Submitted:
07 February 2025
Posted:
10 February 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Possibility of Molecular Computing
2.1. Inorganic Properties of Molecular Computing
2.1.1. Redox and Catalytic Reactions
2.1.2. Magnetic Effects
2.1.3. Photonic Effects
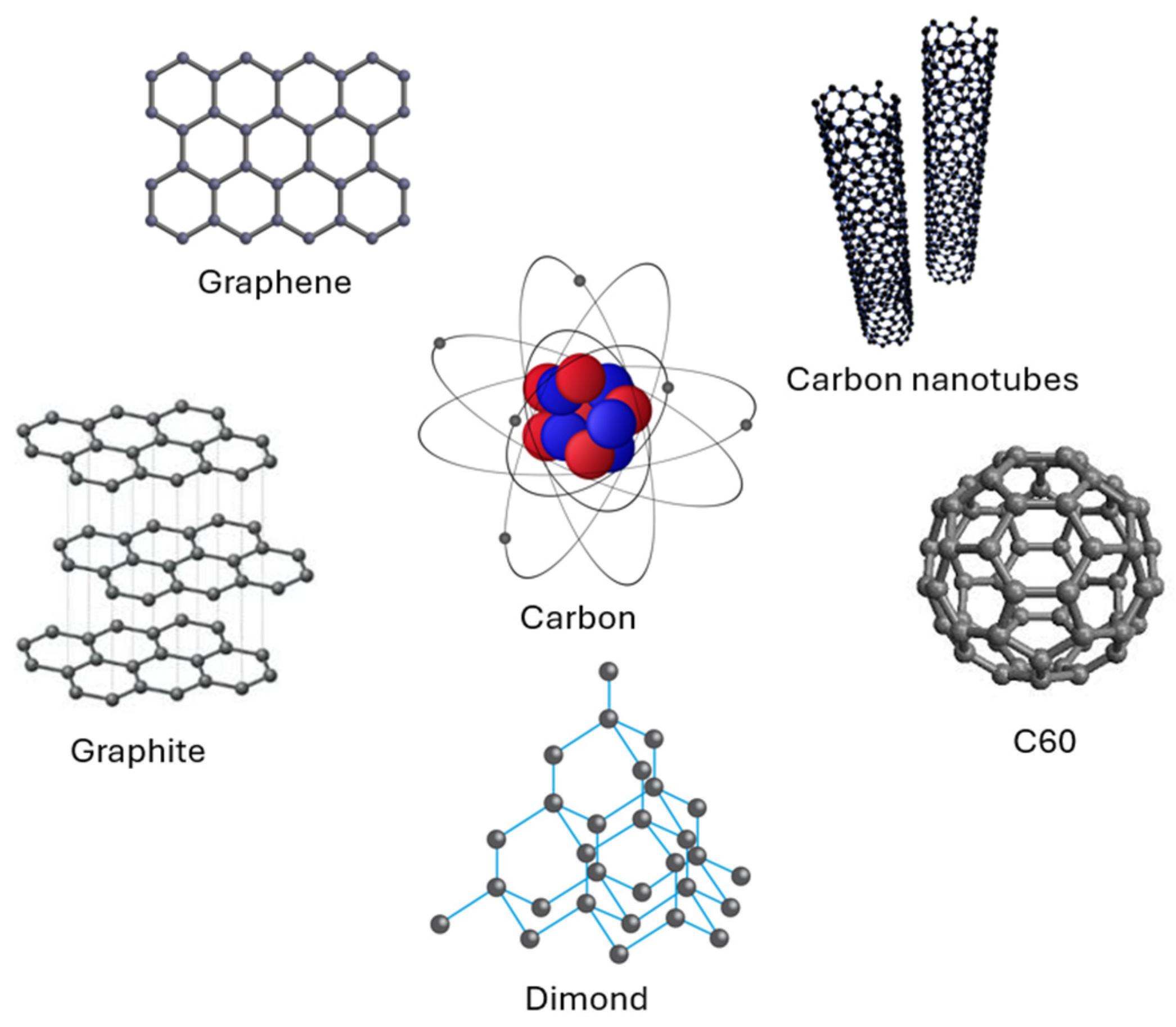
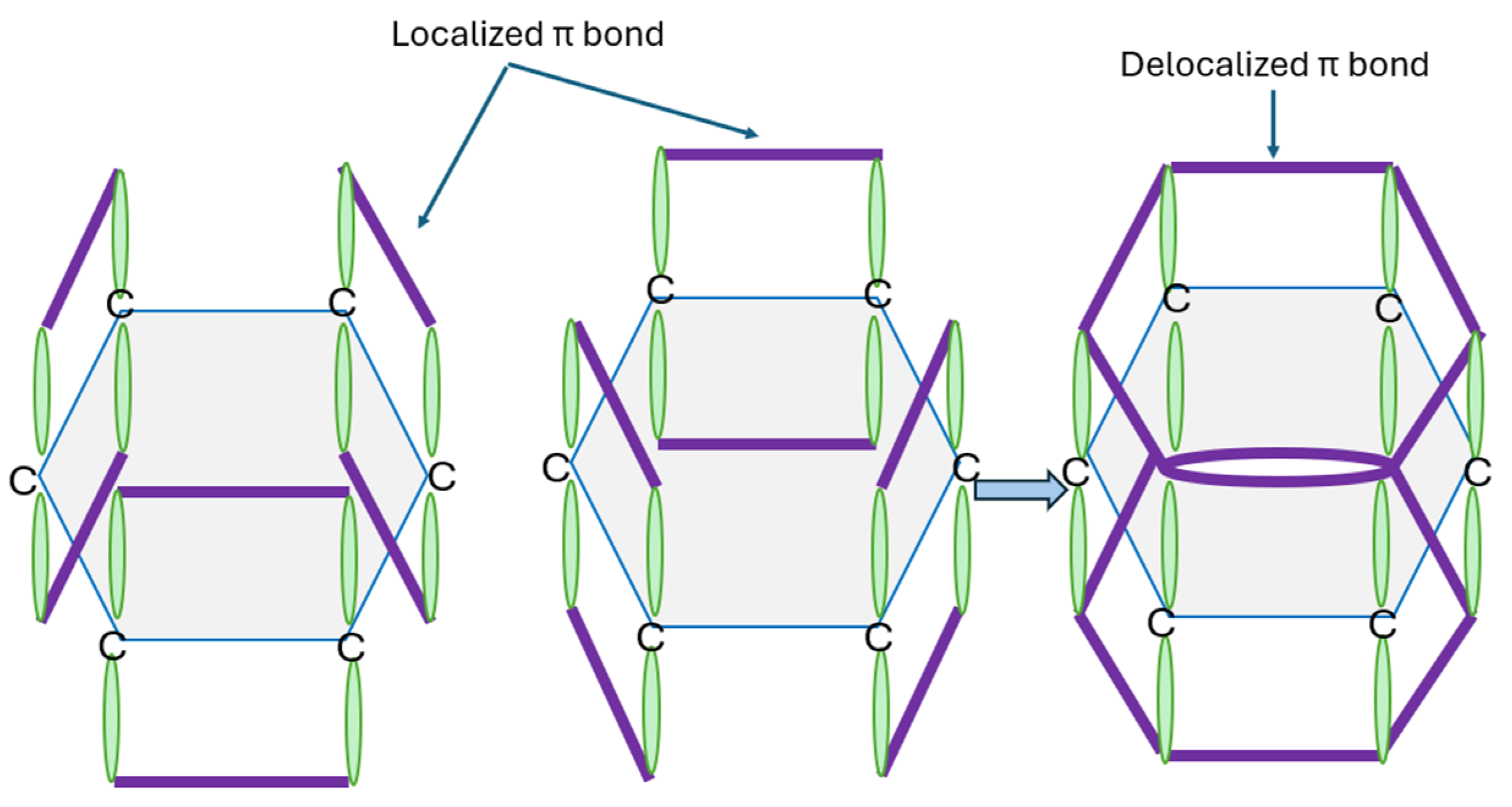
2.2. Carbon-Based Inorganic Molecular Computing
2.2.1. Rotaxanes and Catenanes
2.2.2. Photochromic Dyes
2.2.3. Photochromic Dyes
2.2.4. Photosensitive and Luminescent Molecules
2.2.5. Carbon Triboelectric Nanogenerators (TENGs)
2.3. Organic Molecular Computing
2.3.1. DNA and RNA Molecular Computing
2.3.2. Organic Carbon-Based Computing, Proteins and Lipids
2.3.3. Cell-Based Biological Computing
2.4. Hybrids and CMOL Devices
3. Applications for CMOL Systems in Healthcare
3.1. Biosensors in Healthcare
3.1.1. Nucleic Acid-Based Biosensors
3.1.2. Protein-Based Biosensors
3.1.3. Tissue-Based Biosensors
3.1.4. Cell-Based Biosensing Systems
3.1.5. Biomimetic Sensors
3.2. Targeted Drug Delivery
3.2.1. Mechanism of Release
3.2.2. Mechanism of Action
3.2.3. Type of Targeting Mechanism
3.2.4. Type of Carrier System
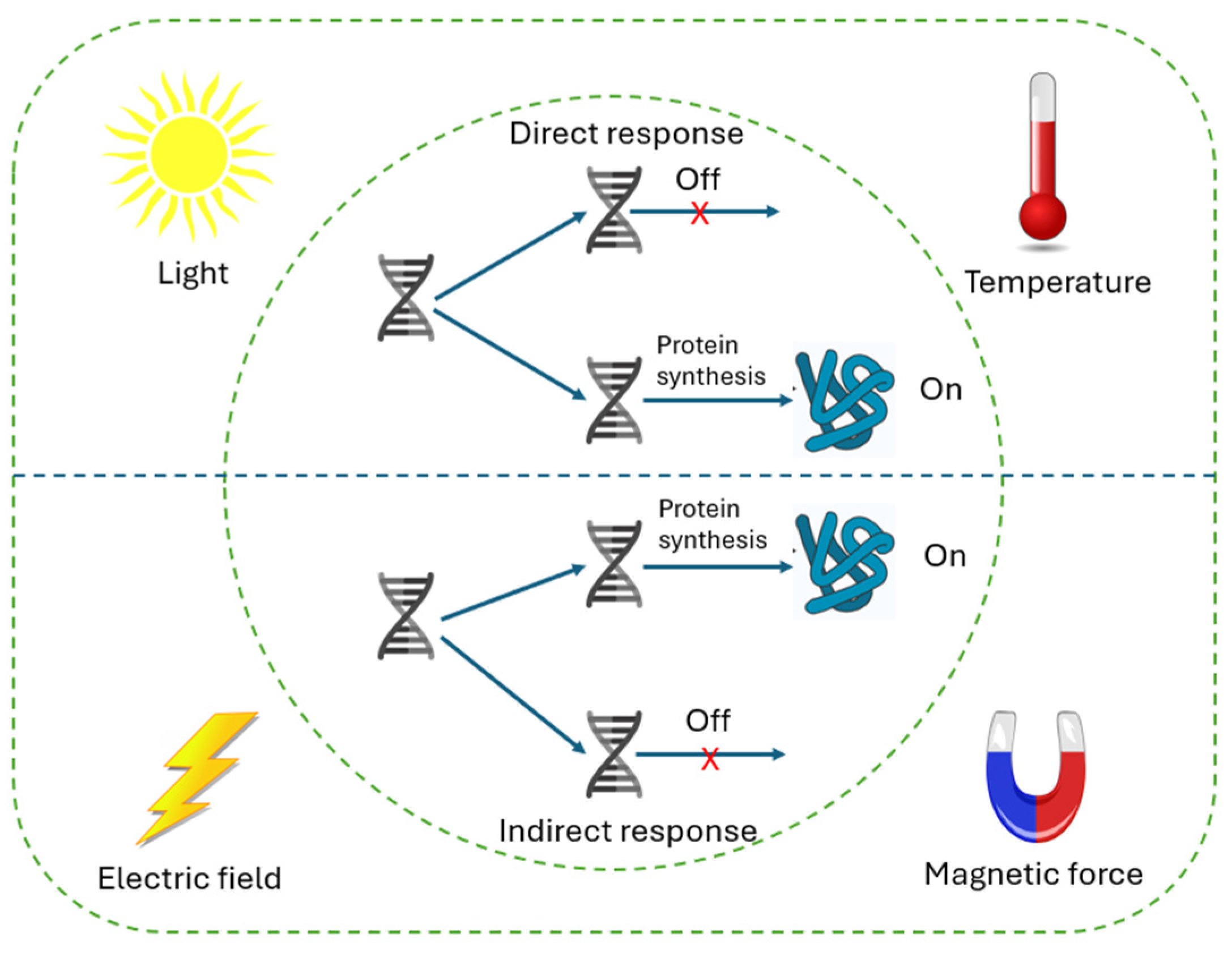
3.3. Gene Expression Physical Control Systems
3.4. Implantable Bioelectronic Devices
| Device Type |
CMOS/Biomolecular/Nanoparticle components |
Activity/ Mechanism of Action |
Target Tissue/ Organ |
Conditions Treated |
| Vagus Nerve Stimulator (VNS) | CMOS pulse generator, conductive polymers (PEDOT), Magnetic nanoparticles |
Electrical stimulation modulating vagus nerve signaling |
Vagus nerve | Epilepsy, Depression, PTSD |
|
Deep Brain Stimulator (DBS) |
CMOS microelectrode arrays, carbon nanotubes (CNTs), conductive hydrogels |
High-frequency pulses modulating deep brain activity |
Basal ganglia, thalamus |
Parkinson’s, Essential Tremor, OCD |
|
Spinal Cord Stimulator (SCS) |
CMOS pulse generator, graphene-coated electrodes, ion-sensitive polymers |
Blocks pain signals by modulating dorsal column activity |
Spinal cord, peripheral nerves | Chronic Pain, Neuropathy |
| Retinal Prosthesis (e.g., Argus II) | CMOS photodiodes, opsins, plasmonic nanoparticles |
Converts light into electrical signals for visual restoration |
Retina | Retinitis Pigmentosa, Macular Degeneration |
| Cochlear Implant | CMOS processor, flexible electrode array, conductive polymers | Converts sound into electrical impulses for auditory nerve stimulation |
Cochlea, Auditory Nerve | Sensorineural Hearing Loss, Deafness |
| Sacral Nerve Stimulator (SNS) | CMOS pulse generator, conductive nanowires, PEDOT-based electrodes |
Modulates sacral nerve activity to control bowel/bladder function | Sacral plexus, spinal cord |
Urinary and Fecal Incontinence |
| Gastric Electrical Stimulator (GES) | CMOS chip, gold nanoparticles, polymer-based electrodes | Electrical Stimulation of stomach muscles for motility control |
Stomach, Digestive Tract | Gastroparesis, Obesity |
|
Cardiac Pacemaker |
CMOS pulse generator, carbon nanotube electrodes, ion-sensitive gels |
Regulates heart rate via electrical impulses | Heart muscle |
Bradycardia, Arrhythmias |
|
Renal Nerve Stimulator; Bioartificial Kidney |
CMOS pulse generator, bioelectronic electrodes, CNT-modified probes; renal tissue |
Electrical stimulation for renal denervation; renal filtration |
Renal artery, Kidney |
Hypertension, Chronic Kidney Disease |
| Bone Regeneration Implant | CMOS microcurrent stimulator, hydroxyapatite-coated electrodes, BMP proteins |
Electrical stimulation promoting osteoblast activity |
Bone (Femur, Tibia, Spine) | Osteoporosis, Bone Fracture Healing |
| Peripheral Nerve Stimulator (PNS) | CMOS electrodes, gold nanoparticle-functionalized microelectrodes |
Electrical stimulation for pain and movement control |
Peripheral nerves | Chronic Pain, Phantom Limb Pain |
|
Bladder Neuromodulator |
CMOS pulse generator, bioelectronic hydrogel electrodes |
Electrical stimulation to modulate bladder activity |
Bladder, Pelvic Nerves | Overactive Bladder, Incontinence |
|
Pancreatic Stimulator |
CMOS electrode array, gold nanoparticle-coated electrodes | Electrical stimulation of pancreatic beta cells for insulin modulation |
Pancreas | Diabetes Mellitus |
|
Dermal Bioelectronic Implant |
CMOS microcurrent device, silver nanoparticles, conductive polymers |
Electrical stimulation for skin regeneration and wound healing |
Skin, Epidermis, Dermis |
Chronic Wounds, Diabetic Ulcers, Burns |
3.5. Real-Time Cellular Imaging
| Method Type | Mechanism of Action | Applications for Pathologies | Tissue/Organ | Cell Types |
| Fluorescence Imaging with CMOS Sensors | Fluorescent protein markers (e.g., GFP) captured using CMOS imaging chips | Cancer imaging, inflammation monitoring | Tumors, lymph nodes | Cancer cells, immune cells |
| Bioluminescence CMOS Imaging | Luminescent proteins (e.g., luciferase) emitting light detected by CMOS sensors | Metastatic cancer tracking, infection studies | Tumours, liver, brain | Cancer cells, hepatocytes |
|
Two-Photon Fluorescence Imaging |
Non-linear light absorption for deep tissue imaging | Brain mapping, neurodevelopment disorders | Brain, heart | Neurons, cardiomyocytes |
| FRET-Based CMOS Imaging | Förster Resonance Energy Transfer (FRET) for protein interaction analysis |
Cancer signalling pathways, protein aggregation diseases | Breast tissue, brain | Cancer cells, neurons |
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Muñoz, J. Rational Design of Stimuli-Responsive Inorganic 2D Materials via Molecular Engineering: Toward Molecule-Programmable Nanoelectronics. Adv. Mater. 2024, 36, 2305546. [Google Scholar] [CrossRef]
- Zauner, K.P. Molecular Information Technology. Crit. Rev. Solid State Mater. Sci. 2005, 30, 33–69. [Google Scholar] [CrossRef]
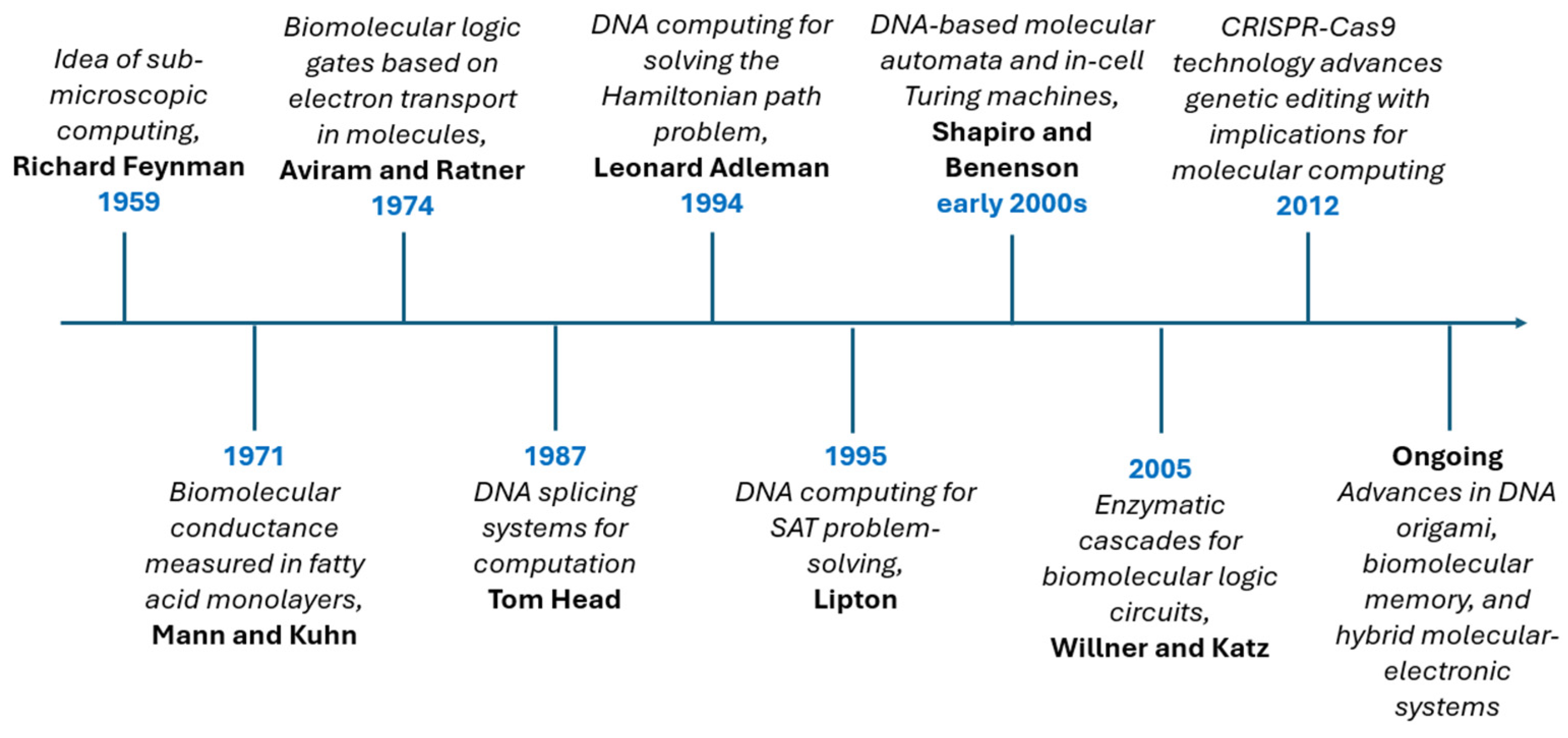
- Adleman, L.M. Molecular Computation of Solutions to Combinatorial Problems. Science 1994, 266, 1021–1024. [Google Scholar] [CrossRef] [PubMed]
- Feynman, R. There’s Plenty of Room at the Bottom. In Feynman and Computation; CRC Press: Boca Raton, FL, USA, 2018; pp. 63–76. [Google Scholar]
- Daley, M.J.; Kari, L. DNA Computing: Models and Implementations. Comments Theor. Biol. 2002, 7, 177–198. [Google Scholar] [CrossRef]
- Lipton, R.J. DNA Solution of Hard Computational Problems. Science 1995, 268, 542–545. [Google Scholar] [CrossRef]
- Shapiro, E.; Benenson, Y. Bringing DNA Computers to Life. Sci. Am. 2006, 294, 44–51. [Google Scholar] [CrossRef]
- Katz, E. Boolean Logic Gates Realized with Enzyme-Catalyzed Reactions—Unusual Look at Usual Chemical Reactions. ChemPhysChem 2019, 20, 9–22. [Google Scholar] [CrossRef]
- Ball, P. CRISPR: Implications for Materials Science. MRS Bull. 2016, 41, 832–835. [Google Scholar] [CrossRef]
- Agrawal, D.K.; Dolan, E.M.; Hernandez, N.E.; Blacklock, K.M.; Khare, S.D.; Sontag, E.D. Mathematical Models of Protease-Based Enzymatic Biosensors. ACS Synth. Biol. 2019, 9. [Google Scholar] [CrossRef]
- Mougkogiannis, P.; Adamatzky, A. Proto–Neural Networks from Thermal Proteins. Biochem. Biophys. Res. Commun. 2024, 149725. [Google Scholar] [CrossRef]
- Garduño-Juárez, R.; Tovar-Anaya, D.O.; Perez-Aguilar, J.M.; Lozano-Aguirre Beltran, L.F.; Zubillaga, R.A.; Alvarez-Perez, M.A.; Villarreal-Ramirez, E. Molecular Dynamic Simulations for Biopolymers with Biomedical Applications. Polymers 2024, 16, 1864. [Google Scholar] [CrossRef]
- Stojanovic, M.N.; Stefanovic, D.; Rudchenko, S. Exercises in Molecular Computing. Acc. Chem. Res. 2014, 47, 1845–1852. [Google Scholar] [CrossRef] [PubMed]
- Erbas-Cakmak, S.; Kolemen, S.; Sedgwick, A.C.; Gunnlaugsson, T.; James, T.D.; Yoon, J.; Akkaya, E.U. Molecular Logic Gates: The Past, Present and Future. Chem. Soc. Rev. 2018, 47, 2228–2248. [Google Scholar] [CrossRef] [PubMed]
- Tsutsui, M.; Taniguchi, M. Single Molecule Electronics and Devices. Sensors 2012, 12, 7259–7298. [Google Scholar] [CrossRef]
- Li, J.; Speyer, G.; Sankey, O.F. Conduction Switching of Photochromic Molecules. Phys. Rev. Lett. 2004, 93, 248302. [Google Scholar] [CrossRef]
- Fioravanti, G.; Haraszkiewicz, N.; Kay, E.R.; Mendoza, S.M.; Bruno, C.; Marcaccio, M.; Wiering, P.G.; Paolucci, F.; Rudolf, P.; Brouwer, A.M.; Leigh, D.A. Three-State Redox-Active Molecular Shuttle That Switches in Solution and on a Surface. J. Am. Chem. Soc. 2008, 130, 2593–2601. [Google Scholar] [CrossRef]
- Reif, J.H. Parallel Molecular Computation. In Proceedings of the Seventh Annual ACM Symposium on Parallel Algorithms and Architectures, Santa Barbara, CA, USA; 1995; pp. 213–223. [Google Scholar]
- Coronado, E. Molecular Magnetism: From Chemical Design to Spin Control in Molecules, Materials and Devices. Nat. Rev. Mater. 2020, 5, 87–104. [Google Scholar] [CrossRef]
- Joachim, C.; Gimzewski, J.K.; Aviram, A. Electronics Using Hybrid-Molecular and Mono-Molecular Devices. Nature 2000, 408, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Symes, M.D.; Cronin, L. The Crystal Computer—Computing with Inorganic Cellular Frameworks and Nets. Int. J. Nanotechnol. Mol. Comput. 2011, 3, 24–34. [Google Scholar] [CrossRef]
- Fuller, E.J.; Li, Y.; Bennet, C.; Keene, S.T.; Melianas, A.; Agarwal, S.; Marinella, M.J.; Salleo, A.; Talin, A.A. Redox Transistors for Neuromorphic Computing. IBM J. Res. Dev. 2019, 63, 9–1. [Google Scholar] [CrossRef]
- Anderson, C.M.; Jain, S.S.; Silber, L.; Chen, K.; Guha, S.; Zhang, W.; McLaughlin, E.C.; Hu, Y.; Tanski, J.M. Synthesis and Characterization of Water-Soluble, Heteronuclear Ruthenium (III)/Ferrocene Complexes and Their Interactions with Biomolecules. J. Inorg. Biochem. 2015, 145, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Dittmann, R.; Strachan, J.P. Redox-Based Memristive Devices for New Computing Paradigm. APL Mater. 2019, 7, 11. [Google Scholar] [CrossRef]
- Wedege, K.; Azevedo, J.; Khataee, A.; Bentien, A.; Mendes, A. Direct Solar Charging of an Organic–Inorganic, Stable, and Aqueous Alkaline Redox Flow Battery with a Hematite Photoanode. Angew. Chem. Int. Ed. 2016, 55, 7142–7147. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.L.; Doty, D.; Soloveichik, D. Deterministic Function Computation with Chemical Reaction Networks. Nat. Comput. 2014, 13, 517–534. [Google Scholar] [CrossRef]
- Matysik, J.; Długosz, O.; Banach, M. Development of Nanozymatic Characteristics in Metal-Doped Oxide Nanomaterials. J. Phys. Chem. B 2024, 128, 8007–8016. [Google Scholar] [CrossRef] [PubMed]
- Roztocki, K.; Bon, V.; Senkovska, I.; Matoga, D.; Kaskel, S. A Logic Gate Based on a Flexible Metal–Organic Framework (JUK-8) for the Concomitant Detection of Hydrogen and Oxygen. Chem.–Eur. J. 2022, 28, e202202255. [Google Scholar] [CrossRef]
- Camarero, J.; Coronado, E. Molecular vs. Inorganic Spintronics: The Role of Molecular Materials and Single Molecules. J. Mater. Chem. 2009, 19, 1678–1684. [Google Scholar] [CrossRef]
- Hirohata, A.; Takanashi, K. Future Perspectives for Spintronic Devices. J. Phys. D Appl. Phys. 2014, 47, 193001. [Google Scholar] [CrossRef]
- Abdullah, R.M.; Vick, A.J.; Murphy, B.A.; Hirohata, A. Spin-Current Signal Amplification by a Geometrical Ratchet. J. Phys. D Appl. Phys. 2014, 47, 482001. [Google Scholar] [CrossRef]
- Suzuki, Y.; Kato, Y. Spin Relaxation, Diffusion, and Edelstein Effect in Chiral Metal Surface. Phys. Rev. B 2023, 107, 115305. [Google Scholar] [CrossRef]
- da Rocha, J.D.G.; Cechinel, M.A.P.; Rocha, L.F.; Riella, H.G.; Padoin, N.; Soares, C. Exploring the Potential of Rare Earth Doped Carbon Dots: Concepts and Applications. Chem. Eng. J. Adv. 2024, 100583. [Google Scholar] [CrossRef]
- Gonçalves, R.A.; Toledo, R.P.; Joshi, N.; Berengue, O.M. Green Synthesis and Applications of ZnO and TiO₂ Nanostructures. Molecules 2021, 26, 2236. [Google Scholar] [CrossRef]
- González-Tudela, A.; Reiserer, A.; García-Ripoll, J.J.; García-Vidal, F.J. Light–Matter Interactions in Quantum Nanophotonic Devices. Nat. Rev. Phys. 2024, 6, 166–179. [Google Scholar] [CrossRef]
- Sharkawy, A.; Shi, S.; Prather, D.W.; Soref, R.A. Electro-Optical Switching Using Coupled Photonic Crystal Waveguides. Opt. Express 2002, 10, 1048–1059. [Google Scholar] [CrossRef] [PubMed]
- Charbonnière, L.J.; Hildebrandt, N. Lanthanide Complexes and Quantum Dots: A Bright Wedding for Resonance Energy Transfer. Eur. J. Inorg. Chem. 2008, 2008, 3241–3251. [Google Scholar] [CrossRef]
- Teunissen, A.J.; Pérez-Medina, C.; Meijerink, A.; Mulder, W.J. Investigating Supramolecular Systems Using Förster Resonance Energy Transfer. Chem. Soc. Rev. 2018, 47, 7027–7044. [Google Scholar] [CrossRef] [PubMed]
- Laurila, T.; Sainio, S.; Caro, M.A. Hybrid Carbon-Based Nanomaterials for Electrochemical Detection of Biomolecules. Prog. Mater. Sci. 2017, 88, 499–594. [Google Scholar] [CrossRef]
- Sauvage, J.P. From Chemical Topology to Molecular Machines (Nobel Lecture). Angew. Chem. Int. Ed. 2017, 56, 11080–11093. [Google Scholar] [CrossRef]
- Ellis, E.; Moorthy, S.; Chio, W.I.K.; Lee, T.C. Artificial Molecular and Nanostructures for Advanced Nanomachinery. Chem. Commun. 2018, 54, 4075–4090. [Google Scholar] [CrossRef]
- Wilson, B.H.; Abdulla, L.M.; Schurko, R.W.; Loeb, S.J. Translational Dynamics of a Non-Degenerate Molecular Shuttle Imbedded in a Zirconium Metal–Organic Framework. Chem. Sci. 2021, 12, 3944–3951. [Google Scholar] [CrossRef]
- Köttner, L.; Dube, H. Path-Independent All-Visible Orthogonal Photoswitching for Applications in Multi-Photochromic Polymers and Molecular Computing. Angew. Chem. Int. Ed. 2024, 63, e202409214. [Google Scholar] [CrossRef] [PubMed]
- Pérez, E.M.; Martín, N. π–π Interactions in Carbon Nanostructures. Chem. Soc. Rev. 2015, 44, 6425–6433. [Google Scholar] [CrossRef]
- Burghard, M.; Klauk, H.; Kern, K. Carbon-Based Field-Effect Transistors for Nanoelectronics. Adv. Mater. 2009, 21, 2586–2600. [Google Scholar] [CrossRef]
- Watanabe, K.; Miura, N.; Taguchi, H.; Komatsu, T.; Aratake, A.; Makita, T.; Tanabe, M.; Wakimoto, T.; Kumagai, S.; Okamoto, T.; Watanabe, S. All-Carbon-Based Complementary Integrated Circuits. Adv. Mater. Technol. 2024, 9, 2301673. [Google Scholar] [CrossRef]
- Hartmann, M.; Hermann, S.; Marsh, P.F.; Rutherglen, C.; Wang, D.; Ding, L.; Peng, L.M.; Claus, M.; Schröter, M. CNTFET Technology for RF Applications: Review and Future Perspective. IEEE J. Microw. 2021, 1, 275–287. [Google Scholar] [CrossRef]
- Gervasi, B. Will Carbon Nanotube Memory Replace DRAM? IEEE Micro 2019, 39, 45–51. [Google Scholar] [CrossRef]
- Blaudeck, T.; Preuß, A.; Scharf, S.; Notz, S.; Kossmann, A.; Hartmann, S.; Kasper, L.; Mendes, R.G.; Gemming, T.; Hermann, S.; Lang, H. Photosensitive Field-Effect Transistors Made from Semiconducting Carbon Nanotubes and Non-Covalently Attached Gold Nanoparticles. Phys. Status Solidi A 2019, 216, 1900030. [Google Scholar] [CrossRef]
- Ogawa, K. Two-Photon Absorbing Molecules as Potential Materials for 3D Optical Memory. Appl. Sci. 2014, 4, 1–18. [Google Scholar] [CrossRef]
- Dias, G.G.; Souto, F.T. Architecture of Molecular Logic Gates: From Design to Application as Optical Detection Devices. Organics 2024, 5, 114–162. [Google Scholar] [CrossRef]
- Chrystie, R.S. A Review on 1-D Nanomaterials: Scaling-Up with Gas-Phase Synthesis. Chem. Rec. 2023, 23, e202300087. [Google Scholar] [CrossRef] [PubMed]
- Prestopino, G.; Orsini, A.; Barettin, D.; Arrabito, G.; Pignataro, B.; Medaglia, P.G. Vertically Aligned Nanowires and Quantum Dots: Promises and Results in Light Energy Harvesting. Materials 2023, 16, 4297. [Google Scholar] [CrossRef]
- Ramaswamy, S.H.; Kondo, R.; Chen, W.; Fukushima, I.; Choi, J. Development of Highly Durable Sliding Triboelectric Nanogenerator Using Diamond-Like Carbon Films. Tribol. Online 2020, 15, 89–97. [Google Scholar] [CrossRef]
- Dos Reis, G.S.; de Oliveira, H.P.; Candido, I.C.M.; Freire, A.L.; Molaiyan, P.; Dotto, G.L.; Grimm, A.; Mikkola, J.P. Supercapacitors and Triboelectric Nanogenerators Based on Electrodes of Greener Iron Nanoparticles/Carbon Nanotubes Composites. Sci. Rep. 2024, 14, 11555. [Google Scholar] [CrossRef] [PubMed]
- Gervasio, J.H.D.B.; da Costa Oliveira, H.; da Costa Martins, A.G.; Pesquero, J.B.; Verona, B.M.; Cerize, N.N.P. How Close Are We to Storing Data in DNA? Trends Biotechnol. 2024, 42, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Takiguchi, S.; Takeuchi, N.; Shenshin, V.; Gines, G.; Genot, A.J.; Nivala, J.; Rondelez, Y.; Kawano, R. [Full reference details needed].
- Stojanovic, M.N.; Stefanovic, D.; LaBean, T.; Yan, H. Computing with Nucleic Acids. In Bioelectronics: From Theory to Applications; Wiley-VCH: Weinheim, Germany, 2005; pp. 427–455. [Google Scholar]
- Faulhammer, D.; Cukras, A.R.; Lipton, R.J.; Landweber, L.F. Molecular Computation: RNA Solutions to Chess Problems. Proc. Natl. Acad. Sci. USA 2000, 97, 1385–1389. [Google Scholar] [CrossRef] [PubMed]
- Benenson, Y. RNA-Based Computation in Live Cells. Curr. Opin. Biotechnol. 2009, 20, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, S.; Ono, H.; Kawasaki, S.; Kuang, Y.; Fujita, Y.; Saito, H. Synthetic RNA-Based Logic Computation in Mammalian Cells. Nat. Commun. 2018, 9, 4847. [Google Scholar] [CrossRef]
- Buckhout-White, S.; Spillmann, C.M.; Algar, W.R.; Khachatrian, A.; Melinger, J.S.; Goldman, E.R.; Ancona, M.G.; Medintz, I.L. Assembling Programmable FRET-Based Photonic Networks Using Designer DNA Scaffolds. Nat. Commun. 2014, 5, 5615. [Google Scholar] [CrossRef]
- Tregubov, A.A.; Nikitin, P.I.; Nikitin, M.P. Advanced Smart Nanomaterials with Integrated Logic-Gating and Biocomputing: Dawn of Theranostic Nanorobots. Chem. Rev. 2018, 118, 10294–10348. [Google Scholar] [CrossRef]
- Allard, S.; Forster, M.; Souharce, B.; Thiem, H.; Scherf, U. Organic Semiconductors for Solution-Processable Field-Effect Transistors (OFETs). Angew. Chem. Int. Ed. 2008, 47, 4070–4098. [Google Scholar] [CrossRef]
- Ariga, K. Materials Nanoarchitectonics for Advanced Devices. Materials 2024, 17, 5918. [Google Scholar] [CrossRef]
- Grant, B.; Bandera, Y.; Foulger, S.H.; Vilčáková, J.; Sáha, P.; Pfleger, J. Boolean and Elementary Algebra with a Roll-To-Roll Printed Electrochemical Memristor. Adv. Mater. Technol. 2022, 7, 2101108. [Google Scholar] [CrossRef]
- Giordano, G.; Carlotti, M.; Mazzolai, B. A Perspective on Cephalopods Mimicry and Bioinspired Technologies Toward Proprioceptive Autonomous Soft Robots. Adv. Mater. Technol. 2021, 6, 2100437. [Google Scholar] [CrossRef]
- Picci, G.; Montis, R.; Gilchrist, A.M.; Gale, P.A.; Caltagirone, C. Fluorescent and Colorimetric Sensors for Anions: Highlights from 2020 to 2022. Coord. Chem. Rev. 2024, 501, 215561. [Google Scholar] [CrossRef]
- Katz, E.; Privman, V. Enzyme-Based Logic Systems for Information Processing. Chem. Soc. Rev. 2010, 39, 1835–1857. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Iwabuchi, S.; Izumi, K.; Takiguchi, S.; Yamaji, M.; Fujita, S.; Suzuki, H.; Kambara, F.; Fukasawa, G.; Cooney, A.; Di Michele, L. Lipid Vesicle-Based Molecular Robots. Lab Chip 2024. [CrossRef]
- TerAvest, M.A.; Li, Z.; Angenent, L.T. Bacteria-Based Biocomputing with Cellular Computing Circuits to Sense, Decide, Signal, and Act. Energy Environ. Sci. 2011, 4, 4907–4916. [Google Scholar] [CrossRef]
- Goñi-Moreno, Á. Biocomputation: Moving Beyond Turing with Living Cellular Computers. Commun. ACM 2024, 67, 70–77. [Google Scholar] [CrossRef]
- Sorenson, C.; Adamala, K.P. Laws of Thought in Living Cells. Cell 2024, 187, 4830–4832. [Google Scholar] [CrossRef]
- Krauhausen, I.; Coen, C.T.; Spolaor, S.; Gkoupidenis, P.; van de Burgt, Y. Brain-Inspired Organic Electronics: Merging Neuromorphic Computing and Bioelectronics Using Conductive Polymers. Adv. Funct. Mater. 2024, 34, 2307729. [Google Scholar] [CrossRef]
- Strukov, D.B.; Likharev, K.K. Prospects for the Development of Digital CMOL Circuits. In Proceedings of the 2007 IEEE International Symposium on Nanoscale Architectures, San Diego, CA, USA; 2007; pp. 109–116. [Google Scholar]
- Jabegu, T.; Li, N.; Okmi, A.; Tipton, B.; Vlassiouk, I.; Xiao, K.; Urazhdin, S.; Yao, Y.; Lei, S. Interfacial Momentum Matching for Ohmic Van Der Waals Contact Construction. Adv. Electron. Mater. 2024, 2400397. [Google Scholar] [CrossRef]
- Kim, B.J.; Bonacchini, G.E.; Ostrovsky-Snider, N.A.; Omenetto, F.G. Bimodal Gating Mechanism in Hybrid Thin-Film Transistors Based on Dynamically Reconfigurable Nanoscale Biopolymer Interfaces. Adv. Mater. 2023, 35, 2302062. [Google Scholar] [CrossRef]
- Abbott, J.; Ye, T.; Park, H.; Ham, D. CMOS Interface with Biological Molecules and Cells: Invited Review Paper. In ESSDERC 2019-49th European Solid-State Device Research Conference (ESSDERC), Cracow, Poland, 2019; pp. 13–16.
- Jorgsson, V.; Kumar, R.; Ahmed, M.; Yung, M.; Pattnayak, A.; Sridhar, S.P.; Varma, V.; Ponnambalam, A.R.; Weidlich, G.; Pinotsis, D. AI-Driven Physics-Informed Bio-Silicon Intelligence System: Integrating Hybrid Systems, Biocomputing, Neural Networks, and Machine Learning for Advanced Neurotechnology. arXiv Prepr. 2024, arXiv:2407.11939.
- Pedro, F. Advances and Challenges in Closed Loop Therapeutics: From Signal Selection to Optogenetic Techniques. J. Biomed. Sustain. Healthc. Appl. 2024, 73. [Google Scholar] [CrossRef]
- Hoffmann, C.; Wang, J.; Ali, R.P.; D'Souza, R.S. Neuromodulation Guide for the Non-Neuromodulator Clinician: What It Is and How It Can Benefit Patients? Biomol. Biomed. 2024. [CrossRef] [PubMed]
- Puccetti, M.; Pariano, M.; Schoubben, A.; Giovagnoli, S.; Ricci, M. Biologics, Theranostics, and Personalized Medicine in Drug Delivery Systems. Pharmacol. Res. 2024, 107086. [Google Scholar] [CrossRef]
- Skottvoll, F.S.; Escobedo-Cousin, E.; Mielnik, M.M. The Role of Silicon Technology in Organ-On-Chip: Current Status and Future Perspective. Adv. Mater. Technol. 2024, 2401254. [Google Scholar] [CrossRef]
- Byrne, R.; Carrico, A.; Lettieri, M.; Rajan, A.K.; Forster, R.J.; Cumba, L.R. Bioinks and Biofabrication Techniques for Biosensors Development: A Review. Mater. Today Bio 2024, 101185. [Google Scholar] [CrossRef]
- Agiba, A.M.; Elsayyad, N.; ElShagea, H.N.; Metwalli, M.A.; Mahmoudsalehi, A.O.; Beigi-Boroujeni, S.; Lozano, O.; Aguirre-Soto, A.; Arreola-Ramirez, J.L.; Segura-Medina, P.; Hamed, R.R. Advances in Light-Responsive Smart Multifunctional Nanofibers: Implications for Targeted Drug Delivery and Cancer Therapy. Pharmaceutics 2024, 16, 1017. [Google Scholar] [CrossRef]
- Radulescu, D.M.; Andronescu, E.; Vasile, O.R.; Ficai, A.; Vasile, B.S. Silk Fibroin-Based Scaffolds for Wound Healing Applications with Metal Oxide Nanoparticles. J. Drug Deliv. Sci. Technol. 2024, 105689. [Google Scholar] [CrossRef]
- Kang, K.; Ye, S.; Jeong, C.; Jeong, J.; Ye, Y.S.; Jeong, J.Y.; Kim, Y.J.; Lim, S.; Kim, T.H.; Kim, K.Y.; Kim, J.U. Bionic Artificial Skin with a Fully Implantable Wireless Tactile Sensory System for Wound Healing and Restoring Skin Tactile Function. Nat. Commun. 2024, 15, 10. [Google Scholar] [CrossRef]
- Moeinfard, T.; Ghafar-Zadeh, E.; Magierowski, S. CMOS Point-of-Care Diagnostics Technologies: Recent Advances and Future Prospects. Micromachines 2024, 15, 1320. [Google Scholar] [CrossRef]
- Stuber, A.; Nakatsuka, N. Aptamer Renaissance for Neurochemical Biosensing. ACS Nano 2024, 18, 2552–2563. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, M.; Cramer, A.; Webb, S.; Goorskey, E.; Chushak, Y.; Mirau, P.; Arroyo-Currás, N.; Chávez, J.L. Rational Approach to Optimizing Conformation-Switching Aptamers for Biosensing Applications. ACS Sens. 2024, 9, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Klebes, A.; Ates, H.C.; Verboket, R.D.; Urban, G.A.; von Stetten, F.; Dincer, C.; Früh, S.M. Emerging Multianalyte Biosensors for the Simultaneous Detection of Protein and Nucleic Acid Biomarkers. Biosens. Bioelectron. 2024, 244, 115800. [Google Scholar] [CrossRef] [PubMed]
- Chaisupa, P.; Wright, R.C. State-of-the-Art in Engineering Small Molecule Biosensors and Their Applications in Metabolic Engineering. SLAS Technol. 2024, 29, 100113. [Google Scholar]
- Cristea, C.; Florea, A.; Tertiș, M.; Săndulescu, R. Immunosensors. In Biosensors-Micro and Nanoscale Applications; IntechOpen: London, UK, 2015. [Google Scholar]
- Del Giovane, S.; Bagheri, N.; Di Pede, A.C.; Chamorro, A.; Ranallo, S.; Migliorelli, D.; Burr, L.; Paoletti, S.; Altug, H.; Porchetta, A. Challenges and Perspectives of CRISPR-Based Technology for Diagnostic Applications. TrAC Trends Anal. Chem. 2024, 117594. [Google Scholar] [CrossRef]
- Mahr, R.; Frunzke, J. Transcription Factor-Based Biosensors in Biotechnology: Current State and Future Prospects. Appl. Microbiol. Biotechnol. 2016, 100, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Selivanovitch, E.; Ostwalt, A.; Chao, Z.; Daniel, S. Emerging Designs and Applications for Biomembrane Biosensors. Annu. Rev. Anal. Chem. 2024, 17, 339–366. [Google Scholar] [CrossRef] [PubMed]
- Doryab, A.; Schmid, O. Towards a Gold Standard Functional Readout to Characterize In Vitro Lung Barriers. Eur. J. Pharm. Sci. 2022, 179, 106305. [Google Scholar] [CrossRef]
- Deguchi, S.; Takayama, K. State-of-the-Art Liver Disease Research Using Liver-on-a-Chip. Inflamm. Regen. 2022, 42, 62. [Google Scholar] [CrossRef] [PubMed]
- Danku, A.E.; Dulf, E.H.; Braicu, C.; Jurj, A.; Berindan-Neagoe, I. Organ-on-a-Chip: A Survey of Technical Results and Problems. Front. Bioeng. Biotechnol. 2022, 10, 840674. [Google Scholar] [CrossRef] [PubMed]
- Dobres, S.; Mula, G.; Sauer, J.; Zhu, D. Applications of 3D Printed Chimeric DNA Biomaterials. Eng. Regen. 2022, 3, 13–23. [Google Scholar] [CrossRef]
- Gheorghiu, M. A Short Review on Cell-Based Biosensing: Challenges and Breakthroughs in Biomedical Analysis. J. Biomed. Res. 2020, 35, 255. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Hu, Q.; San, S.; Kasputis, T.; Splinter, M.G.D.; Yin, K.; Chen, J. CRISPR-Based Biosensors for Human Health: A Novel Strategy to Detect Emerging Infectious Diseases. TrAC Trends Anal. Chem. 2023, 117342. [Google Scholar] [CrossRef] [PubMed]
- Popgeorgiev, N.; Gil, C.; Berthenet, K.; Bertolin, G.; Ichim, G. Shedding Light on Mitochondrial Outer-Membrane Permeabilization and Membrane Potential: State-of-the-Art Methods and Biosensors. Semin. Cell Dev. Biol. 2024, 156, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, P. Cell-Based Biosensors: Principles and Applications; Artech House: London, UK, 2009. [Google Scholar]
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-Dimensional Cell Culture Systems and Their Applications in Drug Discovery and Cell-Based Biosensors. Assay Drug Dev. Technol. 2014, 12, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Bräuer, B.; Unger, C.; Werner, M.; Lieberzeit, P.A. Biomimetic Sensors to Detect Bioanalytes in Real-Life Samples Using Molecularly Imprinted Polymers: A Review. Sensors 2021, 21, 5550. [Google Scholar] [CrossRef]
- Sligar, S.G.; Denisov, I.G. Nanodiscs: A Toolkit for Membrane Protein Science. Protein Sci. 2021, 30, 297–315. [Google Scholar] [CrossRef]
- Gabriele, F.; Palerma, M.; Ippoliti, R.; Angelucci, F.; Pitari, G.; Ardini, M. Recent Advances on Affibody- and DARPin-Conjugated Nanomaterials in Cancer Therapy. Int. J. Mol. Sci. 2023, 24, 8680. [Google Scholar] [CrossRef]
- Kim, K.N.; Sung, M.J.; Park, H.L.; Lee, T.W. Organic Synaptic Transistors for Bio-Hybrid Neuromorphic Electronics. Adv. Electron. Mater. 2022, 8, 2100935. [Google Scholar] [CrossRef]
- Reddy, K.T.K.; Reddy, A.S. Recent Breakthroughs in Drug Delivery Systems for Targeted Cancer Therapy: An Overview. Cell. Mol. Biomed. Rep. 2025, 5, 13–27. [Google Scholar] [CrossRef]
- Park, H.; Otte, A.; Park, K. Evolution of Drug Delivery Systems: From 1950 to 2020 and Beyond. J. Control. Release 2022, 342, 53–65. [Google Scholar] [CrossRef]
- Mayer, A.M.; Mayer, V.A.; Swanson-Mungerson, M.; Pierce, M.L.; Rodríguez, A.D.; Nakamura, F.; Taglialatela-Scafati, O. Marine Pharmacology in 2019–2021: Marine Compounds with Antibacterial, Antidiabetic, Antifungal, Anti-Inflammatory, Antiprotozoal, Antituberculosis and Antiviral Activities; Affecting the Immune and Nervous Systems, and Other Miscellaneous Mechanisms of Action. Mar. Drugs 2024, 22, 309. [Google Scholar] [CrossRef] [PubMed]
- Holland, J.F. Holland-Frei Cancer Medicine 8; PMPH-USA: Shelton, CT, USA, 2010; Volume 8. [Google Scholar]
- Gavas, S.; Quazi, S.; Karpiński, T.M. Nanoparticles for Cancer Therapy: Current Progress and Challenges. Nanoscale Res. Lett. 2021, 16, 173. [Google Scholar] [CrossRef]
- Shi, D.; Beasock, D.; Fessler, A.; Szebeni, J.; Ljubimova, J.Y.; Afonin, K.A.; Dobrovolskaia, M.A. To PEGylate or Not to PEGylate: Immunological Properties of Nanomedicine’s Most Popular Component, Polyethylene Glycol and Its Alternatives. Adv. Drug Deliv. Rev. 2022, 180, 114079. [Google Scholar] [CrossRef]
- Morachis, J.M.; Mahmoud, E.A.; Almutairi, A.; Insel, P.A. Physical and Chemical Strategies for Therapeutic Delivery by Using Polymeric Nanoparticles. Pharmacol. Rev. 2012, 64, 505–519. [Google Scholar] [CrossRef]
- Wakaskar, R.R. General Overview of Lipid–Polymer Hybrid Nanoparticles, Dendrimers, Micelles, Liposomes, Spongosomes and Cubosomes. J. Drug Target. 2018, 26, 311–318. [Google Scholar] [CrossRef]
- Naseri, N.; Valizadeh, H.; Zakeri-Milani, P. Solid Lipid Nanoparticles and Nanostructured Lipid Carriers: Structure, Preparation and Application. Adv. Pharm. Bull. 2015, 5, 305. [Google Scholar] [CrossRef] [PubMed]
- Braatz, D.; Cherri, M.; Tully, M.; Dimde, M.; Ma, G.; Mohammadifar, E.; Reisbeck, F.; Ahmadi, V.; Schirner, M.; Haag, R. Chemical Approaches to Synthetic Drug Delivery Systems for Systemic Applications. Angew. Chem. Int. Ed. 2022, 61, e202203942. [Google Scholar] [CrossRef] [PubMed]
- Kofoed Andersen, C.; Khatri, S.; Hansen, J.; Slott, S.; Pavan Parvathaneni, R.; Mendes, A.C.; Chronakis, I.S.; Hung, S.C.; Rajasekaran, N.; Ma, Z.; Zhu, S. Carbon Nanotubes—Potent Carriers for Targeted Drug Delivery in Rheumatoid Arthritis. Pharmaceutics 2021, 13, 453. [Google Scholar] [CrossRef]
- Paul, W.; Sharma, C.P. Inorganic Nanoparticles for Targeted Drug Delivery. In Biointegration of Medical Implant Materials; Elsevier: Amsterdam, Netherlands, 2020; pp. 333–373. [Google Scholar]
- Pierigè, F.; Serafini, S.; Rossi, L.; Magnani, M. Cell-Based Drug Delivery. Adv. Drug Deliv. Rev. 2008, 60, 286–295. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Fussenegger, M. Programming Mammalian Cell Behaviors by Physical Cues. Trends Biotechnol. 2024. [CrossRef] [PubMed]
- Unal, G.; Fussenegger, M. At the Crossroads of Biology and Electronics. Curr. Opin. Biotechnol. 2025, 91, 103249. [Google Scholar] [CrossRef]
- Del Sol-Fernández, S.; Martínez-Vicente, P.; Gomollón-Zueco, P.; Castro-Hinojosa, C.; Gutiérrez, L.; Fratila, R.M.; Moros, M. Magnetogenetics: Remote Activation of Cellular Functions Triggered by Magnetic Switches. Nanoscale 2022, 14, 2091–2118. [Google Scholar] [CrossRef] [PubMed]
- Nims, R.J.; Pferdehirt, L.; Guilak, F. Mechanogenetics: Harnessing Mechanobiology for Cellular Engineering. Curr. Opin. Biotechnol. 2022, 73, 374–379. [Google Scholar] [CrossRef] [PubMed]
- Emiliani, V.; Entcheva, E.; Hedrich, R.; Hegemann, P.; Konrad, K.R.; Lüscher, C.; Mahn, M.; Pan, Z.H.; Sims, R.R.; Vierock, J.; Yizhar, O. Optogenetics for Light Control of Biological Systems. Nat. Rev. Methods Primers 2022, 2, 55. [Google Scholar] [CrossRef] [PubMed]
- Mariello, M.; Kim, K.; Wu, K.; Lacour, S.P.; Leterrier, Y. Recent Advances in Encapsulation of Flexible Bioelectronic Implants: Materials, Technologies, and Characterization Methods. Adv. Mater. 2022, 34, 2201129. [Google Scholar] [CrossRef]
- Soares dos Santos, M.P.; Bernardo, R.M. Bioelectronic Multifunctional Bone Implants: Recent Trends. Bioelectron. Med. 2022, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Bettucci, O.; Matrone, G.M.; Santoro, F. Conductive Polymer-Based Bioelectronic Platforms Toward Sustainable and Biointegrated Devices: A Journey from Skin to Brain Across Human Body Interfaces. Adv. Mater. Technol. 2022, 7, 2100293. [Google Scholar] [CrossRef]
- Nalesso, F.; Garzotto, F.; Cattarin, L.; Bettin, E.; Cacciapuoti, M.; Silvestre, C.; Stefanelli, L.F.; Furian, L.; Calò, L.A. The Future for End-Stage Kidney Disease Treatment: Implantable Bioartificial Kidney Challenge. Appl. Sci. 2024, 14, 491. [Google Scholar] [CrossRef]
- Oh, S.; Jekal, J.; Liu, J.; Kim, J.; Park, J.U.; Lee, T.; Jang, K.I. Bioelectronic Implantable Devices for Physiological Signal Recording and Closed-Loop Neuromodulation. Adv. Funct. Mater. 2024, 34, 2403562. [Google Scholar] [CrossRef]
- Berggren, M.; Głowacki, E.D.; Simon, D.T.; Stavrinidou, E.; Tybrandt, K. In Vivo Organic Bioelectronics for Neuromodulation. Chem. Rev. 2022, 122, 4826–4846. [Google Scholar] [CrossRef]
- Kishore, A.; Varughese, A.M.; Roth, B.; Zeilinger, C. Fabrication of a Low-Cost Benchtop Optical Imager for Quantum Dot Microarray-Based Stress Biomarker Detection. Biomed. Opt. Express 2024, 15, 4147–4161. [Google Scholar] [CrossRef]
- Dal Din, A.; Amin, O.J.; Wadley, P.; Edmonds, K.W. Antiferromagnetic Spintronics and Beyond. npj Spintronics 2024, 2, 25. [Google Scholar] [CrossRef]

| Characteristic | Molecular Computing | MOSFET Computing |
| Scale | Operates at molecular (nanometer to sub-nanometer) scale, enabling extremely high density of computation | Operates at micrometer scale; scaling is limited by lithographic processes and quantum tunnelling effects |
| Energy Efficiency | Potentially very low energy consumption due to chemical and biological interactions | Energy-intensive due to resistive losses and constant power requirements in high-speed circuits |
| Processing Speed | Relatively slow, limited by chemical reaction rates and molecular diffusion | Fast, with switching speeds in the gigahertz range |
| Complexity of Fabrication |
Can be self-assembled using chemical processes; fabrication is still experimental and less mature | Mature and standardized fabrication using well-established semiconductor manufacturing processes |
| Logic Operations | Can perform complex parallel operations inherently due to molecular interactions | Sequential logic operations using binary signals (0 and 1) |
| Re-programmability | Reprogramming requires modifying the molecular environment or sequences; less straightforward | Easily reprogrammable through software and hardware updates |
| Robustness | Susceptible to environmental conditions like temperature, pH, and contamination | Highly robust under controlled conditions; less sensitive to minor environmental changes |
| Integration Density | Extremely high, as molecules are orders of magnitude smaller than transistors | High but limited by physical dimensions of MOSFETs and wiring complexity |
| Cost | Currently high due to its experimental nature; expected to decrease with advances in chemical synthesis | Cost-effective due to economies of scale in semiconductor manufacturing |
| Parallelism | Intrinsically parallel due to simultaneous molecular interactions | Parallelism requires hardware design like multicore processors and is less inherently parallel |
| Error Handling | Error-prone due to the stochastic nature of chemical reactions; requires redundancy or error-correction mechanisms | Mature error-handling techniques are built into hardware and software |
| Power Source | Energy is derived from chemical reactions, light, or molecular interactions | Electrical power is supplied by external power sources |
| Scalability | High potential for scalability to molecular levels; still experimental | Limited scalability as transistor sizes approach physical and quantum limits |
| Type |
Inorganic Molecules |
Carbon-based Inorganic Molecules |
Organic Molecules |
Hybrid Systems |
| Logic Gates | Molecules based on transition metals (e.g., ruthenium complexes) | Molecular switches (e.g., rotaxanes) | DNA logic gates | DNA-metal nanoparticle conjugates |
| Memory Devices | Metal-organic frameworks (MOFs) | Organic thin-film transistors | Protein-based memory systems | DNA-templated nanowires |
| Switches | Photochromic inorganic compounds | Fullerene derivatives (e.g., C60) | Light-activated proteins (e.g., rhodopsin) | DNA-linked quantum dots |
| Computing Systems | Quantum dot arrays | Molecular tweezers or cages | DNA strand displacement systems | DNA-organic molecule complexes |
| Energy Systems |
Semiconductor materials for solar cells | Organic photovoltaics | Photosynthetic proteins | Biohybrid solar cells with photosystems |
| Signal Amplification |
Catalytic systems with metal ions | Amplified chemical reactions | Enzyme cascades | Metal-enzyme hybrids |
| Artificial Neural Networks |
Memristors using transition metal oxides | Organic polymers | Neural networks with bio-mimicking peptides | Organic-inorganic hybrid memristors |
| Molecule/System | Mechanism | Application |
Molecular Examples/Names |
| Rotaxanes | Conformational switching between mechanically interlocked molecular components | Logic gates, molecular memories | Rotaxane, Stoddart-type rotaxanes |
| Photochromic Dyes | Light-induced isomerization between distinct molecular states |
Optical computing, data storage | Azobenzene, Spiropyran, Diarylethenes |
| Semiconductors: Carbon Nanotubes (CNTs) and Fullerenes | Electron conduction via π-π stacking in delocalized π-electron systems | Transistors, flexible electronics | Pentacene, P3HT (Poly(3-hexylthiophene)); Multi-walled CNTs |
| Photosensitive and Luminescent Molecules | Light absorption and emission, or photoswitching between states |
Displays, sensors, molecular probes, carbon LED | Carbon Dots (CDs), Graphene Quantum Dots (GQDs), CNTs |
| Carbon Triboelectric Nanogenerators (TENGs) | Harvesting mechanical energy via triboelectric charge generation and transfer |
Wearable electronics, self-powered sensors |
Graphene, Carbon Nanotubes (CNTs), Carbon Dots (CDs), Diamond-like Carbon (DLC) films |
| Dimensionality | Applications in Molecular Computing | Examples | Mechanisms Utilized |
| 0D (Zero-Dimensional) | - Carbon Quantum Dots for logic operations - Single-electron transistors (SET) |
- Carbon Quantum Dots (CQDs) - Graphene Quantum Dots (GQDs) - Fullerenes |
Quantum confinement, electron tunnelling, charge storage |
| 1D (One-Dimensional) | - Nanowire-based logic gates. - Molecular interconnects in circuits |
- Carbon Nanotubes (SWCNTs, MWCNTs) - Carbon Nanofibers |
Ballistic electron transport, low resistance conduction |
| 2D (Two-Dimensional) | - High-speed transistors for molecular computing - Flexible logic arrays |
- Graphene - Graphene Oxide (GO) - Reduced Graphene Oxide (rGO) |
High carrier mobility, tunable bandgap (GO/rGO) |
| 3D (Three-Dimensional) | - Neuromorphic computing architectures. - Memory devices with large capacity |
- Diamond - Graphite - Amorphous Carbon - Carbon Aerogels - Carbon Foams |
High surface area for data storage, hierarchical connectivity |
| 3D Hierarchical | - Hybrid logic systems. - Multi-functional computational networks |
- CNT-Graphene Hybrids - Porous Carbon Frameworks - Carbon Nano-onions (CNOs) |
Synergistic properties of combined dimensions |
| System | Molecules Used | Mechanism | Application |
| DNA Strand Displacement | DNA | Hybridization and displacement reactions are where an invading strand displaces a pre-bound strand, releasing it for further reactions | Logic gates, parallel computation, molecular circuits, data storage. |
| DNA Origami Circuits | DNA | DNA self-assembly into nanoscale structures capable of logic operations | Nanorobotics, programmable matters, drug delivery. |
| Ribozymes | RNA | RNA molecules that catalyze specific chemical reactions by folding into unique 3D structures |
Biosensing, RNA-based circuits, gene regulation, synthetic biology |
| CRISPR-Cas Systems | CRISPR RNA, Cas proteins | Sequence-specific DNA targeting for gene editing and programmable logic gates | Gene regulation, synthetic circuits, biological memory |
| Enzyme Cascades | Enzymes (e.g., polymerases, kinases) | Sequential catalytic reactions where the product of one enzymatic step acts as the substrate for the next, amplifying signals |
Biochemical computing, diagnostic tools, metabolic pathway analysis |
| Biomolecular Photon Absorption/Emission | Fluorescent proteins, quantum dots | Absorption or emission of photons for signalling and detection | Bio-imaging, molecular sensors, real-time monitoring |
| Biomolecular Wiring | Conductive polymers, protein nanowires | Conductive pathways are formed by biomolecules for signal transmission | Nanoelectronics, biosensors, molecular-scale circuits |
| Peptide Computing | Peptides, Engineered proteins | Sequence-specific interactions and self-assembly for information processing, Protein-protein interactions and allosteric changes to perform logical operations |
Logic gates, Molecular pattern recognition, targeted drug delivery, Peptide Nucleic Acids (PNAs) |
| Lipid Bilayer Systems | Lipids | Formation of lipid bilayers for compartmentalization and regulation of molecular diffusion and signalling pathways |
Signal transduction in biosensors, microfluidics, synthetic cells |
| Cell-based biological computing, Bacterial Quorum Sensing | Signalling molecules, proteins |
Cell-to-cell communication using diffusible molecules for collective behaviour control. |
Synthetic biology, population control in biosensors. |
| Type of Logic Gate | Type of molecule | Way of Functioning |
| AND | DNA strands with specific sequences | It requires both inputs to be present for the output to form. The DNA strands input have complementary sequences to parts of an output strand. They only form when both inputs bind, creating a new duplex DNA |
| OR | Multiple DNA strands with overlapping sequences | Due to the shared binding regions, each input DNA strand can bind to a segment of the output strand. Thus, the output is produced if either or both inputs are present |
| NOT | DNA with toehold-mediated strand displacement | The output is pre-formed in a complex with another strand. The input strand, when present, displaces the output strand through a 'toehold' (a short single-stranded region that initiates strand displacement), turning off the output by releasing it from the complex |
| NAND | Combination of AND and NOT by DNA strand displacement | AND gate is followed by a NOT gate. DNA strands interact to form an output if both inputs are present (AND), but the other DNA strand displacement reaction then suppresses the output; if both inputs are present (NOT), thus only producing an output if at least one input is missing |
| NOR | Combination of OR and NOT by DNA interactions | Combination of OR gate with NOT gate. Any input produces an output in the OR part, which is negated by a NOT mechanism (strand displacement) if any input is present. Output appears only when no inputs are present |
| XOR | Complex DNA networks or cascades | Complex DNA interactions where outputs vary depending on which inputs are present. It may involve multiple steps where different DNA strands interact in a cascade manner. An output is present only when one but not both inputs are present through the series of strand displacements or catalytic reactions |
| Category | CMOS-Molecular Devices | CMOS-Biomolecular Hybrid Computing |
| Core Components | CMOS transistors, molecular crossbar arrays, nanowires, redox-active molecules, non-crystal molecular layers | CMOS transistors, DNA strands, peptides, enzymes, protein-based gates, vesicules, cells |
| Molecular Elements, examples | Fullerenes, CNTs, CQDs, TCNQ, ruthenium complexes, alkylthiols, ferrocene | DNA, RNA, Peptide Nucleic Acids (PNAs), Enzymes |
| Mechanisms | - Charge transfer & tunnelling; - Single-electron effects; - Resistive switching; - Spin-based logic - Photonic |
- DNA strand displacement for logic operations; - DNA origami patterns - Enzymatic reactions for data storage - Protein-based logic gates - Vesicular or cellular circuits - Photonic |
| Possible Device Architecture | Molecular crossbar array with interface CMOS; 2-layered structures; CMOS interface with different molecular or nanoparts | Biochemical layers (DNA, enzymes, lipids) interfaced with CMOS circuits; CMOS interface |
| Data Storage Mechanism | Molecular charge trapping and resistive switching (ReRAM); photoelectric memory; spintronic-electronic memory | Biochemical reactions storing data as nucleotide sequences; photoelectric memory |
| Advantages | - Ultra-high density memory; - Low power consumption; - Fault-tolerant logic; - Scalable beyond silicon limits |
- Biocompatibility for biomedical applications; - High parallelism; - Self-assembly of biomolecules |
| Challenges | - Precise molecular alignment issues; - Molecular degradation; - Complex fabrication |
- Molecular degradation in non-aqueous conditions; - Limited speed due to biochemical reactions |
|
Possible Applications |
- High-density non-volatile memory (NVM); - Neuromorphic computing - Quantum computing; - AI accelerators |
- Biosensors and medical diagnostics; - DNA-based cryptography; - Biocompatible computing devices |
| Energy Efficiency | Ultra-low power due to single-electron effects and resistive switching | Very low power but slower due to biochemical reaction rates |
| Fault Tolerance | High fault tolerance due to reconfigurable crossbar arrays | Error-prone due to biochemical reaction noise and degradation |
| Fabrication Complexity | High: molecular alignment, nanoscale precision required | Moderate: self-assembly properties of biomolecules |
| Scalability | Ultra-high density, nanoscale compatible | High, but limited by biochemical stability and speed |
| Emerging Research Focus Areas | - 3D CMOL stacks for ultra-dense memory; - Hybrid spintronics |
- DNA-based parallel computing; - Bio-neuromorphic architectures |
| Application | Mechanism of Action | System/Device | Molecular Compound/Method |
|
Biosensing and Diagnostics |
Molecular sensors detect biomarkers and generate electronic signals |
CMOS biochip with molecular sensors | Aptamers, antibodies, DNA probes |
|
Targeted Drug Delivery |
Electronic control triggers drug release via molecular valves or nanocarriers. | Electrostatically controlled nanocarriers | Liposomes, dendrimers, electro-responsive polymers |
| Electrogenetic and optogenetic Control Systems | Electrical signals regulate genes expression through engineered circuits. |
Bioelectronic hybrid with genetic circuits | Electrogenetic switches, CRISPR, redox compounds |
| Implantable Biomodulation | Electrical stimulation modulates nerve activity for therapeutic intervention. | CMOS-integrated nerve stimulator | Conductive polymers, carbon nanotubes |
| Real-Time Cellular Monitoring | Cells tagged with reporters emit signals monitored by CMOS sensors. |
CMOS bioimager with fluorescent reporters | Quantum dots, fluorescent proteins, FRET sensors |
| Targeted Therapy | Molecular compounds respond to electronic stimulation to generate localized heat. |
CMOS-controlled magnetic nanoparticle array | Iron oxide nanoparticles, gold nanoshells, thermoresponsive liposomes |
| Type of Biosensor | Sensing Mechanism | Applications |
| Nucleic Acid-Based Biosensors | DNA, RNA, and PNA probes to detect complementary nucleic acid sequences |
Genetic testing, pathogen identification |
| Antibody-Based Biosensors | Employ antibodies to specifically bind antigens, producing a detectable signal |
Pathogen detection, biomarker diagnostics |
| Cell-Based Biosensors | Use whole cells to monitor cellular responses to toxins, drugs, or environmental changes | Drug testing, toxin detection, cellular research |
| Tissue-Based Biosensors | Use biological tissue slices for broader metabolic sensing and functional assays |
Metabolic studies, experimental biology |
| Biomimetic Sensors | Use synthetic materials or molecular imprints that mimic biological recognition | Environmental monitoring, chemical sensing |
| Classification Type | Subtype | Mechanism Description | Examples |
| Based on Release Mechanism | Diffusion-controlled release | Drug diffuses out based on the concentration |
Reservoir matrix, hydrogel, polymeric nanoparticles |
|
Swelling and/or erosion-controlled release |
Matrix swells and/or degrades, and the drug is released | Biodegradable hydrogels, PLGA microparticles |
|
|
Chemical- controlled release |
Release by bond cleavage or polymer degradation | Drug-polymer conjugates, prodrugs |
|
| Stimuli-responsive release, molecular valve | Triggered by pH, temperature, light, magnetic field | mesoporous silica nanoparticle (MSN), pH-sensitive micelles, thermoresponsive liposomes |
|
| Osmotic pressure-driven release | Water influx creates pressure to push the drug out | Osmotic pumps (e.g., OROS systems) | |
| Enzyme-activated release | Enzymes trigger the release | Enzyme-responsive hydrogels |
|
| Combination Mechanisms | Multiple mechanisms combined for enhanced control | pH and temperature- sensitive liposomes |
|
| Based on mechanism of action |
Receptor agonism or antagonism | Drugs activate or block receptors to modulate cellular signaling pathways |
β-blockers (propranolol), opioids (morphine) |
| Enzyme inhibition | Inhibits specific enzymes, preventing the conversion of substrates into products |
ACE inhibitors (lisinopril), statins (atorvastatin) |
|
|
Ion channel modulation |
Modulates ion channels to alter ion flow, affecting cellular excitability and signaling |
Calcium channel blockers (amlodipine), lidocaine |
|
| Nucleic acid interaction |
Drugs that bind or modify nucleic acids to inhibit transcription; gene expression modulation |
Cisplatin (DNA cross-linker), doxorubicin; siRNA therapy (patisiran), HDAC inhibitors (vorinostat) |
|
| Cytotoxic or cytostatic action |
Induces cell death or inhibits cell proliferation |
Paclitaxel, methotrexate | |
| Hormone modulation or replacement | Modifies hormone levels either by supplementation or inhibition |
Insulin, tamoxifen | |
| Protein binding or sequestration | Binds proteins to inhibit their function or prevent their interaction with other molecules |
TNF Inhibitors (infliximab), paclitaxel |
|
| Based on Type of Targeting | Passive targeting | Utilizes physiological barriers: EPR effect) | Liposomes, polymeric Mmcelles |
| Active targeting | Receptor or ligand binding for specific cell interaction | Antibody-drug conjugates (ADCs) |
|
| Inverse targeting | Avoiding healthy cells interaction | Polyethylene glycol (PEG) coating or PEGylation |
|
| Physical Targeting | External triggers, such as heat, magnetic field, US, Photodynamic therapy (PDT), Photothermal therapy (PTT), electroporation; mechanical |
Magnetic nanoparticles, US-triggered microbubbles or sonoporation; cell apoptosis through the PDT or PTT; microneedles, micro-jets | |
| Based on Type of Carrier System | Nanoparticles | Nano-sized carriers for drug encapsulation | Polymeric Nanoparticles, Lipid Nanoparticles |
|
Microspheres, Microcapsules |
Microscale carriers for larger payloads. | PLGA Microspheres, Calcium Alginate Beads | |
| Liposomes | Lipid bilayer vesicles for drug delivery | Doxil (Doxorubicin Liposome) |
|
|
Polymeric Conjugates |
Drugs chemically linked to polymers. | PEGylated Proteins (PEG-IFN), HPMA Conjugates |
|
|
Hydrogels, Dendrimers |
Water-swollen networks & branched polymers | PAMAM Dendrimers, Smart Hydrogels | |
| Based on Site of Action |
Intracellular Delivery |
Targets drug release inside cells | Liposomes, Antibody-Drug Conjugates |
|
Extracellular Delivery |
Releases drugs outside the cells |
Collagen-Based Drug Systems, Hydrogels | |
|
Organ-Specific Delivery |
Targets specific organs or tissues | Liver-Targeting Nanoparticles, Brain-Targeting Liposomes |
| Type of Carrier System | Description | Applications |
| Liposomes | Spherical vesicles composed of lipid bilayers encapsulating drugs |
Cancer therapy, gene delivery |
| Micelles | Amphiphilic molecules form nanosized spherical structures for hydrophobic drugs |
Cancer treatment, antimicrobial delivery |
| LPNs, SLNs, NLCs | Lipid nanoparticles for encapsulation |
mRNA |
| Nanoparticles, nanotubes | Solid colloidal particles used for controlled drug release and targeting | Tumor targeting, vaccine delivery |
| Dendrimers | Branched macromolecules with controlled architecture for drug conjugation | Gene therapy, anticancer drug delivery |
| Polymeric Carriers | Biodegradable polymers used for sustained and targeted drug release | Chronic disease treatment, cancer therapy |
| Microspheres | Small spherical particles used for controlled drug delivery |
Hormonal therapy, vaccine delivery |
| Cell-based delivery and viral vector | Macrophages and stem cells as drug delivery system; DNA-engineered retrovirus | Oncology, immunology |
| Technique | CMOS/Molecular Components | Mechanism of Action |
Condition/ Organ |
Pathologies/ Conditions Treated |
| Magnetogenetics | Magnetic nanoparticles, magnetically sensitive ion channels (TRPV1), CMOS magnetic field sensors |
Magnetic field activates ion channels for neural stimulation |
Brain, spinal cord, peripheral nerves |
Parkinson’s, depression, chronic pain |
| Thermogenetics | Thermo-responsive proteins (e.g., TRPV1, TRPM8), CMOS thermal sensors, plasmonic nanoparticles | Heat-activated ion channels modulate cell activity | Brain, skin, muscle |
Epilepsy, neuropathic pain, skin cancer |
| Mechanogenetics | Mechanosensitive ion channels (e.g., Piezo1, TREK-1), stretchable CMOS devices, pressure-sensitive nanoparticles |
Mechanical force activates ion channels for cellular control | Muscle, heart, skin, bone | Muscular dystrophy, cardiac arrhythmias, osteoarthritis |
| Sonogenetics | Ultrasound- sensitive proteins (e.g., prestin, TRPA1), CMOS ultrasound transducers, gas vesicle nanoparticles (GVNPs) |
Focused ultrasound activates ion channels for cellular response |
Brain, liver, muscle |
Epilepsy, liver diseases, oncology |
| Electrogenetics | Electrosensitive ion channels (e.g., K2P, NaV), CMOS microelectrode arrays, conductive polymers (PEDOT) | Electrical stimulation induces ion flow and gene expression; DART | Brain, heart, spinal cord |
Epilepsy, cardiac arrhythmias, paralysis, brain disorders |
| Optogenetics | Opsins (e.g., Channelrhodopsin, Halorhodopsin), CMOS micro-LED arrays, light-sensitive proteins | Light stimulation activates opsins for ion flow modulation |
Brain, retina, spinal cord | Parkinson’s, retinal blindness, epilepsy, mood disorders |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





