Submitted:
01 February 2025
Posted:
04 February 2025
You are already at the latest version
Abstract
Keywords:
1. Integration, Innovation and Insight
2. Significance
3. Introduction
3.1. A Combinatorial Problem
3.2. A Possible Solution
3.3. Wnt Signaling and Secretion
4. Methods
5. Time Series Data
6. Design of Experiment
6.1. Pipeline for Time Series Data
7. Results & Discussion
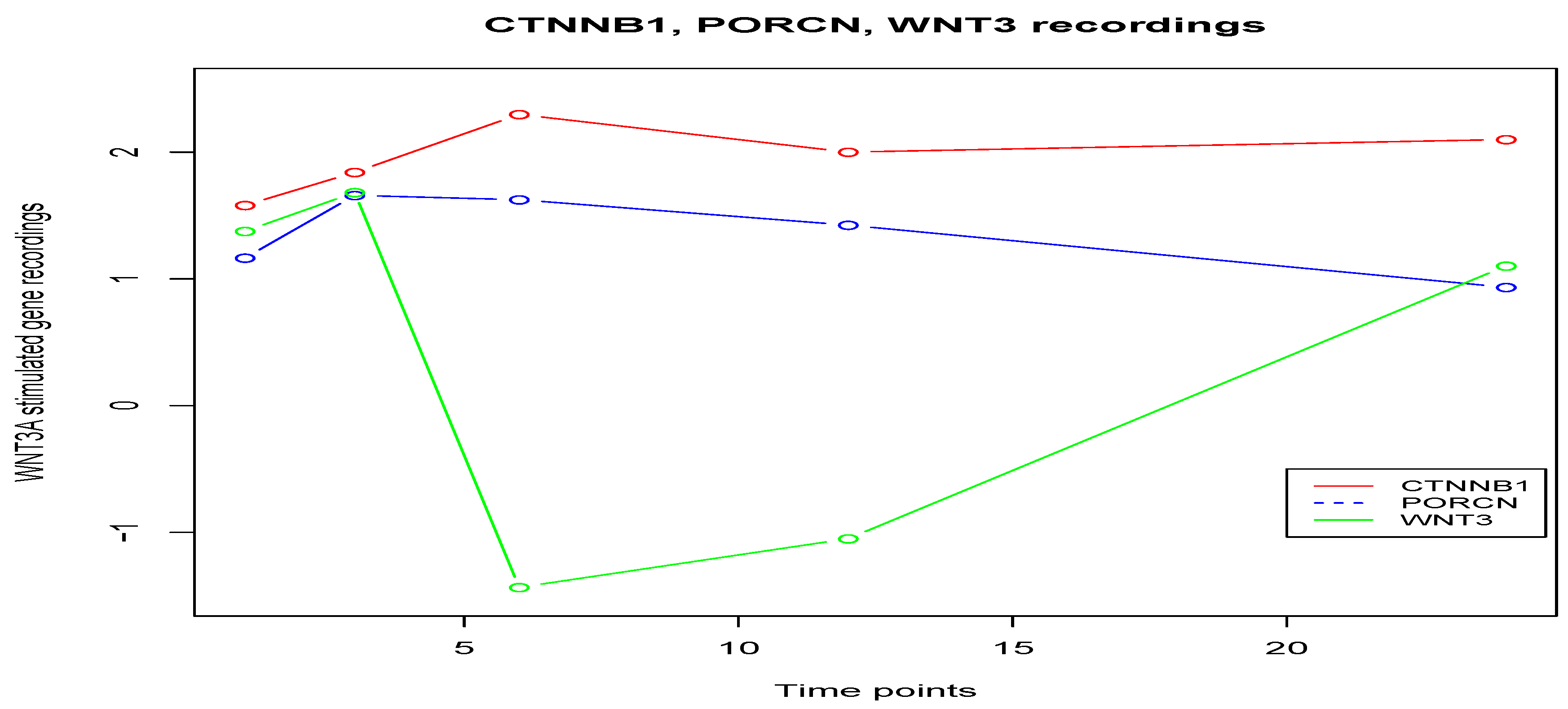
7.1. Time Series Data by Gujral and MacBeath [1]
7.2. Enumeration and Ranking of Combinations from Gujral and MacBeath [1]
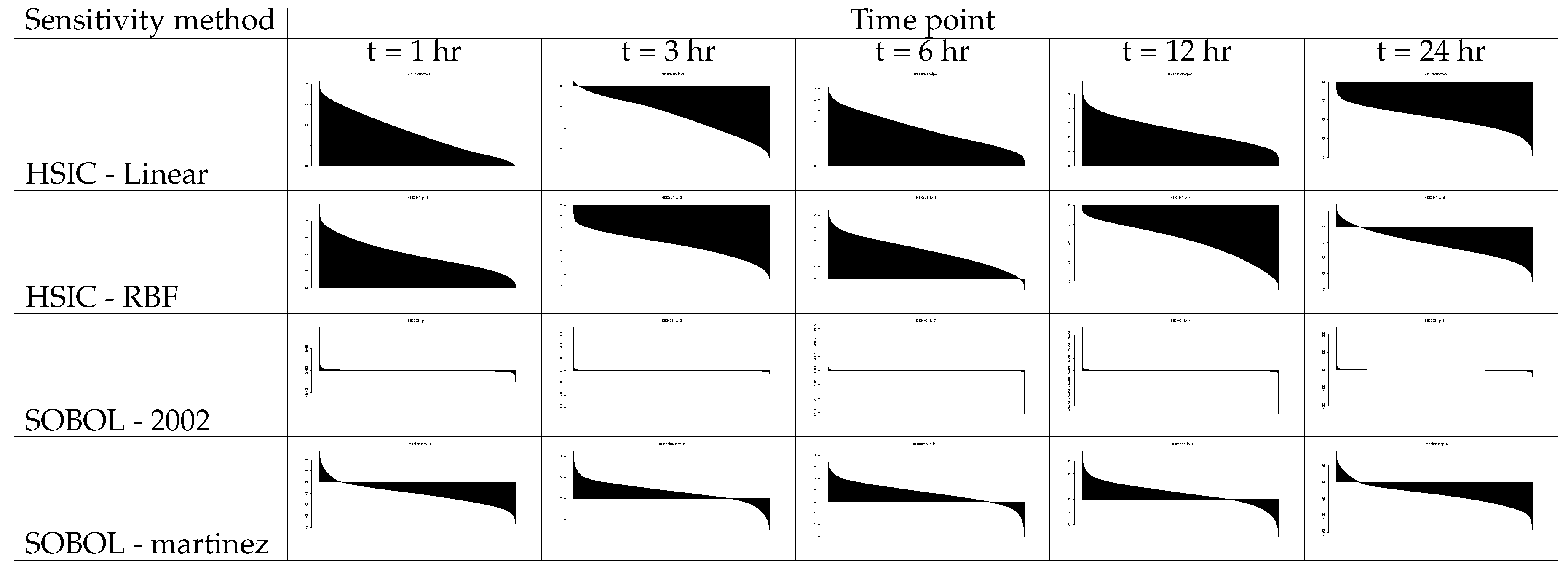
7.3. Conserved Machine Learning Rankings for Tested PORCN-WNT-X Combinations
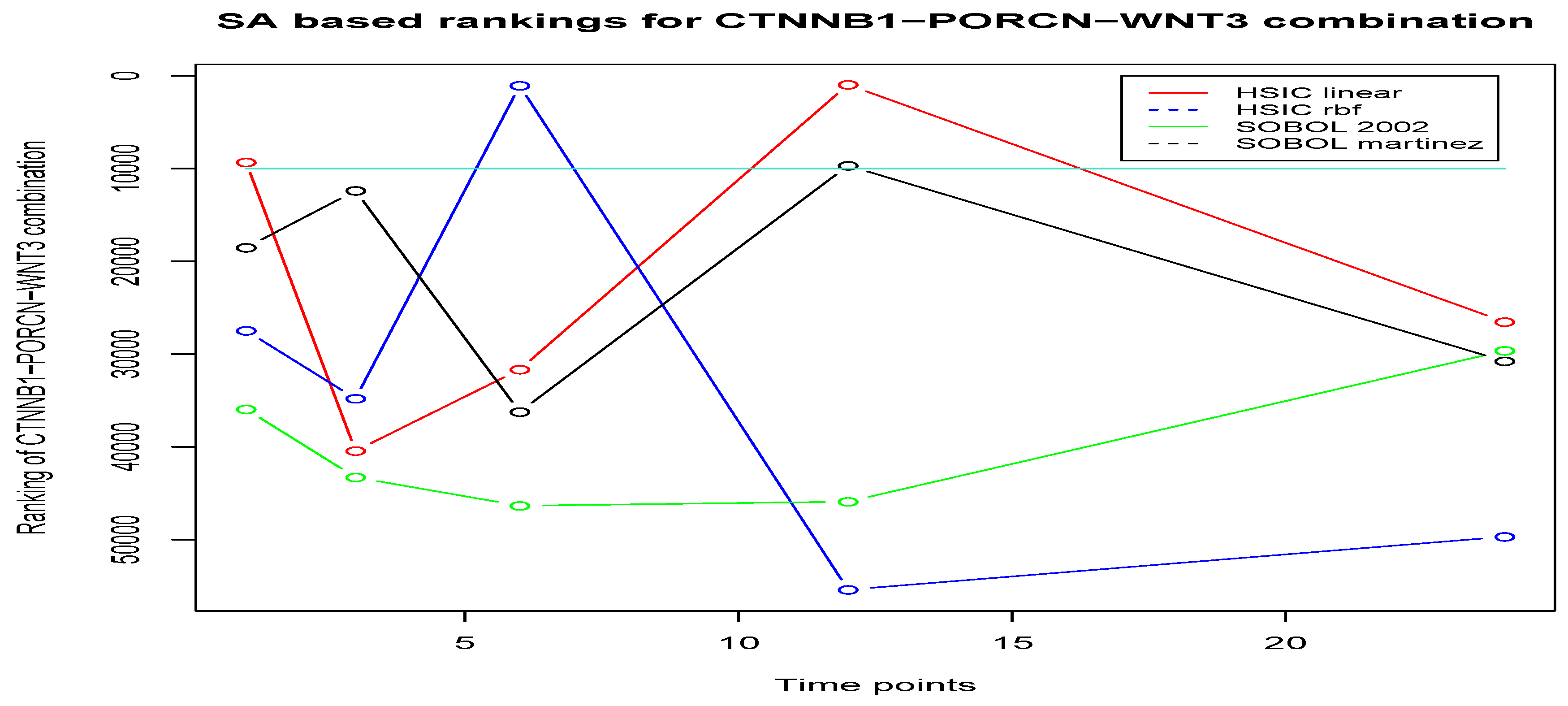
7.3.1. Examining the Behaviour of CTNNB1-PORCN-WNT3 Combination
7.4. Enumeration of top 10, 3rd Order Combinations for WNT3A Stimulated Response Genes
7.4.1. Adenomatosis Polyposis Coli (APC)
7.4.2. V-myc Avian Myelocytomatosis Viral Oncogene Homolog (MYC)
7.4.3. fRizzled Class Receptor (FZD)
7.4.4. Glycogen Synthase Kinase 3 (GSK3)
7.4.5. Dishevelled Segment Polarity Protein (DVL)
7.4.6. Low Density Lipoprotein Receptor-Related Protein (LRP)
7.4.7. C-Terminal Binding Protein (CTBP)
7.4.8. Cyclin D (CCND)
7.4.9. Wnt Family Member (WNT)
8. Conclusion
Competing interests
Author contributions statement
Availability of code
Acknowledgments
Supplementary
References
- Gujral, T.S.; MacBeath, G. A system-wide investigation of the dynamics of Wnt signaling reveals novel phases of transcriptional regulation. PloS one 2010, 5, e10024. [CrossRef]
- Sinha, S. Machine learning ranking of plausible (un) explored synergistic gene combinations using sensitivity indices of time series measurements of Wnt signaling pathway. Integrative Biology 2024, 16, zyae020. [CrossRef]
- Joachims, T. Training linear SVMs in linear time. In Proceedings of the Proceedings of the 12th ACM SIGKDD international conference on Knowledge discovery and data mining. ACM, 2006, pp. 217–226.
- Sharma, R. Wingless a new mutant in Drosophila melanogaster. Drosophila information service 1973, 50, 134–134.
- Thorstensen, L.; Lind, G.E.; Løvig, T.; Diep, C.B.; Meling, G.I.; Rognum, T.O.; Lothe, R.A. Genetic and epigenetic changes of components affecting the WNT pathway in colorectal carcinomas stratified by microsatellite instability. Neoplasia 2005, 7, 99–108. [CrossRef]
- Baron, R.; Kneissel, M. WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nature medicine 2013, 19, 179–192. [CrossRef]
- Clevers, H. Wnt/[β]-catenin signaling in development and disease. Cell 2006, 127, 469–480. [CrossRef]
- Sokol, S. Wnt Signaling in Embryonic Development; Vol. 17, Elsevier, 2011.
- Pinto, D.; Gregorieff, A.; Begthel, H.; Clevers, H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes & development 2003, 17, 1709–1713.
- Zhong, Z.; Ethen, N.J.; Williams, B.O. WNT signaling in bone development and homeostasis. Wiley Interdisciplinary Reviews: Developmental Biology 2014, 3, 489–500. [CrossRef]
- Pećina-Šlaus, N. Wnt signal transduction pathway and apoptosis: a review. Cancer Cell International 2010, 10, 1–5. [CrossRef]
- Kahn, M. Can we safely target the WNT pathway? Nature Reviews Drug Discovery 2014, 13, 513–532. [CrossRef]
- Garber, K. Drugging the Wnt pathway: problems and progress. Journal of the National Cancer Institute 2009, 101, 548–550. [CrossRef]
- Voronkov, A.; Krauss, S. Wnt/beta-catenin signaling and small molecule inhibitors. Current pharmaceutical design 2012, 19, 634. [CrossRef]
- Blagodatski, A.; Poteryaev, D.; Katanaev, V. Targeting the Wnt pathways for therapies. Mol Cell Ther 2014, 2, 28. [CrossRef]
- Curtin, J.C.; Lorenzi, M.V. Drug discovery approaches to target Wnt signaling in cancer stem cells. Oncotarget 2010, 1, 552. [CrossRef]
- Bänziger, C.; Soldini, D.; Schütt, C.; Zipperlen, P.; Hausmann, G.; Basler, K. Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell 2006, 125, 509–522. [CrossRef]
- Bartscherer, K.; Pelte, N.; Ingelfinger, D.; Boutros, M. Secretion of Wnt ligands requires Evi, a conserved transmembrane protein. Cell 2006, 125, 523–533. [CrossRef]
- Kurayoshi, M.; Yamamoto, H.; Izumi, S.; Kikuchi, A. Post-translational palmitoylation and glycosylation of Wnt-5a are necessary for its signalling. Biochemical Journal 2007, 402, 515–523. [CrossRef]
- Gao, X.; Hannoush, R.N. Single-cell imaging of Wnt palmitoylation by the acyltransferase porcupine. Nature chemical biology 2014, 10, 61–68. [CrossRef]
- Sinha, S. Hilbert-Schmidt and Sobol sensitivity indices for static and time series Wnt signaling measurements in colorectal cancer-part A. BMC systems biology 2017, 11, 120. [CrossRef]
- Da Veiga, S. Global sensitivity analysis with dependence measures. Journal of Statistical Computation and Simulation 2015, 85, 1283–1305. [CrossRef]
- Saltelli, A. Making best use of model evaluations to compute sensitivity indices. Computer physics communications 2002, 145, 280–297. [CrossRef]
- Martinez, J. Analyse de sensibilite globale par decomposition de la variance. Presentation in “Journée des GdR Ondes & Mascot 2011, 13, 207.
- Baudin, M.; Boumhaout, K.; Delage, T.; Iooss, B.; Martinez, J.M. Numerical stability of Sobol’indices estimation formula. In Proceedings of the Proceedings of the 8th International Conference on Sensitivity Analysis of Model Output (SAMO 2016), 2016, Vol. 30, pp. 50–51.
- Tanaka, K.; Okabayashi, K.; Asashima, M.; Perrimon, N.; Kadowaki, T. The evolutionarily conserved porcupine gene family is involved in the processing of the Wnt family. European Journal of Biochemistry 2000, 267, 4300–4311. [CrossRef]
- Liu, J.; Xiao, Q.; Xiao, J.; Niu, C.; Li, Y.; Zhang, X.; Zhou, Z.; Shu, G.; Yin, G. Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal transduction and targeted therapy 2022, 7, 3. [CrossRef]
- Willert, K.; Brown, J.D.; Danenberg, E.; Duncan, A.W.; Weissman, I.L.; Reya, T.; Yates III, J.R.; Nusse, R. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 2003, 423, 448–452. [CrossRef]
- Liu, J.; Pan, S.; Hsieh, M.H.; Ng, N.; Sun, F.; Wang, T.; Kasibhatla, S.; Schuller, A.G.; Li, A.G.; Cheng, D.; et al. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proceedings of the National Academy of Sciences 2013, 110, 20224–20229. [CrossRef]
- Madan, B.; Ke, Z.; Harmston, N.; Ho, S.Y.; Frois, A.; Alam, J.; Jeyaraj, D.A.; Pendharkar, V.; Ghosh, K.; Virshup, I.H.; et al. Wnt addiction of genetically defined cancers reversed by PORCN inhibition. Oncogene 2016, 35, 2197–2207. [CrossRef]
- McCrea, P.D.; Turck, C.W.; Gumbiner, B. A homolog of the armadillo protein in Drosophila (plakoglobin) associated with E-cadherin. Science 1991, 254, 1359–1361. [CrossRef]
- Peifer, M.; Rauskolb, C.; Williams, M.; Riggleman, B.; Wieschaus, E. The segment polarity gene armadillo interacts with the wingless signaling pathway in both embryonic and adult pattern formation. Development 1991, 111, 1029–1043. [CrossRef]
- Kemler, R. From cadherins to catenins: cytoplasmic protein interactions and regulation of cell adhesion. Trends in Genetics 1993, 9, 317–321. [CrossRef]
- Kuraguchi, M.; Wang, X.P.; Bronson, R.T.; Rothenberg, R.; Ohene-Baah, N.Y.; Lund, J.J.; Kucherlapati, M.; Maas, R.L.; Kucherlapati, R. Adenomatous polyposis coli (APC) is required for normal development of skin and thymus. PLoS genetics 2006, 2, e146. [CrossRef]
- Wang, X.P.; O’Connell, D.J.; Lund, J.J.; Saadi, I.; Kuraguchi, M.; Turbe-Doan, A.; Cavallesco, R.; Kim, H.; Park, P.J.; Harada, H.; et al. Apc inhibition of Wnt signaling regulates supernumerary tooth formation during embryogenesis and throughout adulthood 2009.
- Yang, B.; Zhang, J.; Wang, J.; Fan, W.; Barbier-Torres, L.; Yang, X.; Justo, M.A.R.; Liu, T.; Chen, Y.; Steggerda, J.; et al. CSNK2A1-mediated MAX phosphorylation upregulates HMGB1 and IL-6 expression in cholangiocarcinoma progression. Hepatology Communications 2023, 7, e00144. [CrossRef]
- Sun, X.X.; Chen, Y.; Su, Y.; Wang, X.; Chauhan, K.M.; Liang, J.; Daniel, C.J.; Sears, R.C.; Dai, M.S. SUMO protease SENP1 deSUMOylates and stabilizes c-Myc. Proceedings of the National Academy of Sciences 2018, 115, 10983–10988. [CrossRef]
- Ghimire, S.R.; Deans, M.R. Frizzled3 and Frizzled6 cooperate with Vangl2 to direct cochlear innervation by type II spiral ganglion neurons. Journal of Neuroscience 2019, 39, 8013–8023. [CrossRef]
- Jaka, O.; Casas-Fraile, L.; Azpitarte, M.; Aiastui, A.; De Munain, A.L.; Saenz, A. FRZB and melusin, overexpressed in LGMD2A, regulate integrin β1D isoform replacement altering myoblast fusion and the integrin-signalling pathway. Expert Reviews in Molecular Medicine 2017, 19, e2. [CrossRef]
- Zhong, Z.; Harmston, N.; Wood, K.C.; Madan, B.; Virshup, D.M.; et al. A p300/GATA6 axis determines differentiation and Wnt dependency in pancreatic cancer models. The Journal of Clinical Investigation 2022, 132. [CrossRef]
- Gan, X.q.; Wang, J.y.; Xi, Y.; Wu, Z.l.; Li, Y.p.; Li, L. Nuclear Dvl, c-Jun, β-catenin, and TCF form a complex leading to stabiLization of β-catenin–TCF interaction. The Journal of cell biology 2008, 180, 1087–1100. [CrossRef]
- Wang, L.; Liu, X.; Gusev, E.; Wang, C.; Fagotto, F. Regulation of the phosphorylation and nuclear import and export of β-catenin by APC and its cancer-related truncated form. Journal of cell science 2014, 127, 1647–1659. [CrossRef]
- Holt, R.J.; Young, R.M.; Crespo, B.; Ceroni, F.; Curry, C.J.; Bellacchio, E.; Bax, D.A.; Ciolfi, A.; Simon, M.; Fagerberg, C.R.; et al. De novo missense variants in FBXW11 cause diverse developmental phenotypes including brain, eye, and digit anomalies. The American Journal of Human Genetics 2019, 105, 640–657. [CrossRef]
- Kim, T.W.; Kwak, S.; Shin, J.; Kang, B.H.; Lee, S.E.; Suh, M.Y.; Kim, J.H.; Hwang, I.Y.; Lee, J.H.; Choi, J.; et al. Ctbp2-mediated β-catenin regulation is required for exit from pluripotency. Experimental & molecular medicine 2017, 49, e385–e385.
- Bao, Y.; Gabrielpillai, J.; Dietrich, J.; Zarbl, R.; Strieth, S.; Schröck, F.; Dietrich, D. Fibroblast growth factor (FGF), FGF receptor (FGFR), and cyclin D1 (CCND1) DNA methylation in head and neck squamous cell carcinomas is associated with transcriptional activity, gene amplification, human papillomavirus (HPV) status, and sensitivity to tyrosine kinase inhibitors. Clinical Epigenetics 2021, 13, 1–18.
- Brandt, M.M.; Van Dijk, C.G.; Chrifi, I.; Kool, H.M.; Bürgisser, P.E.; Louzao-Martinez, L.; Pei, J.; Rottier, R.J.; Verhaar, M.C.; Duncker, D.J.; et al. Endothelial loss of Fzd5 stimulates PKC/Ets1-mediated transcription of Angpt2 and Flt1. Angiogenesis 2018, 21, 805–821. [CrossRef]
| 1 | Aspects of unpublished work presented as poster in the Berkeley Cell Symposyia : Technology, Biology & Data Science, 2016, Berkeley, USA |
 |
| Ranking @ using HSIC - linear | |||||||||||
| 3rd order comb. | 3rd order comb. | ||||||||||
| CXXC4-PORCN-WNT4 | 49 | 6186 | 19448 | 20672 | 51388 | PITX2-PORCN-WNT4 | 177 | 16891 | 32175 | 32123 | 27627 |
| FZD6-PORCN-WNT2B | 379 | 19259 | 55786 | 24330 | 16739 | FZD6-PORCN-WNT4 | 394 | 39861 | 46523 | 785 | 6046 |
| FOSL1-PORCN-WNT4 | 455 | 37570 | 20729 | 10105 | 38487 | PITX2-PORCN-WNT2B | 630 | 42884 | 44910 | 50269 | 15186 |
| FZD5-PORCN-WNT4 | 646 | 40380 | 25866 | 12816 | 56710 | FOSL1-PORCN-WNT2B | 670 | 19667 | 46545 | 40946 | 19709 |
| KREMEN1-PORCN-WNT4 | 693 | 6753 | 23243 | 1864 | 44869 | FZD7-PORCN-WNT3A | 780 | 35610 | 948 | 25632 | 12174 |
| DKK1-PORCN-WNT2B | 1222 | 26601 | 56978 | 25305 | 51044 | KREMEN1-PORCN-WNT3A | 1377 | 25809 | 2588 | 12830 | 26801 |
| BCL9-PORCN-WNT2B | 1394 | 28608 | 33398 | 47599 | 21197 | FZD8-PORCN-WNT4 | 1416 | 27491 | 15836 | 9789 | 36903 |
| NLK-PORCN-WNT4 | 1742 | 49908 | 50644 | 30880 | 6042 | GSK3B-PORCN-WNT4 | 1893 | 29701 | 28162 | 8296 | 19073 |
| FRAT1-PORCN-WNT4 | 1985 | 2835 | 18017 | 16053 | 23730 | FZD6-PORCN-WNT2 | 2024 | 30877 | 41431 | 1558 | 29140 |
| FZD5-PORCN-WNT2B | 2123 | 44490 | 45300 | 33310 | 53770 | FZD6-PORCN-WNT5A | 2165 | 41983 | 26329 | 43017 | 27185 |
| CCND3-PORCN-WNT5A | 2270 | 38617 | 15634 | 39601 | 37699 | CXXC4-PORCN-WNT3 | 2291 | 2240 | 24537 | 6255 | 30135 |
| FZD6-PORCN-WNT3A | 2394 | 56979 | 41510 | 33895 | 6602 | CSNK1D-PORCN-WNT5A | 2899 | 17376 | 34214 | 55252 | 32946 |
| BCL9-PORCN-WNT2 | 2978 | 30124 | 2987 | 21306 | 45043 | DKK1-PORCN-WNT3A | 3027 | 51411 | 42480 | 14175 | 39772 |
| EP300-PORCN-WNT4 | 3047 | 52212 | 12666 | 11882 | 56673 | LRP5-PORCN-WNT5A | 3051 | 25077 | 6874 | 53519 | 30711 |
| FZD8-PORCN-WNT2 | 3084 | 38943 | 6461 | 12608 | 17348 | CSNK1A1-PORCN-WNT4 | 3257 | 26979 | 26665 | 9094 | 28864 |
| DIXDC1-PORCN-WNT2B | 3266 | 11238 | 38406 | 37800 | 51881 | CSNK1G1-PORCN-WNT2 | 3410 | 20366 | 30313 | 12508 | 34531 |
| FOSL1-PORCN-WNT5A | 3631 | 16428 | 9587 | 43895 | 35656 | DAAM1-PORCN-WNT4 | 3680 | 47587 | 5186 | 9818 | 24761 |
| FZD1-PORCN-WNT2 | 3737 | 14464 | 10387 | 26137 | 31141 | EP300-PORCN-WNT2B | 3826 | 37202 | 48507 | 42391 | 54306 |
| DIXDC1-PORCN-WNT5A | 3885 | 26867 | 8379 | 47688 | 52084 | FOSL1-PORCN-WNT3A | 3900 | 15019 | 1518 | 39920 | 12124 |
| FZD2-PORCN-WNT3 | 3994 | 23668 | 24913 | 3556 | 38793 | CSNK1A1-PORCN-WNT2B | 4109 | 24272 | 43637 | 30113 | 14274 |
| BCL9-PORCN-WNT3A | 4125 | 17834 | 1540 | 51723 | 23039 | FOSL1-PORCN-WNT2 | 4144 | 48973 | 6608 | 18938 | 40921 |
| LEF1-PORCN-WNT3 | 4220 | 6502 | 35317 | 5065 | 1556 | LRP6-PORCN-WNT2B | 4288 | 11513 | 52986 | 43798 | 7888 |
| CTNNB1-PORCN-WNT2B | 4381 | 41494 | 54608 | 33159 | 44891 | FZD5-PORCN-WNT3A | 4393 | 44820 | 4226 | 33407 | 54333 |
| GSK3B-PORCN-WNT2B | 4525 | 24848 | 40685 | 28860 | 9721 | FZD2-PORCN-WNT4 | 4576 | 26380 | 31174 | 13954 | 52829 |
| FZD5-PORCN-WNT2 | 4674 | 42137 | 9005 | 17361 | 56340 | EP300-PORCN-WNT5A | 4719 | 30282 | 10559 | 49731 | 55789 |
| FRAT1-PORCN-WNT5A | 4744 | 35332 | 12577 | 52285 | 31329 | FRAT1-PORCN-WNT3A | 4941 | 28915 | 3063 | 37414 | 10241 |
| KREMEN1-PORCN-WNT2 | 4959 | 36712 | 10209 | 2355 | 46390 | CTNNBIP1-PORCN-WNT4 | 5017 | 38878 | 13233 | 4965 | 42568 |
| FZD7-PORCN-WNT2 | 5063 | 48509 | 3534 | 18543 | 37452 | FBXW2-PORCN-WNT5A | 5093 | 9753 | 1927 | 56299 | 23944 |
| CSNK1A1-PORCN-WNT5A | 5243 | 23800 | 15741 | 37419 | 31250 | FRZB-PORCN-WNT5A | 5316 | 15372 | 16217 | 53391 | 42789 |
| FZD2-PORCN-WNT2 | 5319 | 35217 | 4925 | 18448 | 50496 | FZD6-PORCN-WNT3 | 5539 | 17206 | 33991 | 301 | 6205 |
| DVL2-PORCN-WNT2B | 5842 | 46277 | 40915 | 44683 | 55710 | LEF1-PORCN-WNT4 | 5938 | 6340 | 32575 | 15016 | 39684 |
| DIXDC1-PORCN-WNT2 | 6166 | 9846 | 4217 | 17714 | 53792 | DVL1-PORCN-WNT2B | 6171 | 16317 | 38040 | 29254 | 5289 |
| FBXW11-PORCN-WNT2 | 6311 | 39888 | 7951 | 33180 | 19070 | CTNNB1-PORCN-WNT5A | 6382 | 34261 | 18812 | 36691 | 51406 |
| DVL1-PORCN-WNT4 | 6631 | 47068 | 9083 | 4176 | 6383 | FRAT1-PORCN-WNT3 | 6706 | 18258 | 19680 | 8889 | 11973 |
| DAAM1-PORCN-WNT5A | 6758 | 49371 | 3391 | 50318 | 25339 | FBXW11-PORCN-WNT3 | 6762 | 31256 | 9446 | 8712 | 8411 |
| FBXW2-PORCN-WNT2 | 6905 | 25689 | 1063 | 38513 | 34061 | DVL1-PORCN-WNT2 | 7032 | 39563 | 1823 | 5185 | 12571 |
| DKK1-PORCN-WNT3 | 7165 | 12201 | 52957 | 1425 | 47839 | FSHB-PORCN-WNT4 | 7187 | 35341 | 51879 | 43751 | 30960 |
| DIXDC1-PORCN-WNT3A | 7226 | 9557 | 1088 | 34837 | 45504 | APC-PORCN-WNT3 | 7285 | 26324 | 14843 | 5204 | 12325 |
| CSNK2A1-PORCN-WNT2 | 7317 | 28374 | 7626 | 21080 | 31795 | DVL2-PORCN-WNT5A | 7381 | 46344 | 27348 | 46664 | 56984 |
| NLK-PORCN-WNT3 | 7444 | 40963 | 56870 | 11439 | 3041 | GSK3B-PORCN-WNT3A | 7574 | 34492 | 6475 | 23367 | 8516 |
| FRZB-PORCN-WNT3A | 7687 | 8222 | 4498 | 44883 | 17262 | CTNNBIP1-PORCN-WNT5A | 7693 | 15781 | 8108 | 34940 | 32551 |
| CSNK1G1-PORCN-WNT3A | 7824 | 36551 | 31135 | 32075 | 7370 | FZD7-PORCN-WNT3 | 7919 | 50229 | 13866 | 3977 | 19719 |
| FBXW2-PORCN-WNT3A | 8007 | 29570 | 331 | 44149 | 23392 | CTNNBIP1-PORCN-WNT3 | 8175 | 33782 | 15052 | 3217 | 10272 |
| FZD8-PORCN-WNT3 | 8198 | 42890 | 20340 | 2880 | 17514 | DVL2-PORCN-WNT3A | 8315 | 32904 | 22980 | 44056 | 50976 |
| PORCN-SFRP1-WNT2B | 8374 | 33217 | 32367 | 38609 | 43781 | EP300-PORCN-WNT3 | 8497 | 44763 | 16497 | 4191 | 45957 |
| PORCN-SFRP1-WNT5A | 8531 | 30769 | 38133 | 56398 | 46745 | CSNK1A1-PORCN-WNT3 | 8682 | 9887 | 21818 | 2357 | 12384 |
| LRP5-PORCN-WNT3 | 8787 | 10685 | 15101 | 51335 | 9714 | FOSL1-PORCN-WNT3 | 8888 | 41174 | 18861 | 4616 | 20787 |
| AXIN1-PORCN-WNT4 | 9013 | 31986 | 17718 | 11205 | 34131 | PORCN-WNT4-WNT5A | 9093 | 21416 | 44185 | 36030 | 52780 |
| FRAT1-PORCN-WNT2 | 9314 | 30043 | 8901 | 28853 | 31633 | CTNNB1-PORCN-WNT3 | 9325 | 40433 | 31660 | 954 | 26533 |
| GSK3B-PORCN-WNT2 | 9371 | 40431 | 15296 | 6282 | 27880 | AES-PORCN-WNT5A | 9395 | 43267 | 5747 | 17185 | 30918 |
| FRZB-PORCN-WNT3 | 9560 | 6430 | 26600 | 4539 | 19524 | CTBP1-PORCN-WNT3A | 9567 | 4412 | 1206 | 29048 | 13053 |
| MYC-PORCN-WNT2 | 9597 | 42141 | 37704 | 21380 | 33950 | DAAM1-PORCN-WNT2 | 9600 | 49958 | 2043 | 12313 | 45710 |
| CTNNBIP1-PORCN-WNT2 | 9847 | 23180 | 4750 | 9800 | 36378 | FGF4-PORCN-WNT2B | 9991 | 29929 | 44511 | 25174 | 44265 |
| Ranking @ using HSIC - rbf | |||||||||||
| 3rd order comb. | 3rd order comb. | ||||||||||
| CXXC4-PORCN-WNT4 | 12492 | 2278 | 24920 | 46755 | 55842 | PITX2-PORCN-WNT4 | 9247 | 2492 | 36647 | 13332 | 52478 |
| FZD6-PORCN-WNT2B | 47 | 9685 | 16997 | 36922 | 48852 | FZD6-PORCN-WNT4 | 49 | 30454 | 13795 | 53236 | 50505 |
| FOSL1-PORCN-WNT4 | 1429 | 25705 | 30547 | 51837 | 52319 | PITX2-PORCN-WNT2B | 6802 | 25276 | 16938 | 13140 | 54357 |
| FZD5-PORCN-WNT4 | 21655 | 38004 | 30699 | 16505 | 55082 | FOSL1-PORCN-WNT2B | 4296 | 5489 | 1279 | 29512 | 55035 |
| KREMEN1-PORCN-WNT4 | 4572 | 591 | 18551 | 4045 | 46258 | FZD7-PORCN-WNT3A | 7046 | 46922 | 42358 | 23913 | 36667 |
| DKK1-PORCN-WNT2B | 6263 | 7902 | 41853 | 840 | 49894 | KREMEN1-PORCN-WNT3A | 8946 | 29482 | 33980 | 6745 | 37128 |
| BCL9-PORCN-WNT2B | 7648 | 28196 | 20793 | 10962 | 34371 | FZD8-PORCN-WNT4 | 3177 | 7307 | 28634 | 31520 | 31714 |
| NLK-PORCN-WNT4 | 33383 | 45823 | 7542 | 34942 | 49961 | GSK3B-PORCN-WNT4 | 7755 | 14164 | 10378 | 27077 | 53726 |
| FRAT1-PORCN-WNT4 | 6345 | 412 | 40450 | 45568 | 49626 | FZD6-PORCN-WNT2 | 586 | 40987 | 4888 | 4733 | 51712 |
| FZD5-PORCN-WNT2B | 20497 | 40866 | 6253 | 29353 | 51974 | FZD6-PORCN-WNT5A | 88 | 50478 | 30280 | 17366 | 51000 |
| CCND3-PORCN-WNT5A | 12281 | 45137 | 3555 | 19433 | 30263 | CXXC4-PORCN-WNT3 | 37775 | 8521 | 7319 | 54045 | 46168 |
| FZD6-PORCN-WNT3A | 75 | 57089 | 15014 | 7701 | 45470 | CSNK1D-PORCN-WNT5A | 17071 | 12074 | 1312 | 10487 | 50936 |
| BCL9-PORCN-WNT2 | 28083 | 32378 | 4465 | 3529 | 55212 | DKK1-PORCN-WNT3A | 3656 | 53268 | 6994 | 22200 | 48539 |
| EP300-PORCN-WNT4 | 5823 | 49192 | 43675 | 49247 | 45902 | LRP5-PORCN-WNT5A | 4664 | 27857 | 8039 | 7363 | 54523 |
| FZD8-PORCN-WNT2 | 9002 | 35606 | 6653 | 15940 | 32619 | CSNK1A1-PORCN-WNT4 | 22396 | 7881 | 36610 | 47685 | 56275 |
| DIXDC1-PORCN-WNT2B | 20111 | 15273 | 9550 | 18377 | 30375 | CSNK1G1-PORCN-WNT2 | 21543 | 32722 | 24973 | 9534 | 56929 |
| FOSL1-PORCN-WNT5A | 684 | 10157 | 3538 | 9803 | 48095 | DAAM1-PORCN-WNT4 | 35996 | 39470 | 9367 | 45869 | 39406 |
| FZD1-PORCN-WNT2 | 16381 | 31579 | 18868 | 2713 | 56723 | EP300-PORCN-WNT2B | 15830 | 18382 | 7434 | 20043 | 39500 |
| DIXDC1-PORCN-WNT5A | 17412 | 29192 | 9192 | 10144 | 42283 | FOSL1-PORCN-WNT3A | 1761 | 8657 | 47396 | 12988 | 53731 |
| FZD2-PORCN-WNT3 | 3861 | 10371 | 8475 | 41168 | 44534 | CSNK1A1-PORCN-WNT2B | 29063 | 5140 | 20656 | 25842 | 56335 |
| BCL9-PORCN-WNT3A | 14481 | 19317 | 37603 | 22307 | 42095 | FOSL1-PORCN-WNT2 | 8304 | 49480 | 51245 | 9737 | 56572 |
| LEF1-PORCN-WNT3 | 15283 | 16253 | 23588 | 39987 | 45869 | LRP6-PORCN-WNT2B | 9683 | 1526 | 19256 | 22267 | 47876 |
| CTNNB1-PORCN-WNT2B | 3504 | 38996 | 12248 | 21395 | 50262 | FZD5-PORCN-WNT3A | 12410 | 43484 | 27634 | 36130 | 51870 |
| GSK3B-PORCN-WNT2B | 8048 | 11294 | 10179 | 16139 | 54531 | FZD2-PORCN-WNT4 | 695 | 6500 | 28617 | 20769 | 54707 |
| FZD5-PORCN-WNT2 | 36093 | 40927 | 1182 | 2948 | 56700 | EP300-PORCN-WNT5A | 14385 | 21157 | 9773 | 23711 | 50138 |
| FRAT1-PORCN-WNT5A | 9891 | 40070 | 11853 | 25638 | 52101 | FRAT1-PORCN-WNT3A | 2312 | 22937 | 18240 | 22166 | 52333 |
| KREMEN1-PORCN-WNT2 | 11953 | 40138 | 35447 | 19055 | 52764 | CTNNBIP1-PORCN-WNT4 | 8250 | 26459 | 7633 | 44740 | 54344 |
| FZD7-PORCN-WNT2 | 34943 | 49486 | 52615 | 11489 | 44146 | FBXW2-PORCN-WNT5A | 11096 | 281 | 22916 | 22682 | 46584 |
| CSNK1A1-PORCN-WNT5A | 18048 | 10911 | 1449 | 21232 | 54808 | FRZB-PORCN-WNT5A | 4873 | 5304 | 4481 | 15496 | 53540 |
| FZD2-PORCN-WNT2 | 1870 | 34234 | 31607 | 23727 | 56721 | FZD6-PORCN-WNT3 | 9106 | 30120 | 463 | 54159 | 46051 |
| DVL2-PORCN-WNT2B | 6933 | 36546 | 23445 | 39919 | 54053 | LEF1-PORCN-WNT4 | 5168 | 560 | 21906 | 36211 | 39978 |
| DIXDC1-PORCN-WNT2 | 23094 | 19050 | 12525 | 5084 | 37797 | DVL1-PORCN-WNT2B | 11320 | 1449 | 22904 | 39255 | 42653 |
| FBXW11-PORCN-WNT2 | 12778 | 29459 | 19512 | 7578 | 56052 | CTNNB1-PORCN-WNT5A | 9172 | 29306 | 14624 | 17774 | 54309 |
| DVL1-PORCN-WNT4 | 9417 | 29124 | 39214 | 53815 | 39945 | FRAT1-PORCN-WNT3 | 31121 | 16135 | 7261 | 44737 | 43354 |
| DAAM1-PORCN-WNT5A | 12232 | 34746 | 14301 | 11892 | 33603 | FBXW11-PORCN-WNT3 | 21925 | 17070 | 11142 | 36917 | 20820 |
| FBXW2-PORCN-WNT2 | 15457 | 8876 | 5216 | 27560 | 28062 | DVL1-PORCN-WNT2 | 17995 | 21939 | 25504 | 2801 | 52991 |
| DKK1-PORCN-WNT3 | 40172 | 10975 | 8157 | 48246 | 47170 | FSHB-PORCN-WNT4 | 15962 | 23579 | 8654 | 7169 | 56130 |
| DIXDC1-PORCN-WNT3A | 5822 | 6206 | 42682 | 5541 | 30451 | APC-PORCN-WNT3 | 30579 | 10413 | 26221 | 51601 | 50456 |
| CSNK2A1-PORCN-WNT2 | 25929 | 18590 | 35881 | 10625 | 56369 | DVL2-PORCN-WNT5A | 8044 | 40090 | 34866 | 13832 | 54387 |
| NLK-PORCN-WNT3 | 50644 | 29189 | 2713 | 13343 | 42697 | GSK3B-PORCN-WNT3A | 3461 | 14490 | 6838 | 14639 | 54422 |
| FRZB-PORCN-WNT3A | 6749 | 34490 | 49148 | 22906 | 54377 | CTNNBIP1-PORCN-WNT5A | 7305 | 10873 | 2924 | 1222 | 53417 |
| CSNK1G1-PORCN-WNT3A | 21663 | 38683 | 3656 | 2044 | 55146 | FZD7-PORCN-WNT3 | 53596 | 48671 | 6021 | 51770 | 28806 |
| FBXW2-PORCN-WNT3A | 6686 | 29263 | 3513 | 11200 | 19870 | CTNNBIP1-PORCN-WNT3 | 33531 | 24069 | 10483 | 42349 | 51457 |
| FZD8-PORCN-WNT3 | 35456 | 36452 | 36269 | 42228 | 28440 | DVL2-PORCN-WNT3A | 14794 | 20048 | 11512 | 8279 | 52754 |
| PORCN-SFRP1-WNT2B | 4723 | 27655 | 8331 | 26598 | 23511 | EP300-PORCN-WNT3 | 50856 | 39748 | 37310 | 50396 | 40943 |
| PORCN-SFRP1-WNT5A | 8990 | 18239 | 20358 | 7994 | 16383 | CSNK1A1-PORCN-WNT3 | 29342 | 2632 | 19879 | 55053 | 49188 |
| LRP5-PORCN-WNT3 | 24747 | 9932 | 38589 | 30383 | 52298 | FOSL1-PORCN-WNT3 | 18352 | 38735 | 1724 | 45267 | 50576 |
| AXIN1-PORCN-WNT4 | 12916 | 16183 | 30565 | 49994 | 53220 | PORCN-WNT4-WNT5A | 46004 | 7077 | 13157 | 8574 | 11165 |
| FRAT1-PORCN-WNT2 | 7732 | 25639 | 17621 | 15152 | 56257 | CTNNB1-PORCN-WNT3 | 27473 | 34798 | 1060 | 55400 | 49686 |
| GSK3B-PORCN-WNT2 | 7902 | 27843 | 40790 | 20077 | 56514 | AES-PORCN-WNT5A | 44241 | 42535 | 20909 | 4940 | 45583 |
| FRZB-PORCN-WNT3 | 32861 | 8504 | 11671 | 40944 | 46353 | CTBP1-PORCN-WNT3A | 7201 | 6127 | 37568 | 36662 | 54763 |
| MYC-PORCN-WNT2 | 20775 | 49804 | 1366 | 1434 | 56653 | DAAM1-PORCN-WNT2 | 32528 | 49156 | 12339 | 6293 | 46115 |
| CTNNBIP1-PORCN-WNT2 | 13877 | 15718 | 24186 | 3006 | 56598 | FGF4-PORCN-WNT2B | 49418 | 20740 | 4720 | 31683 | 48786 |
| Ranking @ using SOBOL - 2002 | |||||||||||
| 3rd order comb. | 3rd order comb. | ||||||||||
| CXXC4-PORCN-WNT4 | 9600 | 4879 | 1294 | 6463 | 37839 | PITX2-PORCN-WNT4 | 38340 | 23219 | 45866 | 44679 | 30684 |
| FZD6-PORCN-WNT2B | 31884 | 54376 | 34870 | 43757 | 175 | FZD6-PORCN-WNT4 | 12889 | 17968 | 25060 | 5439 | 42744 |
| FOSL1-PORCN-WNT4 | 47380 | 31804 | 36756 | 47696 | 19151 | PITX2-PORCN-WNT2B | 9536 | 5794 | 12824 | 18961 | 17911 |
| FZD5-PORCN-WNT4 | 9207 | 29406 | 23871 | 11381 | 42466 | FOSL1-PORCN-WNT2B | 9686 | 2290 | 2189 | 14059 | 39340 |
| KREMEN1-PORCN-WNT4 | 2994 | 1634 | 5445 | 11732 | 52786 | FZD7-PORCN-WNT3A | 20917 | 23923 | 24750 | 6039 | 49403 |
| DKK1-PORCN-WNT2B | 41383 | 41457 | 40170 | 35729 | 7578 | KREMEN1-PORCN-WNT3A | 36088 | 42706 | 41171 | 37942 | 19003 |
| BCL9-PORCN-WNT2B | 619 | 12023 | 16698 | 20050 | 36808 | FZD8-PORCN-WNT4 | 4950 | 13451 | 24888 | 5374 | 42530 |
| NLK-PORCN-WNT4 | 3277 | 2846 | 7185 | 12509 | 42889 | GSK3B-PORCN-WNT4 | 1508 | 45949 | 15059 | 14068 | 43601 |
| FRAT1-PORCN-WNT4 | 26095 | 16695 | 23293 | 12549 | 25899 | FZD6-PORCN-WNT2 | 25276 | 2810 | 22247 | 13377 | 56981 |
| FZD5-PORCN-WNT2B | 39424 | 1395 | 52348 | 50158 | 12675 | FZD6-PORCN-WNT5A | 44224 | 38925 | 32092 | 51688 | 14427 |
| CCND3-PORCN-WNT5A | 38273 | 8403 | 32926 | 50822 | 31855 | CXXC4-PORCN-WNT3 | 13 | 8251 | 9046 | 18094 | 39176 |
| FZD6-PORCN-WNT3A | 56095 | 49583 | 38183 | 45187 | 10584 | CSNK1D-PORCN-WNT5A | 33173 | 15133 | 30807 | 39641 | 18354 |
| BCL9-PORCN-WNT2 | 54695 | 186 | 56806 | 50335 | 13657 | DKK1-PORCN-WNT3A | 50877 | 8870 | 41126 | 42802 | 14055 |
| EP300-PORCN-WNT4 | 30024 | 54987 | 52222 | 44421 | 1005 | LRP5-PORCN-WNT5A | 52695 | 9213 | 43304 | 43600 | 2584 |
| FZD8-PORCN-WNT2 | 823 | 26064 | 22707 | 13185 | 36084 | CSNK1A1-PORCN-WNT4 | 18363 | 33232 | 26729 | 4216 | 46046 |
| DIXDC1-PORCN-WNT2B | 3373 | 21084 | 13572 | 18388 | 49569 | CSNK1G1-PORCN-WNT2 | 50216 | 4601 | 38088 | 29252 | 9325 |
| FOSL1-PORCN-WNT5A | 17862 | 53252 | 383 | 11834 | 29054 | DAAM1-PORCN-WNT4 | 16426 | 28262 | 4305 | 18485 | 53571 |
| FZD1-PORCN-WNT2 | 4082 | 13681 | 13674 | 16800 | 40849 | EP300-PORCN-WNT2B | 15324 | 657 | 8664 | 22349 | 56194 |
| DIXDC1-PORCN-WNT5A | 952 | 7968 | 6431 | 9090 | 46718 | FOSL1-PORCN-WNT3A | 9807 | 26119 | 20412 | 9520 | 38142 |
| FZD2-PORCN-WNT3 | 12896 | 43979 | 25222 | 26252 | 20584 | CSNK1A1-PORCN-WNT2B | 32766 | 32478 | 29427 | 48576 | 29309 |
| BCL9-PORCN-WNT3A | 10614 | 7797 | 23824 | 20378 | 37147 | FOSL1-PORCN-WNT2 | 32691 | 53282 | 55078 | 52635 | 15390 |
| LEF1-PORCN-WNT3 | 1897 | 49638 | 17744 | 24203 | 37718 | LRP6-PORCN-WNT2B | 14001 | 48495 | 14927 | 23939 | 48458 |
| CTNNB1-PORCN-WNT2B | 21141 | 13797 | 10811 | 11267 | 27556 | FZD5-PORCN-WNT3A | 36290 | 43622 | 45186 | 48760 | 4106 |
| GSK3B-PORCN-WNT2B | 40100 | 22639 | 43535 | 41745 | 32288 | FZD2-PORCN-WNT4 | 19439 | 3391 | 10054 | 16441 | 30372 |
| FZD5-PORCN-WNT2 | 17667 | 55785 | 4806 | 7011 | 44403 | EP300-PORCN-WNT5A | 20382 | 5030 | 5420 | 2444 | 52850 |
| FRAT1-PORCN-WNT5A | 31003 | 40407 | 33852 | 44607 | 31401 | FRAT1-PORCN-WNT3A | 41935 | 48293 | 37010 | 46134 | 10038 |
| KREMEN1-PORCN-WNT2 | 4395 | 4929 | 2983 | 21310 | 24768 | CTNNBIP1-PORCN-WNT4 | 25654 | 40969 | 4949 | 17529 | 55280 |
| FZD7-PORCN-WNT2 | 29459 | 48233 | 34006 | 44227 | 8832 | FBXW2-PORCN-WNT5A | 34712 | 23368 | 45069 | 53538 | 870 |
| CSNK1A1-PORCN-WNT5A | 38789 | 23969 | 30418 | 52936 | 11242 | FRZB-PORCN-WNT5A | 46306 | 37127 | 38922 | 49415 | 8250 |
| FZD2-PORCN-WNT2 | 19066 | 11867 | 15266 | 13958 | 54152 | FZD6-PORCN-WNT3 | 1060 | 7604 | 18960 | 11967 | 46623 |
| DVL2-PORCN-WNT2B | 29853 | 47974 | 32269 | 46097 | 48598 | LEF1-PORCN-WNT4 | 1022 | 22940 | 10143 | 23171 | 24580 |
| DIXDC1-PORCN-WNT2 | 49186 | 4194 | 44785 | 51431 | 14406 | DVL1-PORCN-WNT2B | 40525 | 45123 | 41427 | 54543 | 4682 |
| FBXW11-PORCN-WNT2 | 23357 | 13473 | 24805 | 6018 | 45404 | CTNNB1-PORCN-WNT5A | 13708 | 4031 | 12259 | 7191 | 26151 |
| DVL1-PORCN-WNT4 | 20685 | 17463 | 11589 | 4589 | 42885 | FRAT1-PORCN-WNT3 | 15232 | 8958 | 20105 | 11048 | 47325 |
| DAAM1-PORCN-WNT5A | 40815 | 29141 | 52832 | 38755 | 3576 | FBXW11-PORCN-WNT3 | 16713 | 4954 | 24116 | 4905 | 32441 |
| FBXW2-PORCN-WNT2 | 21418 | 34494 | 12902 | 15427 | 45695 | DVL1-PORCN-WNT2 | 16681 | 12031 | 15758 | 2629 | 52473 |
| DKK1-PORCN-WNT3 | 6334 | 48097 | 16047 | 14394 | 43015 | FSHB-PORCN-WNT4 | 45867 | 2754 | 36614 | 35679 | 21141 |
| DIXDC1-PORCN-WNT3A | 4460 | 39339 | 11468 | 317 | 55081 | APC-PORCN-WNT3 | 16291 | 16865 | 6866 | 16198 | 2665 |
| CSNK2A1-PORCN-WNT2 | 4020 | 41699 | 1257 | 21148 | 55149 | DVL2-PORCN-WNT5A | 30719 | 33390 | 31230 | 44148 | 37843 |
| NLK-PORCN-WNT3 | 5701 | 24382 | 20672 | 24152 | 44887 | GSK3B-PORCN-WNT3A | 53885 | 36640 | 45918 | 41054 | 36072 |
| FRZB-PORCN-WNT3A | 41182 | 40550 | 43804 | 44778 | 35334 | CTNNBIP1-PORCN-WNT5A | 31467 | 16471 | 52175 | 39654 | 1859 |
| CSNK1G1-PORCN-WNT3A | 5826 | 7485 | 13924 | 24296 | 26101 | FZD7-PORCN-WNT3 | 52083 | 39346 | 38543 | 56059 | 12372 |
| FBXW2-PORCN-WNT3A | 29440 | 19195 | 41476 | 51750 | 21814 | CTNNBIP1-PORCN-WNT3 | 23703 | 12476 | 9340 | 1961 | 39642 |
| FZD8-PORCN-WNT3 | 856 | 9226 | 16578 | 5870 | 42396 | DVL2-PORCN-WNT3A | 41245 | 49749 | 29482 | 36589 | 48546 |
| PORCN-SFRP1-WNT2B | 5304 | 40532 | 1039 | 12715 | 4309 | EP300-PORCN-WNT3 | 41860 | 56501 | 48463 | 34741 | 957 |
| PORCN-SFRP1-WNT5A | 3180 | 7525 | 15911 | 15553 | 17157 | CSNK1A1-PORCN-WNT3 | 23223 | 33035 | 27629 | 15910 | 40581 |
| LRP5-PORCN-WNT3 | 6138 | 18536 | 4914 | 23077 | 47094 | FOSL1-PORCN-WNT3 | 47540 | 54840 | 54984 | 43162 | 17864 |
| AXIN1-PORCN-WNT4 | 23738 | 25185 | 14884 | 7905 | 7566 | PORCN-WNT4-WNT5A | 15387 | 7092 | 1712 | 7474 | 56642 |
| FRAT1-PORCN-WNT2 | 19931 | 10329 | 16043 | 9132 | 55049 | CTNNB1-PORCN-WNT3 | 35945 | 43282 | 46348 | 45906 | 29631 |
| GSK3B-PORCN-WNT2 | 17073 | 34553 | 13631 | 15361 | 24594 | AES-PORCN-WNT5A | 39071 | 41331 | 43943 | 36584 | 10293 |
| FRZB-PORCN-WNT3 | 15867 | 16647 | 13322 | 12391 | 21906 | CTBP1-PORCN-WNT3A | 31316 | 34389 | 34735 | 43964 | 25475 |
| MYC-PORCN-WNT2 | 927 | 8233 | 3947 | 17550 | 45400 | DAAM1-PORCN-WNT2 | 26110 | 40488 | 11965 | 20021 | 55712 |
| CTNNBIP1-PORCN-WNT2 | 15297 | 27154 | 10616 | 7247 | 52470 | FGF4-PORCN-WNT2B | 30643 | 33813 | 53833 | 55121 | 19964 |
| Ranking @ using SOBOL - martinez | |||||||||||
| 3rd order comb. | 3rd order comb. | ||||||||||
| CXXC4-PORCN-WNT4 | 4276 | 14596 | 42774 | 57022 | 45130 | PITX2-PORCN-WNT4 | 9395 | 23474 | 32883 | 37297 | 24846 |
| FZD6-PORCN-WNT2B | 5774 | 40911 | 34822 | 40189 | 42079 | FZD6-PORCN-WNT4 | 199 | 7166 | 3089 | 14140 | 1367 |
| FOSL1-PORCN-WNT4 | 7917 | 37593 | 48481 | 35640 | 51574 | PITX2-PORCN-WNT2B | 32916 | 28214 | 19614 | 42831 | 17840 |
| FZD5-PORCN-WNT4 | 8507 | 37632 | 48747 | 30800 | 4942 | FOSL1-PORCN-WNT2B | 51756 | 54848 | 26032 | 20042 | 1751 |
| KREMEN1-PORCN-WNT4 | 25014 | 55908 | 56360 | 6264 | 40529 | FZD7-PORCN-WNT3A | 601 | 7936 | 6391 | 14753 | 43442 |
| DKK1-PORCN-WNT2B | 13119 | 7914 | 33768 | 34345 | 53916 | KREMEN1-PORCN-WNT3A | 5432 | 49056 | 32287 | 41650 | 15766 |
| BCL9-PORCN-WNT2B | 31320 | 49751 | 15858 | 56923 | 5674 | FZD8-PORCN-WNT4 | 16866 | 18194 | 19414 | 12569 | 12962 |
| NLK-PORCN-WNT4 | 839 | 35865 | 37133 | 52537 | 3763 | GSK3B-PORCN-WNT4 | 30196 | 41020 | 21974 | 56816 | 40470 |
| FRAT1-PORCN-WNT4 | 1204 | 15349 | 9265 | 36000 | 46519 | FZD6-PORCN-WNT2 | 2617 | 25245 | 12529 | 7270 | 8966 |
| FZD5-PORCN-WNT2B | 47626 | 21911 | 38180 | 411 | 51516 | FZD6-PORCN-WNT5A | 8901 | 16659 | 3797 | 41382 | 31216 |
| CCND3-PORCN-WNT5A | 39066 | 29570 | 4018 | 3443 | 56436 | CXXC4-PORCN-WNT3 | 49304 | 25501 | 32130 | 16898 | 50343 |
| FZD6-PORCN-WNT3A | 41626 | 8812 | 22868 | 9411 | 30030 | CSNK1D-PORCN-WNT5A | 22402 | 4216 | 2288 | 18135 | 42734 |
| BCL9-PORCN-WNT2 | 54955 | 22154 | 46038 | 51312 | 37377 | DKK1-PORCN-WNT3A | 34106 | 307 | 37594 | 26694 | 44252 |
| EP300-PORCN-WNT4 | 22842 | 5697 | 24417 | 11816 | 30962 | LRP5-PORCN-WNT5A | 14554 | 31962 | 47440 | 55611 | 32184 |
| FZD8-PORCN-WNT2 | 55228 | 25375 | 48312 | 22930 | 2643 | CSNK1A1-PORCN-WNT4 | 39148 | 56389 | 5371 | 51550 | 11271 |
| DIXDC1-PORCN-WNT2B | 1674 | 21335 | 11428 | 19858 | 16898 | CSNK1G1-PORCN-WNT2 | 44086 | 52040 | 53679 | 46699 | 15576 |
| FOSL1-PORCN-WNT5A | 51271 | 54488 | 32643 | 20512 | 3950 | DAAM1-PORCN-WNT4 | 4715 | 18483 | 25056 | 27842 | 55467 |
| FZD1-PORCN-WNT2 | 10972 | 56715 | 2891 | 54766 | 55230 | EP300-PORCN-WNT2B | 34780 | 53516 | 18975 | 9236 | 50588 |
| DIXDC1-PORCN-WNT5A | 16164 | 11219 | 7718 | 36336 | 22820 | FOSL1-PORCN-WNT3A | 49724 | 53367 | 21610 | 17015 | 8003 |
| FZD2-PORCN-WNT3 | 16891 | 31724 | 13141 | 1509 | 53320 | CSNK1A1-PORCN-WNT2B | 51145 | 2966 | 1646 | 6984 | 51942 |
| BCL9-PORCN-WNT3A | 31080 | 25153 | 42987 | 49977 | 8016 | FOSL1-PORCN-WNT2 | 6373 | 1865 | 49888 | 23145 | 13285 |
| LEF1-PORCN-WNT3 | 53701 | 55441 | 52729 | 42382 | 1529 | LRP6-PORCN-WNT2B | 45427 | 12535 | 54497 | 51518 | 467 |
| CTNNB1-PORCN-WNT2B | 51347 | 1712 | 20034 | 6187 | 37020 | FZD5-PORCN-WNT3A | 44264 | 48407 | 37797 | 1058 | 40037 |
| GSK3B-PORCN-WNT2B | 10354 | 12749 | 19731 | 7003 | 20542 | FZD2-PORCN-WNT4 | 34166 | 42118 | 26790 | 28126 | 260 |
| FZD5-PORCN-WNT2 | 14336 | 48237 | 20369 | 3530 | 1362 | EP300-PORCN-WNT5A | 20231 | 51730 | 21290 | 6094 | 9403 |
| FRAT1-PORCN-WNT5A | 17074 | 33012 | 52 | 35018 | 11028 | FRAT1-PORCN-WNT3A | 56576 | 51355 | 16114 | 376 | 8592 |
| KREMEN1-PORCN-WNT2 | 5726 | 54729 | 16213 | 7855 | 32505 | CTNNBIP1-PORCN-WNT4 | 172 | 7513 | 38333 | 6376 | 48756 |
| FZD7-PORCN-WNT2 | 5898 | 37770 | 3588 | 38329 | 46832 | FBXW2-PORCN-WNT5A | 21222 | 11676 | 43880 | 35992 | 48834 |
| CSNK1A1-PORCN-WNT5A | 54650 | 52431 | 7732 | 2602 | 47701 | FRZB-PORCN-WNT5A | 48028 | 29070 | 4466 | 31611 | 7573 |
| FZD2-PORCN-WNT2 | 5093 | 52807 | 23045 | 18722 | 21837 | FZD6-PORCN-WNT3 | 22555 | 18055 | 16279 | 1996 | 2337 |
| DVL2-PORCN-WNT2B | 34774 | 24150 | 6903 | 4977 | 15208 | LEF1-PORCN-WNT4 | 5611 | 19412 | 56055 | 27914 | 1376 |
| DIXDC1-PORCN-WNT2 | 9916 | 15221 | 5173 | 22635 | 55892 | DVL1-PORCN-WNT2B | 7994 | 25962 | 44952 | 31367 | 51921 |
| FBXW11-PORCN-WNT2 | 4568 | 22339 | 17041 | 40983 | 5016 | CTNNB1-PORCN-WNT5A | 43762 | 3615 | 11416 | 6342 | 43345 |
| DVL1-PORCN-WNT4 | 12806 | 51239 | 3810 | 47619 | 49456 | FRAT1-PORCN-WNT3 | 23204 | 52743 | 26732 | 5703 | 32028 |
| DAAM1-PORCN-WNT5A | 35449 | 37756 | 36278 | 4984 | 18388 | FBXW11-PORCN-WNT3 | 22335 | 39756 | 36822 | 12290 | 202 |
| FBXW2-PORCN-WNT2 | 38497 | 36563 | 7161 | 13719 | 3870 | DVL1-PORCN-WNT2 | 51914 | 20319 | 14179 | 47816 | 43578 |
| DKK1-PORCN-WNT3 | 16889 | 6223 | 12205 | 44615 | 49416 | FSHB-PORCN-WNT4 | 48407 | 41026 | 44326 | 38403 | 10738 |
| DIXDC1-PORCN-WNT3A | 50533 | 13326 | 45796 | 13169 | 18807 | APC-PORCN-WNT3 | 31008 | 7341 | 12508 | 22839 | 3590 |
| CSNK2A1-PORCN-WNT2 | 14811 | 7388 | 23266 | 45334 | 3849 | DVL2-PORCN-WNT5A | 25011 | 50148 | 30926 | 12099 | 23878 |
| NLK-PORCN-WNT3 | 1151 | 51209 | 30649 | 40041 | 1150 | GSK3B-PORCN-WNT3A | 50592 | 9533 | 36087 | 5178 | 11848 |
| FRZB-PORCN-WNT3A | 43274 | 25499 | 6194 | 19063 | 9871 | CTNNBIP1-PORCN-WNT5A | 9002 | 40770 | 21026 | 25896 | 23350 |
| CSNK1G1-PORCN-WNT3A | 5132 | 49722 | 52426 | 55953 | 14853 | FZD7-PORCN-WNT3 | 26004 | 2210 | 1421 | 15585 | 23223 |
| FBXW2-PORCN-WNT3A | 8868 | 7822 | 39246 | 38530 | 37536 | CTNNBIP1-PORCN-WNT3 | 1806 | 45771 | 25025 | 3597 | 38354 |
| FZD8-PORCN-WNT3 | 52647 | 14741 | 21742 | 5188 | 12616 | DVL2-PORCN-WNT3A | 56248 | 46719 | 21547 | 4335 | 32075 |
| PORCN-SFRP1-WNT2B | 11371 | 7274 | 38621 | 3196 | 35856 | EP300-PORCN-WNT3 | 46942 | 10437 | 16688 | 1303 | 25139 |
| PORCN-SFRP1-WNT5A | 594 | 30254 | 46200 | 4810 | 32251 | CSNK1A1-PORCN-WNT3 | 3737 | 47095 | 31808 | 23264 | 22340 |
| LRP5-PORCN-WNT3 | 26284 | 10985 | 49664 | 49226 | 2910 | FOSL1-PORCN-WNT3 | 8415 | 55 | 49545 | 34307 | 41439 |
| AXIN1-PORCN-WNT4 | 3439 | 53381 | 53023 | 47853 | 2095 | PORCN-WNT4-WNT5A | 39548 | 22687 | 24609 | 57067 | 2189 |
| FRAT1-PORCN-WNT2 | 16837 | 44503 | 5217 | 24876 | 44412 | CTNNB1-PORCN-WNT3 | 18524 | 12386 | 36234 | 9685 | 30765 |
| GSK3B-PORCN-WNT2 | 4811 | 27688 | 24895 | 50330 | 3255 | AES-PORCN-WNT5A | 47174 | 30282 | 25126 | 4019 | 12452 |
| FRZB-PORCN-WNT3 | 2049 | 35897 | 28774 | 18126 | 55301 | CTBP1-PORCN-WNT3A | 19331 | 3013 | 43990 | 41658 | 27693 |
| MYC-PORCN-WNT2 | 31464 | 54137 | 55028 | 11498 | 44754 | DAAM1-PORCN-WNT2 | 1260 | 3063 | 21010 | 13636 | 38216 |
| CTNNBIP1-PORCN-WNT2 | 6978 | 14005 | 25169 | 2753 | 40759 | FGF4-PORCN-WNT2B | 6331 | 344 | 43865 | 32471 | 20681 |
| WNT3 stimulated response genes | ||||||
|---|---|---|---|---|---|---|
| Response gene family | Gene family member 3rd order combinations | |||||
| adenomatosis polyposis coli | APC-PITX2-SFRP4 | APC-FZD6-SENP2 | APC-PITX2-SENP2 | APC-DIXDC1-WNT2B | APC-PITX2-TCF7 | APC-FZD6-TLE2 |
| (APC) regulator of WNT signaling pathway | APC-FZD2-TCF7L1 | APC-PITX2-PPP2CA | APC-PORCN-SENP2 | APC-PITX2-WNT4 | ||
| v-myc avian myelocytomatosis | CSNK2A1-MYC-SENP2 | CSNK2A1-MYC-SFRP4 | AES-AXIN1-MYC | CSNK2A1-MYC-TCF7L1 | FRZB-MYC-SENP2 | CSNK2A1-MYC-PPP2CA |
| viral oncogene homolog (MYC) | DKK1-MYC-SENP2 | FZD5-MYC-SENP2 | CSNK2A1-MYC-WNT4 | DVL1-MYC-SENP2 | ||
| frizzled class receptor (FZD) | FZD1 | FZD2 | FZD5 | FZD6 | FZD7 | FZD8 |
| FZD1-NLK-SENP2 | FSHB-FZD2-SENP2 | FZD5-JUN-WNT2 | APC-FZD6-FZD8 | FZD7-NKD1-SENP2 | FZD8-FBXW4-WNT3A | |
| AES-AXIN1-FZD1 | FSHB-FZD2-WNT4 | FZD5-JUN-FBXW4 | APC-FZD6-TLE2 | CXXC4-FZD7-PPP2CA | AES-AXIN1-FZD8 | |
| DVL1-FRZB-FZD1 | FSHB-FZD2-FZD7 | FZD5-CCND2-FBXW11 | AES-AXIN1-FZD6 | FZD7-PPP2CA-SFRP4 | FZD8-LRP6-RHOU | |
| FZD1-FZD7-PPP2CA | FSHB-FZD2-KREMEN1 | FZD5-CCND2-SENP2 | FZD6-PORCN-WNT2B | CXXC4-FZD7-SFRP4 | FZD8-PORCN-SFRP1 | |
| FZD1-FZD7-SFRP4 | DKK1-FZD2-LRP5 | FZD5-CCND2-FRZB | FZD6-PORCN-WNT4 | FZD1-FZD7-PPP2CA | DIXDC1-FOXN1-FZD8 | |
| AES-EP300-FZD1 | APC-FZD2-TCF7L1 | FZD5-CCND3-SENP2 | FZD6-GSK3A-WNT2 | FZD1-FZD7-SFRP4 | FZD8-PORCN-SENP2 | |
| DKK1-DVL2-FZD1 | FSHB-FZD2-TLE2 | FZD5-JUN-WNT5A | CCND1-CTBP1-FZD6 | FZD7-PORCN-SENP2 | CXXC4-FRAT1-FZD8 | |
| CXXC4-FRAT1-FZD1 | FSHB-FZD2-LRP5 | FZD5-PITX2-SENP2 | FZD6-PORCN-TLE2 | CSNK1D-FGF4-FZD7 | BTRC-FOXN1-FZD8 | |
| DVL1-EP300-FZD1 | FRZB-FZD2-SENP2 | FZD5-CCND2-DKK1 | FZD6-PORCN-SENP2 | FSHB-FZD2-FZD7 | FZD8-LRP6-TCF7 | |
| FRAT1-FZD1-SFRP4 | FSHB-FZD2-SFRP4 | FZD5-MYC-SENP2 | FZD6-PORCN-SFRP1 | FZD7-PORCN-FBXW4 | FZD8-GSK3A-KREMEN1 | |
| glycogen synthase kinase 3 (GSK3) | GSK3A | GSK3B | ||||
| FRZB-GSK3A-SENP2 | CSNK1D-FGF4-GSK3B | |||||
| FRZB-GSK3A-PPP2R1A | FBXW11-GSK3B-WNT2B | |||||
| DKK1-GSK3A-LRP5 | FOSL1-FOXN1-GSK3B | |||||
| FZD6-GSK3A-WNT2 | GSK3B-LRP6-SLC9A3R1 | |||||
| BTRC-GSK3A-T | CTBP2-GSK3B-RHOU | |||||
| FRZB-GSK3A-LRP5 | CSNK1G1-GSK3B-SLC9A3R1 | |||||
| FZD5-GSK3A-SENP2 | GSK3B-RHOU-SENP2 | |||||
| DKK1-GSK3A-WNT2B | FSHB-FZD2-GSK3B | |||||
| BTRC-GSK3A-NLK | GSK3B-JUN-TLE1 | |||||
| CSNK1D-GSK3A-LEF1 | CTBP1-FGF4-GSK3B | |||||
| dishevelled segment | ||||||
| polarity protein (DVL) | DVL1 | DVL2 | ||||
| DVL1-EP300-FRZB | DKK1-DVL2-SENP2 | |||||
| AXIN1-DVL1-FBXW2 | DKK1-DVL2-FRZB | |||||
| AXIN1-DVL1-FBXW11 | DVL2-JUN-FBXW4 | |||||
| DVL1-FRZB-FZD1 | DKK1-DVL2-FZD1 | |||||
| DVL1-EP300-GSK3B | DVL2-JUN-WNT3A | |||||
| DVL1-EP300-WNT2B | DVL2-JUN-WNT2B | |||||
| DVL1-FBXW11-SLC9A3R1 | CXXC4-DVL2-FRZB | |||||
| DVL1-EP300-WNT4 | DKK1-DVL2-FBXW11 | |||||
| AXIN1-DVL1-RHOU | DVL2-JUN-TCF7 | |||||
| DVL1-EP300-FZD1 | FZD5-DVL2-FRZB | |||||
| low density lipoprotein receptor | ||||||
| related protein (LRP) | LRP5 | LRP6 | ||||
| DVL1-EP300-LRP5 | FBXW11-LRP6-SENP2 | |||||
| DKK1-JUN-LRP5 | FBXW11-LRP6-TCF7 | |||||
| LRP5-NLK-WNT4 | FBXW11-LRP6-SLC9A3R1 | |||||
| FOXN1-KREMEN1-LRP5 | LRP6-TCF7-WNT2B | |||||
| DKK1-FZD2-LRP5 | FBXW11-LRP6-SFRP4 | |||||
| FZD5-FOXN1-LRP5 | DKK1-LRP6-SENP2 | |||||
| FSHB-FZD2-LRP5 | DAAM1-LRP6-SENP2 | |||||
| DKK1-GSK3A-LRP5 | CCND2-LRP6-RHOU | |||||
| LRP5-SFRP1-WNT2B | DAAM1-LRP6-SLC9A3R1 | |||||
| FRZB-GSK3A-LRP5 | CCND2-LRP6-TCF7 | |||||
| C-terminal binding protein (CTBP) | CTBP1 | CTBP2 | ||||
| CCND1-CTBP1-KREMEN1 | CTBP2-CTNNB1-FOSL1 | |||||
| CSNK2A1-CTBP1-PPP2R1A | CTBP2-CTNNB1-WNT4 | |||||
| CCND1-CTBP1-FZD6 | CTBP2-T-TLE2 | |||||
| CTBP1-FGF4-FBXW4 | CTBP2-FOXN1-SENP2 | |||||
| CTBP1-FGF4-FZD1 | CTBP2-CTNNB1-FBXW4 | |||||
| CTBP1-FGF4-WNT5A | CTBP2-CTNNB1-TLE1 | |||||
| CTBP1-GSK3A-PPP2CA | CTBP2-CTNNB1-WNT2B | |||||
| CTBP1-FGF4-TCF7L1 | CTBP2-T-WNT2B | |||||
| CTBP1-FGF4-FRZB | CTBP2-GSK3B-RHOU | |||||
| CTBP1-FGF4-RHOU | CSNK1A1-CTBP2-KREMEN1 | |||||
| cyclin D (CCND) | CCND1 | CCND2 | CCND3 | |||
| CCND1-FGF4-GSK3B | CCND2-LRP6-SENP2 | CCND3-PORCN-WNT4 | ||||
| CCND1-FGF4-RHOU | FZD5-CCND2-FBXW11 | CCND3-PORCN-FBXW4 | ||||
| CCND1-WIF1-WNT2B | FZD5-CCND2-SENP2 | FZD5-CCND3-SENP2 | ||||
| CCND1-FGF4-SFRP1 | FZD5-CCND2-FRZB | FZD5-CCND3-FRZB | ||||
| CCND1-FGF4-WNT5A | CCND2-LRP6-RHOU | AES-AXIN1-CCND3 | ||||
| CCND1-FGF4-FOSL1 | CCND2-FBXW11-SLC9A3R1 | CCND3-WNT1-WNT4 | ||||
| CCND1-PYGO1-WNT2 | CCND2-LRP6-TCF7 | CCND3-PORCN-SFRP1 | ||||
| CCND1-FGF4-FBXW4 | FZD5-CCND2-DKK1 | CCND3-PORCN-TLE2 | ||||
| CCND1-FGF4-PPP2R1A | CCND2-LRP6-FBXW4 | FZD5-CCND3-CSNK1D | ||||
| CCND1-CTBP1-KREMEN1 | CCND2-WNT1-WNT4 | FZD5-CCND3-SFRP4 | ||||
| Wnt family member (WNT) | WNT1 | WNT2B | WNT3A | WNT4 | WNT5A | |
| TLE2-WNT1-WNT2B | CTNNBIP1-WIF1-WNT2B | CSNK1D-FGF4-WNT3A | CXXC4-PORCN-WNT4 | CSNK1D-FGF4-WNT5A | ||
| AXIN1-WNT1-WNT4 | AES-AXIN1-WNT2B | FZD8-FBXW4-WNT3A | CSNK1D-FGF4-WNT4 | FSHB-T-WNT5A | ||
| AXIN1-WNT1-WNT2 | LRP6-TCF7-WNT2B | DKK1-JUN-WNT3A | LRP5-NLK-WNT4 | CTNNBIP1-WIF1-WNT5A | ||
| CCND2-WNT1-WNT4 | CXXC4-PORCN-WNT2B | AES-AXIN1-WNT3A | PITX2-PORCN-WNT4 | CCND1-FGF4-WNT5A | ||
| TLE1-WNT1-WNT2B | TLE1-WIF1-WNT2B | DVL2-JUN-WNT3A | FSHB-FZD2-WNT4 | FZD5-JUN-WNT5A | ||
| TCF7L1-WNT1-WNT2B | CCND1-WIF1-WNT2B | CCND1-FGF4-WNT3A | AXIN1-WNT1-WNT4 | CCND2-LRP6-WNT5A | ||
| CCND2-WNT1-WNT5A | APC-DIXDC1-WNT2B | FBXW2-WNT3A-WNT4 | DVL1-EP300-WNT4 | PITX2-PORCN-WNT5A | ||
| TCF7L1-WNT1-WNT4 | FBXW11-LRP6-WNT2B | JUN-PYGO1-WNT3A | FSHB-T-WNT4 | CXXC4-PORCN-WNT5A | ||
| TLE1-WNT1-WNT4 | FZD7-NKD1-WNT2B | CTNNBIP1-WIF1-WNT3A | APC-PORCN-WNT4 | LEF1-T-WNT5A | ||
| AXIN1-WNT1-WNT3 | TLE2-WIF1-WNT2B | PPP2CA-WNT1-WNT3A | DKK1-JUN-WNT4 | CXXC4-PORCN-WNT5A | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





