Submitted:
04 February 2025
Posted:
05 February 2025
You are already at the latest version
Abstract
Metadichol expresses all nuclear receptors, the TLR family (1-10), the Sirtuin family (1-7), all 48 nuclear receptors, and all Yamanaka factors: Oct 4, Sox2, KLF4, C-myc, and circadian genes: Per1, CRY1, BMAL1, CLOCK, and PPARGC1A. The transcription factor family Kruppel-like factors (KLFs) is essential for cell proliferation, differentiation, and development. Our tiny chemical, Metadichol, a long-chain lipid alcohol nanoemulsion, modulates KLF family 1-18 expression. Concentration dependent. At 1 ng/ml, 16 of 18 KLF transcription factors are downregulated, save KLF 4 and 18. Small chemical modulators of KLF expression and activity offer new therapeutic approaches for many disorders, including cancer. In immunotherapy, KLF10 inhibition may reduce T regulatory cell numbers or function to boost anti-tumor immunity. Small compounds substituting KLF4 in cellular reprogramming could increase iPSC production efficiency and safety in regenerative medicine. In addition to KLF family expression results, this research will examine the regulation mechanisms of KLFs, nuclear receptors, SIRTs, circadian genes, Toll-like receptors, Klotho, P53, and PPARGC1A in cancer. These protein families form a dynamic web of interconnected signaling networks that affect cancer biology, including the mechanisms of crosstalk between them, the roles of individual members in different cancer types, and the tumor microenvironment's effect on these interactions. Our research reveals that molecular biology, genetics, immunology, and clinical oncology must be integrated to understand cancer's intricate networks. This will allow novel therapeutic techniques to target gene family connections, improving cancer therapy results and tumor biology understanding.
Keywords:
Introduction
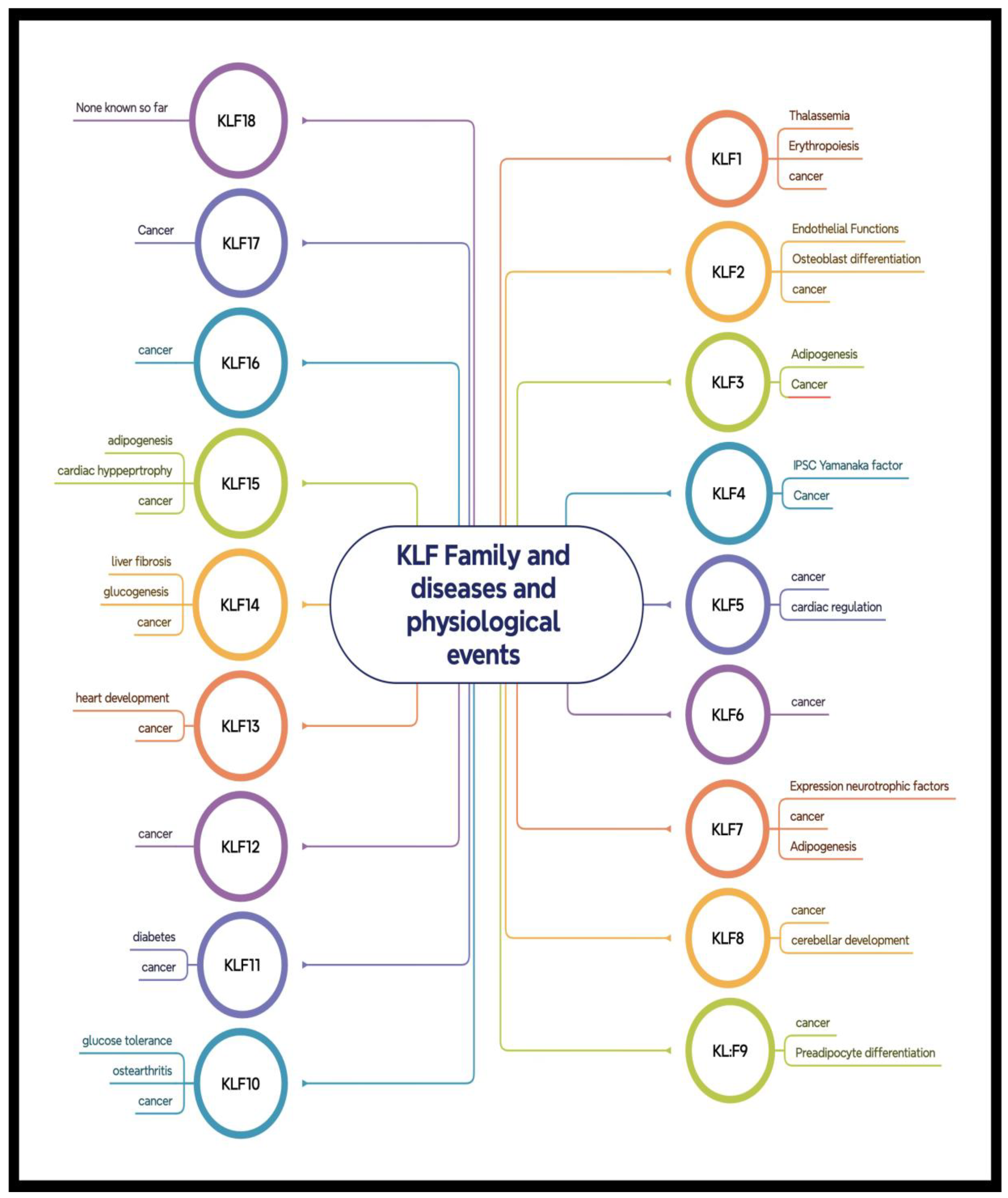
KLF Structure and Classification
KLFs and Development Roles
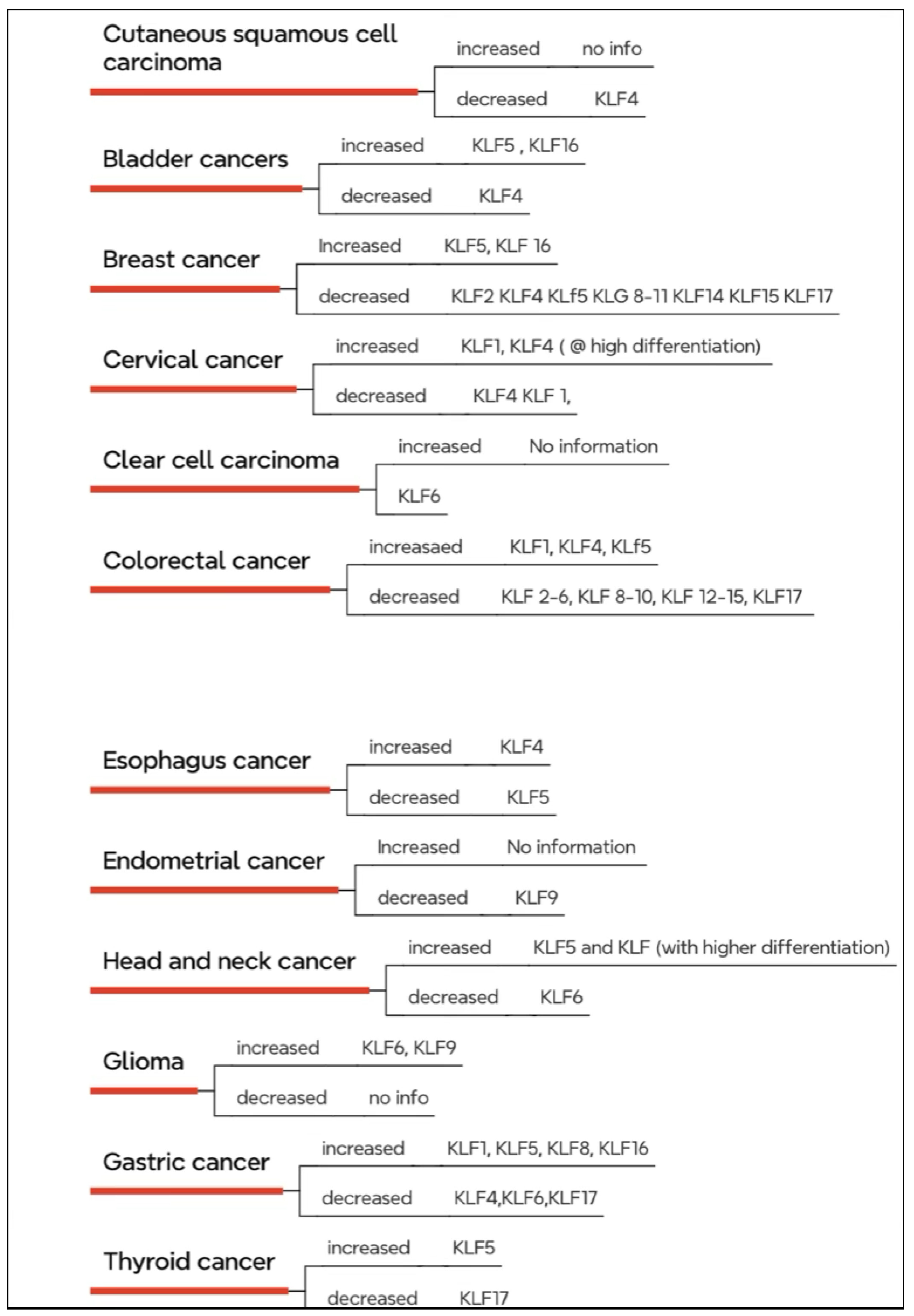
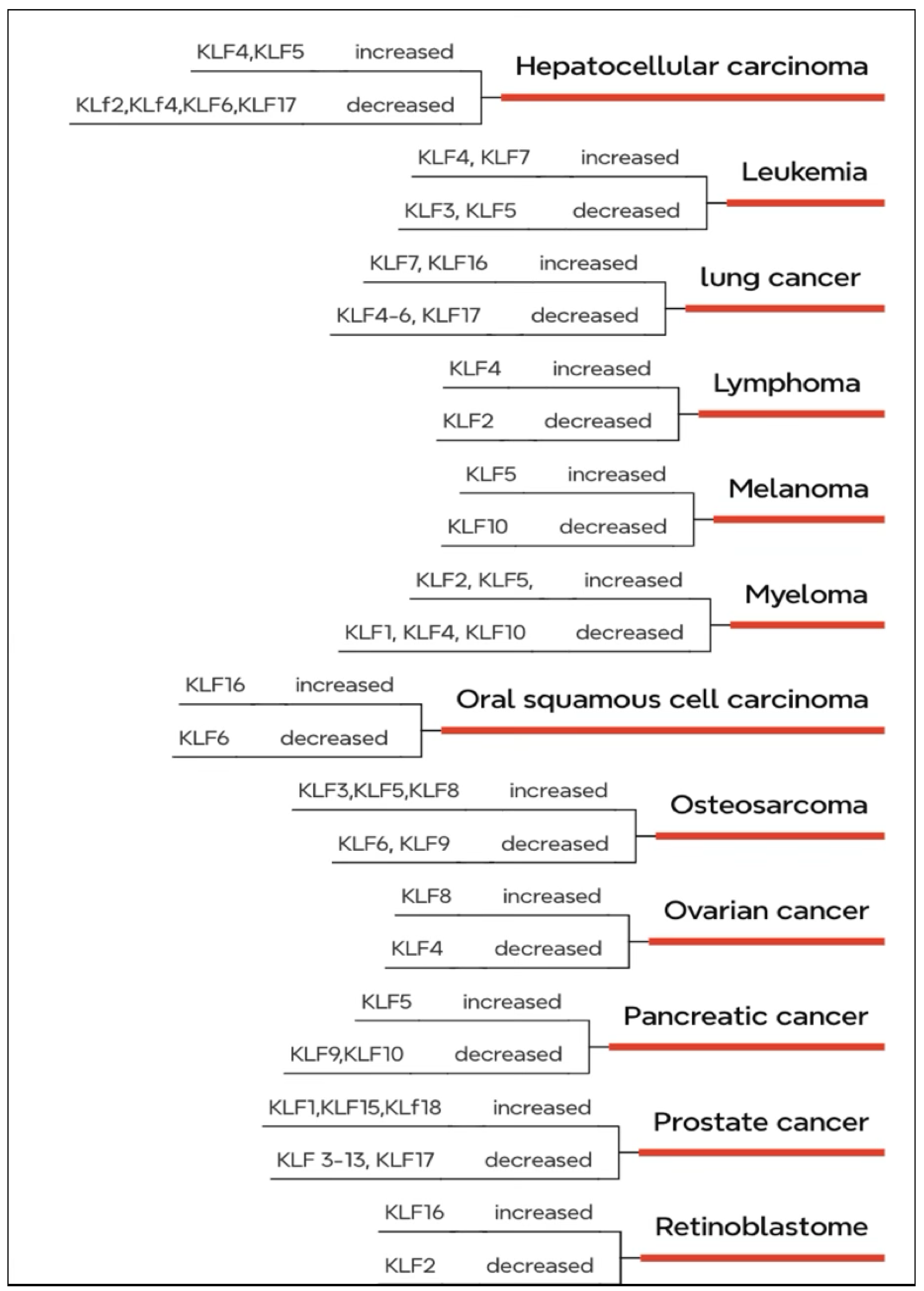
KLFs in Cancer
Immune System and KLFs
Targeting KLFs Therapeutically
KLFs and Cellular Processes
Genetic Regulation via KLFs
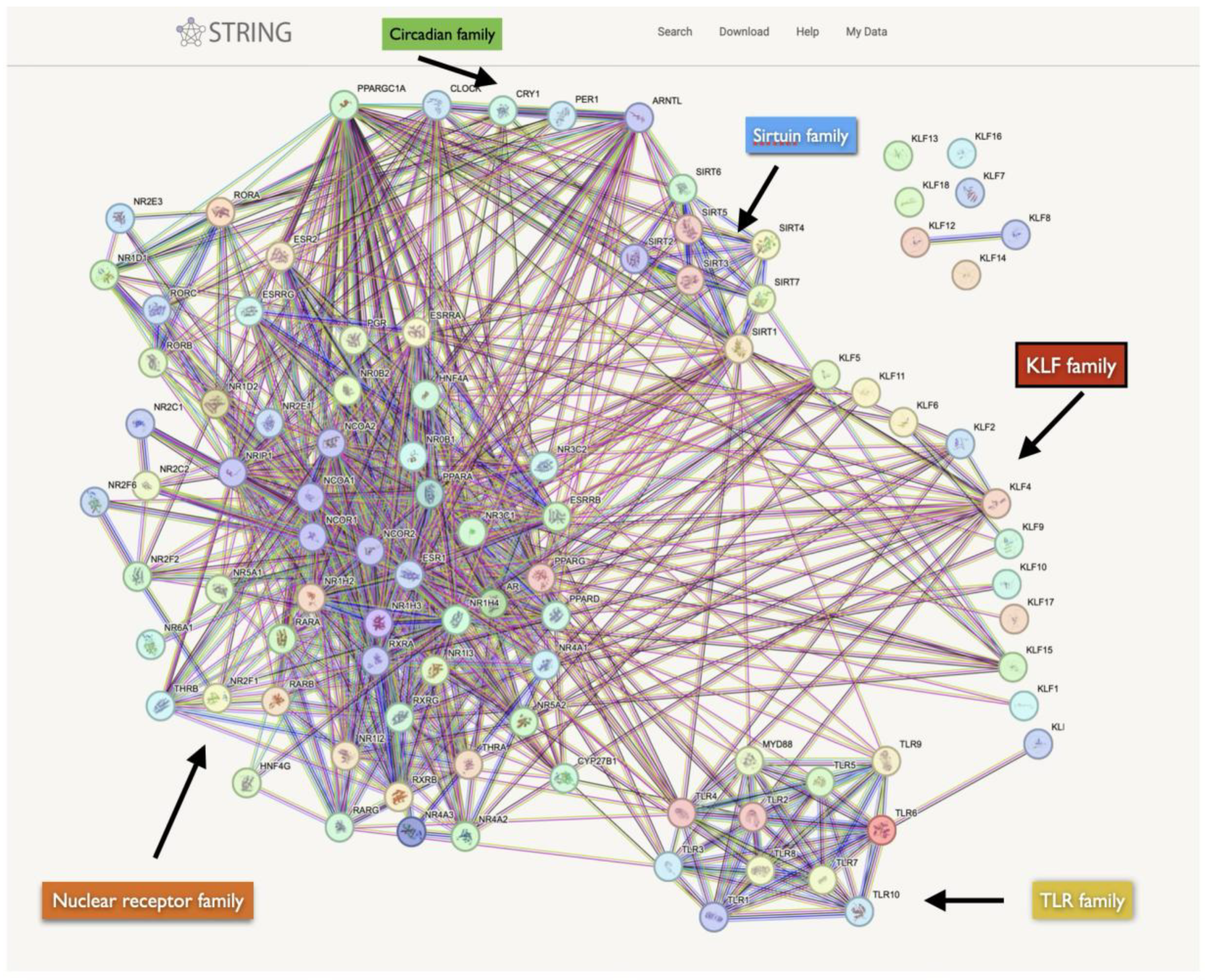
Cross-Regulation and KLF Family Interactions
Experimental
Isolation of Human WBCs by Cell Line and Condition
| Cell line | Sample name | Treatment details |
| Human PBMC | Metadichol | Control |
| 1 pg/ml | ||
| 100 pg/ml | ||
| 1 ng/ml | ||
| 100ng/ml |
Sample Preparation and RNA Isolation
cDNA Production
RT-qPCR Primers and Analysis
Results
Antitumor Synergy
Past Compensatory Mechanisms
Multiple Signaling Pathway Modulation
Addressing Tumor Heterogeneity
Making therapy more effective
Potential for Personalized Medicine
KLF and Regulatory Pathway Interactions
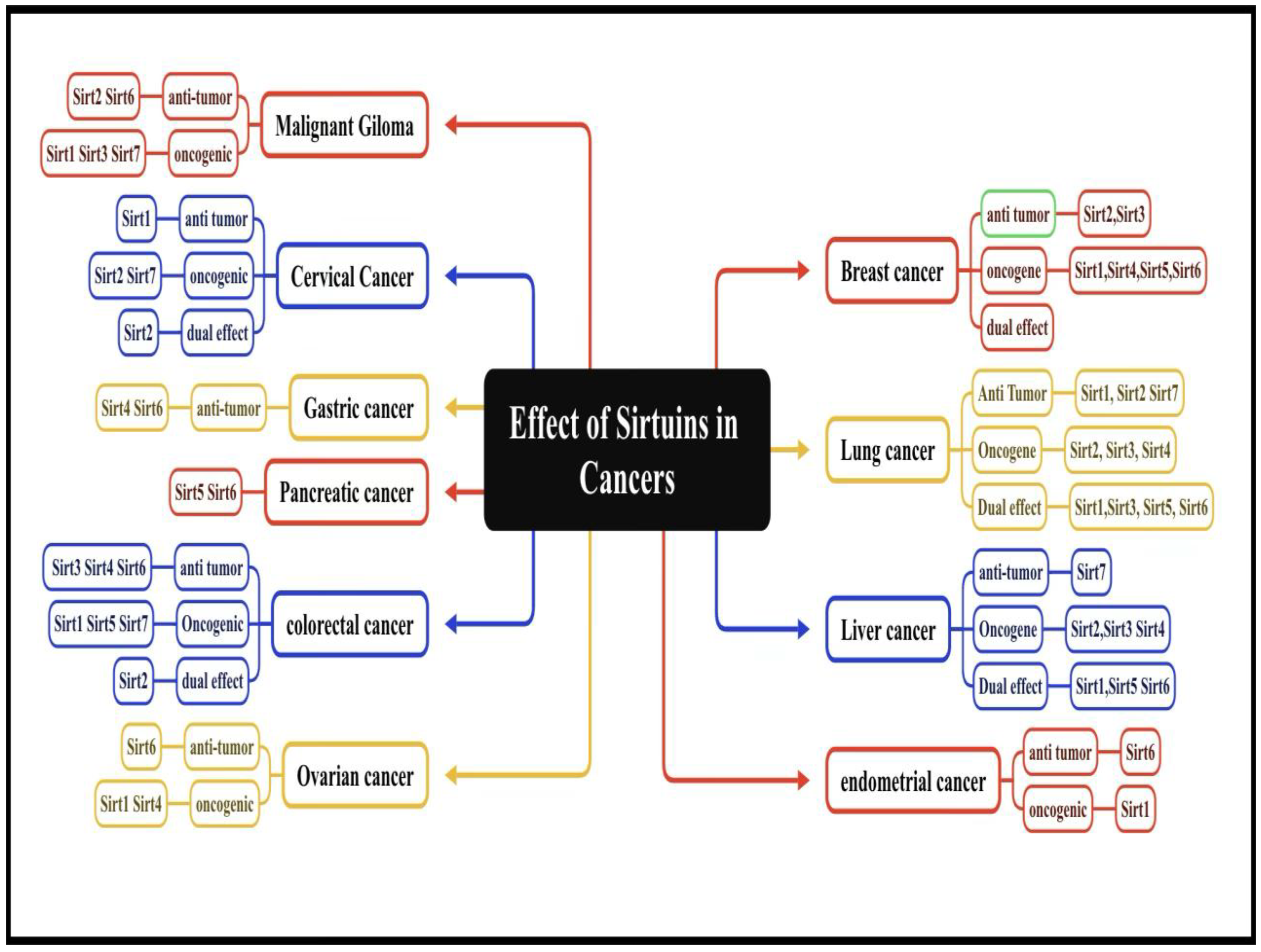
Established KLF-NR Interactions in Cancer
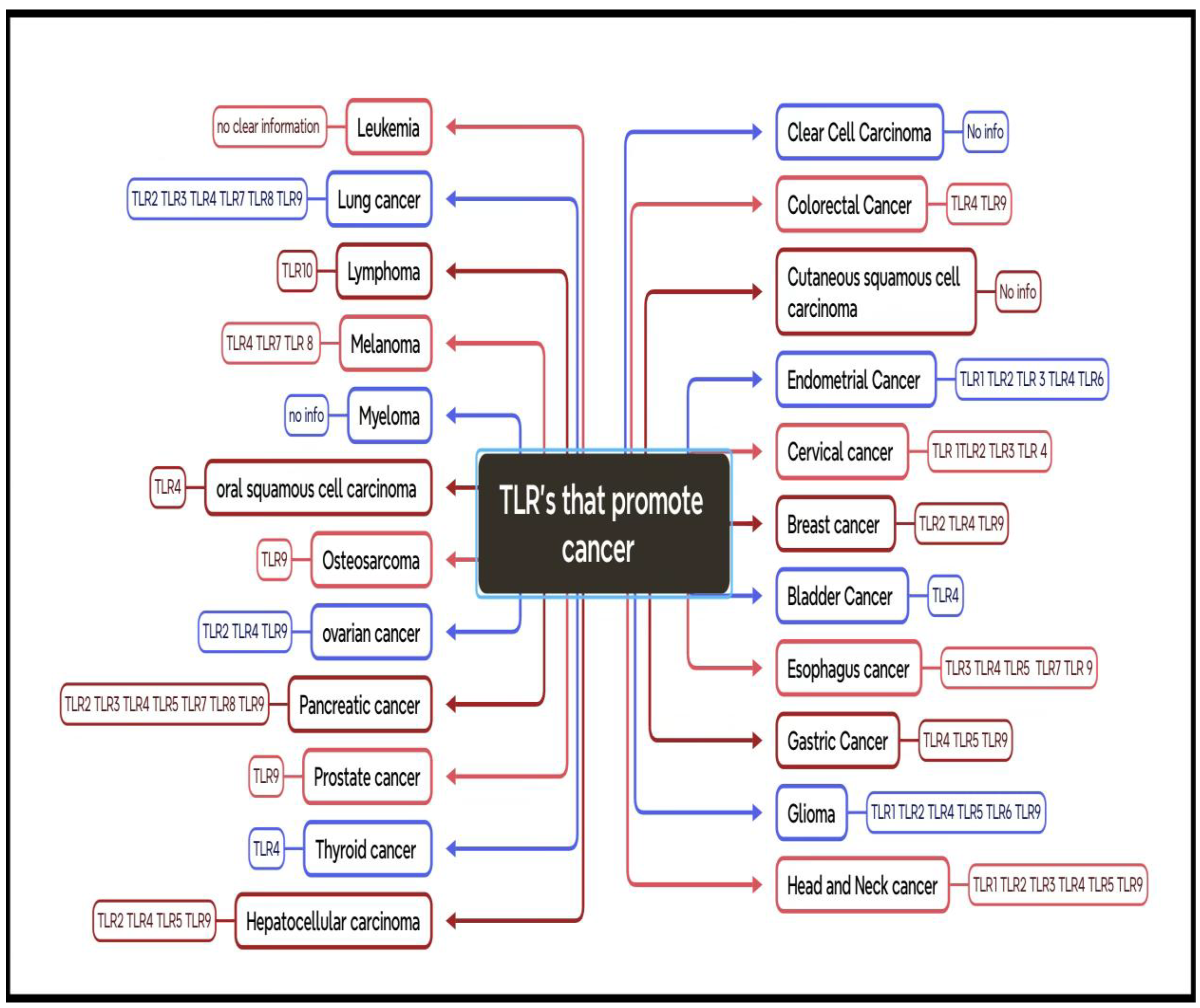
KLF-TLR Interactions in Cancer
Cancer, KLFs, and Circadian Clock
KLF-TP53, TERT, FOXO1, Klotho Interactions in Cancer
Interactions between KLF and FOXO1 in Cancer Pathways Are Complex
Klotho-KLF Interactions in Cancer: An Indirect Influence Network
Synergistic and Antagonistic Effects: Context-Dependent Results

Conclusions
Declarations
Author Contributions
References
- Yuce K, Ozkan AI. The kruppel-like factor (KLF) family, diseases, and physiological events, . Gene. 2024 Feb 15;895:148027. [CrossRef]
- Swamynathan SK. Krüppel-like factors: three fingers in control. Hum Genomics. 2010 Apr;4(4):263-70. [CrossRef]
- Black AR, Black JD, Azizkhan-Clifford J. Sp1 and krüppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001 Aug;188(2):143-60. [CrossRef]
- Gao Y, Cao Q, Lu L, Zhang X, Zhang Z, Dong X, Jia W, Cao Y. Kruppel-like factor family genes are expressed during Xenopus embryogenesis and involved in germ layer formation and body axis patterning. Dev Dyn. 2015 Oct;244(10):1328-46. Epub 2015 Aug 10. PMID: 26198170. [CrossRef]
- Oates AC, Pratt SJ, Vail B, Yan Yl, Ho RK, Johnson SL, Postlethwait JH, Zon LI. The zebrafish KLF gene family. Blood. 2001 Sep 15;98(6):1792-801. PMID: 11535513. [CrossRef]
- Oates AC, Pratt SJ, Vail B, Yan Yl, Ho RK, Johnson SL, Postlethwait JH, Zon LI. The zebrafish KLF gene family. Blood. 2001 Sep 15;98(6):1792-801. PMID: 11535513. [CrossRef]
- Shao M., Ge G., Liu W., Xiao J., Xia H., Fan Y., Zhao F., He B.Chen C. Characterization and phylogenetic analysis of Krüppel-like transcription factor (KLF) gene family in tree shrews (Tupaia belangeri chinensis). Oncotarget. 2017; 8: 16325-16339.
- Shinoda Y, Ogata N, Higashikawa A, Manabe I, Shindo T, Yamada T, Kugimiya F, Ikeda T, Kawamura N, Kawasaki Y, Tsushima K, Takeda N, Nagai R, Hoshi K, Nakamura K, Chung UI, Kawaguchi H. Kruppel-like factor 5 causes cartilage degradation through transactivation of matrix metalloproteinase 9. J Biol Chem. 2008 Sep 5;283(36):24682-9. [CrossRef]
- Mallipattu SK, Estrada CC, He JC. The critical role of Krüppel-like factors in kidney disease. Am J Physiol Renal Physiol. 2017 Feb 1;312(2):F259-F265. [CrossRef]
- Alder JK, Georgantas RW 3rd, Hildreth RL, Kaplan IM, Morisot S, Yu X, McDevitt M, Civin CI. Kruppel-like factor 4 is essential for inflammatory monocyte differentiation in vivo. J Immunol. 2008 Apr 15;180(8):5645-52. [CrossRef]
- Papadakis Konstantinos, Krempski James, P-201 Krüppel-Like Factor KLF10 Regulates Intestinal Macrophages and Innate Immune Colitis, Inflammatory Bowel Diseases, Volume 20, Issue suppl_1, December 2014, Page S106, . [CrossRef]
- Mao A, Zhou X, Liu Y, Ding J, Miao A, Pan G. KLF8 is associated with poor prognosis and regulates glycolysis by targeting GLUT4 in gastric cancer. J Cell Mol Med. 2019 Aug;23(8):5087-5097. [CrossRef]
- Sebzda E, Zou Z, Lee JS, Wang T, Kahn ML. Transcription factor KLF2 regulates the migration of naive T cells by restricting chemokine receptor expression patterns. Nat Immunol. 2008 Mar;9(3):292-300. [CrossRef]
- Tugal D, Jain MK, Simon DI. Endothelial KLF4: crippling vascular injury? J Am Heart Assoc. 2014 Feb 26;3(1):e000769. [CrossRef]
- Liu, C., Shen, M., Tan, W.L.W. et al. Statins improve endothelial function via suppression of epigenetic-driven End MT. Nat Cardiovasc Res 2, 467–485 (2023). [CrossRef]
- Melissa E. Heard, Michael C. Velarde, Linda C. Giudice, Frank A. Simmen, Rosalia C.M. Simmen, Krüppel-Like Factor 13 Deficiency in Uterine Endometrial Cells Contributes to Defective Steroid Hormone Receptor Signaling but Not Lesion Establishment in a Mouse Model of Endometriosis, Biology of Reproduction, Volume 92, Issue 6, 1 June 2015, 140, 1–9, . [CrossRef]
- Athon W. Homeister and Cam Patterson. Zinc Fingers in the Pizza Pie, Circulation Research, 2008, Volume 103, Number 7. /doi.org/10.1161/CIRCRESAHA.108.18576.
- Yang Y, Goldstein BG, Chao HH, Katz JP. KLF4 and KLF5 regulate proliferation, apoptosis and invasion in esophageal cancer cells. Cancer Biol Ther. 2005 Nov;4(11):1216-21. [CrossRef]
- Brown, A. R., Simmen, R., and Simmen, F.. 2011. "Abstract A44: Suppression of insulin-induced fatty acid synthase gene expression and colon cancer cell proliferation by members of the Krppel-like family of transcription factors". Cancer Prev Res 2011;4(10 Suppl):A44.
- Bieker JJ. Krüppel-like factors: three fingers in many pies. J Biol Chem. 2001 Sep 14;276(37):34355-8. [CrossRef]
- Pang CJ, Lemsaddek W, Alhashem YN, Bondzi C, Redmond LC, Ah-Son N, Dumur CI, Archer KJ, Haar JL, Lloyd JA, Trudel M. Kruppel-like factor 1 (KLF1), KLF2, and Myc control a regulatory network essential for embryonic erythropoiesis. Mol Cell Biol. 2012 Jul;32(13):2628-44. [CrossRef]
- Kalra IS, Alam MM, Choudhary PK, Pace BS. Krüppel-like Factor 4 activates HBG gene expression in primary erythroid cells. Br J Haematol. 2011 Jul;154(2):248-59. [CrossRef]
- Vinci, M.; Greco, D.; Treccarichi, S.; Chiavetta, V.; Figura, M.G.; Musumeci, A.; Greco, V.; Federico, C.; Calì, F.; Saccone, S. Bioinformatic Evaluation of KLF13 Genetic Variant: Implications for Neurodevelopmental and Psychiatric Symptoms. Genes 2024, 15, 1056. [CrossRef]
- Blackmore MG, Wang Z, Lerch JK, Motti D, Zhang YP, Shields CB, Lee JK, Goldberg JL, Lemmon VP, Bixby JL. Krüppel-like Factor 7 engineered for transcriptional activation promotes axon regeneration in the adult corticospinal tract. Proc Natl Acad Sci U S A. 2012 May 8;109(19):7517-22. [CrossRef]
- Shaverdashvili K, Padlo J, Weinblatt D, Jia Y, Jiang W, Rao D, Laczkó D, Whelan KA, Lynch JP, Muir AB, Katz JP. KLF4 activates NFκB signaling and esophageal epithelial inflammation via the Rho-related GTP-binding protein RHOF. PLoS One. 2019 Apr 18;14(4):e0215746. [CrossRef]
- Jiang Z, Yu T, Fan Z, Yang H, Lin X. Krüppel-Like Factor 7 is a Marker of Aggressive Gastric Cancer and Poor Prognosis. Cell Physiol Biochem. 2017;43(3):1090-1099. [CrossRef]
- P.R. Raghavan US patents 8,722,093 (2014) and 9,006,292 (2015).
- P.R. Raghavan . Metadichol®-induced expression of Sirtuins 1-7 in somatic and cancer cells, Medical Research Archives, (online), 2024 12(6). [CrossRef]
- P.R. Raghavan . Metadichol-induced expression of Toll receptor family members in peripheral blood mononuclear cells. Medical research archives, 2024. (online) 12(8). [CrossRef]
- P.R. Raghavan. Metadichol® A Nano Lipid Emulsion that Expresses All 49 Nuclear Receptors in Stem and Somatic Cells. Archives of Clinical and Biomedical Research. 7 2023: 524-536. Doi.10.26502/acbr.50170368.
- P.R. Raghavan, Metadichol, a Natural Ligand for the Expression of Yamanaka Reprogramming Factors in Human Cardiac, Fibroblast, and Cancer Cell Lines. Medical Research Archives, (online) 2024, 12(6). [CrossRef]
- P.R. Raghavan PR., 2024. Metadichol®-induced expression of Circadian clock transcription factors in human fibroblasts. Medical Research Archives, online) 2024, 12(6). [CrossRef]
- Tetreault MP, Wang ML, Yang Y, Travis J, Yu QC, Klein-Szanto AJ, Katz JP. Klf4 overexpression activates epithelial cytokines and inflammation-mediated esophageal squamous cell cancer in mice. Gastroenterology. 2010 Dec;139(6):2124-2134.e9. PMID: 20816834; PMCID: PMC3457785. [CrossRef]
- Tarapore RS, Yang Y, Katz JP. Restoring KLF5 in esophageal squamous cell cancer cells activates the JNK pathway leading to apoptosis and reduced cell survival. Neoplasia. 2013 May;15(5):472-80. [CrossRef]
- Lahiri SK, Zhao J. Krüppel-like factor 8 emerges as an important regulator of cancer. Am J Transl Res. 2012;4(3):357-63.
- Li ZY, Zhu YX, Chen JR, Chang X, Xie ZZ. The role of KLF transcription factor in the regulation of cancer progression. Biomed Pharmacother. 2023 Jun;162:114661. [CrossRef]
- McConnell B.B, Yang VW. Mammalian Krüppel-like factors in health and diseases. Physiol Rev. 2010 Oct;90(4):1337-81. [CrossRef]
- Yang, Y. & Katz, J. P. in The Biology of Krüppel-like Factors (eds. Nagai, R. F., Friedman, S. L. & Kasuga, M.) 2009, 67–82 (Springer).
- Tetreault, MP., Yang, Y. & Katz, J. Krüppel-like factors in cancer. Nat Rev Cancer, 2013 13, 701–713 . [CrossRef]
- Zhang Y, Yao C, Ju Z, Jiao D, Hu D, Qi L, Liu S, Wu X, and Zhao C. Krüppel-like actors in tumors: Key regulators and therapeutic avenues. Front. Oncol. 2023, 13:1080720. [CrossRef]
- Rascio F, Spadaccino F, Rocchetti MT, Castellano G, Stallone G, Netti GS, Ranieri E. The Pathogenic Role of PI3K/AKT Pathway in Cancer Onset and Drug Resistance: An Updated Review. Cancers (Basel). 2021 Aug 5;13(16):3949. PMID: 34439105; PMCID: PMC8394096. [CrossRef]
- Zhu B, Liu Q, Han Q, Zeng B, Chen J, Xiao Q. Downregulation of Krüppel-like factor 1 inhibits the metastasis and invasion of cervical cancer cells. Mol Med Rep. 2018 Oct;18(4):3932-3940. [CrossRef]
- Xu R, Chen Y, Wei S, Chen J. Comprehensive Pan-Cancer Analysis of the Prognostic Role of KLF Transcription Factor 2 (KLF2) in Human Tumors. Onco Targets Ther. 2024 Nov 2;17:887-904. [CrossRef]
- Zhu J, Teng H, Zhu X, Yuan J, Zhang Q, Zou Y. Pancancer analysis of Krüppel-like factor 3 and its carcinogenesis in pancreatic cancer. Front Immunol. 2023 Aug 3;14:1167018. [CrossRef]
- He, Z., He, J. & Xie, K. KLF4 transcription factor in tumorigenesis. Cell Death Discov. 2023, 9, 118. [CrossRef]
- Luo Y, Chen C. The roles and regulation of the KLF5 transcription factor in cancers. Cancer Sci. 2021 Jun;112(6):2097-2117. [CrossRef]
- DiFeo A, Martignetti JA, Narla G. The role of KLF6 and its splice variants in cancer therapy. Drug Resist Updat. 2009 Feb-Apr;12(1-2):1-7. [CrossRef]
- Li Z, Liu Q. The Oncogenic Role of KLF7 in Colon Adenocarcinoma and Therapeutic Perspectives. Int J Genomics. 2023 Dec 12;2023:5520926. [CrossRef]
- Lahiri SK, Zhao J. Krüppel-like factor 8 emerges as an important regulator of cancer. Am J Transl Res. 2012;4(3):357-63.
- Ying M, Sang Y, Li Y, Guerrero-Cazares H, Quinones-Hinojosa A, Vescovi AL, Eberhart CG, Xia S, Laterra J. Krüppel-like family of transcription factor 9, a differentiation-associated transcription factor, suppresses Notch1 signaling and inhibits glioblastoma-initiating stem cells. Stem Cells. 2011 Jan;29(1):20-31. [CrossRef]
- Memon A, Lee WK. KLF10 as a Tumor Suppressor Gene and Its TGF-β Signaling. Cancers (Basel). 2018 May 25;10(6):161. [CrossRef]
- Xi Z, Zhang R, Zhang F, Ma S, Feng T. KLF11 Expression Predicts Poor Prognosis in Glioma Patients. Int J Gen Med. 2021 Jun 28;14:2923-2929. [CrossRef]
- Li, Y., Li, S., Shi, X. et al. KLF12 promotes the proliferation of breast cancer cells by reducing the transcription of p21 in a p53-dependent and p53-independent manner. Cell Death Dis, 2023, 14, 313. [CrossRef]
- Chen CC, Xie XM, Zhao XK, Zuo S, Li HY. Krüppel-like Factor 13 Promotes HCC Progression by Transcriptional Regulation of HMGCS1-mediated Cholesterol Synthesis. J Clin Transl Hepatol. 2022 Dec 28;10(6):1125-1137. [CrossRef]
- Wang X, Qu X, Liu X, Wang K, Yang Y, Zhang Y, Wang Z, Fan G, Li Y, Zeng Y, Chen H, Zhu T. KLF14 inhibits tumor progression via FOSL1 in glioma. Biochem Biophys Rep. 2024 Nov 27;41:101885. [CrossRef]
- Kanyomse, Q., Le, X., Tang, J. et al. KLF15 suppresses tumor growth and metastasis in Triple-Negative Breast Cancer by downregulating CCL2 and CCL7. Sci Rep, 2022, 12, 19026. [CrossRef]
- Ma P, Sun CQ, Wang YF, Pan YT, Chen QN, Liu WT, Liu J, Zhao CH, Shu YQ, Li W. KLF16 promotes proliferation in gastric cancer cells by regulating p21 and CDK4. Am J Transl Res. 2017 Jun 15;9(6):3027-3036. PMID: 28670390; PMCID: PMC5489902.
- Jiang X, Shen TY, Lu H, Shi C, Liu Z, Qin H, Wang F. Clinical significance and biological role of KLF17 as a tumor suppressor in colorectal cancer. Oncol Rep. 2019 Nov;42(5):2117-2129. [CrossRef]
- Zhang Y, Yao C, Ju Z, Jiao D, Hu D, Qi L, Liu S, Wu X, Zhao C. Krüppel-like factors in tumors: Key regulators and therapeutic avenues. Front Oncol. 2023 Jan 25;13:1080720. [CrossRef]
- Simmen FA, Alhallak I, Simmen RCM. Krüppel-like Factor-9 and Krüppel-like Factor-13: Highly Related, Multi-Functional, Transcriptional Repressors and Activators of Oncogenesis. Cancers (Basel). 2023 Nov 30;15(23):5667. [CrossRef]
- Fleming-de-Moraes CD, Rocha MR, Tessmann JW, de Araujo WM, Morgado-Diaz JA. Crosstalk between PI3K/Akt and Wnt/β-catenin pathways promote colorectal cancer progression regardless of mutational status. Cancer Biol Ther. 2022 Dec 31;23(1):1-13. [CrossRef]
- Tsai YC, Hsin MC, Liu RJ, Li TW, Ch'ang HJ. Krüppel-like Factor 10 as a Prognostic and Predictive Biomarker of Radiotherapy in Pancreatic Adenocarcinoma. Cancers (Basel). 2023 Oct 30;15(21):5212. PMID: 37958386; PMCID: PMC10648792. [CrossRef]
- P.R. Raghavan. Metadichol, A Modulator that Controls Expression of Toll-Like Receptors in Cancer Cell Lines British Journal of Cancer Research. 2024, 7(3): 720-732. [CrossRef]
- P.R. Raghavan. Metadichol: An Agonist that Expresses the Anti-Aging Gene Klotho in Various Cell Lines. Fortune Journal of Health Sciences. 2023, 6: 357-362. [CrossRef]
- P.R. Raghavan.The Quest for Immortality: Introducing Metadichol®, a Novel Telomerase Activator. Stem Cell Res Ther 2019, 9: 446. [CrossRef]
- Mao A, Zhou X, Liu Y, Ding J, Miao A, Pan G. KLF8 is associated with poor prognosis and regulates glycolysis by targeting GLUT4 in gastric cancer. J Cell Mol Med. 2019 Aug;23(8):5087-5097. [CrossRef]
- Tsai, YiChih, Chen, Suliang, Peng, Shu-Ling, Tsai, Ya-Li, Chang, Zuong-Ming, Chang, Vincent, and Chang, Hui-Ju. "Upregulating sirtuin 6 ameliorates glycolysis, EMT and distant metastasis of pancreatic adenocarcinoma with krppel-like factor 10 deficiency". Springer Nature. [CrossRef]
- Ansari A, Rahman MS, Saha SK, Saikot FK, Deep A, Kim KH. Function of the SIRT3 mitochondrial deacetylase in cellular physiology, cancer, and neurodegenerative disease. Aging Cell. 2017 Feb;16(1):4-16. [CrossRef]
- Limame R, Op de Beeck K, Lardon F, De Wever O, Pauwels P. Krüppel-like factors in cancer progression: three fingers on the steering wheel. Oncotarget. 2014 Jan 15;5(1):29-48. [CrossRef]
- Bureau C, Hanoun N, Torrisani J, Vinel JP, Buscail L, Cordelier P. Expression and Function of Kruppel Like-Factors (KLF) in Carcinogenesis. Curr Genomics. 2009 Aug;10(5):353-60. [CrossRef]
- Zhao E, Hou J, Ke X, Abbas MN, Kausar S, Zhang L, Cui H. The Roles of Sirtuin Family Proteins in Cancer Progression. Cancers (Basel). 2019 Dec 5;11(12):1949. [CrossRef]
- Black AR, Black JD, Azizkhan-Clifford J. Sp1 and krüppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001 Aug;188(2):143-60. [CrossRef]
- Stünkel W, Campbell RM. Sirtuin 1 (SIRT1): the misunderstood HDAC. J Biomol Screen. 2011 Dec;16(10):1153-69. [CrossRef]
- Zhang XL, Zhang D, Michel FJ, Blum JL, Simmen FA, Simmen RC. Selective interactions of Kruppel-like factor 9/basic transcription element-binding protein with progesterone receptor isoforms A and B determine transcriptional activity of progesterone-responsive genes in endometrial epithelial cells. J Biol Chem. 2003 Jun 13;278(24):21474-82. Epub 2003 Apr 2. PMID:. [CrossRef]
- Knoedler JR, Denver RJ. Krüppel-like factors are effectors of nuclear receptor signaling. Gen Comp Endocrinol. 2014 Jul 1;203:49-59. [CrossRef]
- Luo Y, Chen C. The roles and regulation of the KLF5 transcription factor in cancers. Cancer Sci. 2021 Jun;112(6):2097-2117. Epub 2021 May 3. PMID: 33811715; PMCID: PMC8177779. [CrossRef]
- Fan L, Sweet DR, Fan EK, Prosdocimo DA, Madera A, Jiang Z, Padmanabhan R, Haldar SM, Vinayachandran V, Jain MK. Transcription factors KLF15 and PPARδ cooperatively orchestrate genome-wide regulation of lipid metabolism in skeletal muscle. J Biol Chem. 2022 Jun;298(6):101926. [CrossRef]
- Shrestha S, Sewell JA, Santoso CS, Forchielli E, Carrasco Pro S, Martinez M, Fuxman Bass JI. Discovering human transcription factor physical interactions with genetic variants, novel DNA motifs, and repetitive elements using enhanced yeast one-hybrid assays. Genome Res. 2019 Sep;29(9):1533-1544. PMID: 31481462; PMCID: PMC6724672. [CrossRef]
- Giarrizzo M, LaComb JF, Bialkowska AB. The Role of Krüppel-like Factors in Pancreatic Physiology and Pathophysiology. Int J Mol Sci. 2023 May 11;24(10):8589. [CrossRef]
- Yea S, Narla G, Zhao X, Garg R, Tal-Kremer S, Hod E, Villanueva A, Loke J, Tarocchi M, Akita K, Shirasawa S, Sasazuki T, Martignetti JA, Llovet JM, Friedman SL. Ras promotes growth by alternative splicing-mediated inactivation of the KLF6 tumor suppressor in hepatocellular carcinoma. Gastroenterology. 2008 May;134(5):1521-31. [CrossRef]
- Sun H, Gao Y, Lu K, Zhao G, Li X, Li Z, Chang H. Overexpression of Klotho suppresses liver cancer progression and induces cell apoptosis by negatively regulating wnt/β-catenin signaling pathway. World J Surg Oncol. 2015 Oct 24;13:307. [CrossRef]
- Ozkan AD, Eskiler GG, Kazan N, Turna O. Histone deacetylase inhibitor sodium butyrate regulates the activation of toll-like receptor 4/interferon regulatory factor-3 signaling pathways in prostate cancer cells. J Cancer Res Ther. 2023 Oct 1;19(7):1812-1817. [CrossRef]
- Luo, Xinjing, Chen, Jie, Ruan, Jianwei, Chen, Yongfeng, Mo, Xuanrong, Xie, Jiangwen, and Lv, Guoju. 2015. "Krüppel-Like Factor 4 Is a Regulator of Proinflammatory Signaling in Fibroblast-Like Synoviocytes through Increased IL-6 Expression". Mediators of Inflammation.
- Tsai, Kuo-Wang. 2017. "Identify metastasis-associated lnc RNA in triple-negative breast cancer". Journal of Cancer Science & Therapy. [CrossRef]
- Ma L, Feng L, Ding X, Li Y. Effect of TLR4 on the growth of SiHa human cervical cancer cells via the MyD88-TRAF6-TAK1 and NF-κB-cyclin D1-STAT3 signaling pathways. Oncol Lett. 2018 Mar;15(3):3965-3970. [CrossRef]
- Fakhri S, Moradi SZ, Yarmohammadi A, Narimani F, Wallace CE, Bishayee A. Modulation of TLR/NF-κB/NLRP Signaling by Bioactive Phytocompounds: A Promising Strategy to Augment Cancer Chemotherapy and Immunotherapy. Front Oncol. 2022 Mar 1;12:834072. [CrossRef]
- De Paoli F, Staels B, Chinetti-Gbaguidi G. Macrophage phenotypes and their modulation in atherosclerosis. Circ J. 2014;78(8):1775-81. [CrossRef]
- Troutman TD, Bazan JF, Pasare C. Toll-like receptors, signaling adapters and regulation of the pro-inflammatory response by PI3K. Cell Cycle. 2012 Oct 1;11(19):3559-67. Epub 2012 Aug 16. PMID: 22895011; PMCID: PMC3478307. [CrossRef]
- Szebeni GJ, Vizler C, Kitajka K, Puskas LG. Inflammation and Cancer: Extra- and Intracellular Determinants of Tumor-Associated Macrophages as Tumor Promoters. Mediators Inflamm. 2017;2017:9294018. [CrossRef]
- Zou J, Shankar N. Roles of TLR/MyD88/MAPK/NF-κB Signaling Pathways in the Regulation of Phagocytosis and Proinflammatory Cytokine Expression in Response to E. faecalis Infection. PLoS One. 2015 Aug 28;10(8):e0136947. [CrossRef]
- Liao X, Sharma N, Kapadia F, Zhou G, Lu Y, Hong H, Paruchuri K, Mahabeleshwar GH, Dalmas E, Venteclef N, Flask CA, Kim J, Doreian BW, Lu KQ, Kaestner KH, Hamik A, Clément K, Jain MK. Krüppel-like factor 4 regulates macrophage polarization. J Clin Invest. 2011 Jul;121(7):2736-49. [CrossRef]
- Saheed Oluwasina Oseni, Rolando Branly, Mirjana Pavlovic, James Kumi-Diaka; Abstract 5730: Co-targeting toll-like receptor and PI3K survival signaling pathways in stem-like castration resistant prostate cancer cells. Cancer Res 1 July 2018; 78 (13_Supplement): 5730. [CrossRef]
- Schwartz AL, Dickerson E, Dagia N, Malgor R, McCall KD. TLR signaling inhibitor, phenylmethimazole, in combination with tamoxifen inhibits human breast cancer cell viability and migration. Oncotarget. 2016 Jul 1;8(69):113295-113302. PMID: 29371911; PMCID: PMC5768328. [CrossRef]
- Wang H, Flannery SM, Dickhöfer S, Huhn S, George J, Kubarenko AV, Lascorz J, Bevier M, Willemsen J, Pichulik T, Schafmayer C, Binder M, Manoury B, Paludan SR, Alarcon-Riquelme M, Bowie AG, Försti A, Weber ANR. A coding IRAK2 protein variant compromises Toll-like receptor (TLR) signaling and is associated with colorectal cancer survival. J Biol Chem. 2014 Aug 15;289(33):23123-23131. [CrossRef]
- Chu Y, Liu Z, Liu J, Yu L, Zhang D, Pei F. Characterization of lncRNA-Perturbed TLR-Signaling Network Identifies Novel lncRNA Prognostic Biomarkers in Colorectal Cancer. Front Cell Dev Biol. 2020 Jun 18;8:503. PMID: 32626715; PMCID: PMC7314994. [CrossRef]
- Sedighzadeh SS, Khoshbin AP, Razi S, Keshavarz-Fathi M, Rezaei N. A narrative review of tumor-associated macrophages in lung cancer: regulation of macrophage polarization and therapeutic implications. Transl Lung Cancer Res. 2021 Apr;10(4):1889-1916. PMID: 34012800; PMCID: PMC8107755. [CrossRef]
- Yong Zhu, Richard G. Stevens, Aaron E. Hoffman, Liesel M. FitzGerald, Erika M. Kwon, Elaine A. Ostrander, Scott Davis, Tongzhang Zheng, Janet L. Stanford; Testing the Circadian Gene Hypothesis in Prostate Cancer: A Population-Based Case-Control Study. Cancer Res 15 December 2009; 69 (24): 9315–9322. [CrossRef]
- Dierickx P, Van Laake LW, Geijsen N. Circadian clocks: from stem cells to tissue homeostasis and regeneration. EMBO Rep. 2018 Jan;19(1):18-28. [CrossRef]
- Wu Y, Tao B, Zhang T, Fan Y, Mao R. Pan-Cancer Analysis Reveals Disrupted Circadian Clock Associates With T Cell Exhaustion. Front Immunol. 2019 Oct 24;10:2451. PMID: 31708917; PMCID: PMC6821711. [CrossRef]
- Subramaniam M, Hawse JR, Rajamannan NM, Ingle JN, Spelsberg TC. Functional role of KLF10 in multiple disease processes. Biofactors. 2010 Jan-Feb;36(1):8-18. [CrossRef]
- Léveillé M, Besse-Patin A, Jouvet N, Gunes A, Sczelecki S, Jeromson S, Khan NP, Baldwin C, Dumouchel A, Correia JC, Jannig PR, Boulais J, Ruas JL, Estall JL. PGC-1α isoforms coordinate to balance hepatic metabolism and apoptosis in inflammatory environments. Mol Metab. 2020 Apr;34:72-84. [CrossRef]
- Angelousi A, Kassi E, Ansari-Nasiri N, Randeva H, Kaltsas G, Chrousos G. Clock genes and cancer development in particular in endocrine tissues. Endocr Relat Cancer. 2019 Jun;26(6):R305-R317. [CrossRef]
- Sulli G, Lam MTY, Panda S. Interplay between Circadian Clock and Cancer: New Frontiers for Cancer Treatment. Trends Cancer. 2019 Aug;5(8):475-494. Epub 2019 Aug 3. PMID: 31421905; PMCID: PMC7120250. [CrossRef]
- Guillaumond, F., Gréchez-Cassiau, A., Subramaniam, M., Brangolo, S., Peteri-Brünback, B., Staels, B., … Teboul, M.. Krüppel-Like Factor KLF10 Is a Link between the Circadian Clock and Metabolism in Liver. Molecular and Cellular Biology, 2010, 30(12), 3059–3070. [CrossRef]
- Chen P, Hsu WH, Chang A, Tan Z, Lan Z, Zhou A, Spring DJ, Lang FF, Wang YA, DePinho RA. Circadian Regulator CLOCK Recruits Immune-Suppressive Microglia into the GBM Tumor Microenvironment. Cancer Discov. 2020 Mar;10(3):371-381. [CrossRef]
- Kiss Z, Ghosh PM. WOMEN IN CANCER THEMATIC REVIEW: Circadian rhythmicity and the influence of 'clock' genes on prostate cancer. Endocr Relat Cancer. 2016 Nov;23(11):T123-T134. Epub 2016 Sep 22. PMID: 27660402; PMCID: PMC5148656. [CrossRef]
- Zhou S, Tang X, Tang F. Krüppel-like factor 17, a novel tumor suppressor: its low expression is involved in cancer metastasis. Tumour Biol. 2016 Feb;37(2):1505-13. Epub 2015 Dec 12. PMID: 26662959; PMCID: PMC4842221. [CrossRef]
- Yusuf I, Kharas MG, Chen J, Peralta RQ, Maruniak A, Sareen P, Yang VW, Kaestner KH, Fruman DA. KLF4 is a FOXO target gene that suppresses B cell proliferation. Int Immunol. 2008 May;20(5):671-81. [CrossRef]
- Ligumsky H, Rubinek T, Merenbakh-Lamin K, Yeheskel A, Sertchook R, Shahmoon S, Aviel-Ronen S, Wolf I. Tumor Suppressor Activity of Klotho in Breast Cancer Is Revealed by Structure-Function Analysis. Mol Cancer Res. 2015 Oct;13(10):1398-407. [CrossRef]
- Ewendt F, Feger M, Föller M. Role of Fibroblast Growth Factor 23 (FGF23) and αKlotho in Cancer. Front Cell Dev Biol. 2021 Jan 14;8:601006. [CrossRef]
- Melemadathil, Karthika, Udupa, Karthik, Belle, V., Udupa, C. K. K., Pai, Ananth, and Mailankody, S.. 2024. "Role of serum -Klotho level in patients with breast carcinoma.". 2924, Journal of Clinical Oncology. [CrossRef]
- Raj, S.; Ahuja, M. The Versatility of Klotho Protein: Insights into Its Multifaceted Functions in Health and Disease. World Journal of Current Med and Pharm Research2024, 6, 12-17.
- Farrugia MK, Vanderbilt DB, Salkeni MA, Ruppert JM. Kruppel-like Pluripotency Factors as Modulators of Cancer Cell Therapeutic Responses. Cancer Res. 2016 Apr 1;76(7):1677-82. [CrossRef]
- Abramovitz L, Rubinek T, Ligumsky H, Bose S, Barshack I, Avivi C, Kaufman B, Wolf I. KL1 internal repeat mediates klotho tumor suppressor activities and inhibits bFGF and IGF-I signaling in pancreatic cancer. Clin Cancer Res. 2011 Jul 1;17(13):4254-66. [CrossRef]
- Wu Q, Jiang L, Wu J, Dong H, Zhao Y. Klotho Inhibits Proliferation in a RET Fusion Model of Papillary Thyroid Cancer by Regulating the Wnt/β-Catenin Pathway. Cancer Manag Res. 2021 Jun 17;13:4791-4802. PMID: 34168498; PMCID: PMC8216664. [CrossRef]
- Black AR, Black JD, Azizkhan-Clifford J. Sp1 and krüppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001 Aug;188(2):143-60. [CrossRef]
- Simmen RC, Pabona JM, Velarde MC, Simmons C, Rahal O, Simmen FA. The emerging role of Krüppel-like factors in endocrine-responsive cancers of female reproductive tissues. J Endocrinol. 2010 Mar;204(3):223-31. [CrossRef]
- Bureau C, Hanoun N, Torrisani J, Vinel JP, Buscail L, Cordelier P. Expression and Function of Kruppel Like-Factors (KLF) in Carcinogenesis. Curr Genomics. 2009 Aug;10(5):353-60. PMID: 20119532; PMCID: PMC2729999. [CrossRef]
- Mota J, Lima AMM, Gomes JIS, Souza de Andrade M, Brito HO, Silva MMAL, Faustino-Rocha AI, Oliveira PA, Lopes FF, Gil da Costa RM. Klotho in Cancer: Potential Diagnostic and Prognostic Applications. Diagnostics (Basel). 2023 Oct 31;13(21):3357. [CrossRef]
- Mao A, Zhou X, Liu Y, Ding J, Miao A, Pan G. KLF8 is associated with poor prognosis and regulates glycolysis by targeting GLUT4 in gastric cancer. J Cell Mol Med. 2019 Aug;23(8):5087-5097. Epub 2019 May 24. PMID: 31124603; PMCID: PMC6653475. [CrossRef]
- Jiang Z, Yu T, Fan Z, Yang H, Lin X. Krüppel-Like Factor 7 is a Marker of Aggressive Gastric Cancer and Poor Prognosis. Cell Physiol Biochem. 2017;43(3):1090-1099. [CrossRef]
- Wang Y, Chen L, Huang G, He D, He J, Xu W, Zou C, Zong F, Li Y, Chen B, Wu S, Zhao W, Wu J. Klotho sensitizes human lung cancer cell line to cisplatin via PI3k/Akt pathway. PLoS One. 2013;8(2):e57391. [CrossRef]
- Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, Gable AL, Fang T, Doncheva NT, Pyysalo S, Bork P, Jensen LJ, von Mering C. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023 Jan 6;51(D1):D638-D646. PMID: 36370105; PMCID: PMC9825434. [CrossRef]
- Aleman C, Mas R, Hernandez C, Rodeiro I, Cerejido E, Noa M, Capote A, Menendez R, Amor A, Fraga V. A 12-month study of policosanol oral toxicity in Sprague Dawley rats. Toxicol Lett , 1994, 70:77–87.
- Alemán CL, Ferreiro RM, Puig MN, Guerra IR, Ortega CH, Capote A. Carcinogenicity of policosanol in sprague dawley rats: a 24 month study. Teratog Carcinog Mutagen, 1994, 14:239–249.
- Alemán CL, Puig MN, Elias EC, Ortega CH, Guerra IR, Ferreiro RM, Briñis F. Carcinogenicity of policosanol in mice: an 18-month study. Food Chem Toxicol, 1995 33:573–578.

| Test concentrations | |||||
| RNA yield (ng/µl) | 0 | 1 pg/ ml | 100 pg/ ml | 1 ng/ ml | 100 ng/ ml |
| Human PBMC's | 623.120 | 343.123 | 792.123 | 673.111 | 611.123 |
| Primer | Sequence | Amplicon size | Annealing temperature |
| GAPDH | GTCTCCTCTGACTTCAACAGCG | 186 | 60 |
| ACCACCCTGTTGCTGTAGCCAA | |||
| KLF1 | CAGGTGTGATAGCCGAGACC | 111 | 65 |
| TCTTGGTGTAGCTCTTGCCG | |||
| KLF2 | CCAAGAGTTCGCATCTGAAGGC | 131 | 65 |
| CCGTGTGCTTTCGGTAGTGGC | |||
| KLF3 | CTCATGGTCTCCTTATCGGAGG | 131 | 65 |
| TGTCCTCTGTGGTTCGATCCCA | |||
| KLF4 | CCTTCCTGCCCGATCAGATG | 132 | 62 |
| TGAGCATCATCCCGTGTGTC | |||
| KLF5 | GGAGAAACGACGCATCCACTAC | 140 | 65 |
| GAACCTCCAGTCGCAGCCTTC | |||
| KLF6 | GACAGCTCCGAGGAACTTTCT | 156 | 65 |
| CACGCAACCCCACAGTTGA | |||
| KLF7 | CTCACGAGGCACTACAGGAAAC | 135 | 67 |
| TGGCAACTCTGGCCTTTCGGTT | |||
| KLF8 | CCTGAAAGCTCACCGCAGAATC | 113 | 61 |
| TGCTTGCGGAAATGGCGAGTGA | |||
| KLF9 | GGGAAACACGCCTCCGAAAA | 110 | 65 |
| CGTTCACCTGTATGCACTCTGTA | |||
| KLF10 | AGGAGTCACATCTGTAGCCACC | 139 | 67 |
| GAACGGGCAAACCTCCTTTCAC | |||
| KLF11 | ATGGATGCAGCCACACCTGAAC | 115 | 65 |
| GGAGAAACAGGTGTCCTTGTCG | |||
| KLF12 | CCTTTCCATAGCCAGAGCAGTAC | 130 | 65 |
| CTGGCGTCTTGTGCTCTCAATAC | |||
| KLF13 | CAGAGGAAGCACAAGTGCCACT | 137 | 65 |
| CGCGAACTTCTTGTTGCAGTCC | |||
| KLF14 | CATCCAGATATGATCGAGTACCG | 163 | 65 |
| CCTTGAGGGTAAGACTGACAGC | |||
| KLF15 | GTGAGAAGCCCTTCGCCTGCA | 114 | 67 |
| ACAGGACACTGGTACGGCTTCA | |||
| KLF16 | GACTGCGCCAAAGCCTACTACA | 171 | 65 |
| CCTGCCAGTCACAAGCAAAAGG | |||
| KLF17 | GCTGCCCAGGATAACGAGAAC | 128 | 67 |
| ATCTCTGCGCTGTGAGGAAAG | |||
| KLF18 | TCCATGGGCCAGAAAGTGAC | 197 | 67 |
| GGGTGTTCAGCTGGCTACTT |
| Control | 1 pg/ml | 100 pg/ml | 1 ng/ml | 100 ng/ml | |
| KLF1 | 1 | 1.42 | 0.94 | 0.22 | 0.7 |
| KLF2 | 1 | 1.03 | 0.67 | 0.27 | 0.77 |
| KLF3 | 1 | 1.63 | 1.35 | 0.38 | 1.35 |
| KLF4 | 1 | 1.1 | 0.24 | 1.56 | 0.8 |
| KLF5 | 1 | 0.98 | 0.7 | 0.3 | 0.96 |
| KLF6 | 1 | 0.57 | 0.54 | 0.46 | 0.47 |
| KLF7 | 1 | 0.87 | 0.5 | 0.12 | 1.02 |
| KLF8 | 1 | 4.99 | 0.85 | 0.34 | 1.02 |
| KLF9 | 1 | 3.02 | 1.3 | 0.41 | 1.08 |
| KLF10 | 1 | 1.4 | 1.38 | 0.26 | 1.15 |
| KLF11 | 1 | 1.45 | 1.5 | 0.28 | 0.91 |
| KLF12 | 1 | 0.59 | 0.76 | 0.22 | 0.88 |
| KLF13 | 1 | 1.84 | 0.92 | 0.27 | 1.12 |
| KLF14 | 1 | 1.45 | 1.05 | 0.32 | 1.51 |
| KLF15 | 1 | 2.9 | 1.49 | 0.89 | 2.14 |
| KLF16 | 1 | 2.31 | 1.47 | 0.36 | 0.93 |
| KLF17 | 1 | 0.39 | 0.41 | 0.66 | 2.47 |
| KLF18 | 1 | 1.23 | 0.87 | 1.75 | 1.91 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





