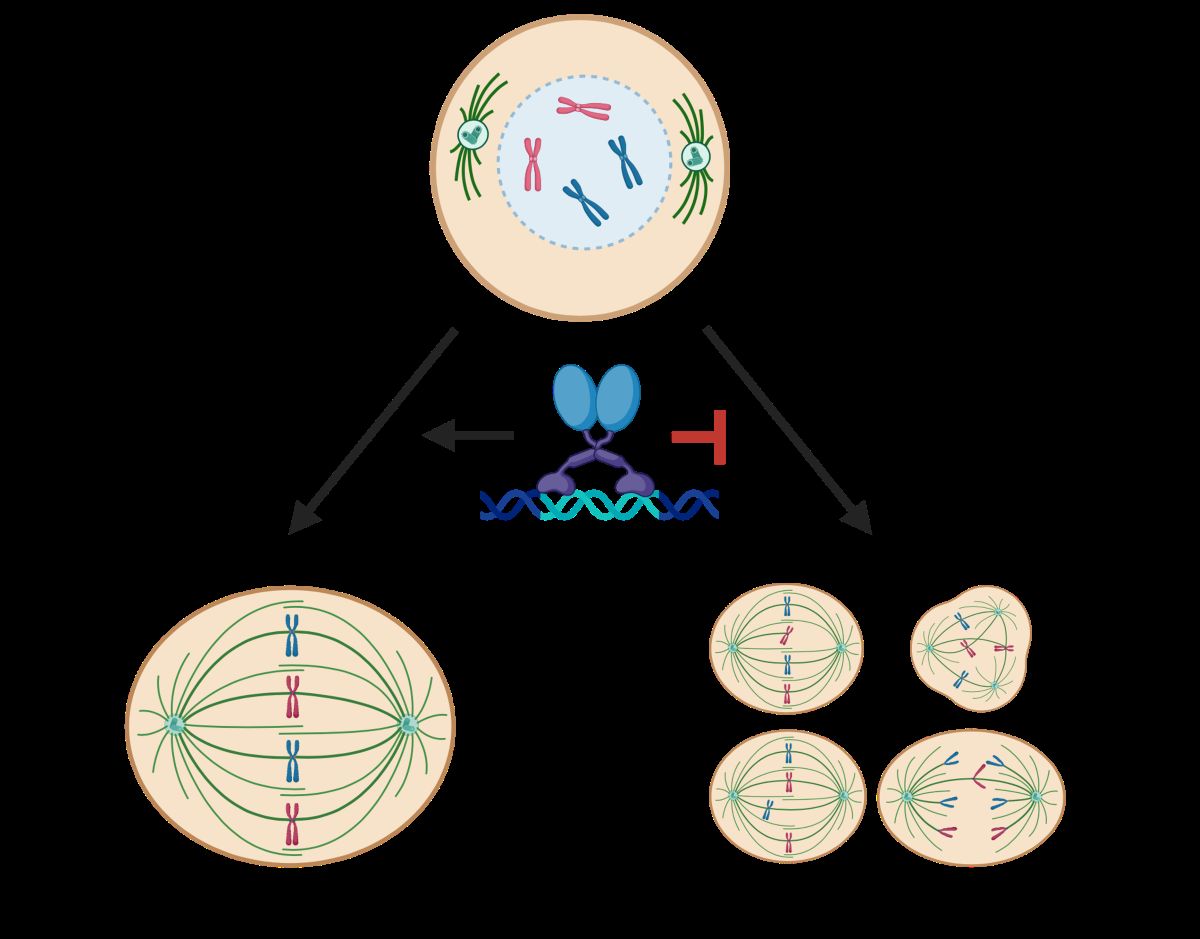
Keywords:
1. Introduction
1.1. Brief Overview of Chromosome Segregation
1.2. Chromosomal Instability (CIN) and Cancer
1.3. Historic Overview of Transcription Factors During Mitosis
1.4. Separating Mitotic Function of Transcription Factors from Their Transcriptional Programs
2. Transcription Factors and Chromosome Condensation
2.1. Overview of Chromosome Condensation
2.2. Chromosome Condensation Is Required for Chromosomal Stability
2.3. Transcription and Condensin Loading
2.4. Transcription Factors and Chromosome Condensation
3. Transcription Factors and Centromeric Transcription
3.1. Overview of Centromere and Kinetochore Assembly and Function
3.2. Centromere Homeostasis Is Required for Chromosomal Stability
3.3. Centromeric Transcription Supports Chromosome Segregation
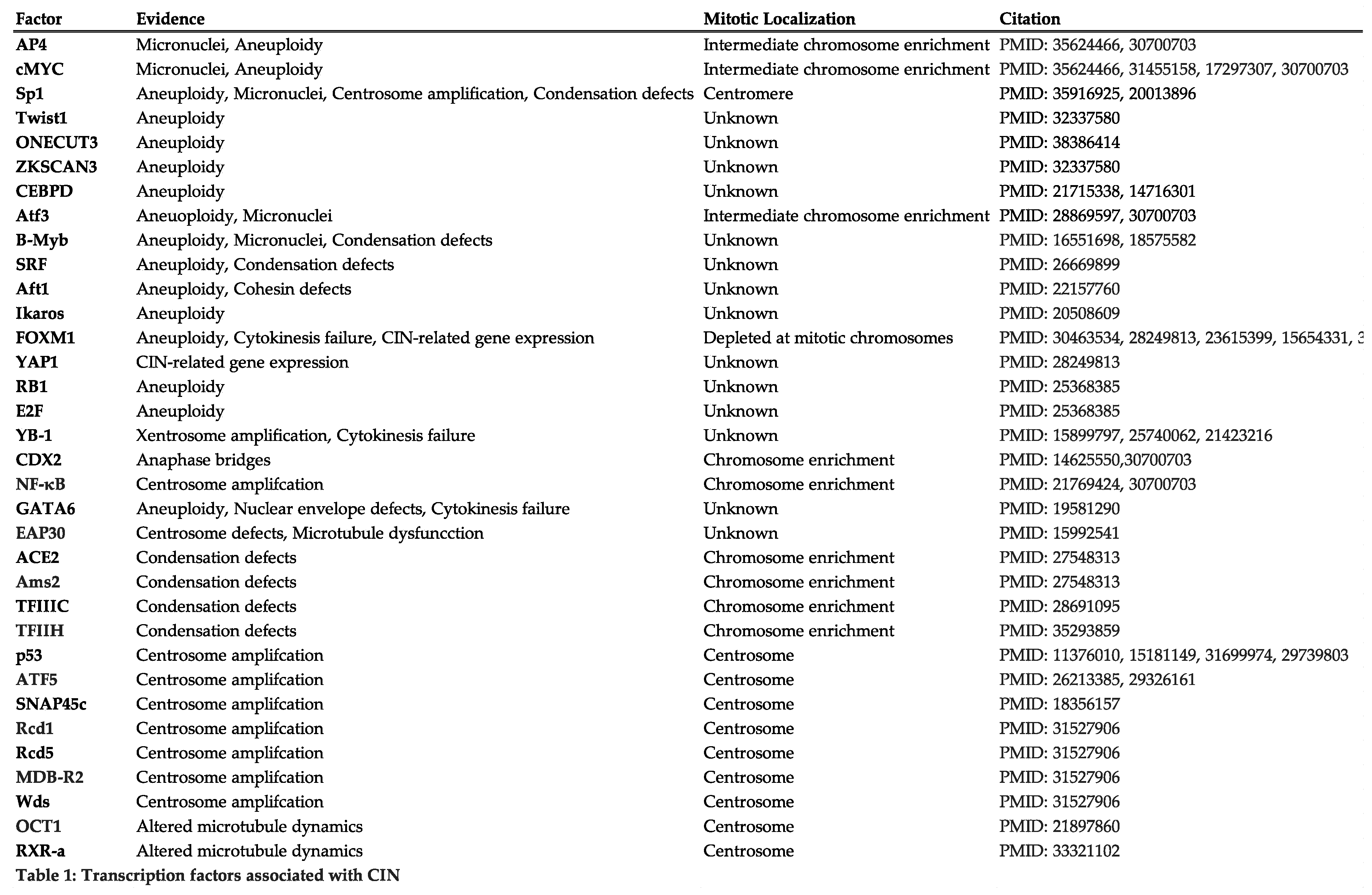
3.4. Regulators of Mitotic Transcription
4. Transcription Factors and Centrosome Biology
4.1. Overview of Centrosome Function
4.2. Centrosome Homeostasis Is Required for Chromosomal Stability
4.3. Transcription Factors Support Centrosome Homeostasis
5. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Milo, R.; Jorgensen, P.; Moran, U.; Weber, G.; Springer, M. BioNumbers--the Database of Key Numbers in Molecular and Cell Biology. Nucleic Acids Res, 2010, 38, D750–3. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, R.A. The Biology of Cancer, Second Edition; Garland Science, 2014.
- Weaver, B.A.; Cleveland, D.W. Does Aneuploidy Cause Cancer? Curr Opin Cell Biol, 2006, 18, 658–667. [Google Scholar] [CrossRef]
- Lee, A.J.X.; Endesfelder, D.; Rowan, A.J.; Walther, A.; Birkbak, N.J.; Futreal, P.A.; Downward, J.; Szallasi, Z.; Tomlinson, I.P.M.; Howell, M.; et al. Chromosomal Instability Confers Intrinsic Multidrug Resistance. Cancer Res, 2011, 71, 1858–1870. [Google Scholar] [CrossRef]
- Thompson, S.L.; Compton, D.A. Examining the Link between Chromosomal Instability and Aneuploidy in Human Cells. Journal of Cell Biology, 2008, 180, 665–672. [Google Scholar] [CrossRef] [PubMed]
- Ramón y Cajal, S.; Sesé, M.; Capdevila, C.; Aasen, T.; De Mattos-Arruda, L.; Diaz-Cano, S.J.; Hernández-Losa, J.; Castellví, J. Clinical Implications of Intratumor Heterogeneity: Challenges and Opportunities. J Mol Med, 2020, 98, 161–177. [Google Scholar] [CrossRef]
- Prescott, D.M.; Bender, M.A. SYNTHESIS OF RNA AND PROTEIN DURING MITOSIS IN MAMMALIAN TISSUE CULTURE CELLS; 1962; Vol. 26.
- Taylor, J.H. NUCLEIC ACID SYNTHESIS IN RELATION TO THE CELL DIVISION CYCLE*. Ann N Y Acad Sci, 1960, 90, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Naumova, N.; Imakaev, M.; Fudenberg, G.; Zhan, Y.; Lajoie, B.R.; Mirny, L.A.; Dekker, J. Organization of the Mitotic Chromosome. Science (1979), 2013, 342, 948–953. [Google Scholar] [CrossRef]
- Martinez-Balbbs, M.A.; Dey, A.; Rabindran, S.K.; Ozato, K.; Wu, C. Displacement of Sequence-Specific Transcription Factors from Mitotic Chromatin; 1995; Vol. 83, pp 29–38.
- Gottesfeld, J.M.; Forbes, D.J. Mitotic Repression of the Transcriptional Machinery. Trends Biochem Sci, 1997, 22, 197–202. [Google Scholar] [CrossRef]
- Kadauke, S.; Udugama, M.I.; Pawlicki, J.M.; Achtman, J.C.; Jain, D.P.; Cheng, Y.; Hardison, R.C.; Blobel, G.A. Tissue-Specific Mitotic Bookmarking by Hematopoietic Transcription Factor GATA1. Cell, 2012, 150, 725–737. [Google Scholar] [CrossRef]
- Caravaca, J.M.; Donahue, G.; Becker, J.S.; He, X.; Vinson, C.; Zaret, K.S. Bookmarking by Specific and Nonspecific Binding of FoxA1 Pioneer Factor to Mitotic Chromosomes. Genes Dev, 2013, 27, 251–260. [Google Scholar] [CrossRef]
- Teves, S.S.; An, L.; Hansen, A.S.; Xie, L.; Darzacq, X.; Tjian, R. A Dynamic Mode of Mitotic Bookmarking by Transcription Factors. Elife, 2016, 5, 1–24. [Google Scholar] [CrossRef]
- Liu, Y.; Pelham-Webb, B.; Di Giammartino, D.C.; Li, J.; Kim, D.; Kita, K.; Saiz, N.; Garg, V.; Doane, A.; Giannakakou, P.; et al. Widespread Mitotic Bookmarking by Histone Marks and Transcription Factors in Pluripotent Stem Cells. Cell Rep, 2017, 19, 1283–1293. [Google Scholar] [CrossRef] [PubMed]
- Pallier, C.; Scaffidi, P.; Chopineau-Proust, S.; Agresti, A.; Nordmann, P.; Bianchi, M.E.; Marechal, V. Association of Chromatin Proteins High Mobility Group Box (HMGB) 1 and HMGB2 with Mitotic Chromosomes. Mol Biol Cell, 2003, 14, 3414–3426. [Google Scholar] [CrossRef]
- Raccaud, M.; Suter, D.M. Transcription Factor Retention on Mitotic Chromosomes: Regulatory Mechanisms and Impact on Cell Fate Decisions. FEBS Lett, 2018, 592, 878–887. [Google Scholar] [CrossRef]
- Raccaud, M.; Friman, E.T.; Alber, A.B.; Agarwal, H.; Deluz, C.; Kuhn, T.; Gebhardt, J.C.M.; Suter, D.M. Mitotic Chromosome Binding Predicts Transcription Factor Properties in Interphase. Nat Commun, 2019, 10, 487. [Google Scholar] [CrossRef] [PubMed]
- Blobel, G.A.; Kadauke, S.; Wang, E.; Lau, A.W.; Zuber, J.; Chou, M.M.; Vakoc, C.R. A Reconfigured Pattern of MLL Occupancy within Mitotic Chromatin Promotes Rapid Transcriptional Reactivation Following Mitotic Exit. Mol Cell. [CrossRef]
- Dey, A.; Nishiyama, A.; Karpova, T.; McNally, J.; Ozato, K. Brd4 Marks Select Genes on Mitotic Chromatin and Directs Postmitotic Transcription. Mol Biol Cell, 2009, 20, 4899–4909. [Google Scholar] [CrossRef] [PubMed]
- Owens, N.D.L.; Gonzalez, I.; Artus, J.; Navarro, P. Mitotic Bookmarking by Transcription Factors and the Preservation of Pluripotency. Stem Cell Epigenet. [CrossRef]
- Kadauke, S.; Blobel, G.A. Mitotic Bookmarking by Transcription Factors. Epigenetics Chromatin, 2013, 6, 6. [Google Scholar] [CrossRef]
- Soares, M.A.F.; Oliveira, R.A.; Castro, D.S. Function and Regulation of Transcription Factors during Mitosis-To-G1 Transition. Open Biol, 2022, 12. [Google Scholar] [CrossRef]
- Budzynśki, M.A.; Wong, A.K.L.; Faghihi, A.; Teves, S.S. A Dynamic Role for Transcription Factors in Restoring Transcription through Mitosis. Biochem Soc Trans, 2024, 52, 821–830. [Google Scholar] [CrossRef]
- Teves, S.S.; An, L.; Bhargava-Shah, A.; Xie, L.; Darzacq, X.; Tjian, R. A Stable Mode of Bookmarking by TBP Recruits RNA Polymerase II to Mitotic Chromosomes. Elife, 2018, 7, 1–22. [Google Scholar] [CrossRef]
- Palozola, K.C.; Donahue, G.; Liu, H.; Grant, G.R.; Becker, J.S.; Cote, A.; Yu, H.; Raj, A.; Zaret, K.S. Mitotic Transcription and Waves of Gene Reactivation during Mitotic Exit. Science (1979), 2017, 358, eaal4671. [Google Scholar] [CrossRef] [PubMed]
- Chan, F.L.; Marshall, O.J.; Saffery, R.; Won Kim, B.; Earle, E.; Choo, K.H.A.; Wong, L.H.; Kim, B.W.; Earle, E.; Choo, K.H.A.; et al. Active Transcription and Essential Role of RNA Polymerase II at the Centromere during Mitosis. PNAS, 2012, 109, 1979–1984. [Google Scholar] [CrossRef]
- Blower, M.D. Centromeric Transcription Regulates Aurora-B Localization and Activation. Cell Rep, 2016, 15, 1624–1633. [Google Scholar] [CrossRef]
- Nishimura, K.; Fukagawa, T.; Takisawa, H.; Kakimoto, T.; Kanemaki, M. An Auxin-Based Degron System for the Rapid Depletion of Proteins in Nonplant Cells. Nat Methods, 2009, 6, 917–922. [Google Scholar] [CrossRef] [PubMed]
- Ottis, P.; Crews, C.M. Proteolysis-Targeting Chimeras: Induced Protein Degradation as a Therapeutic Strategy. ACS Chem Biol, 2017, 12, 892–898. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.M.; Kim, K.B.; Kumagai, A.; Mercurio, F.; Crews, C.M.; Deshaies, R.J. Protacs: Chimeric Molecules That Target Proteins to the Skp1-Cullin-F Box Complex for Ubiquitination and Degradation. Proc Natl Acad Sci U S A, 2001, 98, 8554–8559. [Google Scholar] [CrossRef]
- Clift, D.; So, C.; McEwan, W.A.; James, L.C.; Schuh, M. Acute and Rapid Degradation of Endogenous Proteins by Trim-Away. Nature Protocols 2018 13:10, 2018, 13, 2149–2175. [Google Scholar] [CrossRef]
- Paulson, J.R.; Laemmli, U.K. The Structure of Histone-Depleted Metaphase Chromosomes; 1977; Vol. 12. [CrossRef]
- Marsden, M.P.F.; Laemmli, U.K. Metaphase Chromosome Structure: Evidence for a Radial Loop Model; 1979; Vol. 17. [CrossRef]
- Earnshaw, W.C.; Laemmli, U.K. Architecture of Metaphase Chromosomes and Chromosome Scaffolds. Journal of Cell Biology, 1983, 96, 84–93. [Google Scholar] [CrossRef]
- Ono, T.; Losada, A.; Hirano, M.; Myers, M.P.; Neuwald, A.F.; Hirano, T. Differential Contributions of Condensin I and Condensin II to Mitotic Chromosome Architecture in Vertebrate Cells. Cell, 2003, 115, 109–121. [Google Scholar] [CrossRef]
- Saitoh, N.; Goldberg, I.G.; Wood, E.R.; Earnshaw, W.C. ScII: An Abundant Chromosome Scaffold Protein Is a Member of a Family of Putative ATPases with an Unusual Predicted Tertiary Structure. J Cell Biol, 1994, 127, 303–318. [Google Scholar] [CrossRef]
- Samejima, I.; Matsumoto, T.; Nakaseko, Y.; Beach, D.; Yanagida, M. Identification of Seven New Cut Genes Involved in Schizosaccharomyces Pombe Mitosis. J Cell Sci, 1993, 105, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T.; Mitchison, T.J. A Heterodimeric Coiled-Coil Protein Required for Mitotic Chromosome Condensation In Vitro; 1994; Vol. 79.
- Hirano, T.; Kobayashi, R.; Hirano, M. Condensins, Chromosome Condensation Protein Complexes Containing XCAP- C, XCAP-E and a Xenopus Homolog of the Drosophila Barren Protein. Cell, 1997, 89, 511–521. [Google Scholar] [CrossRef]
- Nasmyth, K. Disseminating the Genome: Joining, Resolving, and Separating Sister Chromatids During Mitosis and Meiosis. Annu Rev Genet, 2001, 35, 673–745. [Google Scholar] [CrossRef] [PubMed]
- Fudenberg, G.; Imakaev, M.; Lu, C.; Goloborodko, A.; Abdennur, N.; Mirny, L.A. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep, 2016, 15, 2038–2049. [Google Scholar] [CrossRef]
- Fudenberg, G.; Abdennur, N.; Imakaev, M.; Goloborodko, A.; Mirny, L.A. Emerging Evidence of Chromosome Folding by Loop Extrusion. Cold Spring Harb Symp Quant Biol. [CrossRef]
- Goloborodko, A.; Imakaev, M.V.; Marko, J.F.; Mirny, L. Compaction and Segregation of Sister Chromatids via Active Loop Extrusion. Elife, 2016, 5, 1–16. [Google Scholar] [CrossRef]
- Kakui, Y.; Rabinowitz, A.; Barry, D.J.; Uhlmann, F. Condensin-Mediated Remodeling of the Mitotic Chromatin Landscape in Fission Yeast. Nat Genet, 2017, 49, 1553–1557. [Google Scholar] [CrossRef] [PubMed]
- Kakui, Y.; Uhlmann, F. SMC Complexes Orchestrate the Mitotic Chromatin Interaction Landscape. Curr Genet, 2018, 64, 335–339. [Google Scholar] [CrossRef]
- Gibcus, J.H.; Samejima, K.; Goloborodko, A.; Samejima, I.; Naumova, N.; Nuebler, J.; Kanemaki, M.T.; Xie, L.; Paulson, J.R.; Earnshaw, W.C.; et al. A Pathway for Mitotic Chromosome Formation. Science (1979), 2018, 359, eaao6135. [Google Scholar] [CrossRef]
- Walther, N.; Julius Hossain, M.; Politi, A.Z.; Koch, B.; Kueblbeck, M.; Ødegård-Fougner, Ø.; Lampe, M.; Ellenberg, J. A Quantitative Map of Human Condensins Provides New Insights into Mitotic Chromosome Architecture. J. Cell Biol, 2018, 217, 2309. [Google Scholar] [CrossRef]
- Hirano, T. Condensin-Based Chromosome Organization from Bacteria to Vertebrates. Cell, 2016, 164, 847–857. [Google Scholar] [CrossRef]
- Lung Chan, K.; Hickson, I.D. Cell Cycle On the Origins of Ultra-Fine Anaphase Bridges. 2009. [CrossRef]
- Martin, C.-A.; Murray, J.E.; Carroll, P.; Leitch, A.; Mackenzie, K.J.; Halachev, M.; Fetit, A.E.; Keith, C.; Bicknell, L.S.; Fluteau, A.; et al. Mutations in Genes Encoding Condensin Complex Proteins Cause Microcephaly through Decatenation Failure at Mitosis. Genes Dev, 2016, 30, 2158–2172. [Google Scholar] [CrossRef] [PubMed]
- Woodward, J.; Taylor, G.C.; Soares, D.C.; Boyle, S.; Sie, D.; Read, D.; Chathoth, K.; Vukovic, M.; Tarrats, N.; Jamieson, D.; et al. Condensin II Mutation Causes T-Cell Lymphoma through Tissue-Specific Genome Instability. Genes Dev, 2016, 30, 2173–2186. [Google Scholar] [CrossRef]
- Ham, M.F.; Takakuwa, T.; Rahadiani, N.; Tresnasari, K.; Nakajima, H.; Aozasa, K. Condensin Mutations and Abnormal Chromosomal Structures in Pyothorax-Associated Lymphoma. Cancer Sci, 2007, 98, 1041–1047. [Google Scholar] [CrossRef] [PubMed]
- Baergen, A.K.; Jeusset, L.M.; Lichtensztejn, Z.; McManus, K.J. Diminished Condensin Gene Expression Drives Chromosome Instability That May Contribute to Colorectal Cancer Pathogenesis. Cancers (Basel), 2019, 11, 1066. [Google Scholar] [CrossRef] [PubMed]
- Je, E.M.; Yoo, N.J.; Lee, S.H. Mutational and Expressional Analysis of SMC2 Gene in Gastric and Colorectal Cancers with Microsatellite Instability. APMIS, 2014, 122, 499–504. [Google Scholar] [CrossRef]
- Kar, S.P.; Beesley, J.; Al Olama, A.A.; Michailidou, K.; Tyrer, J.; Kote-Jarai, Z.S.; Lawrenson, K.; Lindstrom, S.; Ramus, S.J.; Thompson, D.J.; et al. Genome-Wide Meta-Analyses of Breast, Ovarian, and Prostate Cancer Association Studies Identify Multiple New Susceptibility Loci Shared by at Least Two Cancer Types. Cancer Discov, 2016, 6, 1052–1067. [Google Scholar] [CrossRef]
- Baergen, A.K.; Jeusset, L.M.; Lichtensztejn, Z.; McManus, K.J. Diminished Condensin Gene Expression Drives Chromosome Instability That May Contribute to Colorectal Cancer Pathogenesis. Cancers (Basel), 2019, 11, 1066. [Google Scholar] [CrossRef]
- Je, E.M.; Yoo, N.J.; Lee, S.H. Mutational and Expressional Analysis of SMC2 Gene in Gastric and Colorectal Cancers with Microsatellite Instability. APMIS, 2014, 122, 499–504. [Google Scholar] [CrossRef]
- Kar, S.P.; Beesley, J.; Olama, A.A. Al; Michailidou, K.; Tyrer, J.; Kote-Jarai, Z.S.; Lawrenson, K.; Lindstrom, S.; Ramus, S.J.; Thompson, D.J.; et al. Genome-Wide Meta-Analyses of Breast, Ovarian, and Prostate Cancer Association Studies Identify Multiple New Susceptibility Loci Shared by at Least Two Cancer Types. Cancer Discov, 2016, 6, 1052–1067. [Google Scholar] [CrossRef]
- Sundin, O.; Varshavsky, A. Arrest of Segregation Leads to Accumulation of Highly Intertwined Catenated Dimers: Dissection of the Final Stages of SV40 DNA Replication; 1981; Vol. 25.
- Baxter, J.; Sen, N.; López Martínez, V.; Monturus De Carandini, M.E.; Schvartzman, J.B.; Diffley, J.F.X.; Aragón, L. Positive Supercoiling of Mitotic DNA Drives Decatenation by Topoisomerase II in Eukaryotes. Science (1979), 2011, 331, 1328–1332. [Google Scholar] [CrossRef]
- Charbin, A.; Uhlmann, F. Condensin Aids Sister Chromatid Decatenation by Topoisomerase II. [CrossRef]
- Savvidou, E.; Cobbe, N.; Steffensen, S.; Cotterill, S.; Heck, M.M.S.; Marshall, O.; Turnbull, L.; Whitchurch, C.B.; Vagnarelli, P.; Samejima, K.; et al. Drosophila CAP-D2 Is Required for Condensin Complex Stability and Resolution of Sister Chromatids. J Cell Sci, 2005, 118 Pt 11, 2529–2543. [Google Scholar] [CrossRef] [PubMed]
- Shintomi, K.; Takahashi, T.S.; Hirano, T. Reconstitution of Mitotic Chromatids with a Minimum Set of Purified Factors. Nat Cell Biol, 2015, 17, 1014–1023. [Google Scholar] [CrossRef]
- Daniloski, Z.; Bisht, K.K.; Mcstay, B.; Smith, S. Resolution of Human Ribosomal DNA Occurs in Anaphase, Dependent on Tankyrase 1, Condensin II, and Topoisomerase IIα. 2019. [CrossRef]
- Nicklas, R.B.; Koch, C.A. CHROMOSOME MICROMANIPULATION III. Spindle Fiber Tension and the Reorientation of Mal-Oriented Chromosomes. J Cell Biol, 1969, 43, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Akiyoshi, B.; Sarangapani, K.K.; Powers, A.F.; Nelson, C.R.; Reichow, S.L.; Arellano-Santoyo, H.; Gonen, T.; Ranish, J.A.; Asbury, C.L.; Biggins, S. Tension Directly Stabilizes Reconstituted Kinetochore-Microtubule Attachments. Nature, 2010, 468, 576–579. [Google Scholar] [CrossRef]
- Stephens, A.D.; Haase, J.; Vicci, L.; Taylor, R.M.; Bloom, K. Cohesin, Condensin, and the Intramolecular Centromere Loop Together Generate the Mitotic Chromatin Spring. Journal of Cell Biology, 2011, 193, 1167–1180. [Google Scholar] [CrossRef]
- Samoshkin, A.; Arnaoutov, A.; Jansen, L.E.T.; Ouspenski, I.; Dye, L.; Karpova, T.; McNally, J.; Dasso, M.; Cleveland, D.W.; Strunnikov, A. Human Condensin Function Is Essential for Centromeric Chromatin Assembly and Proper Sister Kinetochore Orientation. PLoS One, 2009, 4, e6831. [Google Scholar] [CrossRef] [PubMed]
- Yong-Gonzalez, V.; Wang, B.-D.; Butylin, P.; Ouspenski, I.; Strunnikov, A. Condensin Function at Centromere Chromatin Facilitates Proper Kinetochore Tension and Ensures Correct Mitotic Segregation of Sister Chromatids. [CrossRef]
- Ono, T.; Fang, Y.; Spector, D.L.; Hirano, T. Spatial and Temporal Regulation of Condensins I and II in Mitotic Chromosome Assembly in Human Cells □ D. Mol Biol Cell, 2004, 15, 3296–3308. [Google Scholar] [CrossRef]
- Ribeiro, S.A.; Gatlin, J.C.; Dong, Y.; Joglekar, A.; Cameron, L.; Hudson, D.F.; Farr, C.J.; Mcewen, B.F.; Salmon, E.D.; Earnshaw, W.C.; et al. Condensin Regulates the Stiffness of Vertebrate Centromeres. Mol Biol Cell, 2009, 20, 2371–2380. [Google Scholar] [CrossRef] [PubMed]
- Gerlich, D.; Hirota, T.; Koch, B.; Peters, J.-M.; Ellenberg, J. Condensin I Stabilizes Chromosomes Mechanically through a Dynamic Interaction in Live Cells. Current Biology, 2006, 16, 333–344. [Google Scholar] [CrossRef]
- Oliveira, R.A.; Coelho, P.A.; Sunkel, C.E. The Condensin I Subunit Barren/CAP-H Is Essential for the Structural Integrity of Centromeric Heterochromatin during Mitosis. Mol Cell Biol, 2005, 25, 8971–8984. [Google Scholar] [CrossRef] [PubMed]
- Bernad, R.; Sánchez, P.; Rivera, T.; Rodríguez-Corsino, M.; Boyarchuk, E.; Vassias, I.; Ray-Gallet, D.; Arnaoutov, A.; Dasso, M.; Almouzni, G.; et al. Xenopus HJURP and Condensin II Are Required for CENP-A Assembly. Journal of Cell Biology, 2011, 192, 569–582. [Google Scholar] [CrossRef] [PubMed]
- Barnhart-Dailey, M.C.; Trivedi, P.; Stukenberg, P.T.; Foltz, D.R. HJURP Interaction with the Condensin II Complex during G1 Promotes CENP-A Deposition. Mol Biol Cell, 2017, 28, 54–64. [Google Scholar] [CrossRef]
- Sutani, T.; Sakata, T.; Nakato, R.; Masuda, K.; Ishibashi, M.; Yamashita, D.; Suzuki, Y.; Hirano, T.; Bando, M.; Shirahige, K. Condensin Targets and Reduces Unwound DNA Structures Associated with Transcription in Mitotic Chromosome Condensation. Nat Commun, 2015, 6, 7815. [Google Scholar] [CrossRef]
- Nakazawa, N.; Sajiki, K.; Xu, X.; Villar-Briones, A.; Arakawa, O.; Yanagida, M. RNA Pol II Transcript Abundance Controls Condensin Accumulation at Mitotically Up-Regulated and Heat-Shock-Inducible Genes in Fission Yeast. Genes to Cells, 2015, 20, 481–499. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Zhang, T.; Wong, N.C.; Davidson, N.; Maksimovic, J.; Oshlack, A.; Earnshaw, W.C.; Kalitsis, P.; Hudson, D.F. Condensin I Associates with Structural and Gene Regulatory Regions in Vertebrate Chromosomes. Nat Commun, 2013, 4, 2537. [Google Scholar] [CrossRef] [PubMed]
- Dowen, J.M.; Young, R.A. SMC Complexes Link Gene Expression and Genome Architecture. Current Opinion in Genetics and Development. [CrossRef]
- Lebreton, J.; Colin, L.; Chatre, E.; Bernard, P. RNAP II Antagonizes Mitotic Chromatin Folding and Chromosome Segregation by Condensin. Cell Rep, 2024, 43, 113901. [Google Scholar] [CrossRef] [PubMed]
- Brandão, H.B.; Paul, P.; van den Berg, A.A.; Rudner, D.Z.; Wang, X.; Mirny, L.A. RNA Polymerases as Moving Barriers to Condensin Loop Extrusion. Proc Natl Acad Sci U S A, 2019, 116, 20489–20499. [Google Scholar] [CrossRef]
- Flashner, S.; Swift, M.; Sowash, A.; Fahmy, A.N.; Azizkhan-Clifford, J. Transcription Factor Sp1 Regulates Mitotic Chromosome Assembly and Segregation. Chromosoma, 2022, 131, 175–191. [Google Scholar] [CrossRef]
- Iwasaki, O.; Tanizawa, H.; Kim, K.-D.; Yokoyama, Y.; Corcoran, C.J.; Tanaka, A.; Skordalakes, E.; Showe, L.C.; Noma, K. Interaction between TBP and Condensin Drives the Organization and Faithful Segregation of Mitotic Chromosomes. Mol Cell, 2015, 59, 755–767. [Google Scholar] [CrossRef]
- Kim, K.-D.; Tanizawa, H.; Iwasaki, O.; Noma, K. Transcription Factors Mediate Condensin Recruitment and Global Chromosomal Organization in Fission Yeast. Nat Genet, 2016, 48, 1242–1252. [Google Scholar] [CrossRef]
- Yuen, K.C.; Gerton, J.L. Taking Cohesin and Condensin in Context. PLoS Genet, 2018, 14, e1007118. [Google Scholar] [CrossRef] [PubMed]
- Yuen, K.C.; Slaughter, B.D.; Gerton, J.L. Condensin II Is Anchored by TFIIIC and H3K4me3 in the Mammalian Genome and Supports the Expression of Active Dense Gene Clusters. Sci Adv, 2017, 3, e1700191. [Google Scholar] [CrossRef]
- Xing, H.; Vanderford, N.L.; Sarge, K.D. The TBP–PP2A Mitotic Complex Bookmarks Genes by Preventing Condensin Action. Nat Cell Biol, 2008, 10, 1318–1323. [Google Scholar] [CrossRef]
- Göös, H.; Kinnunen, M.; Salokas, K.; Tan, Z.; Liu, X.; Yadav, L.; Zhang, Q.; Wei, G.H.; Varjosalo, M. Human Transcription Factor Protein Interaction Networks. Nature Communications 2022 13:1, 2022, 13, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Haase, J.; Chen, R.; Parker, W.M.; Bonner, M.K.; Jenkins, L.M.; Kelly, A.E. The TFIIH Complex Is Required to Establish and Maintain Mitotic Chromosome Structure. Elife, 2022, 11. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, O.; Tanizawa, H.; Kim, K.D.; Yokoyama, Y.; Corcoran, C.J.; Tanaka, A.; Skordalakes, E.; Showe, L.C.; Noma, K.I. Interaction between TBP and Condensin Drives the Organization and Faithful Segregation of Mitotic Chromosomes. Mol Cell, 2015, 59, 755–767. [Google Scholar] [CrossRef]
- Kim, K.-D.; Tanizawa, H.; Iwasaki, O.; Noma, K. Transcription Factors Mediate Condensin Recruitment and Global Chromosomal Organization in Fission Yeast. Nat Genet, 2016, 48, 1242–1252. [Google Scholar] [CrossRef]
- Xing, H.; Vanderford, N.L.; Sarge, K.D. The TBP–PP2A Mitotic Complex Bookmarks Genes by Preventing Condensin Action. Nat Cell Biol, 2008, 10, 1318–1323. [Google Scholar] [CrossRef]
- Fachinetti, D.; Diego Folco, H.; Nechemia-Arbely, Y.; Valente, L.P.; Nguyen, K.; Wong, A.J.; Zhu, Q.; Holland, A.J.; Desai, A.; Jansen, L.E.T.; et al. A Two-Step Mechanism for Epigenetic Specification of Centromere Identity and Function. Nat Cell Biol, 2013, 15, 1056–1066. [Google Scholar] [CrossRef]
- Regnier, V.; Vagnarelli, P.; Fukagawa, T.; Zerjal, T.; Burns, E.; Trouche, D.; Earnshaw, W.; Brown, W. CENP-A Is Required for Accurate Chromosome Segregation and Sustained Kinetochore Association of BubR1. Mol Cell Biol, 2005, 25, 3967–3981. [Google Scholar] [CrossRef]
- Liu, S.-T.; Rattner, J.B.; Jablonski, S.A.; Yen, T.J. Mapping the Assembly Pathways That Specify Formation of the Trilaminar Kinetochore Plates in Human Cells. Journal of Cell Biology, 2006, 175, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Black, B.E.; Jansen, L.E.T.; Maddox, P.S.; Foltz, D.R.; Desai, A.B.; Shah, J.V.; Cleveland, D.W. Centromere Identity Maintained by Nucleosomes Assembled with Histone H3 Containing the CENP-A Targeting Domain. Mol Cell, 2007, 25, 309–322. [Google Scholar] [CrossRef]
- Hori, T.; Shang, W.-H.; Takeuchi, K.; Fukagawa, T. The CCAN Recruits CENP-A to the Centromere and Forms the Structural Core for Kinetochore Assembly. J Cell Biol, 2013, 200, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, S.A.; Vagnarelli, P.; Dong, Y.; Hori, T.; McEwen, B.F.; Fukagawa, T.; Flors, C.; Earnshaw, W.C. A Super-Resolution Map of the Vertebrate Kinetochore. Proc Natl Acad Sci U S A, 2010, 107, 10484–10489. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Badger, B.L.; Wan, X.; Deluca, J.G.; Salmon, E.D. The Architecture of CCAN Proteins Creates a Structural Integrity to Resist Spindle Forces and Achieve Proper Intrakinetochore Stretch. [CrossRef]
- Nishino, T.; Rago, F.; Hori, T.; Tomii, K.; Cheeseman, I.M.; Fukagawa, T. CENP-T Provides a Structural Platform for Outer Kinetochore Assembly. EMBO J, 2013, 32, 424–436. [Google Scholar] [CrossRef]
- Cheeseman, I.M.; Niessen, S.; Anderson, S.; Hyndman, F.; Yates, J.R.; Oegema, K.; Desai, A. A Conserved Protein Network Controls Assembly of the Outer Kinetochore and Its Ability to Sustain Tension. Genes Dev, 2004, 18, 2255–2268. [Google Scholar] [CrossRef]
- Cheeseman, I.M.; Chappie, J.S.; Wilson-Kubalek, E.M.; Desai, A. The Conserved KMN Network Constitutes the Core Microtubule-Binding Site of the Kinetochore. [CrossRef]
- Gascoigne, K.E.; Cheeseman, I.M. CDK-Dependent Phosphorylation and Nuclear Exclusion Coordinately Control Kinetochore Assembly State. J Cell Biol, 2013, 201, 23–32. [Google Scholar] [CrossRef]
- Cheeseman, I.M. The Kinetochore. Cold Spring Harb Perspect Biol, 2014, 6. [Google Scholar] [CrossRef]
- Liu, D.; Vader, G.; Vromans, M.J.M.; Lampson, M.A.; Lens, S.M.A. Sensing Chromosome Bi-Orientation by Spatial Separation of Aurora B Kinase from Kinetochore Substrates. Science (1979), 2009, 323, 1350–1353. [Google Scholar] [CrossRef]
- Welburn, J.P.I.; Vleugel, M.; Liu, D.; Yates, J.R.; Lampson, M.A.; Fukagawa, T.; Cheeseman, I.M. Aurora B Phosphorylates Spatially Distinct Targets to Differentially Regulate the Kinetochore-Microtubule Interface. Mol Cell, 2010, 38, 383–392. [Google Scholar] [CrossRef]
- Deluca, J.G.; Gall, W.E.; Ciferri, C.; Cimini, D.; Musacchio, A.; Salmon, E.D. Kinetochore Microtubule Dynamics and Attachment Stability Are Regulated by Hec1. [CrossRef]
- Black, B.E.; Cleveland, D.W. Epigenetic Centromere Propagation and the Nature of CENP-a Nucleosomes. Cell, 2011, 144, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Palmer, D.; O’Day, K.; Wener, M.; Andrews, B.; Margolis, R. A 17-KD Centromere Protein (CENP-A) Copurifies with Nucleosome Core Particles and with Histones. J Cell Biol, 1987, 104, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Kixmoeller, K.; Allu, P.K.; Black, B.E. The Centromere Comes into Focus: From CENP-A Nucleosomes to Kinetochore Connections with the Spindle. Open Biol, 2020, 10, 200051. [Google Scholar] [CrossRef] [PubMed]
- Hooser, A.A. Van; Ouspenski, I.I.; Gregson, H.C.; Starr, D.A.; Yen, T.J.; Goldberg, M.L.; Yokomori, K.; Earnshaw, W.C.; Sullivan, K.F.; Brinkley, B.R. Specification of Kinetochore-Forming Chromatin by the Histone H3 Variant CENP-A. J Cell Sci, 2001, 114, 3529–3542. [Google Scholar] [CrossRef]
- Manuelidis, L. Chromosomal Localization of Complex and Simple Repeated Human DNAs; 1978; Vol. 66.
- McKinley, K.L.; Cheeseman, I.M. The Molecular Basis for Centromere Identity and Function. Nature Reviews Molecular Cell Biology. [CrossRef]
- HFM Peters, A.; Kubicek, S.; Mechtler, K.; O, R.J.; AHA Derijck, A.; Perez-Burgos, L.; Kohlmaier, A.; Opravil, S.; Tachibana, M.; Shinkai, Y.; et al. Partitioning and Plasticity of Repressive Histone Methylation States in Mammalian Chromatin; 2003; Vol. 12.
- Pluta, A.F.; Mackay, A.M.; Ainsztein, A.M.; Goldberg, I.G.; Earnshaw, W.C. The Centromere: Hub of Chromosomal Activities. Science (1979), 1995, 270, 1591–1594. [Google Scholar] [CrossRef]
- Dumont, M.; Gamba, R.; Gestraud, P.; Klaasen, S.; Worrall, J.T.; De Vries, S.G.; Boudreau, V.; Salinas-Luypaert, C.; Maddox, P.S.; Lens, S.M.; et al. Human Chromosome-specific Aneuploidy Is Influenced by DNA -dependent Centromeric Features. EMBO J, 2020, 39. [Google Scholar] [CrossRef]
- Howman, E.V.; Fowler, K.J.; Newson, A.J.; Redward, S.; MacDonald, A.C.; Kalitsis, P.; Choo, K.H. Early Disruption of Centromeric Chromatin Organization in Centromere Protein A (Cenpa) Null Mice. Proc Natl Acad Sci U S A, 2000, 97, 1148–1153. [Google Scholar] [CrossRef]
- Stoler, S.; Keith, K.C.; Curnick, K.E.; Fitzgerald-Hayes, M. A Mutation in CSE4, an Essential Gene Encoding a Novel Chromatin-Associated Protein in Yeast, Causes Chromosome Nondisjunction and Cell Cycle Arrest at Mitosis. Genes Dev, 1995, 9, 573–586. [Google Scholar] [CrossRef]
- Kwon, M.-S.; Hori, T.; Okada, M.; Fukagawa, T. CENP-C Is Involved in Chromosome Segregation, Mitotic Checkpoint Function, and Kinetochore Assembly. https://doi.org/10.1091/mbc.e07-01-0045. [CrossRef]
- Shi, L.; Qalieh, A.; Lam, M.M.; Keil, J.M.; Kwan, K.Y. Robust Elimination of Genome-Damaged Cells Safeguards against Brain Somatic Aneuploidy Following Knl1 Deletion. Nat Commun, 2019, 10. [Google Scholar] [CrossRef]
- Genin, A.; Desir, J.; Lambert, N.; Biervliet, M.; Van der Aa, N.; Pierquin, G.; Killian, A.; Tosi, M.; Urbina, M.; Lefort, A.; et al. Kinetochore KMN Network Gene CASC5 Mutated in Primary Microcephaly. Hum Mol Genet, 2012, 21, 5306–5317. [Google Scholar] [CrossRef]
- Chan, F.L.; Marshall, O.J.; Saffery, R.; Kim, B.W.; Earle, E.; Choo, K.H.A.; Wong, L.H.; Kim, B.W.; Earle, E.; Choo, K.H.A.; et al. Active Transcription and Essential Role of RNA Polymerase II at the Centromere during Mitosis. PNAS, 2012, 109, 1979–1984. [Google Scholar] [CrossRef] [PubMed]
- Ohkuni, K.; Kitagawa, K. Role of Transcription at Centromeres in Budding Yeast. Transcription, 2012, 3, 193. [Google Scholar] [CrossRef] [PubMed]
- Blower, M.D. Centromeric Transcription Regulates Aurora-B Localization and Activation. Cell Rep, 2016, 15, 1624–1633. [Google Scholar] [CrossRef]
- Rošić, S.; Köhler, F.; Erhardt, S. Repetitive Centromeric Satellite RNA Is Essential for Kinetochore Formation and Cell Division. Journal of Cell Biology, 2014, 207, 335–349. [Google Scholar] [CrossRef]
- Molina, O.; Vargiu, G.; Abad, M.A.; Zhiteneva, A.; Jeyaprakash, A.A.; Masumoto, H.; Kouprina, N.; Larionov, V.; Earnshaw, W.C.; Jeyaprakash, A.A.; et al. Epigenetic Engineering Reveals a Balance between Histone Modifications and Transcription in Kinetochore Maintenance. Nat Commun, 2016, 7, 13334. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Guo, Q.; Choi, M.; Liang, Z.; Yuen, K.W.Y. Centromeric and Pericentric Transcription and Transcripts: Their Intricate Relationships, Regulation, and Functions. Chromosoma, 2023, 132, 211–230. [Google Scholar] [CrossRef]
- Liu, H.; Qu, Q.; Warrington, R.; Rice, A.; Cheng, N.; Yu, H.; Bodor, D.L.; Mata, J.F.; Sergeev, M.; David, A.F.; et al. Mitotic Transcription Installs Sgo1 at Centromeres to Coordinate Chromosome Segregation. Mol Cell, 2015, 59, 426–436. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, Q.; Teng, Z.; Liu, H. Centromeric Transcription Maintains Centromeric Cohesion in Human Cells. 2021. [CrossRef]
- Bobkov, G.O.M.M.; Gilbert, N.; Heun, P. Centromere Transcription Allows CENP-A to Transit from Chromatin Association to Stable Incorporation. J. Cell Biol, 2018, 217, 1957. [Google Scholar] [CrossRef]
- Quénet, D.; Dalal, Y. A Long Non-Coding RNA Is Required for Targeting Centromeric Protein A to the Human Centromere. Elife, 2014, 3, e03254. [Google Scholar] [CrossRef]
- McNulty, S.M.; Sullivan, L.L.; Sullivan, B.A. Human Centromeres Produce Chromosome-Specific and Array-Specific Alpha Satellite Transcripts That Are Complexed with CENP-A and CENP-C. Dev Cell, 2017, 42, 226–240.e6. [Google Scholar] [CrossRef]
- Sullivan, B.A.; Karpen, G.H. Centromeric Chromatin Exhibits a Histone Modification Pattern That Is Distinct from Both Euchromatin and Heterochromatin. Nat Struct Mol Biol, 2004, 11, 1076. [Google Scholar] [CrossRef]
- Sullivan, B.A.; Karpen, G.H. Centromeric Chromatin Exhibits a Histone Modification Pattern That Is Distinct from Both Euchromatin and Heterochromatin. Nature Structural & Molecular Biology 2004 11:11, 2004, 11, 1076–1083. [Google Scholar] [CrossRef]
- Gopalakrishnan, S.; Sullivan, B.A.; Trazzi, S.; Della Valle, G.; Robertson, K.D. DNMT3B Interacts with Constitutive Centromere Protein CENP-C to Modulate DNA Methylation and the Histone Code at Centromeric Regions. Hum Mol Genet, 2009, 18, 3178. [Google Scholar] [CrossRef]
- Zhu, J.; Cheng, K.C.L.; Yuen, K.W.Y. Histone H3K9 and H4 Acetylations and Transcription Facilitate the Initial CENP-AHCP-3 Deposition and de Novo Centromere Establishment in Caenorhabditis Elegans Artificial Chromosomes. Epigenetics Chromatin, 2018, 11, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Shang, W.H.; Hori, T.; Westhorpe, F.G.; Godek, K.M.; Toyoda, A.; Misu, S.; Monma, N.; Ikeo, K.; Carroll, C.W.; Takami, Y.; et al. Acetylation of Histone H4 Lysine 5 and 12 Is Required for CENP-A Deposition into Centromeres. Nat Commun, 2016, 7, 13465. [Google Scholar] [CrossRef]
- Bergmann, J.H.; Rodríguez, M.G.; Martins, N.M.C.; Kimura, H.; Kelly, D.A.; Masumoto, H.; Larionov, V.; Jansen, L.E.T.; Earnshaw, W.C. Epigenetic Engineering Shows H3K4me2 Is Required for HJURP Targeting and CENP-A Assembly on a Synthetic Human Kinetochore. EMBO J, 2010, 30, 328. [Google Scholar] [CrossRef]
- Mravinac, B.; Sullivan, L.L.; Reeves, J.W.; Yan, C.M.; Kopf, K.S.; Farr, C.J.; Schueler, M.G.; Sullivan, B.A. Histone Modifications within the Human X Centromere Region. PLoS One, 2009, 4, e6602. [Google Scholar] [CrossRef] [PubMed]
- Hori, T.; Shang, W.H.; Toyoda, A.; Misu, S.; Monma, N.; Ikeo, K.; Molina, O.; Vargiu, G.; Fujiyama, A.; Kimura, H.; et al. Histone H4 Lys 20 Monomethylation of the CENP-A Nucleosome Is Essential for Kinetochore Assembly. Dev Cell, 2014, 29, 740. [Google Scholar] [CrossRef]
- Somma, M.P.; Andreyeva, E.N.; Pavlova, G.A.; Pellacani, C.; Bucciarelli, E.; Popova, J.V.; Bonaccorsi, S.; Pindyurin, A.V.; Gatti, M. Moonlighting in Mitosis: Analysis of the Mitotic Functions of Transcription and Splicing Factors. Cells 2020, Vol. 9, Page 1554, 2020, 9, 1554. [Google Scholar] [CrossRef]
- Nam, H.J.; Naylor, R.M.; van Deursen, J.M. Centrosome Dynamics as a Source of Chromosomal Instability. Trends Cell Biol, 2014, 25, 65. [Google Scholar] [CrossRef]
- Ryniawec, J.M.; Rogers, G.C. Centrosome Instability: When Good Centrosomes Go Bad. Cellular and Molecular Life Sciences 2021 78:21, 2021, 78, 6775–6795. [Google Scholar] [CrossRef]
- Denu, R.A.; Zasadil, L.M.; Kanugh, C.; Laffin, J.; Weaver, B.A.; Burkard, M.E. Centrosome Amplification Induces High Grade Features and Is Prognostic of Worse Outcomes in Breast Cancer. BMC Cancer, 2016, 16, 1–13. [Google Scholar] [CrossRef]
- Singh, C.K.; Denu, R.A.; Nihal, M.; Shabbir, M.; Garvey, D.R.; Huang, W.; Iczkowski, K.A.; Ahmad, N. PLK4 Is Upregulated in Prostate Cancer and Its Inhibition Reduces Centrosome Amplification and Causes Senescence. Prostate, 2022, 82, 957–969. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Ahn, J.S.; Han, H.J.; Kim, H.M.; Hwang, J.; Lee, K.H.; Cha-Molstad, H.; Ryoo, I.J.; Jang, J.H.; Ko, S.K.; et al. Cep131 Overexpression Promotes Centrosome Amplification and Colon Cancer Progression by Regulating Plk4 Stability. Cell Death & Disease 2019 10:8, 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Shinmura, K.; Kurabe, N.; Goto, M.; Yamada, H.; Natsume, H.; Konno, H.; Sugimura, H. PLK4 Overexpression and Its Effect on Centrosome Regulation and Chromosome Stability in Human Gastric Cancer. Mol Biol Rep, 2014, 41, 6635–6644. [Google Scholar] [CrossRef]
- Koutsami, M.K.; Tsantoulis, P.K.; Kouloukoussa, M.; Apostolopoulou, K.; Pateras, I.S.; Spartinou, Z.; Drougou, A.; Evangelou, K.; Kittas, C.; Bartkova, J.; et al. Centrosome Abnormalities Are Frequently Observed in Non-Small-Cell Lung Cancer and Are Associated with Aneuploidy and Cyclin E Overexpression. J Pathol, 2006, 209, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Centrosome Abnormalities in Pancreatic Ductal Carcinoma | Clinical Cancer Research | American Association for Cancer Research https://aacrjournals.org/clincancerres/article/5/5/963/287511/Centrosome-Abnormalities-in-Pancreatic-Ductal (accessed Dec 19, 2024).
- Skyldberg, B.; Fujioka, K.; Hellström, A.C.; Sylvén, L.; Moberger, B.; Auer, G. Human Papillomavirus Infection, Centrosome Aberration, and Genetic Stability in Cervical Lesions. Modern Pathology, 2001, 14, 279–284. [Google Scholar] [CrossRef] [PubMed]
- D’Assoro, A.B.; Lingle, W.L.; Salisbury, J.L. Centrosome Amplification and the Development of Cancer. Oncogene 2002 21:40, 2002, 21, 6146–6153. [Google Scholar] [CrossRef]
- Levine, M.S.; Bakker, B.; Boeckx, B.; Moyett, J.; Lu, J.; Vitre, B.; Spierings, D.C.; Lansdorp, P.M.; Cleveland, D.W.; Lambrechts, D.; et al. Centrosome Amplification Is Sufficient to Promote Spontaneous Tumorigenesis in Mammals. Dev Cell, 2017, 40, 313–322.e5. [Google Scholar] [CrossRef]
- Ciciarello, M.; Mangiacasale, R.; Casenghi, M.; Limongi, M.Z.; D’Angelo, M.; Soddu, S.; Lavia, P.; Cundari, E. P53 Displacement from Centrosomes and P53-Mediated G1 Arrest Following Transient Inhibition of the Mitotic Spindle. Journal of Biological Chemistry, 2001, 276, 19205–19213. [Google Scholar] [CrossRef]
- Tritarelli, A.; Oricchio, E.; Ciciarello, M.; Mangiacasale, R.; Palena, A.; Lavia, P.; Soddu, S.; Cundari, E. P53 Localization at Centrosomes during Mitosis and Postmitotic Checkpoint Are ATM-Dependent and Require Serine 15 Phosphorylation. Mol Biol Cell, 2004, 15, 3751–3757. [Google Scholar] [CrossRef] [PubMed]
- Contadini, C.; Monteonofrio, L.; Virdia, I.; Prodosmo, A.; Valente, D.; Chessa, L.; Musio, A.; Fava, L.L.; Rinaldo, C.; Di Rocco, G.; et al. P53 Mitotic Centrosome Localization Preserves Centrosome Integrity and Works as Sensor for the Mitotic Surveillance Pathway. Cell Death & Disease 2019 10:11, 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Lopes, C.A.M.; Mesquita, M.; Cunha, A.I.; Cardoso, J.; Carapeta, S.; Laranjeira, C.; Pinto, A.E.; Pereira-Leal, J.B.; Dias-Pereira, A.; Bettencourt-Dias, M.; et al. Centrosome Amplification Arises before Neoplasia and Increases upon P53 Loss in Tumorigenesis. Journal of Cell Biology, 2018, 217, 2353–2363. [Google Scholar] [CrossRef]
- Madarampalli, B.; Yuan, Y.; Liu, D.; Lengel, K.; Xu, Y.; Li, G.; Yang, J.; Liu, X.; Lu, Z.; Liu, D.X. ATF5 Connects the Pericentriolar Materials to the Proximal End of the Mother Centriole. Cell, 2015, 162, 580–592. [Google Scholar] [CrossRef]
- Yuan, Y.; Gaither, K.; Kim, E.; Liu, E.; Hu, M.; Lengel, K.; Qian, D.; Xu, Y.; Wang, B.; Knipprath, H.; et al. SUMO2/3 Modification of Activating Transcription Factor 5 (ATF5) Controls Its Dynamic Translocation at the Centrosome. Journal of Biological Chemistry, 2018, 293, 2839–2848. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, M.; Hernandez, N. Mitotic Functions for SNAP45, a Subunit of the Small Nuclear RNA-Activating Protein Complex SNAPc. Journal of Biological Chemistry, 2008, 283, 14845–14856. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, G.A.; Popova, J.V.; Andreyeva, E.N.; Yarinich, L.A.; Lebedev, M.O.; Razuvaeva, A.V.; Dubatolova, T.D.; Oshchepkova, A.L.; Pellacani, C.; Somma, M.P.; et al. RNAi-Mediated Depletion of the NSL Complex Subunits Leads to Abnormal Chromosome Segregation and Defective Centrosome Duplication in Drosophila Mitosis. PLoS Genet, 2019, 15, e1008371. [Google Scholar] [CrossRef]
- Kawaguchi, A.; Asaka, M.N.; Matsumoto, K.; Nagata, K. Centrosome Maturation Requires YB-1 to Regulate Dynamic Instability of Microtubules for Nucleus Reassembly. Scientific Reports 2015 5:1, 2015, 5, 1–8. [Google Scholar] [CrossRef]
- Davies, A.H.; Barrett, I.; Pambid, M.R.; Hu, K.; Stratford, A.L.; Freeman, S.; Berquin, I.M.; Pelech, S.; Hieter, P.; Maxwell, C.; et al. YB-1 Evokes Susceptibility to Cancer through Cytokinesis Failure, Mitotic Dysfunction and HER2 Amplification. Oncogene 2011 30:34, 2011, 30, 3649–3660. [Google Scholar] [CrossRef]
- Bergmann, S.; Royer-Pokora, B.; Fietze, E.; Jürchott, K.; Hildebrandt, B.; Trost, D.; Leenders, F.; Claude, J.C.; Theuring, F.; Bargou, R.; et al. YB-1 Provokes Breast Cancer through the Induction of Chromosomal Instability That Emerges from Mitotic Failure and Centrosome Amplification. Cancer Res, 2005, 65, 4078–4087. [Google Scholar] [CrossRef]
- Kang, J.; Goodman, B.; Zheng, Y.; Tantin, D. Dynamic Regulation of Oct1 during Mitosis by Phosphorylation and Ubiquitination. PLoS One, 2011, 6, e23872. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Zhou, Y.; Tu, X.; Ye, X.; Xu, L.; Xiao, Z.; Wang, Q.; Wang, X.; Du, M.; Chen, Z.; et al. Centrosomal Localization of RXRα Promotes PLK1 Activation and Mitotic Progression and Constitutes a Tumor Vulnerability. Dev Cell, 2020, 55, 707–722.e9. [Google Scholar] [CrossRef]
- Astrinidis, A.; Kim, J.; Kelly, C.M.; Olofsson, B.A.; Torabi, B.; Sorokina, E.M.; Azizkhan-Clifford, J. The Transcription Factor SP1 Regulates Centriole Function and Chromosomal Stability through a Functional Interaction with the Mammalian Target of Rapamycin/Raptor Complex. Genes Chromosomes Cancer, 2010, 49, 282–297. [Google Scholar] [CrossRef]
- Camargo Ortega, G.; Falk, S.; Johansson, P.A.; Peyre, E.; Broix, L.; Sahu, S.K.; Hirst, W.; Schlichthaerle, T.; De Juan Romero, C.; Draganova, K.; et al. The Centrosome Protein AKNA Regulates Neurogenesis via Microtubule Organization. Nature 2019 567:7746, 2019, 567, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Wiegard, A.; Kuzin, V.; Cameron, D.P.; Grosser, J.; Ceribelli, M.; Mehmood, R.; Ballarino, R.; Valant, F.; Grochowski, R.; Karabogdan, I.; et al. Topoisomerase 1 Activity during Mitotic Transcription Favors the Transition from Mitosis to G1. Mol Cell, 2021, 81, 5007–5024.e9. [Google Scholar] [CrossRef] [PubMed]
- Lam, J.C.; Aboreden, N.G.; Midla, S.C.; Wang, S.; Huang, A.; Keller, C.A.; Giardine, B.; Henderson, K.A.; Hardison, R.C.; Zhang, H.; et al. YY1-Controlled Regulatory Connectivity and Transcription Are Influenced by the Cell Cycle. Nature Genetics 2024 56:9, 2024, 56, 1938–1952. [Google Scholar] [CrossRef]
- Zhang, H.; Lam, J.; Zhang, D.; Lan, Y.; Vermunt, M.W.; Keller, C.A.; Giardine, B.; Hardison, R.C.; Blobel, G.A. CTCF and Transcription Influence Chromatin Structure Re-Configuration after Mitosis. Nature Communications 2021 12:1, 2021, 12, 1–16. [Google Scholar] [CrossRef]
- Festuccia, N.; Owens, N.; Papadopoulou, T.; Gonzalez, I.; Tachtsidi, A.; Vandoermel-Pournin, S.; Gallego, E.; Gutierrez, N.; Dubois, A.; Cohen-Tannoudji, M.; et al. Transcription Factor Activity and Nucleosome Organization in Mitosis. Genome Res, 2019, 29, 250–260. [Google Scholar] [CrossRef]
- Chervova, A.; Molliex, A.; Baymaz, H.I.; Coux, R.X.; Papadopoulou, T.; Mueller, F.; Hercul, E.; Fournier, D.; Dubois, A.; Gaiani, N.; et al. Mitotic Bookmarking Redundancy by Nuclear Receptors in Pluripotent Cells. Nature Structural & Molecular Biology 2024 31:3, 2024, 31, 513–522. [Google Scholar] [CrossRef]
- Teves, S.S.; An, L.; Bhargava-Shah, A.; Xie, L.; Darzacq, X.; Tjian, R. A Stable Mode of Bookmarking by TBP Recruits RNA Polymerase II to Mitotic Chromosomes. Elife, 2018, 7. [Google Scholar] [CrossRef]
- Thompson, L.L.; Jeusset, L.M.P.; Lepage, C.C.; McManus, K.J. Evolving Therapeutic Strategies to Exploit Chromosome Instability in Cancer. Cancers (Basel), 2017, 9, 151. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, G.J.; Richmond, P.A.; Bunker, E.N.; Karman, S.S.; Azofeifa, J.; Garnett, A.T.; Xu, Q.; Wheeler, G.E.; Toomey, C.M.; Zhang, Q.; et al. Genome-Wide Dose-Dependent Inhibition of Histone Deacetylases Studies Reveal Their Roles in Enhancer Remodeling and Suppression of Oncogenic Super-Enhancers. Nucleic Acids Res, 2018, 46, 1756–1776. [Google Scholar] [CrossRef] [PubMed]
- Manning, A.L.; Yazinski, S.A.; Nicolay, B.; Bryll, A.; Zou, L.; Dyson, N.J. Suppression of Genome Instability in PRB-Deficient Cells by Enhancement of Chromosome Cohesion. Mol Cell, 2014, 53, 993. [Google Scholar] [CrossRef]
- Swift, M.L.; Azizkhan-Clifford, J. DNA Damage-Induced Sumoylation of Sp1 Induces Its Interaction with RNF4 and Degradation in S Phase to Remove 53BP1 from DSBs and Permit HR. DNA Repair (Amst), 2022, 111, 103289. [Google Scholar] [CrossRef]
- Swift, M.L.; Sell, C.; Azizkhan-Clifford, J. DNA Damage-Induced Degradation of Sp1 Promotes Cellular Senescence. Geroscience, 2021, 44, 683–698. [Google Scholar] [CrossRef] [PubMed]
- Swift, M.L.; Beishline, K.; Azizkhan-Clifford, J. Sp1-Dependent Recruitment of the Histone Acetylase P300 to DSBs Facilitates Chromatin Remodeling and Recruitment of the NHEJ Repair Factor Ku70. DNA Repair (Amst), 2021, 105, 103171. [Google Scholar] [CrossRef]
- Swift, M.L.; Beishline, K.; Flashner, S.; Azizkhan-Clifford, J. DSB Repair Pathway Choice Is Regulated by Recruitment of 53BP1 through Cell Cycle-Dependent Regulation of Sp1. Cell Rep, 2021, 34, 108840. [Google Scholar] [CrossRef]
- Torabi, B.; Flashner, S.; Beishline, K.; Sowash, A.; Donovan, K.; Bassett, G.; Azizkhan-Clifford, J. Caspase Cleavage of Transcription Factor Sp1 Enhances Apoptosis. Apoptosis, 2018, 23, 65–78. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




