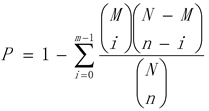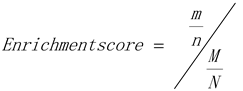Introduction
Asthma is one of the chronic diseases with high morbidity and high mortality worldwide [
1].Asthma is highly heterogeneous, and some patients are complicated with allergic diseases, other respiratory diseases, even gastrointestinal diseases[
2,
3], resulting in poor prognosis and difficult to control of asthma. Our previous clinical work paid attention to patients with asthma complicated with inflammatory bowel disease. The attack of asthma also deteriorated the inflammatory bowel disease. Besides, there were relevant studies suggesting that asthma patients had an increased risk of inflammatory bowel disease[
4,
5]. However, it is not clear the underlying mechanism that IBD affects the pathogenesis of bronchial asthma. Therefore, in-depth study of the pathogenic factors of bronchial asthma combined with IBD can help clarify the pathogenesis of bronchial asthma associated with IBD, which has clinical value for the prevention and treatment of heterogeneous asthma.
Both asthma and IBD are chronic inflammatory diseases with similarities in immune, genetic and environmental factors. This study aimed to explore the role of genetic factors in the pathogenesis of asthma combined with IBD and to identify the differentially expressed genes related to asthma complicated with IBD by Gene Expression Omnibus database (GEO). Due to the lack of data related to patients with asthma combined with IBD in the database, differentially expressed genes were obtained from the datasets of asthma and IBD, respectively. The intersected genes that were differentially expressed in both asthma and IBD were ultimately screened. This study also aimed to analyze the biological functions of intersected genes, especially their roles in immune mechanisms, explore potential regulatory molecular targets, and provide new ideas for the diagnosis and treatment of asthma associated with IBD.
Materials and Method
Data Collection
Inclusion and exclusion criteria: Transcriptome datasets of asthma, inflammatory bowel disease were downloaded from GEO database (
http://www.ucbi.nlm.nih.gov/geo), respectively. ①Inclusion criteria were as follow: datasets from patients with diagnosed asthma associated with inflammatory bowel disease, including blood samples and RNA datasets; ② Exclusion criteria were as follow: age < 18 years old; sputum sample dataset; fecal sample dataset; without control group; DNA dataset.
Asthma: The asthma-related transcriptome dataset (GSE134544) was downloaded from the GEO database after screening. The post-treatment data were removed. Ten cases of asthma (GSM3955526, GSM3955437, GSM3955611, GSM3955462, GSM3955403, GSM3955432, GSM3955511, GSM3955531, GSM3955606, GSM3955486) were randomly selected using the random extraction function of Excel (version V3.8.7.0), as well as 10 cases in the control group (GSM3955623, GSM3955640, GSM3955633, GSM3955625, GSM3955628, GSM3955486). GSM3955636, GSM3955627, GSM3955635, GSM3955621, GSM3955637).
(3)Inflammatory bowel disease: IBD -related transcriptome dataset (GSE169568) was downloaded from the GEO dataset after screening. The post-treatment cohort (German cohort) was removed. Ten cases of IBD(GSM5209527, GSM5209460, GSM5209522, GSM5209540, GSM5209582, GSM5209430, GSM5209482, GSM5209553, GSM5209620, GSM5209613) , as well as 10 cases in the control group (GSM5209605, GSM5209611, GSM5209483, GSM5209500, GSM5209604, GSM 5209498, GSM5209499, GSM5209490, GSM5209601, GSM5209608).
Screening of differentially expressed genes
(1)Dataset processing: ① Normalization: Both two datasets belong to Affymetrix Genechip data. The level of gene expression was reflected by the fluorescence signal. The limma package in R (version 3.6.2) was used to calculate the p value and the RMA method was used for normalization. Finally, the log2 transformation was carried out.
② Principal component analysis (PCA)
The adegenet package in R was used for PCA of gene expression, to investigate the distribution of samples, and to explore the relationship among samples or verify the experimental design.
③ Analysis of differentially expressed genes
The p values of differentiated expressed genes from two group were calculated via t test in R (P < 0.05 is considered as significant difference). The differentiated expressed genes with p<0.05 and |log2 FC|>2 were selected.
④Clustering analysis The cluster package in R was used for unsupervised hierarchical clustering analysis of differentially expressed transcripts. The distance between the pairs of multiple genes was calculated to form the distance matrix. The two classes with the closest distance were merged into a new class, the distance between the new class and the current classes was calculated, and so on until there was only one class. The direct correlation of genes was measured by the expression of the selected differentially expressed transcripts.
Gene functional analysis
① The calculated result will obtain a enrichment p-value (P < 0.05), and a small p value indicates that differentially expressed genes are enriched in the GO entry. The calculation formula is as follows:GO enrichment analysis of differentially expressed genes were performed and their functions were described based on annotation. The number of differentially expressed genes included in each GO entry was counted, and the significance of differentially expressed gene enrichment in each GO entry was calculated by hypergeometric distribution test. The calculation result will return a p value of enrichment significance (P < 0.05). A small p value indicated that differentially expressed genes were enriched in the GO entry. The calculation formula was as follows:
The calculation formula of enrichment score:
Where, N is the number of genes with GO annotation; n is the number of differentially expressed genes with GO annotation in N; M is the number of genes annotated for a specific GO entry; m is the number of differentially expressed genes annotated for a specific GO term. The ggplot2 package in R was used to draw a bar chart for the TOP30 genes.
②KEGG analysis KEGG database was used for Pathway analysis of differentially expressed genes (combined with KEGG annotation), and hypergeometric distribution test was used to calculate the significance of differentially expressed genes enrichment in each Pathway entry. Finally, ggplot2 package in R was used to draw the bubble map for the TOP20 genes.
Analysis of differentially expressed intersected gene and immune cells
Cibersortx (
https://cibersortx.stanford.edu) was used to analyze the relationship between differentially expressed genes and immune cells. CIBERSORT is a web-based tool for deconvolution of the expression matrix of human leukocyte subtypes based on the principle of linear support vector regression, which can estimate the composition and abundance of immune cells in mixed cells based on transcriptome data. In this study, R software was used to analyze the correlation between the previously screened significant differentially expressed genes and immune cells. Spearman was used for correlation calculation, and p less than 0.05 indicated significant difference.
Statistical analysis
All statistical analysis and related visualization were performed using the R software (version 3.6.2).
Results
Asthma
(1)Gene expression analysis : According to the box plot (Figure 1), the median position of the data was in the middle of the box, indicating that the sample data was more concentrated and symmetrical. The median of each group was basically in the same position, and the principal component analysis showed that the samples of the control group were close to that of the case group, indicating a good similarity (Figure 2).
(2) Differential expression analysis: A total of 28402 differentially expressed genes were screened by differential expression analysis, and 157 genes with p value less than 0.05 and | log2FC |>2 were selected, of which 90 were up-regulated and 67 were down-regulated. The differences generated by comparison were reflected in the volcano map, with gray and blue as the genes with non-significant differences. The red is the display of significant differential genes, and the horizontal axis is |log2 FC|>2, and the vertical axis is log10 transform of P (Figure. 3A). In the heat map, the genes with significant differences were clustered according to expression level, with red representing high-expression transcripts and green representing low-expression (Figure. 3B).
(3)Function analysis of differentially expressed genes ①After obtaining the differentially expressed genes, we performed GO enrichment analysis on the differentially expressed genes, described their functions based on the annotation results, counted the number of differentially expressed genes included in each GO entry, and calculated the significance of differentially expressed genes in each GO entry using hypergeometric distribution test method. The results indicated that there were 843 genes with GO annotation with 670 genes with P value < 0.05, of which 547 with high expression and 221 with low expression. Gene function is mainly divided into three categories: biological process, cell composition, and molecular function. GO entries with more than 2 corresponding genes in the three categories are screened, and 10 entries are sorted from largest to smallest according to the -log10 transform of P corresponding to each entry (Figure. 4A). The TOP10 of biological processes included neutrophil downregulation, antimicrobial human response, innate mucosal immune response, anti-gram-negative bacterial response, antibacterial human response regulated by antimicrobial peptides, cell membrane interference in other organs, antibacterial human response, killing cells in other organs, antifungal response, and anti-gram-positive bacterial response. The TOP10 of cell components included specialized granule lumen, cyanophil granule lumen, tertiary granule lumen, extracellular space, extracellular region, phagocytic vesicle cavity, cyanophil granule, specialised granule, tertiary granule membrane and exosomes. The TOP10 of molecular functions included MHC Class I protein binding, protein antigen binding, carbohydrate binding, lipopolysaccharide binding, serine-type endopeptidase activity, heparin binding, transmembrane transport signal transduction receptor activity, chemokine activity, endopeptidase activity.
②KEGG database was used for Pathway analysis of differentially expressed genes. The results indicated that 149 genes were annotated in KEGG database and there were 51 genes with P value < 0.05, among which 41 were highly expressed genes and 76 were lowly expressed genes.The pathway entries with more than 2 corresponding genes were screened. According to the -log10 transform of P corresponding to each entry, the top20 for KEGG enrichment results should be sorted from largest to smallest (Figure. 4B), including Staphylococcus aureus infection, antigen expression and process, cytotoxicity of natural killer cells, graft-versus-host disease, NOD body receptor signaling pathway, transcription mismatch repair in tumors, chemokine signaling pathway, complement and coagulation cascade, phagosome, viral protein and cytokine and cytokine receptor interaction, and cytokine to cytokine receptor interaction Regulation of proteoglycan, actin cytoskeleton, RAS signaling pathway, MAPK signaling pathway, tumor signaling pathway, metabolic pathway in tuberculosis and tumor.
Inflammatory bowel disease
(1) According to the box plot (Figure 1), the median position of the data was in the middle of the box, indicating that the sample data was more concentrated and symmetrical (Figure 5). Principal component analysis showed that the distance between the samples of the control group and that of the case group was far away, indicating a low similarity (Figure. 6).
(2)Differential expression analysis A total of 11,727 differentially expressed genes were screened. Among them, 193 genes with p value less than 0.05 and | log2FC |>2 times were selected, of which 138 were up-regulated and 54 were down-regulated. The differences were reflected in the volcano map (Figure. 7A), with gray and blue genes showing non-significant differences. The red is the display of significant differentially expressed genes, and the horizontal axis is |log2 FC|>2, and the vertical axis is log10 transform of P value. In the heat map (Figure. 7B), the genes with significant differences were clustered according to expression level, with red representing high-expression transcripts and green representing low-expression transcripts.
Function analysis of differentially expressed genes ①After obtaining the differentially expressed genes, we performed GO enrichment analysis on the differentially expressed genes, and calculated the significance of differentially expressed genes in each GO entry using hypergeometric distribution test method. The results indicated that there were 1731 genes with GO annotation with 1269 genes with P value < 0.05, of which 943 with high expression and 623 with low expression. Gene function is mainly divided into three categories: biological process, cell composition, and molecular function. GO entries with more than 2 corresponding genes in the three categories are screened, and 10 entries are sorted from largest to smallest according to the -log10 transform of P corresponding to each entry (Figure. 8A). The top 10 biological processes included neutrophil downregulation, adaptive immune response, immune response, positive regulation of IL-8 secretion, toll-like receptor 4 signaling pathway, innate immune response, positive regulation of calcium ion signaling pathway, negative regulation of T cell receptor signaling pathway, mitochondrial electron transport, cytochrome c to oxygen, and cytokine production; The top 10 of cell components included T cell receptor composition, special granule lumen, tertiary granule membrane, tertiary granule lumen, immune synapses, cytoplasmic pressure particles, intrinsic components of cell membrane, cytoplasmic membrane, blood particles, and exosomes. The TOP10 of molecular functions included RAGE receptor binding, cytochrome c oxidation activity, single strand DNA binding, protein binding, non-transmembrane protein tyrosine kinase activity, transmembrane signal receptor activity, calcium ion dependent receptor activity, manganese ion binding, protein homogeneity binding, cadherin binding.
②KEGG database was used for Pathway analysis of differentially expressed genes, and hypergeometric distribution test was used to calculate the significance of enrichment in each Pathway entry. The results indicated that 176 genes were annotated in KEGG database and there were 47 genes with P value < 0.05, among which 40 were highly expressed genes and 58 were lowly expressed genes.The pathway entries with more than 2 corresponding genes were screened. According to the -log10 transform of P corresponding to each entry, the top20 for KEGG enrichment results should be sorted from largest to smallest (Figure. 8B), including coronary muscle contraction, arrhythmogenic right ventricular cardiomyopathy, PD-L1 expression and PD-L1 checkpoint pathway, hypertrophic cardiomyopathy, tension cardiomyopathy, Toll-like receptor signaling pathway, oxytocin signaling pathway, Th17 cell differentiation, HIV1 infection, Alzheimer's disease, oxidative phosphorylation, Th1 and Th2 cell differentiation, Parkinson's disease, non Alcoholic liver disease, cardiomyocyte adrenergic signaling, hematopoietic cell lineage, PI3K-Akt signaling pathway, cGMP-PKG signaling pathway, thermogenesis, NOD body receptor signaling pathway.
Intersection analysis of asthma and inflammatory bowel disease
Among the 193 differentially expressed genes screened from inflammatory bowel disease and asthma, there were 7 intersected differentially expressed genes, of which 5 genes showed high expression, such as ILMN_1654389, ILMN_1694548, ILMN_1723971, ILMN_1755843, and ILMN_1766896, 2 genes showed low expression, such as ILMN_1699695 and ILMN_2105441 (Figure. 9). The ILMN_1723971 and ILMN_1654389 were not annotated. Thus, it was concluded that differentially expressed genes including ANXA3, SLC26A8, ZDHHC19, JCHAIN, and TNFRSF21 were found in both two diseases. Unfortunately, there was no effective enrichment of the above genes in KEGG. According to GO database analysis, ANXA3 was highly expressed in asthma and IBD. TNFRSF21 showed low expression in asthma and IBD. SLC26A8 was highly expressed in both of asthma and IBD, but SLC26A8 is not in the TOP10 entries in asthma. ZDHHC19 is also highly expressed in both asthma and IBD, but ZDHHC19 is not in the TOP10 entries in both two diseases. JCHAIN showed low expression in asthma and high expression in IBD, but JCHAIN is not in the TOP10 entries in asthma.
Correlation Analysis of Intersected Genes and Immune Cells in Asthma and Inflammatory Bowel Disease
The cibersortx tool was used to analyze the relationship between intersected genes, ANXA3, ZDHHC19, TNFRSF21, including between IBD and asthma and immune cells (Figure 10). ANXA3 is mainly negatively correlated with NK cell dormancy and positively correlated with macrophage M0 phase; TNFRSF21 was mainly positively correlated with NK cell dormancy and negatively correlated with macrophage M0 phase and neutrophils; ZDHHC19 was associated with several immune cell functions, which were positively correlated with naive B cells, memory B cells, plasma cells, CD8+ T cells, Tregs, macrophage M2 polarization, mast cell activation, and eosinophil function, and negatively correlated with naive CD4+ T cells, memory CD4+ T cell dormancy, naive CD4+ T cell activation, γδT cells, monocytes, and neutrophil function.
ANXA3 was negatively associated with the γδT cells and NK cells dormancy, and positively associated with neutrophil function in asthma. TNFRSF21 was positively correlated with plasma cells and negatively correlated with neutrophil function. ZDHHC19 was negatively correlated with γδT cellS and NK cellS dormancy, and positively correlated with neutrophil function. ANXA3 did not show an intersection with immune cells in inflammatory bowel disease; TNFRSF21 was negatively correlated with NK cell activation. ZDHHC19 was negatively associated with activation of memory CD4+T cells
Discussion
Some patients suffered from heterogeneous asthma associated with other types of immune diseases such as inflammatory bowel disease, who often have poor prognosis. It is of great clinical value to investigate the pathogenesis of asthma complicated with IBD. However, there are few studies on the pathogenesis of asthma combined with IBD, and no dataset related to asthma combined with IBD has been found in GEO database.This study aimed to explore the role of genetic factors in the pathogenesis of asthma combined withIBD. Therefore, this study screened the differentially expressed genes in asthma and IBD, respectively, and then identified the intersected genes of two diseases to explore the genes that may affect the development of asthma and IBD. Finally, we found five differentially expressed genes of two diseases through bioinformatics technology, such as ANXA3, SLC26A8, ZDHHC19, JCHAIN, and TNFRSF21.
Annexin A3 (ANXA3), known as lipoprotein 3 and placental anticoagulant protein 3, is a member of the annexin superfamily. Annexin is a kind of proteins that bind to phospholipids in a Ca2+ dependent manner and participate in the regulation of phospholipids and membrane binding.
Annexin is widely found in the cells of mammals, invertebrates, fungi, plants, protists and other organisms[
6]
. Biological function of Annexin family includes regulation of cell proliferation, differentiation and apoptosis, cell membrane and cytoskeleton interaction, inflammatory response et al[
7]
. The Annexin family is divided into A, B, C, D and E classes, of which the Annexin belongs to the Class A[
8],Its main biological functions include participating in cell transport, phosphatizing of cell membranes, conducting mitotic information, anticoagulation, binding with cytoskeletal proteins, mediating leukocyte migration and inflammatory response et al[
9]. In this study, ANXA3 is overexpressed in both asthma and IBD, and is involved in neutrophil down-regulation, exosome release and resistance to bacterial infection in IBD in GO annotation. Since both asthma and IBD are chronic inflammatory diseases, ANXA3 is considered to be involved in the inflammatory responses by regulating neutrophils in asthma and IBD. There are few previous studies on the correlation between ANXA3 and asthma associated with IBD Unfortunately, in this study, KEGG analysis found that ANXA3 was not effectively enriched in both asthma and IBD.Previous studies found that ANXA3 could promote stem cell hepatocellular carcinoma through the PI3K/Akt signaling pathway[
10], breast cancer through the NF-κB signaling pathway[
8]. Over-expression of ANXA3 via JNK signaling pathway plays an important role in promoting hepatocellular liver cancer and inducing stem cell characteristics[
11]. ANXA3 can also be down-regulated by HIF signaling pathway to promote colon cancer [
12], suggesting that ANXA3 is involved in the regulation of the above signaling pathways. Besides, the PI3K/Akt and JNK signaling pathways are related to development of IBD[
13,
14] and asthma[15-17], indicating that ANXA3 may play a pathogenic role in asthma combined with IBD via above signaling pathways, which should be verified in further studies in the future.
Tumor necrosis factor receptor superfamily 21 (TNFRSF21) is a member of the tumor necrosis factor superfamily.TNF is expressed by macrophages, T cells, B cells, NK cells, mast cells, endothelial cells, fibroblasts and neuronal cells[
18], and is involved in inflammatory and immune responses. TNFRSF21, also known as death receptor 6 and a member of the TNF superfamily possessing a structural domain of intracellular death, and plays an important role in regulating transcription factor activation, apoptosis, maturation, and immune responses[
19]. This study found low expression of TNFRSF21 in both asthma and IBD. GO annotation showed TNFRSF21 function in cellular component, which maintains the integrity of the cytoplasmic membrane. Besides, TNFRSF21 is also involved in T-cell receptor signaling pathways, adaptive immune response, and axon function in IBD. Although the KEGG database was not effectively enriched for these two diseases, in terms of GO function, TNFRSF21 mainly maintained the integrity of cells. Down-regulated expression of TNFRSF21 is found in various pathological conditions, which impaired cellular integrity and led to disease. Because the TNF family plays a unique role in immunity and TNFRSF21 function in inflammatory bowel disease involves adaptive immunity and T-cell receptor signaling pathways, it is possible that TNFRSF21 may also be involved in both asthma and IBD via immunology. Gamage et al.[
20] found that overexpression of TNFRSF21 can induce apoptosis and activate the NF-κB and JNK signaling pathways, suggesting that TNFRSF21 may be involved in the pathogenesis of asthma complicated with inflammatory bowel disease by regulating the function of cell membranes through these pathways. Currently, there are few studies on TNFRSF21 in asthma and IBD, and no relevant studies have been found on the role of TNFRSF21in asthma combined with IBD. Therefore, TNFRSF21 may become a new and key target in the pathogenesis of asthma combined with inflammatory bowel disease in the future.
Zinc finger DHHC-Type palmitoyltransferase 19 (ZDHHC19), which has only been studied in glioblastoma, osteosarcoma and other tumors. In the present study, although ZDHHC19 ranked without TOP 10 in GO enrichment, ZDHHC19 was overexpressed in both asthma and IBD. Its functions were enriched in biological processes such as peptidyl-L-cysteine S-palmitoylation, protein cysteine S-palmitoyltransferase activity, and protein-targeting to membranes, suggesting that the function of ZDHHC19 was same in the two diseases. S-palmitoylation is a post-translational protein modification catalyzed by a family of zinc-finger Asp-His-Cys domains containing palmitoyltransferases [21-23].This family contains 23 human DHHC proteins, and the vast majority of these proteins are modified by DHHC family members. DHHC performs S-palmitoylase functions on the cell surface, where they transfer fatty acid acyl groups from acyl coenzyme A to protein substrates[
24]. Current studies have found that abnormal palmitoylation and dysregulation of DHHC were associated with cancer [
25]. FAN[
26] et al found that ZDHHC19 promotes glioblastoma through activation of the TGF-β signaling pathway, it shows that ZDHHC19 could promote glioblastoma by activating the TGF-β signaling pathway. We further found that ZDHHC19 could be involved in leukotriene metabolism and is involved in various immune diseases such as asthma, arthritis and IBD through the LTD4 pathway through the website (
https://www.proteinatlas.org /ENSG00000163958-ZDHHC19/interaction
/Leukotriene+metabolism). Given that ZDHHC19 was not found in the KEGG databases for both diseases in this study, corresponding studies based on the above pathways could be conducted in the future.
In this study,Solute carrier family 26 member 8 (SLC26A8) was overexpressed in both asthma and IBD.SLC26A8 is a member of a multimember family of carriers with transmembrane delivery functions. SLC26A8 is involved mainly transmembrane transport of anions, oxalates and other ionsin in asthma, whereas in IBD, SLC26A8 only acted as a protein-binding molecule. SLC26A8 functions in asthma and IBD do not significantly intersect. It is considered that SLC26A8 may not be an important gene in the pathogenesis of asthma combined with inflammatory bowel disease.
JCHAIN, also known as J-chain immunoglobulin, is an evolutionarily conserved 15 kD protein that links IgA and IgM monomers into pentamers and dimers, respectively. JCHAIN is expressed at all stages of B-lymphocyte differentiation and thymocyte development, and its only known function is to promote secretory IgA production[
27,
28]. In this study,JCHAIN showed low expression level in asthma, with no effective enrichment in GO analysis and KEGG analysis. In inflammatory bowel disease, JCHAIN was associated with innate immunity with high expression level, which was considered to be mainly associated with IgA secretion. Since JCHAIN was not effectively enriched in both GO and KEGG in asthma, suggesting that JCHAIN is only associated with IBD. Thus, it is considered that JCHAIN may not be an important gene contributing to the pathogenesis of asthma combined with IBD.
This study screened three differentially expressed genes in the transcriptome of asthma and IBD, including ANXA3, TNFRSF21 and ZDHHC19. The relationship between the above three genes and the immune cells was analyzed by cibersortx. Results indicated that the roles of these three genes in the immune mechanism are not completely consistent. From the correlation analysis between ANXA3, TNFRSF21, ZDHHC19 and immune cells, it is seen that ZDHHC19 plays more roles in immunity, and there is no obvious functions intersection among three genes.
ANXA3 is negatively correlated with NK cell dormancy and γδT cells, and positively correlated with neutrophil function in asthma. Given that asthma is divided into eosinophilic type and neutrophil type, ANXA3 maybe closely related to neutrophil type, which may be a potential therapeutic target for neutrophil type in the future. However, in IBD, ANXA3 has not been shown to play a role in the immune process. Therefore, in this study, ANXA3 is considered to have no role in the pathogenesis via immunity, and may not be the factor in the pathogenesis of asthma complicated with IBD.
TNFRSF21 in asthma is positively correlated with plasma cells and negatively correlated with neutrophil function. Based on analysis of asthma typing and differential expression, this study found low expression of TNFRSF21 in asthma, which contributes to the over-differentiation of neutrophils and leads to the occurrence of the disease. Thus, it is believed that TNFRSF21 plays an important role in the pathogenesis of asthma. However, in IBD, TNFRSF21 is negatively correlated with the activation of NK cells. In patients with IBD, there are a large number of NK cells infiltrated in the intestinal mucosal tissues, which can directly kill the targeted cells by secreting cytotoxic killing proteins and can secrete pro-inflammatory mediators, participating in the inflammatory damage of intestinal mucosa[
29].Therefore, based on the differential expression analysis, the low expression of TNFRSF21 in IBD led to high expression of NK cells, thus promoting the disease. TNFRSF21 is related to immune cell function in both two diseases, it is believed that the low expression of TNFRSF21 may be a factor leading to immune dysfunction and thus causing the pathogenesis of asthma combined with IBD, which promotes the over-differentiation of neutrophils in respiratory tissues and the high expression of NK cells in the intestinal tissues.
The ZDHHC19 is also negatively associated with NK cell dormancy, γδ T cells, and positively associated with neutrophil function in asthma, which is similar to the ANXA3 in terms of immune function. However, ZDHHC1 is negatively associated with memory CD4+ T cell activation in IBD. Inflammatory bowel disease is mainly T-cell-mediated allergic reaction mainly characterized by single nucleated cell infiltration, in which the role of CD4+ T cells in the pathogenesis of IBD is complex. Th1/Th2 imbalance and increased differentiation of Th17 are related to IBD, and the Treg mediates immune tolerance, which plays an inhibitory role in the pathogenesis of IBD[
30].The current results did not clarify the relationship between the ZDHHC19 and CD4+ T-cell subsets, so the role of ZDHHC19 in immune regulation in IBD still needs to be confirmed by further experiments. Similarly, for asthma combined with IBD, the role of ZDHHC19 in the immune mechanism of pathogenesis remains unclear.
There are also shortcomings in this study. First, due to the small amount of data screened, the results may be biased, and the results may be affected if sample size is sufficient. Second, this study did not perform clinical verification, and we will verify the identified three genes, as well as pathways and mechanisms in subsequent studies.
In this study, ANXA3, TNFRSF21 and ZDHHC19, which are differentically expressed genes in the transcriptomes of asthma associated with inflammatory bowel disease, were screened by bioinformatics technology. These genes may contribute to the disease development via cell membrane function and S-palmitoylation. The low expression of TNFRSF21 may be a factor leading to immune dysfunction and the pathogenesis of asthma associated with inflammatory bowel disease. In the future, more further studies on the pathogenesis of the gene on asthma, inflammatory bowel disease, and asthma associated with inflammatory bowel disease will be conducted.
Author Contributions
(I) Conception and design: Xiaohong Yang and Chao Wu. (II) Administrative support: Liping Chen. (III) Provision of study materials or patients: Yu Tian. (IV) Collection and assembly of data: Yu Tian and Zhichuang Lian. (V) Data analysis and interpretation: Yu Tian and Zhichuang Lian. (VI) Manuscript writing: All authors. (VII) Final approval of manuscript: All authors.
Funding
This study was supported by Tianshan medical and health talents training program of Xinjiang Uygur Autonomous Region(Project No: TSYC202301B136) and Tianshan innovation team program of Xinjiang Uygur Autonomous Region(Project No: 2022D14006).
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable.
Availability of data and materials
All data generated or analyzed during this study are included in this article and supplementary information files.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- Huang, K.; Yang, T.; Xu, J.; Yang, L.; Zhao, J.; Zhang, X.; Bai, C.; Kang, J.; Ran, P.; Shen, H.; et al. Prevalence, risk factors, and management of asthma in China: a national cross-sectional study. Lancet 2019, 394, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Nishida, C.; Tomonaga, T.; Izumi, H.; Wang, K.Y.; Higashi, H.; Ishidao, T.; Takeshita, J.I.; Ono, R.; Sumiya, K.; Fujii, S.; et al. Inflammogenic effect of polyacrylic acid in rat lung following intratracheal instillation. Part Fibre Toxicol 2022, 19, 8. [Google Scholar] [CrossRef] [PubMed]
- Freuer, D.; Linseisen, J.; Meisinger, C. Asthma and the risk of gastrointestinal disorders: a Mendelian randomization study. BMC Med 2022, 20, 82. [Google Scholar] [CrossRef] [PubMed]
- Kuenzig, M.E.; Bishay, K.; Leigh, R.; Kaplan, G.G.; Benchimol, E.I. Co-occurrence of Asthma and the Inflammatory Bowel Diseases: A Systematic Review and Meta-analysis. Clin Transl Gastroenterol 2018, 9, 188. [Google Scholar] [CrossRef] [PubMed]
- Barbiellini Amidei, C.; Zingone, F.; Zanier, L.; Canova, C. Risk of Prevalent Asthma among Children Affected by Inflammatory Bowel Disease: A Population-Based Birth Cohort Study. Int J Environ Res Public Health 2020, 17. [Google Scholar] [CrossRef] [PubMed]
- Dallacasagrande, V.; Hajjar, K.A. Annexin A2 in Inflammation and Host Defense. Cells 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Cesarman, G.M.; Guevara, C.A.; Hajjar, K.A. An endothelial cell receptor for plasminogen/tissue plasminogen activator (t-PA). II. Annexin II-mediated enhancement of t-PA-dependent plasminogen activation. J Biol Chem 1994, 269, 21198–21203. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, A. Role of annexin A3 in breast cancer (Review). Mol Clin Oncol 2022, 16, 111. [Google Scholar] [CrossRef]
- Wang Jiaojiao, Y.K. Research progresses of Annexin A3 in malignant tumors. Med Postgra 2019, 32, 6. [Google Scholar]
- Guo, C.; Li, N.; Dong, C.; Wang, L.; Li, Z.; Liu, Q.; Ma, Q.; Greenaway, F.T.; Tian, Y.; Hao, L.; et al. 33-kDa ANXA3 isoform contributes to hepatocarcinogenesis via modulating ERK, PI3K/Akt-HIF and intrinsic apoptosis pathways. J Adv Res 2021, 30, 85–102. [Google Scholar] [CrossRef] [PubMed]
- Tong, M.; Fung, T.M.; Luk, S.T.; Ng, K.Y.; Lee, T.K.; Lin, C.H.; Yam, J.W.; Chan, K.W.; Ng, F.; Zheng, B.J.; et al. ANXA3/JNK Signaling Promotes Self-Renewal and Tumor Growth, and Its Blockade Provides a Therapeutic Target for Hepatocellular Carcinoma. Stem Cell Reports 2015, 5, 45–59. [Google Scholar] [CrossRef]
- Du, K.; Ren, J.; Fu, Z.; Wu, X.; Zheng, J.; Li, X. ANXA3 is upregulated by hypoxia-inducible factor 1-alpha and promotes colon cancer growth. Transl Cancer Res 2020, 9, 7440–7449. [Google Scholar] [CrossRef] [PubMed]
- Lou, X.; Zhu, H.; Ning, L.; Li, C.; Li, S.; Du, H.; Zhou, X.; Xu, G. EZH2 Regulates Intestinal Inflammation and Necroptosis Through the JNK Signaling Pathway in Intestinal Epithelial Cells. Dig Dis Sci 2019, 64, 3518–3527. [Google Scholar] [CrossRef]
- Larabi, A.; Barnich, N.; Nguyen, H.T.T. New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy 2020, 16, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Athari, S.S.; Mehrabi Nasab, E.; Zhao, L. PI3K/AKT/mTOR and TLR4/MyD88/NF-κB Signaling Inhibitors Attenuate Pathological Mechanisms of Allergic Asthma. Inflammation 2021, 44, 1895–1907. [Google Scholar] [CrossRef]
- Song, Y.; Wang, Z.; Jiang, J.; Piao, Y.; Li, L.; Xu, C.; Piao, H.; Li, L.; Yan, G. DEK-targeting aptamer DTA-64 attenuates bronchial EMT-mediated airway remodelling by suppressing TGF-β1/Smad, MAPK and PI3K signalling pathway in asthma. J Cell Mol Med 2020, 24, 13739–13750. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Su, J.; Zhao, W.; Deng, Z.; Wang, P.; Dong, H.; Zhao, H.; Cai, S. JNK modulates RAGE/β-catenin signaling and is essential for allergic airway inflammation in asthma. Toxicol Lett 2021, 336, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Hayden, M.S.; Ghosh, S. Regulation of NF-κB by TNF family cytokines. Semin Immunol 2014, 26, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Z, L.; X, J.; Y, L. Expression and clinical significance of tumor necrosis factor receptor superfamily 21 gene in hepatocellular carcinoma. Chinese Clinical Oncology 2022, 027. [Google Scholar]
- Gamage, K.K.; Cheng, I.; Park, R.E.; Karim, M.S.; Edamura, K.; Hughes, C.; Spano, A.J.; Erisir, A.; Deppmann, C.D. Death Receptor 6 Promotes Wallerian Degeneration in Peripheral Axons. Curr Biol 2017, 27, 890–896. [Google Scholar] [CrossRef] [PubMed]
- De, I.; Sadhukhan, S. Emerging Roles of DHHC-mediated Protein S-palmitoylation in Physiological and Pathophysiological Context. Eur J Cell Biol 2018, 97, 319–338. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Qiao, H.; Wang, P.; Wang, Y.; Qin, D. ZDHHC19 Is Dispensable for Spermatogenesis, but Is Essential for Sperm Functions in Mice. Int J Mol Sci 2021, 22. [Google Scholar] [CrossRef]
- Liang, S.; Zhang, X.; Li, J. Zinc finger Asp-His-His-Cys palmitoyl -acyltransferase 19 accelerates tumor progression through wnt/β-catenin pathway and is upregulated by miR-940 in osteosarcoma. Bioengineered 2022, 13, 7367–7379. [Google Scholar] [CrossRef]
- Lan, T.; Delalande, C.; Dickinson, B.C. Inhibitors of DHHC family proteins. Curr Opin Chem Biol 2021, 65, 118–125. [Google Scholar] [CrossRef]
- Gottlieb, C.D.; Linder, M.E. Structure and function of DHHC protein S-acyltransferases. Biochem Soc Trans 2017, 45, 923–928. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Fan, J.; Yang, H.; Zhao, C.; Niu, W.; Fang, Z.; Chen, X. Heterogeneity of subsets in glioblastoma mediated by Smad3 palmitoylation. Oncogenesis 2021, 10, 72. [Google Scholar] [CrossRef]
- Zheng, Y.; Lu, P.; Deng, Y.; Wen, L.; Wang, Y.; Ma, X.; Wang, Z.; Wu, L.; Hong, Q.; Duan, S.; et al. Single-Cell Transcriptomics Reveal Immune Mechanisms of the Onset and Progression of IgA Nephropathy. Cell Rep 2020, 33, 108525. [Google Scholar] [CrossRef] [PubMed]
- Meeuwsen, M.H.; Wouters, A.K.; Wachsmann, T.L.A.; Hagedoorn, R.S.; Kester, M.G.D.; Remst, D.F.G.; van der Steen, D.M.; de Ru, A.H.; van Hees, E.P.; Kremer, M.; et al. Broadly applicable TCR-based therapy for multiple myeloma targeting the immunoglobulin J chain. J Hematol Oncol 2023, 16, 16. [Google Scholar] [CrossRef]
- Shen Minqiang, L.Z. NK cell activation and immune response in inflammatory bowel disease. World Chinese Journal of Digestology 2008, 16, 6. [Google Scholar]
- Chao Kang, Z.S. , Chen Minhu. CD4+T cell and inflammatory bowel disease. Chinese Journal of Gastroenterology and Hepatology 2010, 19, 1058–1061. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).